Abstract
MUC4 mucin is well known as an important potential target to overcome pancreatic cancer. Three unique domains (NIDO, AMOP, and vWD) with unclear roles only present in MUC4 but are not found in other membrane-bound mucins. Our previous studies first reported that its splice variant, MUC4/Y can be a model of MUC4 (MUC4 gene fragment is more than 30KB, too huge to clone and eukaryotic express) in pancreatic cancer. More importantly, based on MUC4/Y with the appropriate length of gene sequence, it is easy to construct the unique domain-lacking models of MUC4/Y (MUC4) for research. The present study focuses on investigation of the respective role of the unique NIDO, AMOP, and vWD domain or their synergistic effect on MUC4(MUC4/Y)-mediated functions and mechanisms by series of in vitro assays, sequence-based transcriptome analysis, validation of qRT-PCR & Western blot, and systematic comparative analysis. Our results demonstrate: 1) NIDO, AMOP, and vWD domain or their synergy play significant roles on MUC4/Y-mediated malignant function of pancreatic cancer, downstream of molecule mechanisms, particularly MUC4/Y-triggered malignancy-related positive feedback loops, respectively. 2) The synergistic roles of three unique domains on MUC4/Y-mediated functions and mechanisms are more prominent than the respective domain because the synergy of three domain plays the more remarkable effects on MUC4/Y-mediated signaling hub. Thus, to improve reversed effects of domain-lacking and break the synergism of domains will contribute to block MUC4/Y(MUC4) triggering various oncogenic signaling pathways.
Keywords: NIDO, AMOP, vWD, synergy, MUC4/Y
INTRODUCTION
As a high-molecular-weight member of the transmembrane mucin family [1, 2], MUC4 mucin plays important roles in the carcinogenesis and malignant progression of human pancreatic cancer [1, 3–5]. MUC4 is not expressed on in normal parts of the pancreas, but aberrantly overexpressed in pancreatic ductal adenocarcinoma (PDAC) and precancerous pancreatic intraepithelial neoplasias(PanIN) [6–9]. The levels of MUC4 expression rise consistently with the PanIN-PDAC progression model, and correlate significantly with poor prognosis of PDAC [7, 9, 10]. As our and other studies in the literature have reported, pancreatic cancer cells can exploit the multi-functions of MUC4 to trigger malignant activities including proliferation [11, 12, 14, 16], resistance to apoptosis [14, 16], motility [16], invasiveness & neural invasion [15, 16, 17], angiogenesis [12, 16], metastasis [11–13, 15, 16], supressing immune [18], and chemoresistance [19–21]. Thus MUC4 is an important potential target to overcome pancreatic cancer.
MUC4 has been mapped to chromosome 3 in the q29 region [22], which was cloned from the human tracheobronchial chromosomal DNA library and a human pancreatic tumor cell line [2, 23, 24]. The full-length MUC4 gene (abbr. FL-MUC4. NCBI Reference Sequence: NM_018406.6) contains 26 exons, which encode various functional domains from the amino end to carboxyl end, in order as follows: a 27 residue signal peptide, serine & threonine-enriched imperfect repetition motifs, a centrally located large tandem repeat (TR) domain, nidogen (NIDO)-like domain, adhesion-associated domain (AMOP; present in MUC4 and other proteins), von Willebrand factor (vWD; type D domain), and three EGF-like domains [2, 22–25], hydrophobic transmembrane region by which MUC4 is anchored to the cell surface, followed by a short cytoplasmic tail of 22 amino acids. Among them, it has been proved that the EGF-like domains present in MUC4 interact with ErbB2 and ErbB3 receptors to trigger intrinsic protein–tyrosine kinase activity and further activate intracellular signaling pathways (e.g., mitogen-activated protein kinase [MAPK], phosphatidylinositol-3-kinase [PI3K]–Akt, protein kinase C [PKC] pathways), and coactivate transcription of the downstream effector molecules to mediate malignant functions of tumor, including pancratic cancer [15, 16, 26–30]. Notably, the unique domains present in the MUC4 mucin but not found in other membrane-bound mucins are NIDO, AMOP, and vWD domains [31]. A homology analysis between MUC4 proteins in different vertebrate species reveals NIDO, AMOP, and vWD domains are conservative and important motifs [3, 23]. However, the role or effect of the three unique domains on MUC4-mediated functions and mechanisms is unclear, especially when in different tumor microenvironment.
In our previous studies [14, 16], we first proved that its splice variant, MUC4/Y (NCBI Reference Sequence: NM_004532.5) can be as a model of MUC4 (MUC4 gene fragment is more than 30KB, too huge to clone and eukaryotic express) in pancreatic cancer for function and mechanism research, as shown in Additional File 4 (Supplementary Figure S1). More importantly, based on MUC4/Y with the appropriate lenth of gene sequence, it is easy to construct the unique domain-lacking models of MUC4/Y (MUC4) for research. Thus, the present study aimed to investigate the respective role of the unique NIDO, AMOP, and vWD domain or their synergistic effect on MUC4(MUC4/Y)-mediated functions and mechanisms: 1) Series of stable PANC-1 cell strains transfected by the MUC4/Y gene without or with domain-lacking were established and consistent overexpression quantity of target genes were verified for comparison of different quality caused by domain-lacking. 2) Series of in vitro assays were conducted to detect the changes of malignant activities of PANC-1 cells caused by domain-lacking. 3) sequence-based transcriptome analysis, validation of qRT-PCR & Western blot, and systematic comparative analysis were carried out to find the effect afforded by domain-lacking on MUC4-mediated molecule mechanisms, particularly the impact on MUC4/Y(MUC4)-triggered malignancy-related positive feedback loops. 4) Comparison and induction were done to illustrate the universality, individuality and synergism of the role of these three unique domains, which can contribute to provide potential targets for overcome pancreatic cancer.
RESULTS
Establishment of series of stable PANC-1 cell strains with consistent expression quantity of target genes for comparison of different quality caused by domain-lacking
To investigate the function change of MUC4/Y gene without or with domain-lacking in pancreatic cancer, PANC-1 cells, which do not express endogenous MUC4 [32], were stable transfected by target genes, and selected using 10% DMEM containing puromycin (2.0 μg/mL), respectively.
Figure 1A depicts the design of the MUC4/Y gene without or with domain-lacking, corresponding amino acids sequences of domains denoted by different color in Additional File 1. The stable PANC-1 cell clones (N△, A△, V△ and NAV△) with over-expression of target genes in mRNA and protein level were verified separately. Figure 1B shows that the expression and subcellular localization of MUC4/Y gene with domain-lacking (MUC4/Y-NIDO△, MUC4/Y-AMOP△, MUC4/Y-vWD△, MUC4/Y-NIDO△-AMOP△-vWD△) was same as that of MUC4/Y, which was both membranous and cytoplasmic staining, indicating similar protein processing in these clones after transfected by target genes, respectively. These distributions were as same as the pancreatic cancer cell line BXPC-3 which is wild-type MUC4 positive-expression as positive control [33]. Cell membrane surface expression was determined by FCM analysis. Figure 1C shows that the frequency of expression of the phenotypic marker MUC4 was 99.37% in BXPC-3 cell line, 99.42% in Y cell clone, 99.50% in N△ cell clone, 99.94% in A△ cell clone, 99.68% in V△cell clone, 98.97% in NAV△ cell clone, respectively, indicating high purities of the series stable PANC-1 cell clones and stable overexpression of target genes. These results demonstrated that stable PANC-1 cell strains of overexpression of MUC4/Y gene without or with domain-lacking were established, and they were consistent with each other in expression quantity. Thus the different quality between different groups lies in domain-lacking.
Figure 1. Design and identification of stable consistent expression of the MUC4/Y gene without or with domain-lacking.
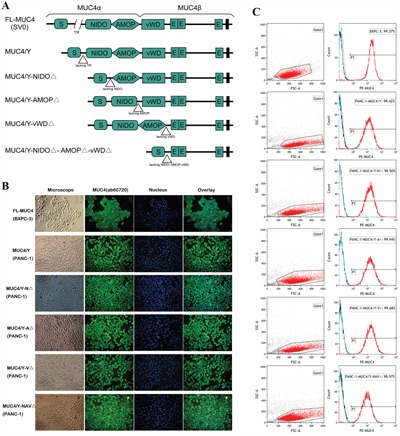
A. Schematic representation of the design strategy. B. IF demonstrating the expression and subcellular localization of MUC4/Y gene with domain-lacking (MUC4/Y-NIDO△, MUC4/Y-AMOP△, MUC4/Y-vWD△, MUC4/Y-NIDO△-AMOP△-vWD△) was same as that of MUC4/Y and wild-type MUC4. C. FCM analysis confirmation of target gene expression on cell membrane surface and high purities of the series stable PANC-1 cell clones with overexpression of target gene, respectively.
Roles of MUC4/Y’s unique domains in cell proliferation, DNA replication, cell cycle and anti-apoptosis under low-nutritional-pressure
Figure 2A and 2B shows that under stress from low nutritional status (1% serum), the percentage of EdU-positive cells of NAV△ group decreased significantly (P = 0.0004), and cell proliferation rate of NAV△ group decreased significantly at 72h, 96h, 120 h (P = 0.038, 0.005, 0.000, respectively), compared to the PANC-1-MUC4/Y control group. These consistent results suggests the simultaneous lack of NIDO, AMOP and vWD domains can lower significantly the ability of cell proliferation and DNA replication in compared to the control (MUC4/Y gene without domain-lacking) under stress from low nutritional status.
Figure 2. Domain-lacking weakened the role of MUC4/Y on malignant activities of PANC-1 cell.
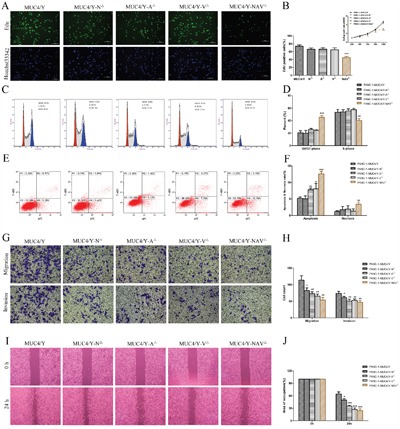
A and B. The altered DNA replication ability was determined by the EdU incorporation assay at various MUC4/Y-domains-lacking groups and the control group. The cells growth rates were determined by CCK-8 proliferation assays at various time points. Cell growth rate = point-in-time of the absorbance at 450 nm(A450) / Mean of A450 in 24h. C and D. Cell cycle distribution was analyzed by using flow cytometry with PI staining. The percentages of cells at the G0/G1 & S & G2/M phase were plotted. E and F. Representative templates of FCM analysis showing the proportion of cells positive for annexin V(-APC) and 7-AAD (top right quadrant) representing the percentage of necrotic cells; the proportion of cells that were annexin V(-APC)–positive and 7-AAD–negative (bottom right quadrant) represented the percentage of apoptotic cells. Bar denotes the percentage of apoptotic and necrotic cells in PANC-1–derived clones. G and H. Metastatic potential in vitro was detected by matrigel migration and invasion assay. Bar graph shows the number of PANC-1–derived clones that had migrated or invaded through the Matrigel. I and J. Migration capability was detected by wound healing assay. Wound closure was delayed in different groups at 24 h. Occupied area of wound closure by migrated cells was calculated and is depicted in the bar chart. “*”(P<0.05), “**”(P<0.01), and “***”(P< 0.001) indicate a significant difference from PANC-1-MUC4/Y control cells.
We further detected cell cycle distribution cultured in low-serum (1% FBS) medium by flow cytometry. As shown in Figure 2C&2D, compared with PANC-1-MUC4/Y control groups, cells transfected with MUC4/Y-NAV△ presented a prominent accumulation of cells in the G0/G1 phase (P = 0.0006) and a decrease in the S phase (P = 0.007), indicating that the simultaneous lack of NIDO, AMOP and vWD domains can reduce cell proliferation rate by facilitating cell cycle arrest at G0/G1 phase in compared to the control under stress from low nutritional status.
Moreover, as shown in Figure 2E&2F, apoptosis assay showed under stress from low nutritional status (1% serum), compared to the PANC-1-MUC4/Y control cells, there were significant increases of apoptosis rate in A△, V△, NAV△ group (P = 0.002, 0.045, 0.0001, respectively), and there was the significant increase of necrosis rate in PANC-1-MUC4/Y-NAV△ cells (P = 0.005), indicating that the lack of AMOP or vWD, and the simultaneous lack of NIDO, AMOP and vWD domains (NAV△) can restore the cellular response to apoptotic stimuli caused by stress from low nutritional status.
Altogether, these results show that synergistic effect of NIDO, AMOP and vWD domains on proliferation, DNA replication, cell cycle and anti-apoptosis of PANC-1 cells is significant, AMOP or vWD domain of MUC4/Y also has significant role on anti-apoptosis of PANC-1 cells.
Roles of MUC4/Y’s unique domains in cell migration and invasion
We used Transwells without or with Matrigel-coated membranes to examine cell migration and invasion, respectively, in vitro. Figure 2G&2H shows that compared to the PANC-1-MUC4/Y control cells, there were significant decreases of average number of migrating cells in N△, A△, V△, NAV△ groups (P =0.025, 0.008, 0.005, 0.003, respectively), and there were significant decreases of average number of cells invading through the Matrigel in N△, A△, V△, NAV△ groups (P = 0.034, 0.008, 0.008, 0.005, respectively). Consistently, as shown in Figure 2I&2J, wound healing assays showed that the migration capability of N△, A△, V△, NAV△ groups was less than that of control cells (P = 0.013, 0.0007, 0.0005, 0.0004, respectively). These data suggest that the lack of domains can significantly down regulate the effects of MUC4/Y on pancreatic cancer cells migration and invasion, and indicate that NIDO, AMOP, vWD, or synergism of them play roles in migration and invasion of PANC-1 cells.
DEG screening and functional annotation for global mRNA analysis of MUC4/Y’s unique domains triggered signatures
We have revealed 1575 differentially expressed genes (DEGs) of MUC4/Y over-expressing PANC-1 cells compared with the blank and negative controls in earlier research [16]. Here domain-lacking triggered signatures were analyzed. The filtering condition of DEGs was intersection sets beween DEGs of Domain-lacking triggering and DEGs of MUC4/Y gene triggering, absolute value of log2 ratio ≥ 1. Comparative analyses of Domain-lacking groups vs control group (PANC-1-MUC4/Y) revealed 932 DEGs for N△ vs Y, 990 DEGs for A△ vs Y, 1033 DEGs for V△ vs Y, 1214 DEGs for NAV△ vs Y, respectively, as shown in Supplementary Table S1-S4 (Additional File 2).
As shown in Supplementary Table S5-S10 (Additional File 3), DEGs annotated against GO and KEGG databases were enriched to identify significant GO biological process terms and pathways, respectively, and adjusted with corrected P ≤ 0.05 for GO analysis and pathways, as follows: 1) N△ vs Y: The significant GO biological process terms were cell projection, synaptic vesicle (P = 0.03743, 0.00213, successively); None of pathways were enriched to identify significantly. 2) A△ vs Y: The significant GO biological process terms were cell projection, membrane, membrane part, intrinsic to membrane, extracellular region, extracellular region part, neuron projection (P = 4.74e-05, 0.00100, 0.00207, 0.00385, 0.01009, 0.01542, 0.00213, successively) in “Cellular Component”subdirectory of GO, and signaling, signal transmission (P = 0.02800, 0.04822, successively) in “Biological Process”subdirectory of GO; None of pathways were enriched to identify significantly. 3) V△ vs Y: The significant GO biological process terms were cell projection, neuron projection, membrane, membrane part, intrinsic to membrane, extracellular region (P = 1.08e-07, 2.56e-05, 8.63e-05, 0.00054, 0.00105, 0.03525, successively) in “Cellular Component”subdirectory of GO, and signaling, signal transmission, signaling process, neurogenesis, generation of neurons, nervous system development, neuron differentiation, regulation of multicellular organismal development, signal transduction (P = 8.42e-05, 0.00016, 0.00020, 0.00121, 0.00659, 0.00860, 0.01776, 0.01919, 0.02644, successively) in “Biological Process”subdirectory of GO; The significant pathway-enriched terms were MAPK signaling pathway, Complement and coagulation cascades, Chemokine signaling pathway, Calcium signaling pathway, Cytokine-cytokine receptor interaction (P = 0.01527118, 0.03150312, 0.03150312, 0.04294926, 0.04324608, successively). 4) NAV△ vs Y: The significant GO biological process terms were cell projection, membrane, intrinsic to membrane, membrane part, integral to membrane (P = 0.00033, 0.00138, 0.00502, 0.01920, 0.04597, successively) in “Cellular Component”subdirectory of GO, GTPase regulator activity (P =0.04792) in “Molecular Function”subdirectory of GO, and signaling, signal transmission, signaling process (P = 2.52e-06, 3.68e-05, 4.65e-05, successively) in “Biological Process”subdirectory of GO; The significant pathway-enriched terms were MAPK signaling pathway (P = 0.000554462).
Altogether, the above enrichment results and analyses of GO function and the KEGG pathway show the global function triggered by NIDO(N), or AMOP(A), or vWD(V), or synergism of NIDO, AMOP and vWD domain(NAV) of MUC4/Y: 1) The universality of N, A, V, NAV is the function about “cell projection”; The common characters of A, V, NAV are function about “membrane” and “signaling”; The similar characters of A, V are function about “extracellular region” and “neuron projection”. 2) The individuality is the function about “synaptic vesicle” in N, “nervous system generation, development, differentiation” and “regulation of multicellular organismal development” and “effect on Immune system” in V, “GTPase regulator activity” in NAV. 3) As shown in Supplementary Table S1-S4 (Additional File 2), comparative analyses between [domain-lacking groups vs control group(PANC-1-MUC4/Y)] and [MUC4/Y over-expressing groups vs control group(PANC-1 cells transfected with empty lentiviral vectors were designated PANC-1-EV. Wild-type PANC-1 and PANC-1-EV cells were used as blank and negative control groups, respectively)] revealed that the expression levels (in transcripts per million, TPM) of most DEGs of MUC4/Y triggering were reversed afforded by domain-lacking to a variable extent. Altered gene expression was then confirmed by carrying out qRT–PCR and Western blotting as follows, which results in variation trends were consistent with sequence-based transcriptome analysis.
QPCR validation of roles of MUC4/Y’s unique domains on MUC4/Y-mediated mechanisms
To verify that the expression levles of most DEGs of MUC4/Y triggering can be reversed by the absence of unique MUC4/Y domains, we selected a batch of DEG molecules of MUC4/Y triggering for QPCR validation, and we focused on MUC4/Y-mediated key mechanisms. Additional File 4 (Supplementary Figure S1-S2) list the summary of representative downstream effector molecules of MUC4/Y to activate malignant functions, trigger the positive feedback regulatory loops, and relate with energy metabolism, protein synthesis & modification.
These selected molecules were classified as twelve feature subsets, i.e. group1-12, as shown in Figure 3. The twelve feature subsets (Group1-12) were “MUC4/EGF-ERBB2-ERBB3 signaling hub”, “PKC and AKT signaling molecules”, “SHC-Grb2-SOS-ERK pathway molecules”, “RALGDS-RAC-JNK pathway molecules”, “Endonuclear Transcription Factors”, “Extracellular Growth Factors & Membrane Receptors”, “Crucial Factors Mediating Actin Dynamics & Migration”, “Crucial Factors Involved in Proliferation & Anti-apotosis”, “Crucial Factors Involved in Metastasis”, “Crucial Factors Involved in Energy Metabolism & Mitochondrial function”, “Crucial Factors Involved in Protein Synthesis & Modification”, “Crucial Factors Involved in Glycosylated Modification”, successively.
Figure 3. The expression of most DEGs of MUC4/Y triggering was reversed in the absence of unique MUC4/Y domains(NIDO, AMOP, vWD, or synergy).
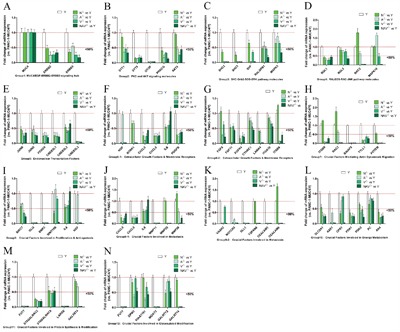
Representative downstream effector molecules of MUC4/Y (i.e. DEGs of MUC4/Y over-expressing PANC-1 cells compared with controls in earlier research [16], as shown in Additional File 4) were classified as twelve feature subsets, i.e. Group1-12. QPCR validation results of altered gene expression of these molecules in lacking-domains groups (N△, A△, V△, NAV△) and control group (MUC4/Y-overexpression). The expression quantity in controls is defined as 1.0, so under the line of “1.0” represents mean expression quantity of experimental groups below that of control group, and under the line of “0.5” represents mean expression quantity of experimental group less than 50% expression of control group.
To investigate the role of MUC4/Y’s domains (N, A, V, NAV) on MUC4/Y-mediated mechanisms, altered gene expression of the lacking-domains groups (N△, A△, V△, NAV△) vs. control group (MUC4/Y-overexpression) was validated by independent qRT-PCR test. The results showed the expression levels of most DEGs of MUC4/Y triggering were reversed by the absence of unique MUC4/Y domains. And the expression levels of representative 68 molecules were all reversed in V△ group (vs Y). Only small number of molecules expression levels were not reversed, as follows: 1) N△ vs Y: 8 molecules which failed to reverse expression were RAC2, MAPK10, FGF5, ITGB8, MMP25, MMP28, TSPAN8, CEACAM6 (Figure 3D, 3G, 3H, 3J, 3K, successively) 2) A△ vs Y: 2 molecules which failed to reverse expression were MAPK10, WNT10B (Figure 3D, 3I, successively) 3) NAV△ vs Y: one molecule which failed to reverse expression was IL8 (Figure 3F, 3I, 3J, successively). These results were also consistent with sequence-based transcriptome analysis, indicating that our sequencing approach and analytical pipeline were reliable.
Altogether, these results show that MUC4/Y’s unique domains (i.e. NIDO, AMOP, vWD, or synergism of them) have roles in MUC4/Y-mediated downstream of molecule mechanisms.
WB validation of effects of MUC4/Y’s unique domains on MUC4/Y-mediated signaling pathways
To investigate the effect of MUC4/Y’s domains (N, A, V, NAV) on MUC4/Y-mediated main signaling pathways, the expression changes of the key nodes in the signal path, especially in protein phosphorylation, were verified and compared between the lacking-domains groups (N△, A△, V△, NAV△) and control group (MUC4/Y-overexpression) by western blotting and measurement of optical density value.
Additional File 4 (Supplementary Figure S1) also shows the main signaling pathways mediated by MUC4/Y, which include MUC4/Y-dependent increase of ERBB2 and ERBB3 phosphorylation activation, paralleled by the upregulation of key molecules (Ras, Src, focal adhesion kinase [FAK], extracellular signal–regulated kinase [ERK], c-Jun amino-terminal kinase [JNK], AKT, nuclear factor κB [NFκB], inhibitor of NFκB [IκBα], c-Jun), as either total or phosphorylated proteins, in the downstream signaling pathways.
As shown in Figure 4, domain-lacking reduced protein expression levels of key nodes of MUC4/Y-mediated signal path in the majority. And the expression levels of representative 20 molecules were all reversed in NAV△ group (vs Y). Non- reverse effects were in the minority, as follows: 1) N△ vs Y: Molecules which failed to lower 6 proteins expression were ERBB2, Src, FAK, ERK, Phospho-ERK, Phospho-c-JUN. 2) A△ vs Y: Molecules which failed to reduce 2 proteins expression were ERBB3, Phospho-NFκB. 3) V△ vs Y: Molecules which failed to down- regulate 2 proteins expression were ERBB3, c-JUN. So, these results indicate that MUC4/Y’s unique domains have effects on MUC4/Y-mediated downstream of signaling pathways, consistently with sequence-based transcriptome analysis and QPCR validation.
Figure 4. Domain-lacking reduced protein expression level of key nodes of MUC4/Y-mediated signal path in the majority.
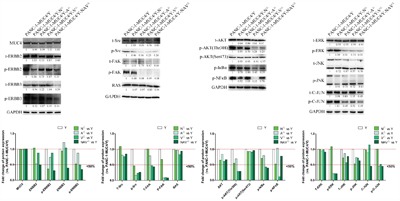
Representative signaling pathways mediated by MUC4/Y were described as earlier research [16] (Additional File 4, Supplementary Figure S1). WB validation results of the expression changes of the key nodes in the signal path and measurement of optical density value by Image J software. The expression quantity in controls is defined as 1.0, so under the line of “1.0” represents mean expression quantity of experimental groups below that of control group, and under the line of “0.5” represents mean expression quantity of experimental group less than 50% expression of control group.
Systematic comparative analysis of FDR-corrected P-values of the 18 sigificant enriched functional categories of MUC4/Y and reverse effects triggered by domain-lacking
As mentioned above, on the whole, loss of MUC4/Y’s unique domains weakened the roles of MUC4/Y on malignant activities, and impaired expression levels of downstream effector molecules of MUC4/Y triggering, demonstrating that MUC4/Y’s unique domains have significant roles in MUC4/Y-mediated malignant function of pancreatic cancer, downstream of molecule mechanisms, including signaling pathways, respectively. To illustrate the separate feature (of N, A, V, NAV) and reverse-effect rate or extent (of N△, A△, V△, NAV△) to MUC4/Y functional categories, systematic comparative analysis were carried out, as shown in Figure 5A-5H.
Figure 5. Systematic comparative analysis of FDR-corrected P-values of the 18 sigificant enriched functional categories of MUC4/Y and reverse effects triggered by domain-lacking.
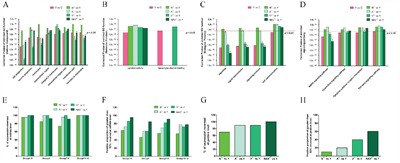
A. The comparison of corrected P-value of enriched GO function (Cellular Component). B. The comparison of corrected P-value of enriched GO function (Molecular Function). Three Groups (N△ vs Y, A△ vs Y, NAV△ vs Y) failed to enriched on the functional subset “lipopolysaccharide binding”, so they lacked the P-value. C. The comparison of corrected P-value of enriched GO function (Biological Process). D. The comparison of corrected P-value of enriched signaling pathway. To (A-D), under the red line of “0.5”, or labeled with red asterisk, *, represents FDR-corrected P-values, ≤0.05. E. Positive proportion of genes (verified by QPCR) number of expression-reversed at different groups to compare reverse-effect rate. F. Positive proportion of greater than 50% reversal rate (verified by QPCR) at different groups to compare reverse-effect extent. To (E&F), Group1-5 on behalf of the feature subset “crucial factors involved in signaling hub”. Group 6 on behalf of the feature subset “extracellular growth factors & membrane receptors”. Group7-9 on behalf of the feature subset “crucial factors involved in oncogenic function”. Group10-12 on behalf of the feature subset “crucial factors involved in energy metabolism (Mitochondrial function) and protein synthesis & modification including glycosylated modification (Golgi function)”. G. Positive proportion of genes (verified by WB) number of expression-reversed at different groups to compare reverse-effect rate. H. Positive proportion of greater than 50% reversal rate (verified by WB) at different groups to compare reverse-effect extent.
Figure 5A-5D show that the validated sigificant enrichment of GO function and the KEGG pathway of MUC4/Y overexpression in PANC-1 cells focuses on 18 functional categories [16] (marked with pink, under the red line of “0.5” represented FDR-corrected P-values, ≤0.05), as follows: cell projection, neuron projection, membrane, extracellular region, integral to membrane, extracellular region part, membrane part, intrinsic to membrane, cytokine activity, lipopolysaccharide binding, signaling, signal transmission, signaling process, cell communication, MAPK signaling pathway, Chemokine signaling pathway, Cytokine-cytokine receptor interaction, TGF-beta signaling pathway.
Compared with MUC4/Y overexpression in PANC-1 cells, DEGs of separate domain-lacking groups with reversed expression level were also enriched significantly (labeled with red asterisk, *, Figure 5A-5D), as follows: N△ vs Y (cell projection), A△ vs Y(cell projection, neuron projection, membrane, extracellular region, extracellular region part, membrane part, intrinsic to membrane, signaling, signal transmission), V△ vs Y (cell projection, neuron projection, membrane, extracellular region, membrane part, intrinsic to membrane, signaling, signal transmission, signaling process, MAPK signaling pathway, Chemokine signaling pathway, Cytokine-cytokine receptor interaction), NAV△ vs Y (cell projection, membrane, integral to membrane, membrane part, intrinsic to membrane, signaling, signal transmission, signaling process, MAPK signaling pathway).
The comparison of FDR-corrected P-values of the above18 functional categories (of MUC4/Y) showed that the least P-values of “extracellular region”and “extracellular region part” were acquired by A△ vs Y, suggesting that the two functional categories of MUC4/Y are the most highly correlated to the function or role of AMOP domain.
The least P-values of “cell projection”, “neuron projection”, “membrane”, “membrane part”, “intrinsic to membrane”, “Chemokine signaling pathway”, and “Cytokine-cytokine receptor interaction” were acquired by V△ vs Y, suggesting that the seven functional categories of MUC4/Y are the most highly correlated to the function or role of vWD domain.
The least P-values of “signaling”, “signal transmission”, “signaling process”, “MAPK signaling pathway” were acquired by NAV△ vs Y, suggesting that the four functional categories are the most highly correlated to the synergistic role of unique domains.
Figure 5E-5F show the reverse-effect rate or extent of different domains-lacking at mRNA level verified by QPCR. As shown in Figure 3, representative downstream effector molecules of MUC4/Y were classified as twelve feature subsets, i.e. group1-12. Group1-5 can be incorporated as a feature subset of crucial factors involved in signaling hub. Group 6 is a feature subset of extracellular growth factors & membrane receptors. Group7-9 can be incorporated as a feature subset of crucial factors involved in oncogenic function. Group10-12 can be incorporated as a feature subset of crucial factors involved in energy metabolism (Mitochondrial function) and protein synthesis & modification including glycosylated modification (Golgi function). Figure 5E shows positive proportion of genes number of expression-reversed at group N△, A△, V△, NAV△, as follows: 1) Group1-5: 95.45% (21/22), 95.45% (21/22), 100% (22/22), 100% (22/22), respectively. 2) Group 6: 84.61% (11/13), 100% (13/13), 100% (13/13), 92.31% (12/13), respectively. 3) Group7-9: 73.91% (17/23), 95.65% (22/23), 100% (23/23), 91.30% (21/23), respectively. 4) Group10-12: 100% (18/18), 100% (18/18), 100% (18/18), 100% (18/18), respectively. Figure 5F shows positive proportion of greater than 50% reversal rate at group N△, A△, V△, NAV△, as follows: 1) Group1-5: 63.64% (14/22), 72.73% (16/22), 86.36% (19/22), 95.45% (21/22), respectively. 2) Group 6: 46.15% (6/13), 61.54% (8/13), 61.54% (8/13), 84.62% (11/13), respectively. 3) Group7-9: 56.53% (13/23), 69.57% (16/23), 82.61% (19/23), 91.30% (21/23), respectively. 4) Group10-12: 55.56% (10/18), 77.78% (14/18), 72.22% (13/18), 77.78% (14/18), respectively.
Figure 5G-5H show the reverse-effect rate or extent of different domains-lacking at protein level confirmed by WB. As shown in Figure 4, the expression of representative key nodes in the signal pathways mediated by MUC4/Y were detected. Figure 5G shows positive proportion of genes number of expression-reversed at group N△, A△, V△, NAV△ was 70% (14/20), 90% (18/20), 90% (18/20), 100% (20/20), respectively. Figure 5H shows positive proportion of greater than 50% reversal rate at group N△, A△, V△, NAV△ was 10% (2/20), 20% (4/20), 40% (8/20), 60% (12/20), respectively.
Altogether, these results indicate that the lack of three unique domains (N△, A△, V△) plays the definite effects on reversing MUC4/Y-mediated malignant function and downstream of molecule mechanisms, among which N△ has the weakest effects on that. Notably, the simultaneous lack of three unique domains (NAV△) plays the most significant effects on reversing MUC4/Y-mediated downstream of signal pathways than the respective lack groups (N△, A△, V△), which is consistent with above mentioned that four functional categories of MUC4/Y (signaling, signal transmission, signaling process, MAPK signaling pathway) were the most highly correlated to the synergistic role of unique domains.
DISCUSSION
In the present study, we focus on investigate the respective role of the unique NIDO, AMOP, and vWD domain or their synergistic effect on MUC4/Y(MUC4)-mediated functions and mechanisms. Based on MUC4/Y, we initially constructed homologous genes lacking unique domains present in MUC4, i.e., MUC4/Y-NIDO△, MUC4/Y-AMOP△, MUC4/Y-vWD△, MUC4/Y-NIDO△-AMOP△-vWD△. We also constructed series of stable PANC-1 cell strains transfected by the MUC4/Y gene without or with domain-lacking, which with consistent forced gene expression rate and subcellular localization, for comparison of different quality caused by domain-lacking.
On the whole, the results of function assays in vitro, sequence-based transcriptome analysis and confirmatory testing were similar in variation trends, as follows: 1) Domain-lacking weakened the roles of MUC4/Y on malignant activities of PANC-1 cell in vitro, which include the significant decrease of cell proliferation and DNA replication, consistently followed by the cell cycle arrest at G0/G1 phase, significant increase of apoptosis & necrosis rate under stress from low nutritional status, which also include significant decrease of the capabilities of migration and invasion. 2) Domain-lacking reversed the expression levels of the most of differentially expressed genes (DEGs) caused by MUC4/Y over-expressing PANC-1 cells compared with the blank and negative controls. Firstly, sequence-based transcriptome analysis revealed that among 1575 DEGs of MUC4/Y-overexpression triggered, the expression levels (in transcripts per million, TPM) of 932 genes were reversed 2 folds or more than by NIDO-lacking triggered, 990 genes by AMOP-lacking triggered, 1033 genes by vWD-lacking triggered, 1214 genes by three domains-simultaneously-lacking triggered, as shown in Additional File 2 (Supplementary Table S1-S4). Secondly, QPCR validation of representative downstream effector molecules of MUC4/Y (which to trigger malignancy-related positive feedback regulatory loops, and relate with energy metabolism, protein synthesis & modification) revealed that the expression levels of representative 68 molecules were all reversed by vWD-lacking with 100% reversed rate, NIDO-lacking with 88.24% (60/68) reversed rate, AMOP-lacking with 97.06% (66/68) reversed rate, three domains-simultaneously-lacking with 98.53% (67/68) reversed rate. Thirdly, WB validation of the expression changes (causesd by domain-lacking) of the key nodes on MUC4/Y-mediated main signaling pathways revealed that the expression levels of representative 20 molecules were all reversed by three domains-simultaneously-lacking with 100% reversed rate, NIDO-lacking with 70% (14/20) reversed rate, AMOP-lacking with 90% (18/20) reversed rate, vWD-lacking with 90% (18/20) reversed rate. Altogether, these results demonstrate that MUC4/Y’s unique domains have significant roles in MUC4/Y-mediated malignant function of pancreatic cancer, downstream of molecule mechanisms, particularly MUC4/Y-triggered malignancy-related positive feedback loops, respectively.
Notably, the simultaneous lack of three unique domains (NAV△) had the most significant effects on reversing MUC4/Y-mediated functions in vitro (as shown in Figure 2). That was consistent with the most significant effects on reversing MUC4/Y-mediated molecule mechanisms than the respective lack groups (N△, A△, V△), as shown in Figure 5F & 5H, NAV△ group ranked the highest overall on positive proportion of greater than 50% reversal rate (95.45% in Group1-5 at mRNA level, 84.62% in Group6 at mRNA level, 91.30% in Group7-9 at mRNA level, 77.78% in Group10-12 at mRNA level, and 60% at protein level). Coincidentally, the above two points are consistent with the results of systematic comparative analysis, which revealed that the four enriched functional categories of MUC4/Y (signaling, signal transmission, signaling process, MAPK signaling pathway) were the most highly correlated to the synergy of three unique domains (NAV). Thus, we conclude that the synergistic roles of NIDO, AMOP and vWD domains on MUC4/Y-mediated functions and mechanisms are more prominent than the respective domain because the synergy of three domain plays the more remarkable effects on MUC4/Y-mediated signaling hub.
In addition, the results and analyses of GO function and the KEGG pathway show that the functional category “GTPase regulator activity” was enriched or triggered only by the synergy of three unique domains (NAV). The functional category “GTPase regulator activity” plays roles in the growth control, regulating the organization and remodelling of the actin cytoskeleton, regulating cell migration, regulating cancer metastasis via modulation of GTPases and GTP hydrolysis [34, 35]. Consistently, our results showed the simultaneous lack of NIDO, AMOP and vWD domains (NAV△) weakened the roles of MUC4/Y on malignant activities of PANC-1 cell in vitro, including cancer metastasis-related capabilities. And NAV△ reversed batches of crucial factors involved in oncogenic function (crucial factors mediating proliferation, anti-apotosis, actin dynamics & migration, metastasis, etc). Thus, finding paths to break the synergy of three unique domains (NAV) will be helpful to weaken capabilities of pancreatic cancer metastasis, specially to the patients with MUC4 positive expression.
In contrast to previous studies [3, 23, 36, 40–44], this article is not confined to a piont. Instead, based on transcriptome analysis with big data and accurate statistic mathematic model, systematic validation and comparison, we found and noted that the respective roles of the unique NIDO, AMOP, and vWD domain or their synergistic effects on MUC4/Y (MUC4)-mediated mechanisms were with complex features, even the overlapping in different dimension, in accordance with MUC4/Y (MUC4)-mediated mechanisms.
Our previous studies [16] have detailed the enormity of the potential regulatory circuitry in pancreatic cancer afforded by MUC4/Y (MUC4), which is with remarkable features, as follows: 1) The malignancy-related positive feedback loops work on the ring circuit path, i.e., triggering(MUC4/EGF-ERBB2-ERBB3 signaling hub)- activating(main MAPK signaling pathways)- transmitting(endonuclear transcription factors)- producing or upregulating(cytokines, growth factors, extracellular matrix, integrins, membrane receptors)- activating(main MAPK signaling pathways)- transmitting(endonuclear transcription factors)- sustained upregulating (MUC4/EGF-ERBB2-ERBB3 signaling hub). 2) Producing or upregulating crucial factors involved in oncogenic function is for malignant activities of pancreatic cancer. 3) Producing or upregulating crucial factors involved in energy metabolism, protein synthesis & modification is for supplying energy and survial material. 4) Producing or upregulating cytokines, growth factors, and adhesion molecules is for pancreatic cancer cell to affect the tumor milieu by cell–ECM and cell–cell interplay. 5) Complex interplays between several signaling pathways form network. So, MUC4 overexpression correlates significantly with poor prognosis of PDAC, not only because it plays important roles in the carcinogenesis and malignant progression of human pancreatic cancer, but also its triggered malignancy-related positive feedback loops are the root of resistance to chemotherapy and molecular targeted therapy.
Thus, in light of MUC4/Y (MUC4)-mediated complex mechanisms, batches of MUC4/Y triggering DEG molecules which can represent above mentioned five remarkable features were selected for QPCR and western blotting validation of changes afforded by different domains-lacking, followed by systematic comparison to effect extent of different domains-lacking on two levels. Importantly, the present study is the first to demonstrate that NIDO, AMOP, and vWD domain or their synergy play significant roles on MUC4/Y (MUC4)- triggered malignancy-related positive feedback loops of pancreatic cancer to a variable extent. Excitingly, we find and verify that the absence of the unique domains (NIDO, AMOP, vWD) respectively or simultanously contributes to weaken MUC4-mediated malignant activities, cut off MUC4/Y-triggered malignancy-related positive feedback loops, and down-regulating transcription of the cascading downstream effectors in pancreatic cancer. These mean that in order to defeat the refractory and drug-resistant pancreatic cancer with MUC4 expression, some pathways can be explored besides repressing MUC4 transcription [45, 46], as follows: 1) To improve reversed effects of domain-lacking. 2) To break the synergism of domains of MUC4. 3) To disrupt MUC4/EGF-ERBB2-ERBB3 signaling hub. 4) Above three remedy combined chemotherapy.
Additionally, in this paper, the results of sequence-based functional annotation for global mRNA analysis also are in agreement with the results of QPCR validation, and are helpful for illustrating the universality and individuality of the role of these three unique domains. For example, the enrichment analyses of GO function [47] and the KEGG pathway [48] show that the common function of NIDO(N), or AMOP(A), or vWD(V), or the synergism of NIDO, AMOP and vWD domain(NAV) were enriched in the feature set “cell projection”, consistently with the structural features of MUC4 and MUC4/Y, i.e., MUC4 or MUC4/Y is anchored to the cell surface by hydrophobic transmembrane region, which locates carboxyl terminal of NIDO, AMOP and vWD domains, so the three domains protrude from the cell surface. The common characters of A, V, NAV were enriched in two function-sets “membrane” and “signaling”, suggesting that A, V, NAV have the more significant role on MUC4/Y-mediate membrane-related molecules expression and signal activation and transmission, which contribute to MUC4/Y-triggered malignancy-related positive feedback loops. The common characters of A, V were enriched in two function-sets “extracellular region” and “neuron projection”, suggesting that both of the domains play more prominent roles in cell-cell interaction, adhesion to the extracellular matrix, and specialized features of neuron. Furthermore, systematic comparative analysis revealed that the two enriched functional categories of MUC4/Y (extracellular region, extracellular region part) were the most highly correlated to AMOP domain.
Interestingly, GO function and the KEGG pathway show that the function on “nervous system generation, development, differentiation” and “regulation of multicellular organismal development” and “effect on Immune system”was enriched or triggered only by vWD domain, which is consistent with systematic comparative analysis results that the seven enriched functional categories of MUC4/Y (cell projection, neuron projection, membrane, membrane part, intrinsic to membrane, chemokine signaling pathway, cytokine-cytokine receptor interaction) were the most highly correlated to vWD domain. Altogether, these individuality of vWD domain suggests MUC4-vWD domain may be involved in various biological function owing to its structure characteristics, i.e., vWD carry the putative GDPH cleavage site in its N-terminal region [23] and adjacent to MUC4-EGF domains with the the nearest distance [3, 23, 31], suggesting vWD domain may be the most potential target for blocking MUC4-triggered malignancy-related regulatory circuitry. Thus, further studies on vWD domain is ongoing in our laboratory.
MATERIALS AND METHODS
Establishment and identification of series of PANC-1 cell strains expressing MUC4/Y with domain-lacking stablely
The PANC-1 pancreatic cancer cell line was obtained from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. As described earlier [16], we estalished series of PANC-1 cell strains expressing MUC4/Y with domain-lacking stably, major workflow as follows: 1) Series of cDNA fragments encoding the MUC4/Y gene with domain-lacking were designed (Additional File 1), including the lack of NIDO, or AMOP, or vWD, or the simultaneous lack of NIDO, AMOP and vWD domain, named MUC4/Y-NIDO△, MUC4/Y-AMOP△, MUC4/Y-vWD△, MUC4/Y-NIDO△-AMOP△-vWD△ correspondingly. A Kozak sequence (GCCACC) before the ATG initiation codon for optimal translation and with unique restriction sites present in the multiple clone site (MCS) of the lentiviral vector but absent from the target cDNA sequence was contained, respectively. These target sequences were synthesized and cloned in a pUC57vector (GenScript). 2) The series of target cDNA sequences were subcloned into the lentiviral vector pCDH-CMV-MCS-EF1-Puro (Cat.#CD510B-1, System Biosciences, USA) respectively, and lentiviral supernatant were produced by 293T cells which were transiently transfected with pCDH-CMV-MCS-EF1-Puro/ target-gene and the pPACKH1 Lentivector Packaging Kit (Cat. #LV500A-1, System Biosciences, USA) with Lipofectamine 2000 (Invitrogen LifeTechnology) according to the manufacturer’s instructions. 3) PANC-1 cells were transfected by series of target genes by carrying out at 20 multiplicity of infection with the lentivirus and using polybrene (8 μg/mL; Sigma-Aldrich) to augment infection efficiency. Stable clones were then selected in medium containing puromycin (2 μg/mL; Sigma-Aldrich). 4) As described previously [16], stable transfected PANC-1 cells overexpressing the MUC4/Y gene were designated PANC-1-MUC4/Y(abbr. Y). Correspondingly, stable transfected PANC-1 cells overexpressing the series of MUC4/Y with domain-lacking genes AMOP were designated PANC-1- MUC4/Y-NIDO△(abbr. N△), PANC-1- MUC4/Y-AMOP△(abbr. A△), PANC-1- MUC4/Y-vWD△(abbr. V△), PANC-1- MUC4/Y-NIDO△-AMOP△-vWD△(abbr. NAV△), respectively.
To verify these stable clones and identify the characteristic of expression and localization of target genes, a variety of detection methods were used, including immunofluorescence (IF), flow cytometry (FCM), quantitative real-time PCR (QPCR), and western blotting (WB).
We used anti-MUC4 mouse monoclonal antibody (#ab60720, Abcam, Cambridge, UK) to detect specific protein expression because this specific antibody is against N-terminal amino acids 79-189 of Human MUC4 (http://www.abcam.com/muc4-antibody-ab60720.html), which is existing consistently in the sequence of MUC4/Y, MUC4/Y-NIDO△, MUC4/Y-AMOP△, MUC4/Y-vWD△, MUC4/Y-NIDO△-AMOP△-vWD△, respectively. Detailed methods of IF, FCM and WB have been described earlier [16, 18, 44].
As described previously [16], the specific primers (forward: 5′-TGGGTGTCCCTGAGCTGC-3′, reverse: 5′-TGATGTGGCTGTGCGTCTC-3′) and TaqMan probe (5′-ATGTGGTCCCAGGAATGACAACACCGT-3′) designed for MUC4/Y, also lie in target Sequences of MUC4/Y-NIDO△, MUC4/Y-AMOP△, MUC4/Y-vWD△, and MUC4/Y-NIDO△-AMOP△-vWD△, so the specific primers and TaqMan probe were used to detect the expression of MUC4/Y with domain-lacking genes in mRNA level. Each quantification PCR was performed in triplicate and repeated thrice independently.
Proliferation assays and Edu retention assays
A Cell Counting Kit-8 (#C0038; Beyotime) cell proliferation assay was performed according to the manufacturer’s instructions. Cells were grown in low-serum (1% FBS) medium as described earlier [16]. Cell growth rate = point-in-time of the absorbance at 450 nm(A450) / Mean of A450 in 24h. Edu retention assays were performed to examine DNA replication. Dissociated cells were exposed to 25 μM of 5-ethynyl-2′-deoxyuridine (Edu, RiboBio, Guangzhou, China) for 2 hr at 37°C, and then the cells were fixed in 4% paraformal-dehyde. After permeabilization with 0.5% Triton-X, the cells were reacted with 1× Apollo reaction cocktail (RiboBio) for 30 min. Subsequently, the DNA contents of the cells were stained with Hoechst 33342 for 30 min and visualized under a fluorescence microscope. The experiments were repeated thrice independently.
Cell cycle analysis
Cells were treated with trypsin at 72 hours after incubation with low-serum (1% FBS) medium and fixed in 70% ethanol for 2 hours at 4°C. After being washed twice with phosphate-buffered saline, the cells were incubated with 0.1 mg/mL RNase A (Sigma-Aldrich Co., St Louis, MO, USA) at 37°C for 30 minutes. The cells were then resuspended in 0.05 mg/mL of propidium iodide (Keygen Biotech, Nanjing, People’s Republic of China) at 4°C for 30 minutes while being protected from light. Finally, the processed cells were analyzed in a FACSort flow cytometer (BD Biosciences, San Jose, CA, USA). Evaluation of the data was performed by CellQuest software (BD Biosciences). The experiments were repeated thrice independently.
Apoptosis assay
Following 48-h treatment with low-serum (1% FBS) medium, PANC-1–derived clones were collected in PBS for apoptosis assay and flow cytometric analysis as described previously [16]. The experiments were repeated thrice independently.
In vitro migration and invasion assays
We used modified 24-well Boyden chambers for the cell migration and invasion assays as described earlier [16]. For the in vitro wound-healing assay, a cell-free area of the culture medium was wounded by scratching with a 200-μL pipette tip. Cell migration into the wound area was monitored in serum-free medium and photographed under a fluorescence microscope at 0 and 24 h. The experiments were repeated thrice independently.
Sequence-based digital gene expression analysis, DEG Gene Ontology functional enrichment and pathway enrichment analysis
For the five groups of cells: PANC-1-MUC4/Y, PANC-1-MUC4/Y-N△, PANC-1-MUC4/Y-A△, PANC-1-MUC4/Y-V△, PANC-1-MUC4/Y-NAV△, same protocol was carried out, as described previously [16], mainly including extracting total RNA from different groups of cells, confirming RNA integrity, transcriptome analysis, Illumina sequencing, screening of differentially expressed genes (DEGs), and functional annotation through the in-house bioinformatics analysis pipeline. DEGs annotated against the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were enriched to identify significant GO biological process terms and pathways, respectively, and adjusted with corrected P ≤ 0.05 for GO and pathways analysis.
Validation of altered gene expression of different groups by QPCR
Altered gene expression of the lacking- domains groups vs. MUC4/Y-overexpression group were validated by Quantitative real-time PCR (QPCR), which was performed using standard procedures using a SYBR Premix Ex Taq Kit (TaKaRa, China) with specific primers as described previously [16]. cDNA was generated using an iScript cDNA Synthesis Kit (Bio-Rad). Ct values were normalized to the 18S gene and a relative quantitative method (ΔΔCt) was used to evaluate quantitative variation. The relative expression level (defined as the fold change) of target genes were calculated to the relative expression detected in the corresponding control cells, which was defined as 1.0.
Validation of altered expression of crucial factors involved in signaling hub in different groups by WB
Altered expression of crucial factors involved in signaling hub in different groups were validated by western blotting (WB), which was performed using standard procedures as described previously [16]. Briefly, cell lysates were prepared as described previously [49]. After the concentrations were determined using the Bradford assay, proteins (30 μg/lane) were resolved on 4-20% Mini-PROTEAN TGX precast gels (#456-1093; Bio-Rad). The resolved proteins were transferred onto polyvinylidene difluoride membranes, blocked with 5% non-fat milk in phosphate-buffered saline (PBS) for 2 h, and immunoblotted with a primary antibody. After incubation with a secondary antibody, blots were visualized by enhanced chemiluminescence (Millipore, Billerica, MA). GADPH was used as the loading control.
Statistical analysis
Statistical analysis was performed using SPSS 20.0 (IBM SPSS Inc.). The results were confirmed by conducting at least three independent experiments in the present study. All data presented are the mean ± standard deviation (SD) of n independent measurements unless noted otherwise. Statistical analysis was performed with one-way ANOVA for multiple groups and the unpaired Student t-test for individual groups. P<0.05 was considered statistically significant.
SUPPLEMENTARY DATA
Footnotes
CONFLICTS OF INTEREST
All authors have reviewed the final version of the manuscript and approved it for publication. The authors have no conflicts of interest to declare.
GRANT SUPPORT
This work was supported by grants from the National Natural Science Foundation of China (81272712, 81572337 and 81672471), Talents planning of six summit fields of Jiangsu Province (WSW-032), the Innovation Capability Development Project of Jiangsu Province (BM2015004), the Research Innovation Program for College Graduates of Jiangsu Province (KYLX15_0954) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801).
REFERENCES
- 1.Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nature reviews Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 2.Porchet N, Nguyen VC, Dufosse J, Audie JP, Guyonnet-Duperat V, Gross MS, Denis C, Degand P, Bernheim A, Aubert JP. Molecular cloning and chromosomal localization of a novel human tracheo-bronchial mucin cDNA containing tandemly repeated sequences of 48 base pairs. Biochemical and biophysical research communications. 1991;175:414–422. doi: 10.1016/0006-291x(91)91580-6. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi P, Singh AP, Batra SK. Structure, evolution, and biology of the MUC4 mucin. FASEB journal. 2008;22:966–981. doi: 10.1096/fj.07-9673rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bafna S, Singh AP, Moniaux N, Eudy JD, Meza JL, Batra SK. MUC4, a multifunctional transmembrane glycoprotein, induces oncogenic transformation of NIH3T3 mouse fibroblast cells. Cancer research. 2008;68:9231–9238. doi: 10.1158/0008-5472.CAN-08-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh AP, Chaturvedi P, Batra SK. Emerging roles of MUC4 in cancer: a novel target for diagnosis and therapy. Cancer research. 2007;67:433–436. doi: 10.1158/0008-5472.CAN-06-3114. [DOI] [PubMed] [Google Scholar]
- 6.Andrianifahanana M, Moniaux N, Schmied BM, Ringel J, Friess H, Hollingsworth MA, Buchler MW, Aubert JP, Batra SK. Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clinical cancer research. 2001;7:4033–4040. [PubMed] [Google Scholar]
- 7.Swartz MJ, Batra SK, Varshney GC, Hollingsworth MA, Yeo CJ, Cameron JL, Wilentz RE, Hruban RH, Argani P. MUC4 expression increases progressively in pancreatic intraepithelial neoplasia. American journal of clinical pathology. 2002;117:791–796. doi: 10.1309/7Y7N-M1WM-R0YK-M2VA. [DOI] [PubMed] [Google Scholar]
- 8.Jhala N, Jhala D, Vickers SM, Eltoum I, Batra SK, Manne U, Eloubeidi M, Jones JJ, Grizzle WE. Biomarkers in Diagnosis of pancreatic carcinoma in fine-needle aspirates. American journal of clinical pathology. 2006;126:572–579. doi: 10.1309/cev30be088cbdqd9. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Zhang JJ, Zhu R, Zhu Y, Liang WB, Gao WT, Yu JB, Xu ZK, Miao Y. The increase in the expression and hypomethylation of MUC4 gene with the progression of pancreatic ductal adenocarcinoma. Medical oncology. 2011;28:S175–184. doi: 10.1007/s12032-010-9683-0. [DOI] [PubMed] [Google Scholar]
- 10.Saitou M, Goto M, Horinouchi M, Tamada S, Nagata K, Hamada T, Osako M, Takao S, Batra SK, Aikou T, Imai K, Yonezawa S. MUC4 expression is a novel prognostic factor in patients with invasive ductal carcinoma of the pancreas. Journal of clinical pathology. 2005;58:845–852. doi: 10.1136/jcp.2004.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh AP, Moniaux N, Chauhan SC, Meza JL, Batra SK. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer research. 2004;64:622–630. doi: 10.1158/0008-5472.can-03-2636. [DOI] [PubMed] [Google Scholar]
- 12.Zhi X, Tao J, Xie K, Zhu Y, Li Z, Tang J, Wang W, Xu H, Zhang J, Xu Z. MUC4-induced nuclear translocation of beta-catenin: a novel mechanism for growth, metastasis and angiogenesis in pancreatic cancer. Cancer letters. 2014;346:104–113. doi: 10.1016/j.canlet.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 13.Senapati S, Chaturvedi P, Chaney WG, Chakraborty S, Gnanapragassam VS, Sasson AR, Batra SK. Novel INTeraction of MUC4 and galectin: potential pathobiological implications for metastasis in lethal pancreatic cancer. Clinical cancer research. 2011;17:267–274. doi: 10.1158/1078-0432.CCR-10-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie K, Zhi X, Tang J, Zhu Y, Zhang J, Li Z, Tao J, Xu Z. Upregulation of the splice variant MUC4/Y in the pancreatic cancer cell line MIA PaCa-2 potentiates proliferation and suppresses apoptosis: new insight into the presence of the transcript variant of MUC4. Oncology reports. 2014;31:2187–2194. doi: 10.3892/or.2014.3113. [DOI] [PubMed] [Google Scholar]
- 15.Rachagani S, Macha MA, Ponnusamy MP, Haridas D, Kaur S, Jain M, Batra SK. MUC4 potentiates invasion and metastasis of pancreatic cancer cells through stabilization of fibroblast growth factor receptor 1. Carcinogenesis. 2012;33:1953–1964. doi: 10.1093/carcin/bgs225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Zhang JJ, Xie KL, Tang J, Liang WB, Zhu R, Zhu Y, Wang B, Tao JQ, Zhi XF, Li Z, Gao WT, Jiang KR, Miao Y, Xu ZK. Specific-detection of clinical samples, systematic functional investigations, and transcriptome analysis reveals that splice variant MUC4/Y contributes to the malignant progression of pancreatic cancer by triggering malignancy-related positive feedback loops signaling. Journal of translational medicine. 2014;12:309. doi: 10.1186/s12967-014-0309-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Zhi X, Zhu Y, Zhang Q, Wang W, Li Z, Tang J, Wang J, Wei S, Li B, Zhou J, Jiang J, Yang L, et al. MUC4-promoted neural invasion is mediated by the axon guidance factor Netrin-1 in PDAC. Oncotarget. 2015;6:33805–33822. doi: 10.18632/oncotarget.5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Zhang JJ, Liang WB, Zhu R, Wang B, Miao Y, Xu ZK. Pancreatic cancer counterattack: MUC4 mediates Fas-independent apoptosis of antigen-specific cytotoxic T lymphocyte. Oncology reports. 2014;31:1768–1776. doi: 10.3892/or.2014.3016. [DOI] [PubMed] [Google Scholar]
- 19.Bafna S, Kaur S, Momi N, Batra SK. Pancreatic cancer cells resistance to gemcitabine: the role of MUC4 mucin. British journal of cancer. 2009;101:1155–1161. doi: 10.1038/sj.bjc.6605285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skrypek N, Duchene B, Hebbar M, Leteurtre E, van Seuningen I, Jonckheere N. The MUC4 mucin mediates gemcitabine resistance of human pancreatic cancer cells via the Concentrative Nucleoside Transporter family. Oncogene. 2013;32:1714–1723. doi: 10.1038/onc.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimeault M, Johansson SL, Senapati S, Momi N, Chakraborty S, Batra SK. MUC4 down-regulation reverses chemoresistance of pancreatic cancer stem/progenitor cells and their progenies. Cancer letters. 2010;295:69–84. doi: 10.1016/j.canlet.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross MS, Guyonnet-Duperat V, Porchet N, Bernheim A, Aubert JP, Nguyen VC. Mucin 4 (MUC4) gene: regional assignment (3q29) and RFLP analysis. Annales de genetique. 1992;35:21–26. [PubMed] [Google Scholar]
- 23.Moniaux N, Nollet S, Porchet N, Degand P, Laine A, Aubert JP. Complete sequence of the human mucin MUC4: a putative cell membrane-associated mucin. The Biochemical journal. 1999;338:325–333. [PMC free article] [PubMed] [Google Scholar]
- 24.Khorrami AM, Choudhury A, Andrianifahanana M, Varshney GC, Bhattacharyya SN, Hollingsworth MA, Kaufman B, Batra SK. Purification and characterization of a human pancreatic adenocarcinoma mucin. Journal of biochemistry. 2002;131:21–29. doi: 10.1093/oxfordjournals.jbchem.a003073. [DOI] [PubMed] [Google Scholar]
- 25.Escande F, Lemaitre L, Moniaux N, Batra SK, Aubert JP, Buisine MP. Genomic organization of MUC4 mucin gene. Towards the characterization of splice variants. European journal of biochemistry / FEBS. 2002;269:3637–3644. doi: 10.1046/j.1432-1033.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 26.Carraway KL, Price-Schiavi SA, Komatsu M, Idris N, Perez A, Li P, Jepson S, Zhu X, Carvajal ME, Carraway CA. Multiple facets of sialomucin complex/MUC4, a membrane mucin and erbb2 ligand, in tumors and tissues (Y2K update) Frontiers in bioscience. 2000;5:D95–D107. doi: 10.2741/carraway. [DOI] [PubMed] [Google Scholar]
- 27.Arango ME, Li P, Komatsu M, Montes C, Carraway CA, Carraway KL. Production and localization of Muc4/sialomucin complex and its receptor tyrosine kinase ErbB2 in the rat lacrimal gland. Investigative ophthalmology & visual science. 2001;42:2749–2756. [PubMed] [Google Scholar]
- 28.Komatsu M, Jepson S, Arango ME, Carothers Carraway CA, Carraway KL. Muc4/sialomucin complex, an intramembrane modulator of ErbB2/HER2/Neu, potentiates primary tumor growth and suppresses apoptosis in a xenotransplanted tumor. Oncogene. 2001;20:461–470. doi: 10.1038/sj.onc.1204106. [DOI] [PubMed] [Google Scholar]
- 29.Kozloski GA, Carraway CA, Carraway KL. Mechanistic and signaling analysis of Muc4-ErbB2 signaling module: new insights into the mechanism of ligand-independent ErbB2 activity. Journal of cellular physiology. 2010;224:649–657. doi: 10.1002/jcp.22163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakshmanan I, Seshacharyulu P, Haridas D, Rachagani S, Gupta S, Joshi S, Guda C, Yan Y, Jain M, Ganti AK, Ponnusamy MP, Batra SK. Novel HER3/MUC4 oncogenic signaling aggravates the tumorigenic phenotypes of pancreatic cancer cells. Oncotarget. 2015;6:21085–21099. doi: 10.18632/oncotarget.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desseyn JL, Tetaert D, Gouyer V. Architecture of the large membrane-bound mucins. Gene. 2008;410:215–222. doi: 10.1016/j.gene.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Yamada N, Nishida Y, Tsutsumida H, Goto M, Higashi M, Nomoto M, Yonezawa S. Promoter CpG methylation in cancer cells contributes to the regulation of MUC4. British journal of cancer. 2009;100:344–351. doi: 10.1038/sj.bjc.6604845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choudhury A, Moniaux N, Winpenny JP, Hollingsworth MA, Aubert JP, Batra SK. Human MUC4 mucin cDNA and its variants in pancreatic carcinoma. Journal of biochemistry. 2000;128:233–243. doi: 10.1093/oxfordjournals.jbchem.a022746. [DOI] [PubMed] [Google Scholar]
- 34.Mansour M, Haupt S, Chan AL, Godde N, Rizzitelli A, Loi S, Caramia F, Deb S, Takano EA, Bishton M, Johnstone CN, Monahan B, Levav-Cohen Y, Jiang YH, Yap AS, Fox SB, et al. E3 ligase E6AP represses breast cancer metastasis via regulation of ECT2-Rho signalling. Cancer research. 2016 doi: 10.1158/0008-5472.CAN-15-1553. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Yang SH, Li CF, Chu PY, Ko HH, Chen LT, Chen WW, Han CH, Lung JH, Shih NY. Overexpression of regulator of G protein signaling 11 promotes cell migration and associates with advanced stages and aggressiveness of lung adenocarcinoma. Oncotarget. 2016;7:31122–31136. doi: 10.18632/oncotarget.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duraisamy S, Ramasamy S, Kharbanda S, Kufe D. Distinct evolution of the human carcinoma-associated transmembrane mucins, MUC1, MUC4 AND MUC16. Gene. 2006;373:28–34. doi: 10.1016/j.gene.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Timpl R. Structure and biological activity of basement membrane proteins. European journal of biochemistry / FEBS. 1989;180:487–502. doi: 10.1111/j.1432-1033.1989.tb14673.x. [DOI] [PubMed] [Google Scholar]
- 38.Mayer U, Kohfeldt E, Timpl R. Structural and genetic analysis of laminin-nidogen interaction. Annals of the New York Academy of Sciences. 1998;857:130–142. doi: 10.1111/j.1749-6632.1998.tb10113.x. [DOI] [PubMed] [Google Scholar]
- 39.Ciccarelli FD, Doerks T, Bork P. AMOP, a protein module alternatively spliced in cancer cells. Trends in biochemical sciences. 2002;27:113–115. doi: 10.1016/s0968-0004(01)02049-7. [DOI] [PubMed] [Google Scholar]
- 40.Colombatti A, Bonaldo P, Doliana R. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix. 1993;13:297–306. doi: 10.1016/s0934-8832(11)80025-9. [DOI] [PubMed] [Google Scholar]
- 41.Ruggeri ZM, Ware J. von Willebrand factor. FASEB journal. 1993;7:308–316. doi: 10.1096/fasebj.7.2.8440408. [DOI] [PubMed] [Google Scholar]
- 42.Aksoy N, Akinci OF. Mucin macromolecules in normal, adenomatous, and carcinomatous colon: evidence for the neotransformation. Macromolecular bioscience. 2004;4:483–496. doi: 10.1002/mabi.200300099. [DOI] [PubMed] [Google Scholar]
- 43.Senapati S, Gnanapragassam VS, Moniaux N, Momi N, Batra SK. Role of MUC4-NIDO domain in the MUC4-mediated metastasis of pancreatic cancer cells. Oncogene. 2012;31:3346–3356. doi: 10.1038/onc.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang J, Zhu Y, Xie K, Zhang X, Zhi X, Wang W, Li Z, Zhang Q, Wang L, Wang J, Xu Z. The role of the AMOP domain in MUC4/Y-promoted tumour angiogenesis and metastasis in pancreatic cancer. Journal of experimental & clinical cancer research : CR. 2016;35:91. doi: 10.1186/s13046-016-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang JJ, Zhu Y, Zhang XF, Liang WB, Xie KL, Tao JQ, Peng YP, Xu ZK, Miao Y. Transcriptional regulation of human MUC4 gene: identification of a novel inhibitory element and its nuclear binding protein. Molecular biology reports. 2013;40:4913–4920. doi: 10.1007/s11033-013-2591-6. [DOI] [PubMed] [Google Scholar]
- 46.Zhang JJ, Zhu Y, Xie KL, Peng YP, Tao JQ, Tang J, Li Z, Xu ZK, Dai CC, Qian ZY, Jiang KR, Wu JL, Gao WT, Du Q, Miao Y. Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Molecular cancer. 2014;13:130. doi: 10.1186/1476-4598-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic acids research. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma C, Zhang J, Durrin LK, Lv J, Zhu D, Han X, Sun Y. The BCL2 major breakpoint region (mbr) regulates gene expression. Oncogene. 2007;26:2649–2657. doi: 10.1038/sj.onc.1210069. [DOI] [PubMed] [Google Scholar]
- 50.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, Kim CJ, Kusanovic JP, Romero R. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


