Abstract
Tetraspanin CD151, also known as PETA-3 or SFA-1, has been reported to predict prognosis in various solid tumors. Yet, the results of these studies remained inconclusive. Here, we performed this meta-analysis of relevant studies published on the topic to quantitatively evaluate the clinicopathological significance of CD151 in solid tumors. The relevant articles were identified via searching the PubMed, Web of Science and Embase database. The pooled hazard ratios (HRs) and corresponding 95% confidence intervals (CI) of overall survival (OS) and disease-free survival (DFS) were calculated to evaluate the prognostic value of CD151 expression in patients with solid tumors. A total of 19 studies involving 4, 270 participants were included in the study, we drew the conclusion that CD151 overexpression was associated with statistically significant poor OS (pooled HR = 1.498, 95% CI = 1.346-1.667, P<0.001) and poor DFS (pooled HR = 1.488, 95% CI = 1.314-1.685, P<0.001). Furthermore, the subgroup analysis revealed that the associations between CD151 overexpression and the outcome endpoints (OS or TTP) were significant within the Asian region and European, as well in patients with breast cancer or gastric cancer. Taken together, the incorporative HR showed CD151 overexpression was associated with poor survival in human solid tumors. CD151 could be a valuable prognosis biomarker or a potential therapeutic target of solid tumors.
Keywords: CD151, solid tumors, prognosis, overall survival, disease-free survival
INTRODUCTION
Tetraspanins CD151, a known transmembrane 4 superfamily protein, contains several different structure domains. Expression of CD151 was observed in different cell types [1]. CD151 and other tetraspanins participate in a variety of key cellular functions [1–3]. CD151 is the first characterized member of the tetraspanin family. It is also known as glycoprotein-27 (GP-27), Red blood cell antigen MER 2 (MER 2), platelet-endothelial tetraspan antigen-3 (PETA-3), Raph blood group antigen (RAPH)/Membrane Glycoprotein SFA-1 (SFA-1) and tetraspanin-24 (TSPAN-24), was located on chromosome 11p15.5 [4, 5]. Its expression was found in a number of cell types [6]. Altered expression of CD151 was involved in cancer growth [7], progression [8], motility, invasion, and metastasis [9]. Existing evidences have demonstrated that CD151 overexpression was observed in several malignancies [10–23]. These striking evidences on the role of CD151 in cancer suggest the tetraspanin would be further considered as a novel potential oncotarget of cancer patients.
An increasing number of studies showed that increased CD151 expression in tumor tissue was associated with cancer patients’ poor survival [10–13, 15, 17–27]. However, in invasive lobular breast cancer [15] and endometrial cancer [16], overexpression of CD151 might have diverse, even opposing roles, correlating with better clinical outcomes. The results of those individual studies were inconsistent. Thus, this comprehensive meta-analysis was performed to clarify the possible prognostic role of CD151 expression in solid tumors.
RESULTS
Demographic characteristics
125 articles were retrieved from the different databases (PubMed, Embase, and Web of Science). As showed in the search flow diagram (Figure 1), 125 records were initially retrieved through the pre-defined search strategy. Because of repeated data, 65 records were removed. After browsing the retrieved titles and abstracts, 39 records were excluded due to no relevant endpoint provided. The remaining 26 records were downloaded as full-text and accessed very carefully. Among them, another 18 studies were excluded, including 1 study that was experimental study, 7 studies that were without prognosis data. After selection, 18 published studies including 4, 270 patients were finally selected for this meta-analysis. The median sample-size was 145, with a wide range from 30 to 886. Among all cohorts, Asian (n = 14) became the major source region of literatures, followed by Austria (n = 1), Poland (n = 2), USA(n = 1), UK (n = 1). Newcastle-Ottawa Scale (NOS) were applied to analyze these studies. The quality scores of these studies ranged from 6 to 9, indicating that the methodological quality was high.
Figure 1. The flow chart of the selection process in our meta-analysis.
As for the cancer type, two studies evaluated non-small cell lung cancer; Two studies focused on glioblastoma; Three studies evaluated breast cancer; One study evaluated gallbladder carcinoma; Three studies evaluated gastric carcinomas; One study evaluated CCRCC or clear cell renal cell carcinoma; One study evaluated esophageal squamous cell carcinoma; One study evaluated pancreatic cancer; Two studies evaluated liver cancer; One study evaluated prostate cancer; One study evaluated colon cancer, and one study evaluated endometrial cancer. Of all the studies, 19 of them focused on OS, other 10 studies focused on DFS.
Evidence synthesis
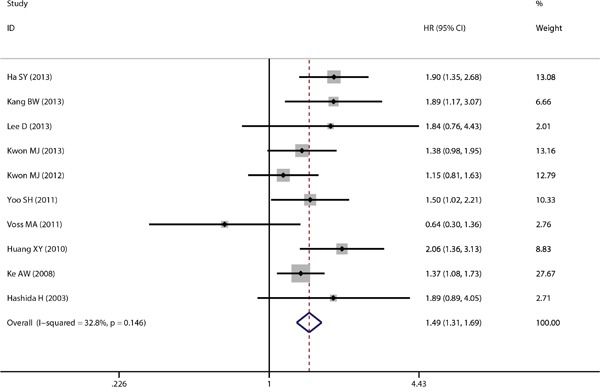
As described, this meta-analysis was based on two outcome endpoints: OS and DFS. 19 studies were included in the current meta-analysis of OS. A fixed-effects model was utilized to calculate the pooled hazard ratio (HR) along with the 95% confidence interval (CI). Heterogeneity test reported a P value of 0.054 and an I2 value of 36.9%. The results indicated that CD151 overexpression was significantly associated with cancer patients’ poor OS (pooled HR =1.498, 95% CI = 1.346-1.667, P<0.001) (Figure 2). 10 studies were included in this current meta-analysis of DFS. Due to the fact that the heterogeneity test reported a P value of 0.146 and an I2 value of 32.8%, a fixed-effects model was again applied. The results demonstrated again a significant association between CD151 expression and DFS (pooled HR = 1.488, 95% CI = 1.314-1.685, P<0.001) (Figure 3). Subgroup study was then performed, the results indicated that the associations between CD151 overexpression and patients’ poor OS and patients poor DFS were also significant in Mongoloid patients (OS: pooled HR =1.511, 95% CI =1.348-1.693, P<0.001; DFS: pooled HR =1.524, 95% CI =1.343-1.729, P<0.001), as well in Caucasian (OS: pooled HR =1.400, 95% CI 1.018–1.924, P=0.038). The significant association was also detected between CD151 overexpression and poor OS in patients with breast cancer (OS: pooled HR = 1.399, 95% CI =1.022-1.915, P=0.036) and gastric cancer (OS: pooled HR = 1.498, 95% CI =1.188-1.890, P=0.001).
Figure 2. The correlation between CD151 expression and overall survival in solid tumor.
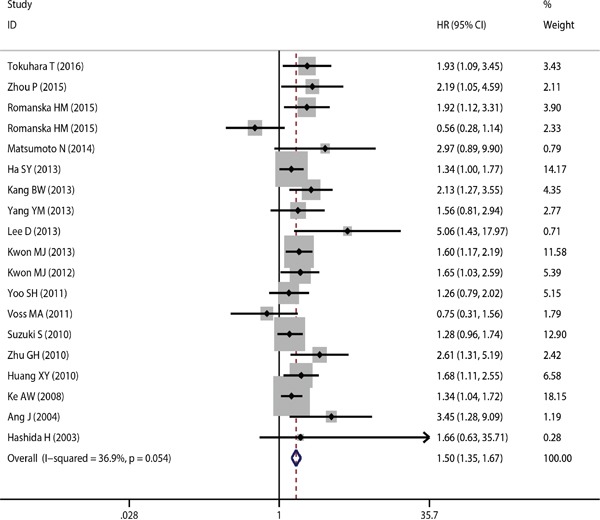
Figure 3. The correlation between CD151 expression and disease-free survival in solid tumor.
Analysis of possible publication bias and sensitivity analysis
In this study, we utilized the Begg's funnel plot as well as the Egger's test to evaluate the possible publication bias. As demonstrated, the funnel plots shapes for the OS and DFS had no obvious heterogeneity (Figure 4), and Egger's tests revealed only slight publication bias concerning OS (P=0.097), but not DFS (P=0.839). Therefore, we performed trim and fill method to make pooled HR more reliable, and the pooled HR P value was also less than 0.01 (Figure not shown). Next, we performed the sensitivity analysis to determine the robustness of the above results. Not a single study was identified to dominate the current meta-analysis, and deletion of each single study had no significant impact on the general conclusions (Figure 5).
Figure 4. Begg's funnel plots for the studies involved in the meta-analysis of CD151 expression and the prognosis of patients with solid tumors.
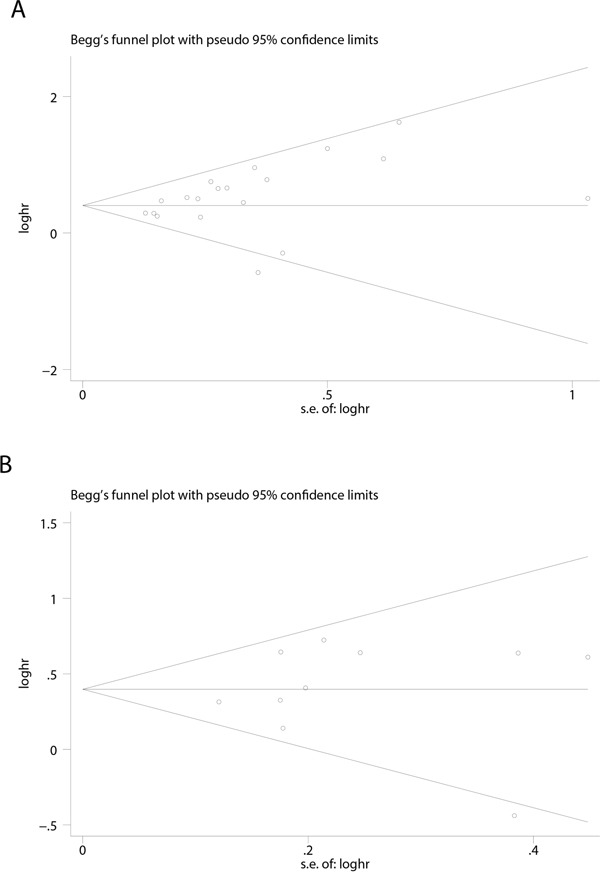
A. Overall survival. B. disease-free survival(DFS). Abbreviations: loghr, logarithm of hazard ratios; s.e., standard error.
Figure 5. Sensitivity analysis of the meta-analysis.
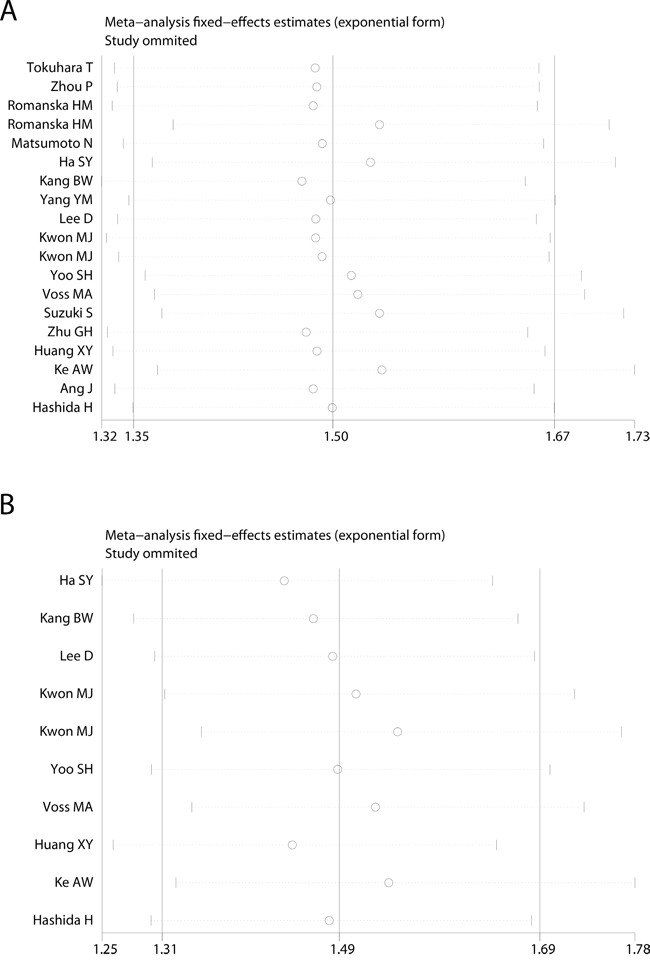
A. Overall survival(OS). B. disease-free survival(DFS).
DISCUSSION
Tetraspanin CD151 is composed of various structure domains [4]. Site-directed mutagenesis studies have revealed that CD151 could be palmitoylated at multiple sites, which is vital for the assemble of tumor endothelial marker (TEM) network [28, 29]. For instance, CD151 interacts with α3β1 and α6β4 integrins via intracellular N-terminal and C-terminal cysteine palmitoylation [28]. Studies have demonstrated that integrin subunits α3 and α6 may directly associate with CD151 at QRD194-196 site [30].
As the first identified member of the tetraspanin family, CD151 was shown to participate in tumor cell behaviors, including cell growth, proliferation, motility and invasiveness, possibly via interacting with a number of different proteins. CD151 silence could decrease above cancer cell behaviors [31–36]. Existing evidences have also shown that CD151 expression is also important for tumor neoangiogenesis and epithelial-tomesenchymal transition [35, 37].
In addition, clinical studies had investigated the potential prognostic value of CD151. Most of these studies, however, include only limited number of patients, and the results are inconclusive. CD151 overexpression often predicts unfavorable outcome in many cancer, such as gastric [11, 14, 24], prostate [10] and non-small cell lung cancer [18, 27]. On the other hand, it is a favorable prognostic indicator in invasive lobular breast cancer [15] and endometrial cancer [16]. To our knowledge, the present study is the first and most full-scale meta-analysis systemically exploring the possible prognostic role of CD151 up-regulation in solid malignancies.
We systematically evaluated survival data of 4, 270 solid tumor patients included in 19 different studies. Overall, these results clearly show that high CD151 expression could be a poor prognostic factor of various solid tumors, with both results of poor OS (pooled HR =1.498, 95% CI = 1.346-1.667, P<0.001) and poor DFS (pooled HR = 1.488, 95% CI = 1.314-1.685, P<0.001). Similarly, further subgroup analysis demonstrated the associations between CD151 overexpression and poor OS and DFS were significant within Mongoloid patients, as well poor OS in Caucasian. The results showed that there was no difference OS or DFS HR among different descent populations. When data was stratified according to cancer type, associations between CD151 overexpression and poor OS were also significant in breast cancer and gastric cancer. Together, our quantitative results strongly supported the current mainstream viewpoint that an undesirable impact of CD151 redundancy was correlated with not only the overall survival, but also disease-free survival. Additionally, Several important implications in this meta-analysis were displayed. High CD151 expression is very likely a general cancer's poor prognostic marker. We included a total of twelve different cancer types [10–27]. The pooled results showed that high CD151 expression was associated with patients’ poor OS and DFS. These conclusion could possibly be extended to all the solid tumors. Finally, it foreshadow the potential of CD151 developing a valuable therapeutic target and prognostic biomarker for solid tumor.
In conclusion, the significant association between CD151 over-expression and cancer patients’ poor survival was clearly demonstrated in the present meta-analysis. These results indicate that CD151 could be a potential prognostic biomarker and a promising therapeutic oncotarget for solid tumors.
MATERIALS AND METHODS
Publication search
This present meta-analysis was performed with the guidelines from Preferred Reporting Items for Systematics Reviews and Meta-Analyses [38]. Databases including PubMed, EMBASE and Web of Science were searched from their incipiency to June, 2016 using the search terms: ‘CD151’, ‘Tetraspanin’ and ‘cancer or tumor or neoplasm or carcinoma’ and ‘prognosis or prognostic or survival or predict or outcome or alive’ and the following limits: Human, article in English. All potentially eligible studies were retrieved. The bibliographies in these studies were also carefully scanned to identify other possible eligible studies and extra studies. At a situation when multiple studies of the same patient population were identified, the published report with the largest sample size was included.
Inclusion criteria
To be eligible for selection of this meta-analysis, studies: (a) should test CD151 expression for prognostic value in cancer; (b) CD151 expression was tested by immunohistochemistry (IHC) or quantitative real-time polymerase chain reaction (qRT-PCR); (c) should have hazard ratios (HRs) with 95% confidence intervals (CIs), or should enable calculation of these statistics from the data presented; (d) should classify CD151 expression as “high” and “low” or “positive” and “negative”; (e) should be written in English.
Exclusion criteria
Exclusion criteria were: (a) literatures such as letters, editorials, abstracts, reviews, case reports and expert opinions were excluded; (b) experiments were performed in vitro or in vivo, but were not associated with patients; (c) articles showing on HRs with 95% CI of OS, or the Kaplan-Meier curves which had inadequate survival data for further analysis; (d) The duration of the follow-up study was less than three years.
Data extraction
Two outcome endpoints were analyzed: overall survival (OS) and disease-free survival (DFS). Parameters of these literatures were extracted from each single paper, including the first author's surname, publication year, country origin, number of total patients, antibodies utilized, types of measurements, score for CD151 assessment, CD151's cut-off values, as well as OS and DFS. The main features of these selected studies were summarized in Table 1.
Table 1. Characteristics of studies included in the meta-analysis.
| author | year | country | case | disease | antibody | method | cutoff value | outcome | NOS |
|---|---|---|---|---|---|---|---|---|---|
| endpoints | score | ||||||||
| Tokuhara T [27] | 2016 | Japan | 145 | non-small cell lung cancer | anti-CD151 monoclonal antibody SFA1.2B4 | ICH | >50% of tumor cells stained | OS | 7 |
| Zhou P [26] | 2015 | USA | 88 | glioblastoma | anti-CD151 monoclonal antibodies (5C11) | ICH | ≥15%cells positive | OS | 7 |
| Romanska HM(a) [15] | 2015 | Poland | 182 | invasive ductal breast carcinoma | mouse anti-human; 1 : 100; Novocastra, Newcastle upon Tyne, UK | ICH | scores of 2+ and 3+ | OS | 8 |
| Romanska HM(b) [15] | 2015 | Poland | 117 | invasive lobular breast cancer | mouse anti-human; 1 : 101; Novocastra, Newcastle upon Tyne, UK | ICH | scores of 2+ and 3+ | OS | 8 |
| Matsumoto N [20] | 2014 | Japan | 45 | gallbladder carcinoma | mouse monoclonal antibody against CD151 (ab33315; Abcam Inc, Cambridge, UK) | ICH | H score≥100 (range of 0-900) | OS | 7 |
| Ha SY [11] | 2013 | Korea | 491 | gastric carcinomas | mouse monoclonal anti-CD151 (NCL-CD151, 1:300 dilutio, Novocastra, Newcastle upon Tyne, UK) | ICH | score of 2+or 3+ | OS, DFS | 7 |
| Kang BW [14] | 2013 | Korea | 159 | gastric cancer | mouse monoclonal anti-CD151 (NCL-CD151; 1:300 dilution, Novocastra, Newcastle upon Tyne, UK) | ICH | grade 2 and 3 | OS, DFS | 7 |
| Yang YM [24] | 2013 | China | 76 | gastric cancer | Mouse anti-human CD151(11G5a, 1:200; Serotec, UK) | ICH | >50% of the tumor section | OS | 9 |
| Lee D [19] | 2013 | Korea | 36 | glioblastoma | mouse monoclonal anti-human CD151 (NCL-CD151, 1:300 dilution; Novocastra, Newcastle-upon-Tyne, UK) | ICH | grades 2 and 3 | OS, DFS | 6 |
| Kwon MJ [18] | 2013 | Korea | 380 | non-small cell lung cancer | monoclonal mouse anti-human CD151 antibody(1:100 dilution, RLM30, Novocastra, Newcastle uponTyne, UK) | ICH | scores of 2+ and 3+ | OS, DFS | 8 |
| Kwon MJ [25] | 2012 | Korea | 886 | breast cancer | monoclonal mouse anti-human CD151 antibody(1:100 dilution, RLM31, Novocastra, Newcastle uponTyne, UK) | ICH | score of 3+ | OS, DFS | 8 |
| Yoo SH [22] | 2011 | Korea | 489 | clear cell renal cell carcinoma | Monoclonal mouse anti-human CD151 antibody (1:100, Novocastra, Newcastle upon Tyne, UK) | ICH | diffuse moderate or diffuse strong | OS, DFS | 8 |
| Voss MA [16] | 2011 | UK | 131 | endometrial cancer | Mouse anti-CD151 monoclonal antibody (mAb) (NCL-CD151, 1:50, Novocastra, Newcastle, Upon Tyne, UK) | ICH | H score≥100 (range of 0-300) | OS, DFS | 8 |
| Suzuki S [21] | 2010 | Japan | 138 | esophageal squamous cell carcinoma | mouse anti-CD151 monoclonal antibody (RLM30; 1:50, Novocastra, Newcastle, UK) | ICH | 10% of tumor cells | OS | 6 |
| Zhu GH [23] | 2010 | China | 71 | Pancreatic cancer | mouse anti-human CD151 monoclonal antibody (sc-80715, 1:100 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA) | ICH | 3-7points | OS | 8 |
| Huang XY [13] | 2010 | China | 140 | intrahepatic cholangiocarcinoma | Monoclonal mouse anti-human CD151 (11G5a, 1:199; Serotec, UK) | ICH | >median | OS, DFS | 8 |
| Ke AW [17] | 2008 | China | 520 | hepatocellular carcinoma | Monoclonal mouse anti-human CD151 (11G5a, 1:200; Serotec, UK) | ICH | >50% of tumor section | OS, DFS | 8 |
| Ang J [10] | 2004 | Australia | 30 | prostate cancer | monoclonal mouse anti-human CD151 antibody (11B1, purified immunoglobulin IgG2a, 4 Ag/mL working concentration; ref. 10). | ICH | >the third quartile (17.52) | OS | 8 |
| Hashida H [12] | 2003 | Japan | 146 | colon cancer | anti-CD151 MAb SFA1.2B4 | ICH | score≥120 (range of 0-300) | OS, DFS | 8 |
ICH : Immunohistochemistry; NOS: Newcastle-Ottawa Scale;
There were two parts of data (a and b) in the study Romanska HM [15].
Cancer-specific survival, disease-specific survival and five-year OS were combined into OS. On the other hand, cumulative recurrence and progression-free survival were combined into DFS. OS was expressed as time (in months) from cancer diagnosis to death. To assess prognostic value of CD151 expression, multivariate hazard ratio (HR) was extracted. For the articles in which prognosis was plotted only as the Kaplan-Meier curves, the Engauge Digitizer V4.1 was then applied to extract survival data, and the Tierney's was utilized to calculate the HRs and 95% CIs [39].
Statistical analysis
Pooled HRs and 95% CIs for two outcome endpoints (OS and DFS) were calculated. Statistical heterogeneity was assessed through the Chisquare test and I-square test, which was checked through the Q test, and a P value >0.10 indicated a lack of heterogeneity. We also quantified the effect of heterogeneity via I2 =100%×(Q - df)/Q. I2 values of <25% could be considered “low”, values of about 50% could be considered “moderate”, and values of over 75% could be considered “high” [40]. According to the absence or presence of heterogeneity, random-effects model or fixed-effects model was applied to merge the RR, respectively. Without statistical heterogeneity, a fixed-effects model was employed to calculate the pooled HRs, otherwise the random-effects model was performed [41]. Funnel plots and the Egger's test were utilized to determine the possible publication bias [26]. Sensitivity analysis was also tested. Statistical analyses were performed via the Stata 14.0 (StataCorp, College Station, TX). P values for all comparisons were two-tailed.
Footnotes
CONFLICTS OF INTEREST
The authors have declared that no competing interests exist.
GRANT SUPPORT
This work is supported by the National Natural Science Foundation (81472786, 81472305), The Six Talents Peak Project of Jiangsu Province (2012-WSN-012, 2014-WSW-061).
REFERENCES
- 1.Hemler ME. Tetraspanin functions and associated microdomains. Nature reviews Molecular cell biology. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 2.Saad RS, Liu Y, Han H, Landreneau RJ, Silverman JF. Prognostic significance of HER2/neu, p53, and vascular endothelial growth factor expression in early stage conventional adenocarcinoma and bronchioloalveolar carcinoma of the lung. Modern pathology. 2004;17:1235–1242. doi: 10.1038/modpathol.3800171. [DOI] [PubMed] [Google Scholar]
- 3.Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nature reviews Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 4.Fitter S, Tetaz TJ, Berndt MC, Ashman LK. Molecular cloning of cDNA encoding a novel platelet-endothelial cell tetra-span antigen, PETA-3. Blood. 1995;86:1348–1355. [PubMed] [Google Scholar]
- 5.Hasegawa H, Kishimoto K, Yanagisawa K, Terasaki H, Shimadzu M, Fujita S. Assignment of SFA-1 (PETA-3), a member of the transmembrane 4 superfamily, to human chromosome 11p15.5 by fluorescence in situ hybridization. Genomics. 1997;40:193–196. doi: 10.1006/geno.1996.4563. [DOI] [PubMed] [Google Scholar]
- 6.Geary SM, Cambareri AC, Sincock PM, Fitter S, Ashman LK. Differential tissue expression of epitopes of the tetraspanin CD151 recognised by monoclonal antibodies. Tissue antigens. 2001;58:141–153. doi: 10.1034/j.1399-0039.2001.580301.x. [DOI] [PubMed] [Google Scholar]
- 7.Novitskaya V, Romanska H, Dawoud M, Jones JL, Berditchevski F. Tetraspanin CD151 regulates growth of mammary epithelial cells in three-dimensional extracellular matrix: implication for mammary ductal carcinoma in situ. Cancer research. 2010;70:4698–4708. doi: 10.1158/0008-5472.CAN-09-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nankivell P, Williams H, McConkey C, Webster K, High A, MacLennan K, Senguven B, Rabbitts P, Mehanna H. Tetraspanins CD9 and CD151, epidermal growth factor receptor and cyclooxygenase-2 expression predict malignant progression in oral epithelial dysplasia. British journal of cancer. 2013;109:2864–2874. doi: 10.1038/bjc.2013.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohno M, Hasegawa H, Miyake M, Yamamoto T, Fujita S. CD151 enhances cell motility and metastasis of cancer cells in the presence of focal adhesion kinase. International journal of cancer. 2002;97:336–343. doi: 10.1002/ijc.1605. [DOI] [PubMed] [Google Scholar]
- 10.Ang J, Lijovic M, Ashman LK, Kan K, Frauman AG. CD151 protein expression predicts the clinical outcome of low-grade primary prostate cancer better than histologic grading: a new prognostic indicator? Cancer epidemiology, biomarkers & prevention. 2004;13:1717–1721. [PubMed] [Google Scholar]
- 11.Ha SY, Do IG, Lee J, Park SH, Park JO, Kang WK, Choi MG, Lee JH, Bae JM, Kim S, Kim KM, Sohn TS. CD151 overexpression is associated with poor prognosis in patients with pT3 gastric cancer. Annals of surgical oncology. 2014;21:1099–1106. doi: 10.1245/s10434-013-3339-1. [DOI] [PubMed] [Google Scholar]
- 12.Hashida H, Takabayashi A, Tokuhara T, Hattori N, Taki T, Hasegawa H, Satoh S, Kobayashi N, Yamaoka Y, Miyake M. Clinical significance of transmembrane 4 superfamily in colon cancer. British journal of cancer. 2003;89:158–167. doi: 10.1038/sj.bjc.6601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang XY, Ke AW, Shi GM, Ding ZB, Devbhandari RP, Gu FM, Li QL, Dai Z, Zhou J, Fan J. Overexpression of CD151 as an adverse marker for intrahepatic cholangiocarcinoma patients. Cancer. 2010;116:5440–5451. doi: 10.1002/cncr.25485. [DOI] [PubMed] [Google Scholar]
- 14.Kang BW, Lee D, Chung HY, Han JH, Kim YB. Tetraspanin CD151 expression associated with prognosis for patients with advanced gastric cancer. Journal of cancer research and clinical oncology. 2013;139:1835–1843. doi: 10.1007/s00432-013-1503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romanska HM, Potemski P, Krakowska M, Mieszkowska M, Chaudhri S, Kordek R, Kubiak R, Speirs V, Hanby AM, Sadej R, Berditchevski F. Lack of CD151/integrin alpha3beta1 complex is predictive of poor outcome in node-negative lobular breast carcinoma: opposing roles of CD151 in invasive lobular and ductal breast cancers. British journal of cancer. 2015;113:1350–1357. doi: 10.1038/bjc.2015.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voss MA, Gordon N, Maloney S, Ganesan R, Ludeman L, McCarthy K, Gornall R, Schaller G, Wei W, Berditchevski F, Sundar S. Tetraspanin CD151 is a novel prognostic marker in poor outcome endometrial cancer. British journal of cancer. 2011;104:1611–1618. doi: 10.1038/bjc.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke AW, Shi GM, Zhou J, Wu FZ, Ding ZB, Hu MY, Xu Y, Song ZJ, Wang ZJ, Wu JC, Bai DS, Li JC, Liu KD, Fan J. Role of overexpression of CD151 and/or c-Met in predicting prognosis of hepatocellular carcinoma. Hepatology. 2009;49:491–503. doi: 10.1002/hep.22639. [DOI] [PubMed] [Google Scholar]
- 18.Kwon MJ, Seo J, Kim YJ, Kwon MJ, Choi JY, Kim TE, Lee DH, Park S, Shin YK, Han J, Choi YL. Prognostic significance of CD151 overexpression in non-small cell lung cancer. Lung cancer. 2013;81:109–116. doi: 10.1016/j.lungcan.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Lee D, Suh YL, Park TI, Do IG, Seol HJ, Nam DH, Kim ST. Prognostic significance of tetraspanin CD151 in newly diagnosed glioblastomas. Journal of surgical oncology. 2013;107:646–652. doi: 10.1002/jso.23249. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto N, Morine Y, Utsunomiya T, Imura S, Ikemoto T, Arakawa Y, Iwahashi S, Saito Y, Yamada S, Ishikawa D, Takasu C, Miyake H, Shimada M. Role of CD151 expression in gallbladder carcinoma. Surgery. 2014;156:1212–1217. doi: 10.1016/j.surg.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki S, Miyazaki T, Tanaka N, Sakai M, Sano A, Inose T, Sohda M, Nakajima M, Kato H, Kuwano H. Prognostic significance of CD151 expression in esophageal squamous cell carcinoma with aggressive cell proliferation and invasiveness. Annals of surgical oncology. 2011;18:888–893. doi: 10.1245/s10434-010-1387-3. [DOI] [PubMed] [Google Scholar]
- 22.Yoo SH, Lee K, Chae JY, Moon KC. CD151 expression can predict cancer progression in clear cell renal cell carcinoma. Histopathology. 2011;58:191–197. doi: 10.1111/j.1365-2559.2011.03752.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu GH, Huang C, Qiu ZJ, Liu J, Zhang ZH, Zhao N, Feng ZZ, Lv XH. Expression and prognostic significance of CD151, c-Met, and integrin alpha3/alpha6 in pancreatic ductal adenocarcinoma. Digestive diseases and sciences. 2011;56:1090–1098. doi: 10.1007/s10620-010-1416-x. [DOI] [PubMed] [Google Scholar]
- 24.Yang YM, Zhang ZW, Liu QM, Sun YF, Yu JR, Xu WX. Overexpression of CD151 predicts prognosis in patients with resected gastric cancer. PloS one. 2013;8:e58990. doi: 10.1371/journal.pone.0058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon MJ, Park S, Choi JY, Oh E, Kim YJ, Park YH, Cho EY, Kwon MJ, Nam SJ, Im YH, Shin YK, Choi YL. Clinical significance of CD151 overexpression in subtypes of invasive breast cancer. British journal of cancer. 2012;106:923–930. doi: 10.1038/bjc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P, Erfani S, Liu Z, Jia C, Chen Y, Xu B, Deng X, Alfaro JE, Chen L, Napier D, Lu M, Huang JA, Liu C, et al. CD151-alpha3beta1 integrin complexes are prognostic markers of glioblastoma and cooperate with EGFR to drive tumor cell motility and invasion. Oncotarget. 2015;6:29675–29693. doi: 10.18632/oncotarget.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokuhara T, Hasegawa H, Hattori N, Ishida H, Taki T, Tachibana S, Sasaki S, Miyake M. Clinical significance of CD151 gene expression in non-small cell lung cancer. Clinical cancer research. 2001;7:4109–4114. [PubMed] [Google Scholar]
- 28.Yang X, Claas C, Kraeft SK, Chen LB, Wang Z, Kreidberg JA, Hemler ME. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Molecular biology of the cell. 2002;13:767–781. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berditchevski F, Odintsova E, Sawada S, Gilbert E. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. The Journal of biological chemistry. 2002;277:36991–37000. doi: 10.1074/jbc.M205265200. [DOI] [PubMed] [Google Scholar]
- 30.Kazarov AR, Yang X, Stipp CS, Sehgal B, Hemler ME. An extracellular site on tetraspanin CD151 determines alpha 3 and alpha 6 integrin-dependent cellular morphology. The Journal of cell biology. 2002;158:1299–1309. doi: 10.1083/jcb.200204056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devbhandari RP, Shi GM, Ke AW, Wu FZ, Huang XY, Wang XY, Shi YH, Ding ZB, Xu Y, Dai Z, Fan J, Zhou J. Profiling of the tetraspanin CD151 web and conspiracy of CD151/integrin beta1 complex in the progression of hepatocellular carcinoma. PloS one. 2011;6:e24901. doi: 10.1371/journal.pone.0024901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franco M, Muratori C, Corso S, Tenaglia E, Bertotti A, Capparuccia L, Trusolino L, Comoglio PM, Tamagnone L. The tetraspanin CD151 is required for Met-dependent signaling and tumor cell growth. The Journal of biological chemistry. 2010;285:38756–38764. doi: 10.1074/jbc.M110.145417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klosek SK, Nakashiro K, Hara S, Shintani S, Hasegawa H, Hamakawa H. CD151 forms a functional complex with c-Met in human salivary gland cancer cells. Biochemical and biophysical research communications. 2005;336:408–416. doi: 10.1016/j.bbrc.2005.08.106. [DOI] [PubMed] [Google Scholar]
- 34.Takeda Y, Li Q, Kazarov AR, Epardaud M, Elpek K, Turley SJ, Hemler ME. Diminished metastasis in tetraspanin CD151-knockout mice. Blood. 2011;118:464–472. doi: 10.1182/blood-2010-08-302240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ke AW, Shi GM, Zhou J, Huang XY, Shi YH, Ding ZB, Wang XY, Devbhandari RP, Fan J. CD151 amplifies signaling by integrin alpha6beta1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology. 2011;140:1629–1641 e1615. doi: 10.1053/j.gastro.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Hong IK, Jin YJ, Byun HJ, Jeoung DI, Kim YM, Lee H. Homophilic interactions of Tetraspanin CD151 up-regulate motility and matrix metalloproteinase-9 expression of human melanoma cells through adhesion-dependent c-Jun activation signaling pathways. The Journal of biological chemistry. 2006;281:24279–24292. doi: 10.1074/jbc.M601209200. [DOI] [PubMed] [Google Scholar]
- 37.Shi GM, Ke AW, Zhou J, Wang XY, Xu Y, Ding ZB, Devbhandari RP, Huang XY, Qiu SJ, Shi YH, Dai Z, Yang XR, Yang GH, Fan J. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology. 2010;52:183–196. doi: 10.1002/hep.23661. [DOI] [PubMed] [Google Scholar]
- 38.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. International journal of surgery. 2010;8:336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]


