Abstract
Purpose
mRNA degradation is an important regulatory step for controlling gene expression and cell functions. Genetic abnormalities of the genes involved in mRNA degradation were found to be associated with cancer risks. Therefore, we systematically investigated the roles of genetic variants of genes in the general mRNA degradation pathway in lung cancer risk.
Experimental design
Meta-analyses were conducted in six lung cancer genome-wide association studies (GWASs) from the Transdisciplinary Research in Cancer of the Lung and additional two GWASs from Harvard University and deCODE in the International Lung Cancer Consortium. Expression quantitative trait loci analysis (eQTL) was used for in silico functional validation of the identified significant susceptibility loci.
Results
This pathway-based analysis included 4,603 single nucleotide polymorphisms (SNP) in 68 genes in 14,463 lung cancer cases and 44,188 controls, of which 20 SNPs were found to be associated with lung cancer risk with a false discovery rate threshold of <0.05. Among the 11 newly identified SNPs in CNOT6, which were in high linkage disequilibrium, the rs2453176 with a RegulomDB score “1f” was chosen as the tag SNP for further analysis. We found that the rs2453176 T allele was significantly associated with lung cancer risk (odds ratio=1.11, 95% confidence interval=1.04–1.18, P=0.001) in the eight GWASs. In the eQTL analysis, we found that levels of CNOT6 mRNA expression were significantly correlated with the rs2453176 T allele, which provided additional biological basis for the observed positive association.
Conclusion
The CNOT6 rs2453176 SNP may be a new functional susceptible locus for lung cancer risk.
Keywords: lung cancer risk, pathway analysis, molecular epidemiology
Introduction
Lung cancer is one of the most frequently diagnosed cancers with about 1.8 million new lung cancer cases reported in 2012 worldwide, accounting for about 13% of total cancer diagnoses [1]. In the United States, 224,390 new lung cancer cases are estimated to occur in 2016 [2]. In addition to other factors, such as occupational and environmental carcinogens, cigarette smoking is the major risk factor for lung cancer [3,4], but not all smokers develop lung cancer, which suggests that genetic predisposition play an essential role in the lung carcinogenesis [5].
In recent years, some genome-wide association studies (GWASs) of lung cancer have been conducted, and a number of genetic variants, i.e., single nucleotide polymorphisms (SNPs), have been found to be associated with lung cancer risk. For example, the significant susceptibility loci associated with lung cancer risk include 5p15.3 (rs401681, rs4975616 and rs472010 in CLPTM1L and rs2736100 in TERT) [6–11], 6p21.3 (rs3117582 in BAG6 or APOM and rs2395185 in HLA-DRB5 or HLA-DRB9) [6,8,9,11], 6q22.1 (rs9387478 in RAP1BP3 or DCBLD1) [11] and 15q25.1 (rs8034191 in HYKK and rs1051730 in CHRNA3) [6,8,9,12–15]. Among these SNPs, rs1051370, rs3117582 and rs2731600 were found to be specifically associated with risk of lung adenocarcinoma (AD) [9], whereas rs12296850 (mapped to 12q23.1) in SLC17A8 or NR1H4 was found to be a susceptibility locus for risk of squamous cell carcinoma (SC) [16]. Interestingly, the vast majority of the SNPs identified by GWASs are in introns or intergenic regions, and their functional evidence is limited. In the present study, we employed the pathway-based strategy that dramatically decreases the number of SNPs to be analyzed and thus significantly reduced multiple testing with the aim to identify possible lung cancer risk-associated functional SNPs that may have not been revealed by previous lung cancer GWASs.
The degradation of mRNA is an important regulatory step for controlling gene expression and cell functions [17,18]. The general cytoplasmic mRNA decay pathway usually begins with the deadenylation, which removes the poly(A) tail Ccr4-Not complex [19], followed by degradation of mRNA proceeding in two directions of 5’-3’ or 3’-5’. The 5’-3’ mRNA degradation initiates with decapping N7-methylguanosine (m7G) cap mainly by DCP1/DCP2 proteins and subsequently degraded by the exoribonuclease Xrn1, while the 3’-5’ mRNA degradation is mainly catalyzed by 10–12 subunit exosome [20].
Some studies suggest that genetic abnormalities of genes involved in the general mRNA degradation pathway may be associated with lung cancer. For example, various genetic variants in LSM2-LSM8, which encode cofactors for mRNA decapping, were recently found in lung cancer cell lines [21]. Therefore, we hypothesize that genetic variants of the general mRNA degradation pathway are associated with lung cancer risk. To test the hypothesis, we conducted the comprehensive meta-analysis of the eight published lung cancer GWASs from the ILCCO (International Lung Cancer Consortium)-TRICL (Transdisciplinary Research in Cancer of the Lung) consortia, focusing on the SNPs of the genes in the general mRNA degradation pathway.
Materials and Methods
Study populations
The first part of the study populations came from the TRICL consortium, which included 12,160 lung cancer cases and 16,838 controls (all Europeans) of six previously published GWASs from: the MD Anderson Cancer Center (MDACC), the Institute of Cancer Research (ICR), the National Cancer Institute (NCI), the International Agency for Research on Cancer (IARC), Toronto study from Samuel Lunenfeld Research Institute study (Toronto), and the German Lung Cancer Study (GLC) [22]. The second part of the study populations included GWASs of European ancestry from Harvard Lung Cancer Study (984 cases and 970 controls) [23] and Icelandic Lung Cancer Study (deCODE) (1,319 cases and 26,380 controls) [15] of the ILCCO. Written informed consents were achieved for all participants, and the present study was approved by each institutional review board of the participating institutions.
GWAS genotyping and imputation
Genotyping in the eight GWASs was performed by Illumina HumanHap 317, 317+240S, 370Duo, 550, 610 or 1M arrays. The imputation was conducted by IMPUTE2 v2.1.1 or MaCH v1.0 software using the reference panel from the 1000 Genomes Project (phase I integrated release 3, March 2012). Standard quality control on samples was performed on all scans in the analysis, excluding any participants with low call rate (< 90%), extremely high or low heterozygosity (P < 1.0×10−4), non-European (with the HapMap phase II CEU, JPT/CHB and YRI populations as a reference) and imputed SNPs with an information score < 0.40 in IMPUTE2 or r2 < 0.30 in MaCH.
Gene and SNP selection
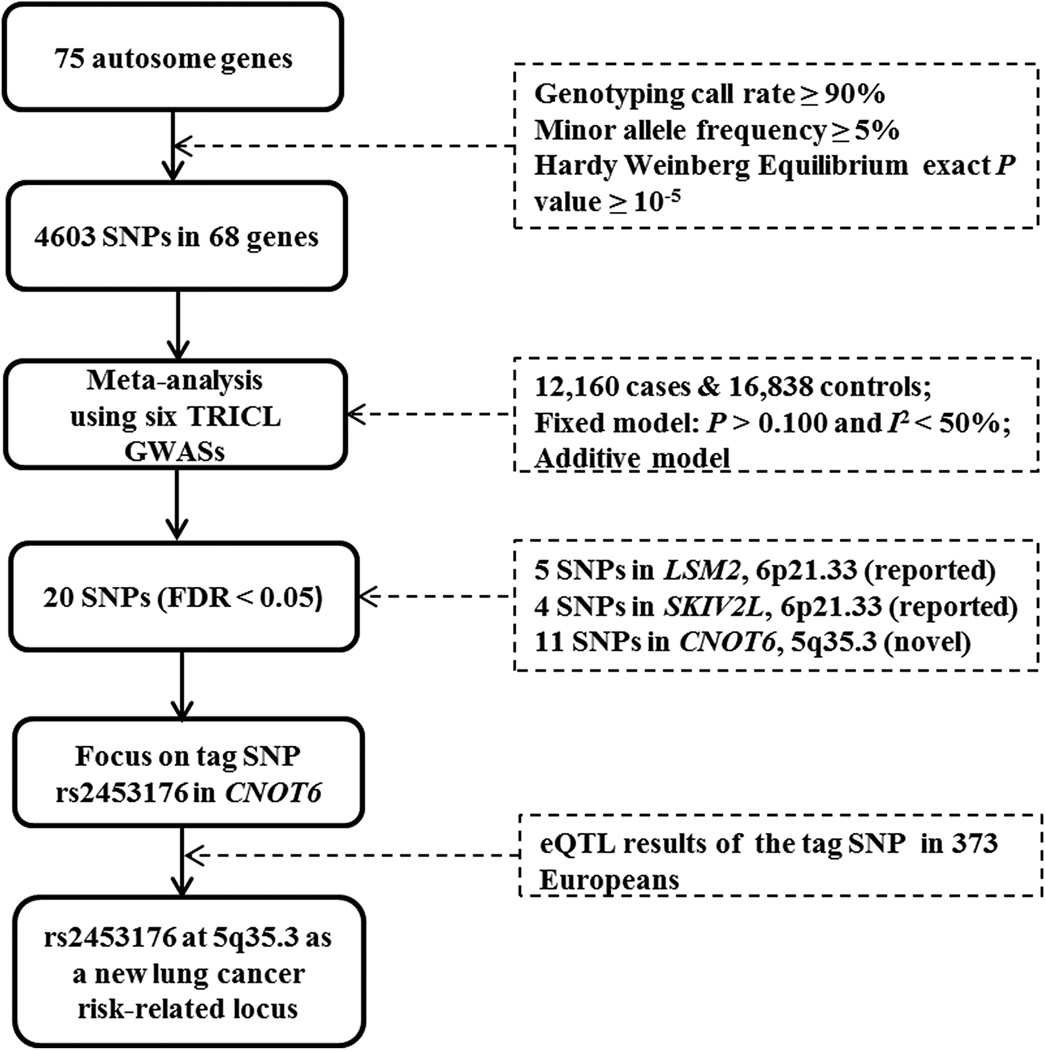
Genes in the general mRNA degradation pathway were identified from the Molecular Signatures Database [24] and the literature [19]. Overall, 75 genes located on autosomal chromosomes were selected. Among them, seven genes were pseudogenes or duplicates or withdrawn from updated NCBI. After removal of these genes, genotypes of 68 genes were abstracted from the GWAS datasets (detailed in Supplementary Table S1). The final meta-analysis contained 4,603 SNPs with the following standards: genotyping rate ≥ 90%, minor allele frequency ≥ 5%, and Hardy Weinberg Equilibrium exact P value ≥ 10−5. The overall workflow is shown in Figure 1.
Figure 1. Study workflow.
SNP: single nucleotide polymorphism; FDR: false discovery rate; TRICL: Transdisciplinary Research in Cancer of the Lung; GWAS: genome-wide association study; eQTL: expression quantitative trait loci.
In silico functional validation
Two in silico tools, SNPinfo (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm) [25], RegulomeDB (http://regulomedb.org/) [26], were used to predict potential functions. Expression quantitative trait loci (eQTL) analysis was performed by using the expression data of lymphoblastoid cell lines from 373 Europeans available in the 1000 Genomes Project (http://www.1000genomes.org/category/frequently-asked-questions/gene-expression) [27] and The Cancer Genome Atlas (TCGA)( https://tcga-data.nci.nih.gov/tcga/)[28].
Statistical analysis
Logistic regression model was used to calculate the odds ratios (ORs) and their 95% confidence intervals (CIs) in an additive genetic model with PLINK (v1.06) software. A meta-analysis with the inverse variance method was employed on the 4,603 SNPs with Stata software (v12, State College, Texas, US). Cochran's Q statistic was applied to test for heterogeneity and the I2 statistic for the proportion of the total variation in the meta-analysis [29]. The fixed-effects model was used when there was no heterogeneity among GWASs (Q-test P > 0.100 and I2 < 50%); otherwise, the random-effects model was used. Multiple testing correction was conducted with false discovery rate (FDR) with a threshold < 0.050 [30]. A linear regression model was also performed to evaluate the correlation between SNPs and mRNA expression levels of the corresponding genes. A paired t-test was used to compare the mRNA expression levels of genes in the lung cancer and normal adjacent tissue from the TCGA database. LocusZoom (http://locuszoom.sph.umich.edu/locuszoom/) was applied to construct regional association plots using Europeans from the 1000 Genomes Project as the reference (phase I integrated release 3, March 2012) [31]. Haploview v4.2 was used to generate the Manhattan plot and LD plots [32]. All analyses were conducted with SAS (version 9.4; SAS Institute, Cary, NC, USA) except for those specified otherwise.
Results
Associations of the SNPs with lung cancer risk
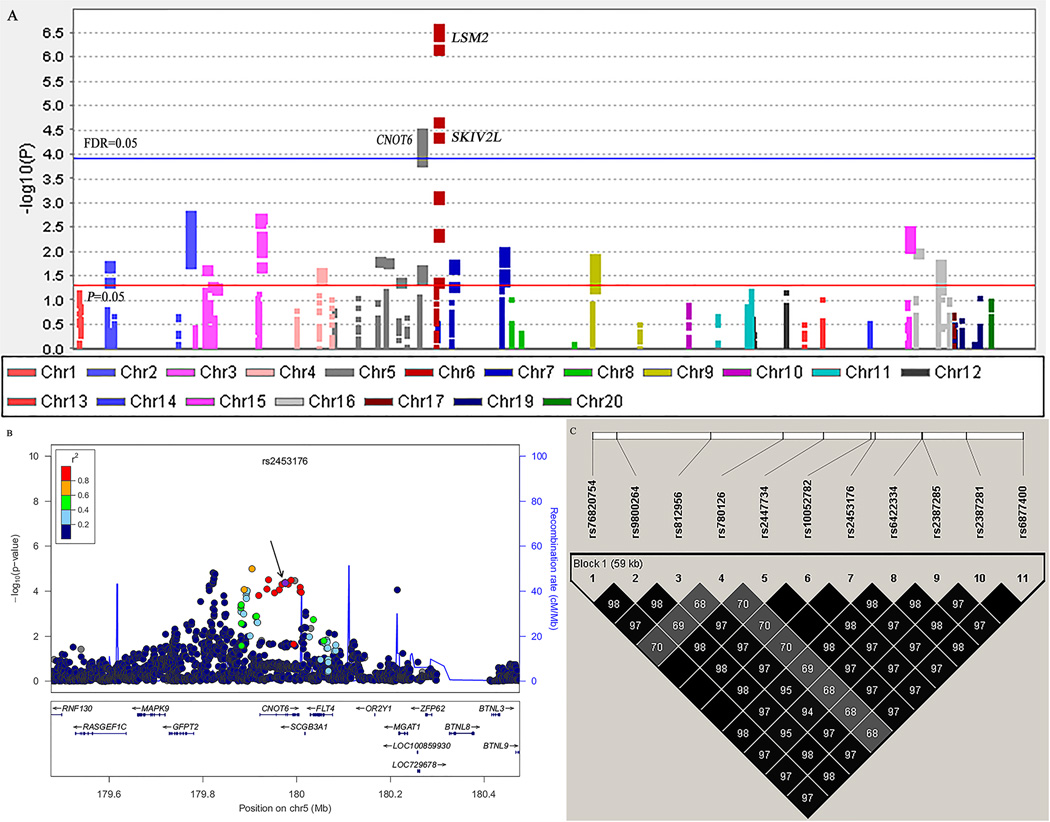
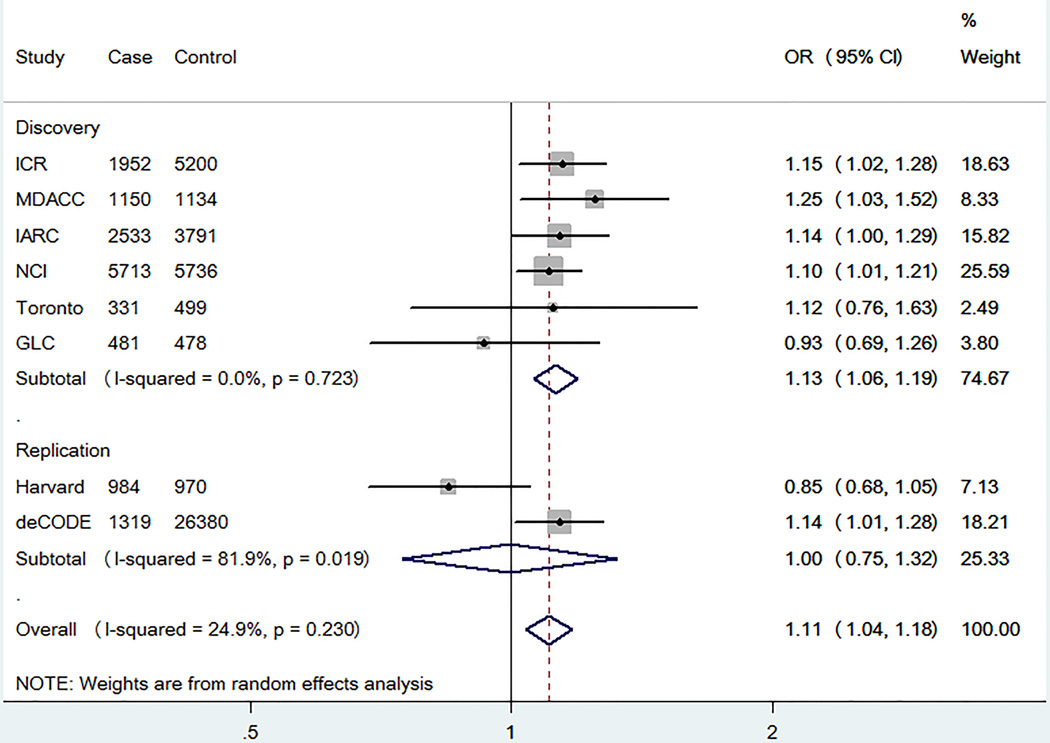
We first performed a meta-analysis in the TRICL database consisted of six previously published GWAS datasets with 12,160 cases and 16,838 controls. The basic information of these six studies is presented in Supplemental Table S2. A total of 4,603 SNPs in the pathway were extracted, of which 318 SNPs were associated with lung cancer risk at P < 0.05 in the additive model and 20 SNPs on LSM2SKIV2L and CNOT6 remained significantly associated with lung cancer risk with FDR < 0.05 after multiple testing corrections (Figure 2A and Table 1). Among these SNPs, we excluded those of LSM2 and SKIV2L, because they were mapped to and in high LD with previously GWAS-reported locus at 6p21.33 [6,8]. As a result, 11 SNPs of CNOT6 located at 5q35.3 were left for further analysis. In the LD analysis, these 11 SNPs shared moderate to high LD (r2 ≥ 0.60, Figure 2B and 2C). We finally chose rs2453176 as the tag SNP, because it was significantly associated with lung cancer risk (OR = 1.13, 95% CI = 1.06–1.19, P = 4.33×10−5) (Table 1) and potentially functional according to function prediction and its imputation quality was the best among the 11 SNPs (Table 2). We used the forest plot to illustrate the association between rs2453176 and lung cancer risk in the six GWASs (Figure 3), and the rs2453176 T allele was associated with an increased lung cancer risk in five GWASs, except for the GLC GWAS.
Figure 2. Screening of SNPs in the general mRNA degradation pathway.
A, Manhattan Plot of genome-wide association results from the general mRNA degradation pathway in TRICL. The x-axis shows SNPs’ positions on each chromosome. The y-axis shows the association P values with lung cancer risk (as –log10 P values). The FDR threshold of 0.05 was shown by a horizontal blue line. The P value of 0.05 was shown by a horizontal red line. B, Regional association plot for SNP rs2453176 in 500 kb up- and downstream region. The left-hand y-axis shows P values of the SNPs, which are transformed as −log10 (P) against chromosomal base pair positions. The right-hand y-axis shows the recombination rate estimated from HapMap Data Rel 22/phase II European population; C, The linkage disequilibrium plots of 11 SNPs in CNOT6. The value within each diamond represents the pairwise correlation between SNPs (measured as r2) defined by the upper left and the upper right sides of the diamond.
Table 1.
Associations between SNPs in the general mRNA degradation pathway and lung cancer risk with FDR < 0.050 in TRICL GWASs
| SNP | Gene | Chr. | Position (hg19) | Allelea | EAF | Qb | I2 | Effectc | OR (95% CI) | P | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs115834633 | LSM2 | 6 | 31765984 | G/A | 0.11 | 0.200 | 30.79 | ++++++ | 1.20 (1.14–1.27) | 3.92E-11 | <.0001 |
| rs114312980 | LSM2 | 6 | 31768799 | A/C | 0.11 | 0.230 | 26.77 | ++++++ | 1.20 (1.14–1.27) | 4.39E-11 | <.0001 |
| rs115801685 | LSM2 | 6 | 31772093 | C/A | 0.11 | 0.220 | 27.36 | ++++++ | 1.20 (1.14–1.27) | 4.54E-11 | <.0001 |
| rs115489726 | LSM2 | 6 | 31766660 | C/T | 0.11 | 0.240 | 25.69 | ++++++ | 1.20 (1.14–1.27) | 8.76E-11 | <.0001 |
| rs114637560 | LSM2 | 6 | 31765864 | T/A | 0.15 | 0.260 | 22.42 | +−++++ | 1.14 (1.08–1.20) | 3.04E-07 | 0.0003 |
| rs114984862 | SKIV2L | 6 | 31936668 | C/T | 0.27 | 0.290 | 18.64 | ++++++ | 1.09 (1.05–1.13) | 1.87E-05 | 0.013 |
| rs9800264 | CNOT6 | 5 | 179940091 | G/A | 0.10 | 0.750 | 0.00 | +++++− | 1.13 (1.07–1.19) | 3.12E-05 | 0.013 |
| rs2387281 | CNOT6 | 5 | 179988283 | T/C | 0.10 | 0.743 | 0.00 | +++++− | 1.13 (1.07–1.19) | 3.28E-05 | 0.013 |
| rs6877400 | CNOT6 | 5 | 179996111 | T/C | 0.10 | 0.747 | 0.00 | +++++− | 1.13 (1.07–1.19) | 3.41E-05 | 0.013 |
| rs116188106 | SKIV2L | 6 | 31927342 | G/A | 0.27 | 0.298 | 17.82 | ++++++ | 1.09 (1.04–1.13) | 3.70E-05 | 0.013 |
| rs114011334 | SKIV2L | 6 | 31928799 | C/T | 0.27 | 0.297 | 17.92 | ++++++ | 1.09 (1.04–1.13) | 3.77E-05 | 0.013 |
| rs115002281 | SKIV2L | 6 | 31929014 | C/A | 0.27 | 0.297 | 17.95 | ++++++ | 1.08 (1.04–1.13) | 3.79E-05 | 0.013 |
| rs10052782 | CNOT6 | 5 | 179975104 | C/T | 0.10 | 0.723 | 0.00 | +++++− | 1.13 (1.06–1.19) | 4.09E-05 | 0.013 |
| rs6422334 | CNOT6 | 5 | 179982151 | C/T | 0.10 | 0.734 | 0.00 | +++++− | 1.13 (1.06–1.19) | 4.14E-05 | 0.013 |
| rs2453176 | CNOT6 | 5 | 179975792 | C/T | 0.10 | 0.723 | 0.00 | +++++− | 1.13 (1.06–1.19) | 4.33E-05 | 0.013 |
| rs2387285 | CNOT6 | 5 | 179982278 | A/G | 0.10 | 0.700 | 0.00 | +++++− | 1.12 (1.06–1.19) | 4.92E-05 | 0.014 |
| rs2447734 | CNOT6 | 5 | 179968674 | G/C | 0.10 | 0.720 | 0.00 | +++++− | 1.12 (1.06–1.19) | 4.97E-05 | 0.014 |
| rs76820754 | CNOT6 | 5 | 179936737 | G/A | 0.10 | 0.735 | 0.00 | +++++− | 1.12 (1.06–1.19) | 8.06E-05 | 0.021 |
| rs780126 | CNOT6 | 5 | 179963034 | C/T | 0.13 | 0.758 | 0.00 | +++++− | 1.12 (1.06–1.18) | 8.67E-05 | 0.021 |
| rs812956 | CNOT6 | 5 | 179953048 | G/C | 0.10 | 0.653 | 0.00 | +++++− | 1.12 (1.06–1.18) | 1.18E-04 | 0.027 |
SNP: single nucleotide polymorphism; FDR: false discovery rate; TRICL: Transdisciplinary Research in Cancer of the Lung; GWAS: genome-wide association study; Chr.: chromosome; EAF: effect allele frequency; OR: odds ratio; CI: confidence interval;
Reference allele/effect allele;
Fixed effect models were used when no heterogeneity was found between studies (Q-test P > 0.100 and I2 < 50.0%); otherwise, random effect models were used;
“+” means a positive association, and “−“ means a negative association.
Table 2.
Linkage disequilibrium between the 11 SNPs of CNOT6 in European populations included in the 1000 Genomes Project and imputation quality scores
| SNP | Position (hg19) |
D' | r2 | Function prediction | Imputation quality | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPinfoa | Regulome DBb | Info ICR |
Rsq MDACC |
Rsq IARC |
Info NCI |
Info Toronto |
Rsq GLC |
||||
| rs2453176 | 179975792 | -- | 1f | 1.000 | 0.999 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| rs780126 | 179963034 | 1.00 | 0.71 | -- | -- | 0.874 | 0.751 | 0.703 | 0.859 | 0.857 | 0.784 |
| rs2387281 | 179988283 | 1.00 | 0.97 | -- | -- | 0.998 | 0.972 | 0.969 | 0.996 | 0.990 | 0.967 |
| rs6877400 | 179996111 | 1.00 | 0.97 | Splicing site | 5 | 0.998 | 0.964 | 0.965 | 0.996 | 0.990 | 0.953 |
| rs2387285 | 179982278 | 1.00 | 0.97 | -- | 4 | 0.990 | 0.966 | 0.923 | 0.988 | 0.981 | 0.964 |
| rs812956 | 179953048 | 1.00 | 0.97 | -- | 6 | 0.991 | 0.961 | 0.962 | 0.988 | 0.978 | 0.976 |
| rs9800264 | 179940091 | 1.00 | 0.99 | -- | -- | 0.999 | 0.970 | 0.977 | 0.998 | 0.993 | 1.000 |
| rs6422334 | 179982151 | 1.00 | 0.99 | -- | 5 | 0.999 | 0.982 | 0.976 | 0.997 | 0.994 | 0.979 |
| rs10052782 | 179975104 | 1.00 | 1.00 | -- | 6 | 1.000 | 0.998 | 1.000 | 1.000 | 1.000 | 1.000 |
| rs76820754 | 179936737 | 1.00 | 1.00 | -- | 6 | 1.000 | 0.970 | 0.971 | 0.999 | 0.998 | 0.999 |
| rs2447734 | 179968674 | 1.00 | 1.00 | -- | -- | 1.000 | 0.994 | 0.999 | 0.999 | 0.997 | 0.999 |
SNP: single nucleotide polymorphism;
Imputation quality: Rsq: MaCH r-squared; Info: IMPUTE2 information score;
Figure 3. Forest plots for associations between CNOT6 rs2453176 and lung cancer risk for all participants.
(P = 0.0013).
We expanded our analysis to include additional two independent lung cancer GWASs (Supplemental Table S2). The deCODE GWAS validated our result of the CNOT6 rs2453176 tag SNP (OR = 1.14, 95% CI = 1.01–1.28, P = 0.032), while the GWAS from Harvard University displayed the same trend as the GLC GWAS (OR = 0.85, 95% CI = 0.68–1.05, P = 0.133) (Figure 3 and Table 3).
Table 3.
Associations between of CNOT6 rs2453176 (C >T) and lung cancer risk stratified by histologic types in all eight lung cancer GWASs from ILCCO-TRICL
| Study | Overall | AD | SC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | P | Case | Control | OR (95% CI) | P | Case | Control | OR (95% CI) | P | |
| ICR | 1952 | 5200 | 1.15 (1.02–1.28) | 0.020 | 465 | 5200 | 1.38 (1.12–1.70) | 0.002 | 611 | 5200 | 1.14 (0.95–1.38) | 0.158 |
| MDACC | 1150 | 1134 | 1.25 (1.03–1.52) | 0.027 | 619 | 1134 | 1.06 (0.92–1.47) | 0.206 | 306 | 1134 | 1.45 (1.08–1.94) | 0.013 |
| IARC | 2533 | 3791 | 1.14 (1.00–1.29) | 0.053 | 517 | 2824 | 1.13 (0.90–1.42) | 0.301 | 911 | 2968 | 1.18 (0.98–1.41) | 0.081 |
| NCI | 5713 | 5736 | 1.10 (1.01–1.21) | 0.025 | 1841 | 5736 | 1.17 (1.03–1.33) | 0.016 | 1447 | 5736 | 1.04 (0.91–1.20) | 0.543 |
| Toronto | 331 | 499 | 1.12 (0.76–1.63) | 0.057 | 90 | 499 | 1.48 (0.83–2.64) | 0.186 | 50 | 499 | 1.00 (0.44–2.25) | 0.998 |
| GLC | 481 | 478 | 0.93 (0.69–1.26) | 0.064 | 186 | 478 | 1.25 (0.86–1.83) | 0.240 | 97 | 478 | 0.90 (0.52–1.54) | 0.695 |
| Discovery combined | 12160 | 16838 | 1.13 (1.06–1.19) | 4.33E-05 | 3818 | 15871 | 1.21 (1.11–1.32) | 2.04E-05 | 3424 | 16015 | 1.13 (1.03–1.23) | 0.009 |
| Harvard | 984 | 970 | 0.85 (0.68–1.05) | 0.133 | 597 | 970 | 0.79 (0.62–1.01) | 0.130 | 216 | 970 | 1.08 (0.75–1.56) | 0.678 |
| deCODE | 1319 | 26380 | 1.14 (1.01–1.28) | 0.032 | 547 | 26380 | 1.02 (0.85–1.21) | 0.858 | 259 | 26380 | 1.11 (0.86–1.43) | 0.436 |
| Replication combined | 2303 | 27350 | 1.00 (0.75–1.32) | 0.098 | 1144 | 27350 | 0.91 (0.71–1.16) | 0.449 | 475 | 27350 | 1.10 (0.89–1.36) | 0.381 |
| Overall | 14463 | 44188 | 1.11 (1.04–1.18) | 0.001 | 4862 | 43221 | 1.13 (1.00–1.27) | 0.050 | 3897 | 43365 | 1.12 (1.03–1.22) | 0.006 |
GWAS: genome-wide association study; ILCCO: International Lung Cancer Consortium; TRICL: Transdisciplinary Research in Cancer of the Lung; AD, adenocarcinoma; SC, squamous cell carcinoma; OR: odds ratio; CI: confidence interval
Homogeneity tests suggest that there is no heterogeneity between the subgroups of AD and SC in each GWAS and overall result (P > 0.05).
As we combined all the data from the eight GWASs, the functional CNOT6 rs2453176 tag SNP was found to be significantly associated with an increased risk of lung cancer (OR = 1.11, 95% CI = 1.04–1.18, P = 0.001) after the FDR correction (Figure 3 and Table 3).
Stratified analyses by lung cancer histology
Since lung cancer has different histological types that could have distinct biological behaviors, we performed AD and SC subgroup analysis and found that the rs2453176 T allele was associated with a borderlinely increased risk in AD (OR = 1.13, 95% CI = 1.00–1.27, P = 0.050, Table 3), but it was significantly associated with SC risk (OR = 1.12, 95% CI = 1.03–1.22, P = 0.006, Table 3). Because smoking is a major risk factor for lung cancer, we further stratified the data into smokers and non-smokers and found that that the rs2453176 T allele was associated with a significantly increased risk in smokers (OR = 1.09, 95% CI = 1.02–1.17, P = 0.011, Table 4), while the allele was not statistically significant in non-smokers (OR = 1.10, 95% CI = 0.89–1.36, P = 0.363, Table 4). Homogeneity tests suggested that there was no heterogeneity between strata either in subgroups of histologic types or smoking status (Table 3 and Table 4, all P > 0.05).
Table 4.
Associations between of CNOT6 rs2453176 (C >T) and lung cancer risk stratified by smoking status in six lung cancer GWASs from ILCCO-TRICL Consortia
| Study | Smoker | Non-smoker | ||||||
|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | P | Case | Control | OR (95% CI) | P | |
| MDACC | 1150 | 1134 | 1.25 (1.03–1.52) | 0.027 | ||||
| IARC | 2367 | 2508 | 1.12 (0.97–1.29) | 0.131 | 159 | 1253 | 1.40 (0.94–2.09) | 0.096 |
| NCI | 5342 | 4336 | 1.10 (1.00–1.22) | 0.058 | 350 | 1379 | 0.99 (0.72–1.37) | 0.972 |
| Toronto | 236 | 272 | 1.14 (0.70–1.86) | 0.606 | 95 | 217 | 1.13 (0.61–2.11) | 0.702 |
| GLC | 433 | 258 | 1.00 (0.67–1.49) | 0.995 | 35 | 220 | 1.64 (0.71–3.82) | 0.250 |
| Harvard | 892 | 809 | 0.87 (0.70–1.09) | 0.221 | 92 | 161 | 0.71 (0.38–1.33) | 0.288 |
| Overall | 10420 | 9317 | 1.09 (1.02–1.17) | 0.011 | 731 | 3230 | 1.10 (0.89–1.36) | 0.363 |
GWAS: genome-wide association study; ILCCO: International Lung Cancer Consortium; TRICL: Transdisciplinary Research in Cancer of the Lung; OR: odds ratio; CI: confidence interval.
Homogeneity tests suggest there is no heterogeneity between the subgroups of smoker and non-smoker in each GWAS and overall result (P > 0.05).
Functional validation by eQTL analysis
Because the CNOT6 rs2453176 SNP was predicted with a score of "1f", suggesting the most confident functional annotation by regulomeDB [26], we further explored the underlying molecular mechanism by performing the eQTL analysis. With mRNA expression data of lymphoblastoid cell lines from 373 Europeans available from the 1000 Genomes Project, We found that expected mRNA expression levels of CNOT6 were significantly decreased with an increased number of the rs2453176 T allele in both the additive (P = 0.008) (Figure 4A) and dominant (P = 0.007) (Figure 4B) models but not the recessive model (Figure 4C). We also used the 105 normal adjacent tissue samples in the TCGA to further explore the correlation between the rs2453176 genotypes and their corresponding mRNA expression levels, but we did not observe a statistical significance (P > 0.05) (Supplemental Figure S1A-S1C). We also compared the mRNA expression level of CNOT6 in the 107 paired samples and did not find a statistically significant difference (P > 0.05) (Supplemental Figure S1D).
Figure 4. The eQTL analysis of CNOT6 mRNA expression for rs2453176 with lymphoblastoid cell data of 373 Europeans from 1000 Genomes Project.
A. additive model, P = 0.008; B. dominant model, P = 0.007; C. recessive model, P = 0.634.
Discussion
In the present study, we found that a novel potentially functional susceptibility locus rs2453176 C>T of CNOT6 in the general mRNA degradation pathway was associated with an increased lung cancer risk in 14,463 cases and 44,188 controls. This association was further supported by a significant correlation between a decreased mRNA expression level and an increasing number of the A allele in the eQTL analysis.
Gene expression disorder is one of cancer hallmarks, and instability of mRNA may result in altered transcprit/protein levels of oncogenes and tumor repressor genes [33]. The degradation of mRNA is a key step in controlling the expression of genes related to cell proliferation. For example, the CCR4-Not complex consists of highly conserved exoribonucleases and adaptor proteins that hydrolyze and shorten the poly(A) tail, which starts the initial and the rate-limiting step of mRNA degradation [19]. Located at 5q35.3, CNOT6 encodes a protein that has a 3'-5' RNase activity and acts as a catalytic subunit of the CCR4-Not deadenylation complex [34]. Although it remains unclear how the catalytic subunit works during the deadenylation process, some studies reported that its expression level was associated with carcinogenesis or prognosis. For example, one study of lung cancer found that the CNOT6 overexpression in lung SC predicted a significantly less metastasis [33]. Another study of acute leukemia discovered that CNOT6 had a significantly lower expression in patients than in controls [35]. These two studies suggest that high expression levels of CNOT6 may promote the degradation of mRNA of some oncogenes and the suppression of cell proliferation in carcinogenesis.
In the present study, we identified that the CNOT6 rs2453176 T allele was associated with an increased risk of lung cancer, which was supported by the association of CNOT6 rs2453176 T allele with a decreased mRNA expression level in lymphoblastoid cell lines from 373 Europeans. This finding is consistent with the role of CNOT6 in lung cancer prognosis as previously described [33]. The ENCODE project data from University of California Santa Cruz show that the CNOT6 rs2453176 locus is located at the DNase I hypersensitive region (Supplemental Figure S2). Usually such an area has a loose chromatin structure and renders it a region with a high affinity for transcription factors (TFs). As a result, some TFs, including MAFK and MAFF, bond to this region in many cell types (Supplemental Figure S2). For example, MAFK and MAFF were found to form heterodimers with a series of TFs and suppressed gene transcriptions [36,37]. Based on these, we speculate that the rs2453176 T allele may have a relatively high affinity with MAFK or MAFF and thus leads to the decreased mRNA expression of CNOT6. It is likely that a reduced quantity of CNOT6 may not be optimal in the mRNA degradation of some aberrant genes, which may in turn increases lung cancer risk, but these speculations need to be further investigated.
In the stratification analysis, rs2453176 was associated with lung cancer risk in both AD and SC subtypes, which is not surprising, because smoking has been established as a predominant risk factor for developing lung cancer, regardless histologic types [38]. Genetic susceptibility to smoking-related lung cancer risk may determine smoking behavior and tobacco metabolism [39]. Indeed, we found that the rs2453176 T allele was associated with a higher risk of lung cancer in smokers than in non-smokers. One study reported that smoking would enhance the activity of the GATA family [40], and another study reported that nicotine would increase the expression of EP300 and promote the lung cancer growth [41]. From the Supplemental Figure S2, GATA1, GATA2 and EP300 are the TFs that bind to the rs2453176 locus, possibly explaining why carriers of the rs2453176 T allele may have an increased risk of lung cancer in smokers than non-smokers.
There are some limitations in the present study. First, we employed the gene set enrichment analysis with a collection of annotated gene sets to define the general mRNA degradation pathway to be investigated, but we may have missed some newly discovered genes in the pathway. However, we searched the literatures and added genes as many as possible. Second, due to the data limitation, we had no access to family history and others factors that may have an impact on lung cancer risk. Third, we used the eQTL analyses from lymphoblastoid cell lines and normal adjacent tissue in TCGA database to validate the risk association. Although the results from the cell lines support our identified association, they may only reflect the baseline or genetically determined expression levels without exposure to smoking. The gene expressions in the normal adjacent lung tissues may be in some degree different from the normal lung tissue and did not support the association.
Overall, the present study of eight published GWASs identified a novel CNOT6 rs2453176 SNP in the general mRNA degradation pathway to be significantly associated with lung cancer risk in European populations, and the risk was more evident in smokers than in non-smokers. Although we used the publically available gene expression database from blood to confirm the biological significance of the variant, further functional evaluations in normal lung tissue are warranted to validate our findings.
Supplementary Material
A. additive model, P = 0.491, B. dominant model, P = 0.990, C. recessive model, P = 0.667; D, The mRNA expression of CNOT6 in the 107 paired lung cancer and normal adjacent tissue samples from the TCGA database (P = 0.237).
Acknowledgments
As Duke Cancer Institute members, QW, KO and NR acknowledge support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236). QW was also supported by the start-up funds from Duke Cancer Institute, Duke University Medical Center.
TRICL
This work was supported by the Transdisciplinary Research in Cancer of the Lung (TRICL) Study and, U19-CA148127 on behalf of the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network. The Toronto study was supported by Canadian Cancer Society Research Institute (020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award to RH The ICR study was supported by Cancer Research UK (C1298/A8780 andC1298/A8362—Bobby Moore Fund for Cancer Research UK) and NCRN, HEAL and Sanofi-Aventis. Additional funding was obtained from NIH grants (5R01CA055769, 5R01CA127219, 5R01CA133996, and 5R01CA121197). The Liverpool Lung Project (LLP) was supported by The Roy Castle Lung Cancer Foundation, UK. The ICR and LLP studies made use of genotyping data from the Wellcome Trust Case Control Consortium 2 (WTCCC2); a full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Sample collection for the Heidelberg lung cancer study was in part supported by a grant (70–2919) from the Deutsche Krebshilfe. The work was additionally supported by a Helmholtz-DAAD fellowship (A/07/97379 to MNT) and by the NIH (U19CA148127). The KORA Surveys were financed by the GSF, which is funded by the German Federal Ministry of Education, Science, Research and Technology and the State of Bavaria. The Lung Cancer in the Young study (LUCY) was funded in part by the National Genome Research Network (NGFN), the DFG (BI576/2-1; BI 576/2-2), the Helmholtzgemeinschaft (HGF) and the Federal office for Radiation Protection (BfS: STSch4454). Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum Muenchen. Support for the Central Europe, HUNT2/Tromsø and CARET genome-wide studies was provided by Institut National du Cancer, France. Support for the HUNT2/Tromsø genome-wide study was also provided by the European Community (Integrated Project DNA repair, LSHG-CT- 2005–512113), the Norwegian Cancer Association and the Functional Genomics Programme of Research Council of Norway. Support for the Central Europe study, Czech Republic, was also provided by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101). Support for the CARET genome-wide study was also provided by grants from the US National Cancer Institute, NIH (R01 CA111703 and UO1 CA63673), and by funds from the Fred Hutchinson Cancer Research Center. Additional funding for study coordination, genotyping of replication studies and statistical analysis was provided by the US National Cancer Institute (R01 CA092039). The lung cancer GWAS from Estonia was partly supported by a FP7 grant (REGPOT245536), by the Estonian Government (SF0180142s08), by EU RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure’s Roadmap and by University of Tartu (SP1GVARENG). The work reported in this paper was partly undertaken during the tenure of a Postdoctoral Fellowship from the IARC (for MNT). The Environment and Genetics in Lung Cancer Etiology (EAGLE), the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), and the Prostate, Lung, Colon, Ovary Screening Trial (PLCO)studies and the genotyping of ATBC, the Cancer Prevention Study II Nutrition Cohort (CPS-II) and part of PLCO were supported by the Intramural Research Program of NIH, NCI, Division of Cancer Epidemiology and Genetics. ATBC was also supported by US Public Health Service contracts (N01-CN-45165, N01-RC-45035 and N01-RC-37004) from the NCI. PLCO was also supported by individual contracts from the NCI to the University of Colorado Denver (NO1-CN-25514), Georgetown University(NO1-CN-25522), Pacific Health Research Institute (NO1-CN-25515), Henry Ford Health System (NO1-CN-25512), University of Minnesota(NO1-CN-25513), Washington University(NO1-CN-25516), University of Pittsburgh (NO1-CN-25511), University of Utah (NO1-CN-25524), Marshfield Clinic Research Foundation (NO1-CN-25518), University of Alabama at Birmingham (NO1-CN-75022, Westat, Inc. NO1-CN-25476), University of California, Los Angeles (NO1-CN-25404). The Cancer Prevention Study II Nutrition Cohort was supported by the American Cancer Society. The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 andRO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C) and study coordination at the GENEVA Coordination Center (U01 HG004446) for EAGLE and part of PLCO studies. Funding for the MD Anderson Cancer Study was provided by NIH grants (P50 CA70907, R01CA121197, R01CA127219, U19 CA148127, R01 CA55769, and K07CA160753) and CPRIT grant (RP100443). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is funded through a federal contract from the NIH to The Johns Hopkins University (HHSN268200782096C). The Harvard Lung Cancer Study was supported by the NIH (National Cancer Institute) grants CA092824, CA090578, and CA074386.
deCODE
The project was funded in part by GENADDICT: LSHMCT-2004-005166), the National Institutes of Health (R01-DA017932)
Abbreviations
- AD
Adenocarcinoma
- CI
confidence interval
- eQTL
expression quantitative trait loci
- FDR
false discovery rate
- GWAS
genome-wide association study
- ILCCO
International Lung Cancer Consortium
- LD
linkage disequilibrium
- OR
odds ratio
- SC
squamous cell carcinoma
- SNP
single nucleotide polymorphisms
- TCGA
The Cancer Genome Atlas
- TRICL
Transdisciplinary Research in Cancer of the Lung
Footnotes
Conflict of interest:
The authors disclose no potential conflicts of interest.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Field RW, Withers BL. Occupational and environmental causes of lung cancer. Clinics in chest medicine. 2012;33(4):681–703. doi: 10.1016/j.ccm.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schottenfeld D, Fraumeni JF. Cancer epidemiology and prevention. xviii. Oxford ; New York: Oxford University Press; 2006. p. 1392. [Google Scholar]
- 5.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nature reviews Cancer. 2007;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nature genetics. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nature genetics. 2008;40(12):1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer research. 2009;69(16):6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. American journal of human genetics. 2009;85(5):679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z, Wu C, Shi Y, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nature genetics. 2011;43(8):792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 11.Lan Q, Hsiung CA, Matsuo K, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nature genetics. 2012;44(12):1330–1335. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature genetics. 2008;40(5):616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. Journal of the National Cancer Institute. 2008;100(18):1326–1330. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 15.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, Jin G, Wu C, et al. Genome-wide association study identifies a novel susceptibility locus at 12q23.1 for lung squamous cell carcinoma in han chinese. PLoS genetics. 2013;9(1):e1003190. doi: 10.1371/journal.pgen.1003190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess HM, Mohr I. Cellular 5'-3' mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell host & microbe. 2015;17(3):332–344. doi: 10.1016/j.chom.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brewer G. Messenger RNA decay during aging and development. Ageing research reviews. 2002;1(4):607–625. doi: 10.1016/s1568-1637(02)00023-5. [DOI] [PubMed] [Google Scholar]
- 19.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochimica et biophysica acta. 2012;1819(6):593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Molecular cell. 2008;32(5):605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young JH, Peyton M, Kim HS, et al. Computational Discovery of Pathway-Level Genetic Vulnerabilities in Non-Small-Cell Lung Cancer. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nature genetics. 2014;46(7):736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su L, Zhou W, Asomaning K, et al. Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis. 2006;27(5):1024–1029. doi: 10.1093/carcin/bgi283. [DOI] [PubMed] [Google Scholar]
- 24.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu ZL, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lappalainen T, Sammeth M, Friedlander MR, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodgers K Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma (vol 511, pg 543,2014) Nature. 2014;514(7521) doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 31.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Maragozidis P, Papanastasi E, Scutelnic D, et al. Poly(A)-specific ribonuclease and Nocturnin in squamous cell lung cancer: prognostic value and impact on gene expression. Molecular cancer. 2015;14(1):187. doi: 10.1186/s12943-015-0457-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittal S, Aslam A, Doidge R, Medica R, Winkler GS. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Molecular biology of the cell. 2011;22(6):748–758. doi: 10.1091/mbc.E10-11-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maragozidis P, Karangeli M, Labrou M, et al. Alterations of deadenylase expression in acute leukemias: evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta haematologica. 2012;128(1):39–46. doi: 10.1159/000337418. [DOI] [PubMed] [Google Scholar]
- 36.Kannan MB, Solovieva V, Blank V. The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochimica et biophysica acta. 2012;1823(10):1841–1846. doi: 10.1016/j.bbamcr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Katsuoka F, Yamamoto M. Small Maf proteins (MafF, MafG, MafK): History, structure and function. Gene. 2016 doi: 10.1016/j.gene.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis DR, Check DP, Caporaso NE, Travis WD, Devesa SS. US lung cancer trends by histologic type. Cancer. 2014;120(18):2883–2892. doi: 10.1002/cncr.28749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shields PG. Molecular epidemiology of smoking and lung cancer. Oncogene. 2002;21(45):6870–6876. doi: 10.1038/sj.onc.1205832. [DOI] [PubMed] [Google Scholar]
- 40.Zhao J, Harper R, Barchowsky A, Di YP. Identification of multiple MAPK-mediated transcription factors regulated by tobacco smoke in airway epithelial cells. American journal of physiology Lung cellular and molecular physiology. 2007;293(2):L480–L490. doi: 10.1152/ajplung.00345.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dasgupta P, Rizwani W, Pillai S, et al. ARRB1-mediated regulation of E2F target genes in nicotine-induced growth of lung tumors. Journal of the National Cancer Institute. 2011;103(4):317–333. doi: 10.1093/jnci/djq541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. additive model, P = 0.491, B. dominant model, P = 0.990, C. recessive model, P = 0.667; D, The mRNA expression of CNOT6 in the 107 paired lung cancer and normal adjacent tissue samples from the TCGA database (P = 0.237).