Abstract
Obesity increases the risk of distant metastatic recurrence and reduces breast cancer (BC) survival. However, the mechanisms behind this pathology and identification of relevant therapeutic targets are poorly defined. Plasma free fatty acids (FFA) levels are elevated in obese individuals. Here we report that TGF-β transiently activates ERK and subsequently phosphorylates SMAD4 at Thr277, which facilitates a SMAD4-USP9x interaction, SMAD4 nuclear retention, and stimulates TGF-β /SMAD3-mediated transcription of Twist and Snail. USP9x inhibited the E3 ubiquitin-protein ligase TIF1γ from binding and monoubiquitinating SMAD4, hence maintaining SMAD4 nuclear retention. FFA further facilitated TGF-β-induced ERK activation, SMAD4 phosphorylation and nuclear retention, promoting TGF-β-dependent cancer progression. Inhibition of ERK and USP9x suppressed obesity-induced metastasis. Additionally, clinical data indicated that phospho-ERK and -SMAD4 levels correlate with activated TGF-β signaling and metastasis in overweight/obese patient BC specimens. Altogether, we demonstrate the vital interaction of USP9x and SMAD4 for governing TGF-β signaling and dyslipidemia-induced, aberrant TGF-β activation during BC metastasis.
Introduction
Breast cancer (BC) is the most common cancer in women and one of the top five cancers causing overall cancer mortality globally, with a continuously rising incidence (1,2). Recent estimates suggest that up to 35% of cases may be avertible via lifestyle and diet alteration (2). More recent studies have revealed obesity to be an established risk factor for BC. It has also been related to increased incidence and mortality, poorer prognosis, a more aggressive tumor phenotype (3,4). Being overweight/obese for a woman diagnosed with BC increases the risk of developing distant metastatic recurrence, and reduces survival irrespective of treatment factors (5). The link between obesity and survival does not vary by menopause or tumor hormone receptor (HR) status. Overweight in rodents is also related to augmented incidence of spontaneous and chemically induced cancers (6). In addition, it has been proposed that BC is associated with consumption of a high fat diet (HFD) (7). A meta-analysis of substantial rodent BC models demonstrated that HFD enhances susceptibility to mammary tumors (8). Although the precise mechanisms remain to be illuminated, dietary factors have been involved in nearly 35% of cancer deaths (9).
Overweight/obesity and HFD in both humans and rodents is characterized by elevated free tatty acids (FFA) levels (10,11). Increasing evidence points to FFA signaling playing an important role in tumorigenesis and BC development and progression. The mean levels of total FFA, two of the saturated fatty acids and one unsaturated fatty acid (palmitic acid C16:0, stearic acid C18:0 and linoleic acid (ω6) C18:2) in the serum are remarkably higher in the BC patients than the benign and the control groups and they have been identified as possible biomarkers for BC (12). As for individual fatty acids, palmitate has been implicated in an increase in BC risk in postmenopausal women cohort studies (13). Furthermore, Shannon et al. (14) reported a significant direct association between erythrocyte palmitic acid and risk of BC. Fatty acid synthase (FAS), which catalyzes the synthesis of palmitic acid, is also commonly overexpressed in BC and other cancers (15,16). Louie et al. (17) demonstrated that cancer cells strongly incorporate and remodel exogenous palmitate into structural and oncogenic glycerophospholipids, sphingolipids, and ether lipids, suggesting that cancer cells are addicted to FFA and utilize exogenous FFA for producing lipids required for proliferation and pro-tumorigenic lipid signaling and energy production. By measuring the membrane lipid composition of BC tissue, Hilvo et al. (18) reveal elevated levels of palmitate-containing phosphatidylcholine species in BC relative to normal adjacent tissue. This trend is in accordance with BC progression, predicted reduced survival, and more prominent in high histological grade cancers. Moreover, BC and other cancer cells can be rescued from the pro-apoptotic effect of fatty acid synthase (FASN) suppression by the exposure to exogenous palmitate (19). In spite of the physiological significance of FFA in the BC biology, the precise molecular mechanisms by which FFA, especially palmitate, might influence cancer development and progression, still remain to be elucidated.
Active transforming growth factor-β (TGF-β) and related factors have various regulatory activities that affect cell proliferation, differentiation, apoptosis, migration, adhesion, survival, development, tissue repair, tumorigenesis, immune defense, and inflammation (20). Hence, TGF-β family members are important in maintaining the homeostasis of adult cells and tissues. Aberrant TGF-β signaling results in many human diseases, e.g. cancer and fibrosis (21,22). Importantly, the TGF-β pathway is involved in various metastatic processes and intensely influence the ability of cancer cells to spread throughout the body (23), nonetheless little is known about its molecular mechanism(s) or regulation. An essential step in TGF-β signal transduction is dependent on the translocation of the SMADs from the cytoplasm to the nucleus (24). Upon binding with ligands, the TGF-β type I receptors are activated and directly phosphorylate the receptor-regulated SMADs (R-SMADs), e.g., SMAD2/SMAD3, which subsequently form complexes with SMAD4, then together accumulate in the nucleus, where they are implicated in the regulation of transcription of target genes (25,26).
As palmitate is the most abundant free saturated fatty acid in human serum and in the diet (27), we examined the effects of palmitate on the TGF-β signaling pathway, which plays crucial roles in the pathogenesis of cancer (28), in human breast cancer cells. Here, we demonstrate that palmitate facilitates TGF-β-induced ERK activation and SAMD4 nuclear retention, thus promoting TGF-β–dependent cancer invasion. Using a HFD-induced obese animal xenograft model of BC, our results further explicated the therapeutic targets of the obesity-induced BC metastasis and exhibited pronounced promise to efficiently thwart obesity-related breast cancer progression. Additionally, tissue microarray analysis of overweight/obese breast cancer patient specimens further substantiated that phospho-ERK levels correlated with activated TGF-β signaling and metastasis. This study not only explains the crucial molecular mechanism by which FFA promotes the TGF-β signaling but also provides molecular characterization of the high FFA-induced BC metastasis.
Materials and methods
Cell culture, treatment and standard assays
Human breast cancer cell lines, MCF-7, MDA-MB-231, BT549 and MDA-MB-468 cells were obtained directly from the American Type Culture Collection (ATCC). MCF7-HER2 and MCF7-neo cells (MCF-7 cells transfected with empty vector) were kind gift from Dr. Kent Osborne (Baylor College of Medicine, Texas). These cells were authenticated by Bio-Synthesis, Inc. (Lewisville, TX), by short tandem repeat (STR) profiling and monitoring cell morphology and biologic behavior, and tested to exclude mycoplasma contamination before experiments. Cells were cultured not more than 3 months after resuscitation. Palmitic acid was added into the cell culture medium as PA-BSA complex as described in our previous work (29). Control groups were incubated with fatty acid-free BSA. Standard cell culture, immunoprecipitation and immunoblotting analysis, immunostaining analysis, quantitative real-time RT-PCR, in vitro invasion assay, wound-healing assay were carried out as described previously (30–33).
Nuclear and cytoplasmic fractionation
Subcellular fractionation was performed as described in our previous work (27). Nuclear and cytoplasmic fractions were assessed by immunoblotting of histone H3 and vinculin, GAPDH or actin, respectively, which were used as loading controls.
Plasmids and shRNAs
Expression plasmids for wild-type SMAD4 and two SMAD4 mutants T277A and T277D were obtained from Shanghai Harmonious One Biotech Co., Ltd. Plasmids were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or FuGENE 6 (Roche, San Francisco, CA). Human USP9x shRNAs and control shRNA lentiviral particles were obtained from Santa Cruz Biotechnology. For optimal shRNA plasmid transfection efficiency, Santa Cruz Biotechnology’s shRNA Plasmid Transfection Reagent and shRNA Plasmid Transfection Medium were used according to manufacturer’s Transfection Protocol.
In vivo model of obesity-induced metastases
Female athymic nude mice (4 weeks of age) were anesthetized and ovariectomized as described previously (34) and allowed to recover for two weeks. Diet-induced obesity was induced by feeding mice with a 45% kcal fat diet containing primarily lard with 17% sucrose (high fat sucrose diet, HFSD) for an indicated time (n=8/group). Control groups were fed with a standard chow-based diet (Chow). The mice were fed the aforementioned diets for six weeks and the body weight was measured every week. The cultured 4TO7 cells labeled with luciferase (4TO7-Luc+ cells) in the logarithmic phase were collected and diluted to 1×107 cells/mL. Then 100 µl cell suspension (in Matrigel) was inoculated into the second mammary fat pad on the right side of mice.
PD0325901 (Sigma-Aldrich) was dissolved originally in DMSO as a stock solution (50 mg/ml) (35). The stock solution was then diluted in water containing 0.05% (hydroxypropyl)methylcellulose and 0.02% Tween 80. The 250 µl PD0325901 formulation was administered to mice (25 mg/kg dose) by gavage three times a week for the duration of each study. Control mice were treated with vehicle by the same route. Tumor-bearing mice were administered with 100 µL of WP1130 suspension (40 mg/kg, i.p.) every other day. Treatments began one week after cell inoculations. Prior to tumor observation, mice were intraperitoneally injected with luciferin (1. 5 mg/10 g). After anesthesia with saturated Avertin (0.18 ml/10 g, i.p.), mice were placed in a Small Animal in Vivo Imaging System and images were acquired. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Wuhan University.
Statistical analysis
Statistical analysis in the current study was calculated with SPSS version 18.0 software (SPSS Inc.). Data were expressed as "mean value ± SD." The significance of mean values between 2 groups was determined by Student’s t test. All differences were two-sided. The significance of the data from patient specimens was analyzed by the χ2 test or the Pearson correlation coefficient test. A P value less than 0.05 was considered statistically significant.
Results
FFA treatment promotes TGF-β-induced ERK activation, SMAD4 nuclear accumulation and gene expression
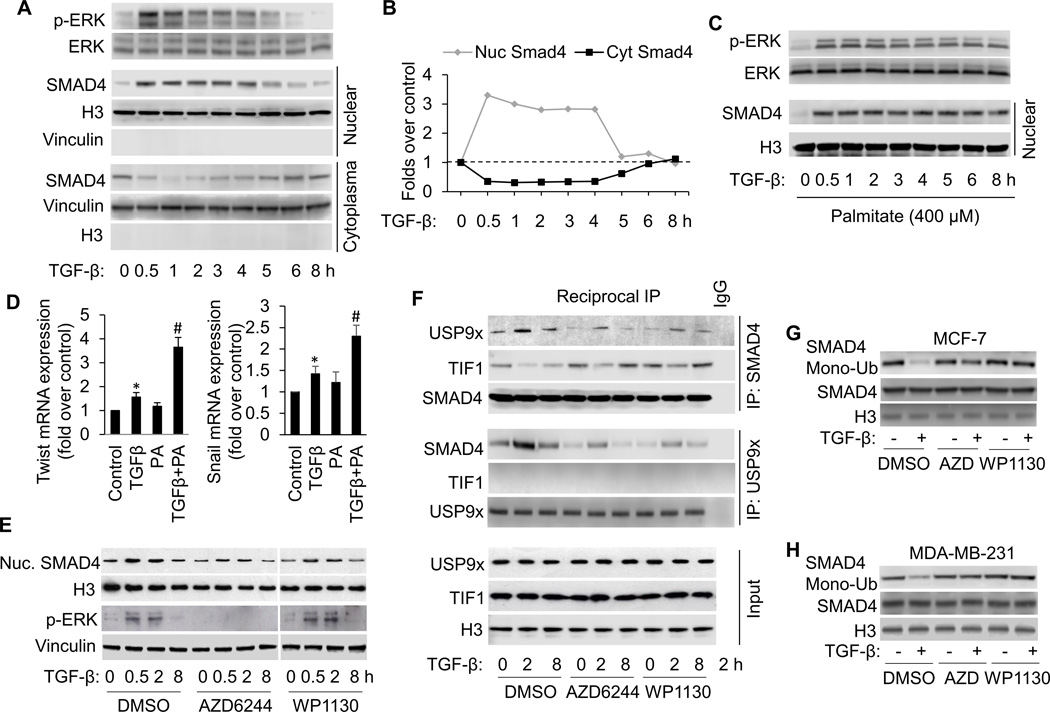
First, we investigated the fate of ERK and SMAD4 after prolonged TGF-β signaling. Nuclear and cytoplasmic extracts were prepared from MCF-7 cells incubated with TGF-β for different times. Nuclear and cytoplasmic fractions were assessed by immunoblotting of histone H3 and vinculin, respectively, which were used as loading controls. Control experiments demonstrated that there was no cross-contamination of nuclei with cytoplasm or vice versa (Fig. 1A and Supplementary Fig. S1A). The levels of p-ERK1/2 are increased at early times after TGF-β treatment and subsided to its basal levels after five hours of treatment. The transient increase induced by TGF-β is coincident with nuclear SMAD4. As shown in Figure 1A and B, SMAD4 levels start to increase in the nuclear fraction after a 30-min treatment with TGF-β. As the SMAD4 levels elevate in the nuclear extracts, it correspondingly drops in the cytoplasmic extracts. After 5 h of TGF-β treatment, the levels of SMAD4 in the nuclear fraction subside to its basal levels. After prolonged TGF-β stimulation (5 and 8 h), levels of SMAD4 in the cytoplasm augment again to approximately the levels observed in unstimulated cells (Fig. 1A and B). Therefore, the dynamics of nuclear SMAD4 could be due to its import into the nucleus from cytoplasm at the early stage of TGF-β stimulation and export of SMAD4 to the cytoplasm after prolonged TGF-β stimulation. The same experiment was performed in MDA-MB-231 cells (Supplemental Fig. S1). The levels of p-ERK and nuclear SMAD4 exhibit basically the same pattern of increase and decrease in MDA-MB-231 cells as it was observed in MCF-7 cells.
Figure 1.
FFA facilitates TGF-β-induced ERK activation, which promotes TGFβ-induced SMAD4 nuclear localization and gene expression by USP9x-SMAD4 interaction and nuclear SMAD4 deubiquitination. A, Western blot analysis of p-ERK and nuclear SMAD4 in MCF-7 cells treated with 3 ng/ml of TGF-β according to the indicated time points. B, The Western blots were quantitated, and the results are presented graphically to show the dynamics of SMAD4 nuclear cytoplasmic shuttling. C, Western blot analysis of p-ERK and nuclear SMAD4 in MCF-7 cells treated with 3 ng/ml of TGF-β in the presence of palmitate (PA, 400 µM) for the shown time periods. D, Induction of Twist and Snail mRNA expression in MCF-7 cells treated with TGFβ1 in the presence or absence of PA for 8 hours analyzed by qRT-PCR. All mRNA are normalized to PUM1 and presented as fold (mean ± SD) over Control based on three experiments. *p<0.05 versus Control; #p < 0.05 versus TGFβ. E, Western blot analysis of p-ERK and nuclear SMAD4 in MCF-7 cells treated with 3 ng/ml of TGF-β in the presence or absence of AZD6244 (1 µM) or WP1130 (5 µM). To detect nuclear USP9x-SMAD4 interaction, TIF1γ-SMAD4 interaction (F) and SMAD4 modifications (G and H), cells were also treated with 15 ng/ml of LMB to block SMAD4 nuclear export. F, Reciprocal co-immunoprecipitations (co-IP) were performed with lysates from MCF-7 cells using anti-SMAD4 and anti-USP9x antibodies and Western blot analysis was performed to detect SMAD4, USP9x and TIF1γ, respectively in IP elutes and in whole cell lysates (input). G and H, Nuclear SMAD4 ubiquitination was detected by transfecting cells with WT-SMAD4 and HA-ubiquitin, immunoprecipitation with anti-Smad4 antibody, followed by immunoblotting with anti-HA antibody.
To investigate the effect of FFA on p-ERK and nuclear SMAD4 levels, we treated MCF-7 cells with 400 µM PA, a concentration that mimics hyperlipidemia condition, for various durations. As shown in Fig. 1C and Supplementary Fig. S1C, incubation with PA further increases p-ERK and nuclear SMAD4 levels and maintain their high levels at the late stage of TGF-β exposure (5–8 h). Our data indicate that PA facilitates and stabilizes ERK activation which may lead to SMAD4 nuclear accumulation in human breast cancer cells.
To determine whether TGF-β target genes are further induced in response to PA, we performed qRT-PCR (Figure 1D). The expression of Twist and Snail mRNA is induced by TGF-β1. PA significantly promotes TGF-β-induced Twist mRNA expression with a maximal induction of 3.7-fold when compared to untreated controls. PA treatment alone does not significantly increase Twist mRNA levels.
ERK activation promotes TGFβ-induced SMAD4 nuclear accumulation via USP9x-SMAD4 interaction and nuclear SMAD4 deubiquitination
Nucleocytoplasmic shuttling of SMADs is a critical regulatory step in TGF-β signaling and plays an important role in controlling gene expression (36). In certain cases this intracellular trafficking is regulated frequently by post-translational modifications such as phosphorylation and ubiquitination (37). Next, we determined whether ERK, an upstream kinase of SMADs (38), and USP9x, a deubiquitinating enzyme essential for TGF-β signaling and SMAD4 monoubiquitination status (39), plays a role in TGF-β-induced SMAD4 nuclear accumulation. As indicated in Fig. 1E and Supplementary Fig. S1E, inhibition of ERK by AZD6244, a potent and selective ERK inhibitor, significantly abolishes PA-induced SMAD4 nuclear accumulation in breast cancer cells. Similar effects were also observed when cells were treated with USP9x DUB activity inhibitor WP1130. These results suggest that ERK and USP9x signalings are upstream events of SMAD4 nuclear accumulation.
To further substantiate mechanisms underlying the nuclear retention of SMAD4 by TGF-β, we examined levels of USP9x-SMAD4 interaction and SMAD4 monoubiquitination. As indicated in Fig. 1F, G and H and Supplementary Fig. S1F, G and H, TGF-β treatment dramatically increases USP9x-SMAD4 interaction and suppresses SMAD4 monoubiquitination in nuclear fraction. These effects are significantly reversed by inhibition of ERK and USP9x with their corresponding inhibitors. Intriguingly, USP9x-SMAD4 interaction negatively correlates with TIF1γ-SMAD4 interaction, implying that USP9x selective binding to SMAD4 in competition with TIF1γ facilitates nuclear SMAD4 deubiquitination and retention.
Activation of ERK is required for FFA promotion of TGF-β-induced nuclear USP9x-SMAD4 interaction, SMAD4 deubiquitination, SMAD3-SMAD4 complex formation and gene expression induction
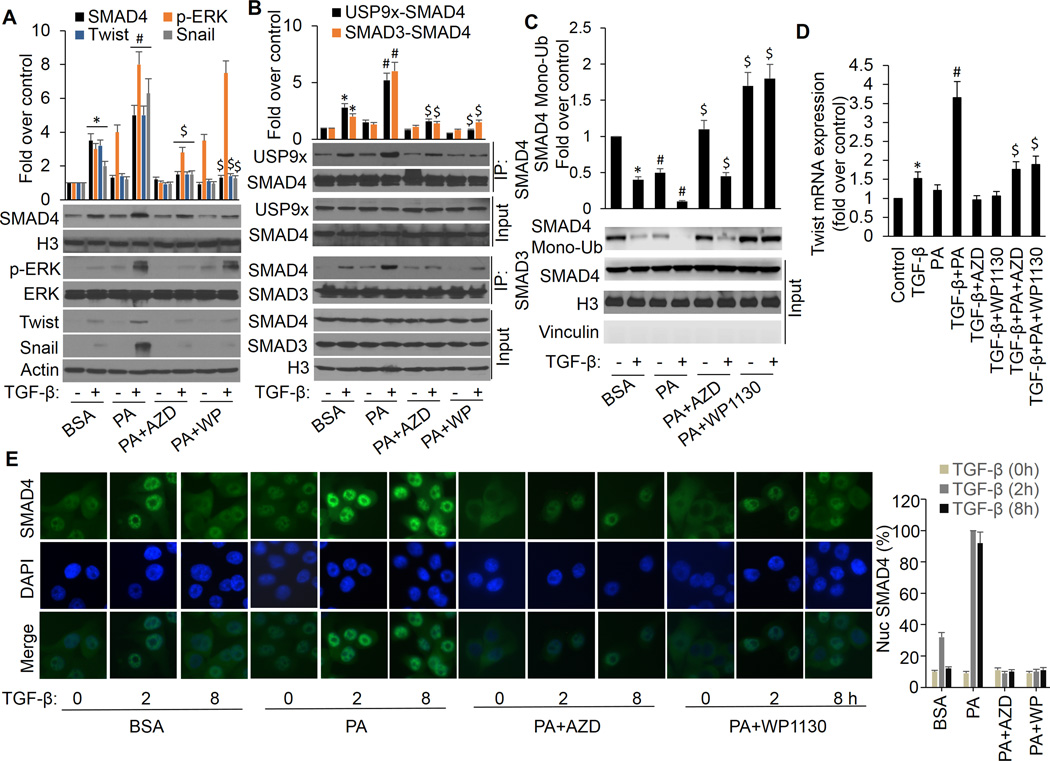
To substantiate the effects of FFA on TGF-β signaling pathway, we show that PA further promotes TGF-β-induced ERK phosphorylation and SMAD4 nuclear retention, and that these effects are completely blocked by AZD6244. In addition, inhibition of USP9x by WP1130 ablates PA promotion of TGF-β-induced SMAD4 nuclear retention (Fig. 2A). These results were further confirmed by immunofluorescence staining that PA further enhances TGF-β-induced SMAD4 nuclear retention and cells co-incubated with PA and AZD6244 or WP1130 exhibited a decrease of nuclear SMAD4 in response to TGF-β (Fig. 2E and Supplementary Fig. S2B). To further explore the molecular mechanism(s) leading to this observation, we analyzed nuclear SMAD4 protein interaction and modification. We demonstrate that PA further increases TGF-β-induced nuclear USP9x-Smad4 interaction, SMAD4 deubiquitination and SMAD3-SMAD4 complex formation, and these effects completely blocked by either AZD6244 or WP1130 (Fig. 2B, C and Supplementary Fig. S2A). Furthermore, PA can further enhance TGF-β-induced expression of target genes, Twist and Snail, at protein and mRNA levels, which can be abolished by inhibition of ERK and USP9x with their pharmaceutical inhibitors (Fig. 2A and D). Taken together, these results suggest that ERK-induced nuclear USP9x-SMAD4 interaction and SMAD4 deubiquitination are affected by PA resulting in the aggravated and sustained activation of TGF-β signaling and higher expression of downstream target genes. Such effects may enhance the risk of breast cancer development and metastasis.
Figure 2.
ERK activation is responsible for FFA promotion of TGF-β-induced USP9x-SMAD4 interaction, SMAD3-SMAD4 complex formation, nuclear SAMD4 retention, and gene expression. MCF-7 cells were treated with BSA or PA in the presence or absence of AZD6244 or WP1130 for 4 h followed by treatment with or without 3 ng/ml of TGF-β for another 4 h. A, Nuclear extracts were made to analyze nuclear SMAD4 levels. Whole-cell extracts were prepared and subjected to Western analysis using p-ERK, ERK, twist, snail and actin antibodies. B, nuclear extracts were made and coimmunoprecipitation of endogenous SMAD4 with USP9x or SMAD3 were performed. Data are presented as mean fold increases (±SD) in treated groups over basal values from three independent experiments. *p<0.01 vs controls; #p<0.01 vs BSA/+TGF-β; $p<0.01 vs PA/+TGF-β. C, nuclear extracts were made and SMAD4 monoubiquitination was detected as described above. *p<0.01 vs controls (BSA/−TGF-β); #p<0.01 PA vs BSA; $p<0.01 PA+AZD vs PA, PA+WP1130 vs PA. D, total RNA was extracted and analyzed for twist mRNA by real-time PCR. *p<0.05 vs controls; #p<0.01 vs TGF-β; $p<0.01 vs PA/+TGF-β. E, MCF-7 cells were treated with TGF-β in the presence or absence of PA, AZD6244 or WP1130 and processed for immunofluorescence with anti-SMAD4 antibody. The same cells were also stained with DAPI to visualize nuclei. Intensity of nuclear SMAD4 among these cells was quantified with Image-Pro Plus 6.0 software. The percentages of nuclear SMAD4 levels illustrated at the right panel represent the mean of three independent experiments, and error bars indicate the SD.
FFA enhances TGF-β-induced invasion and migration by activating ERK and USP9x
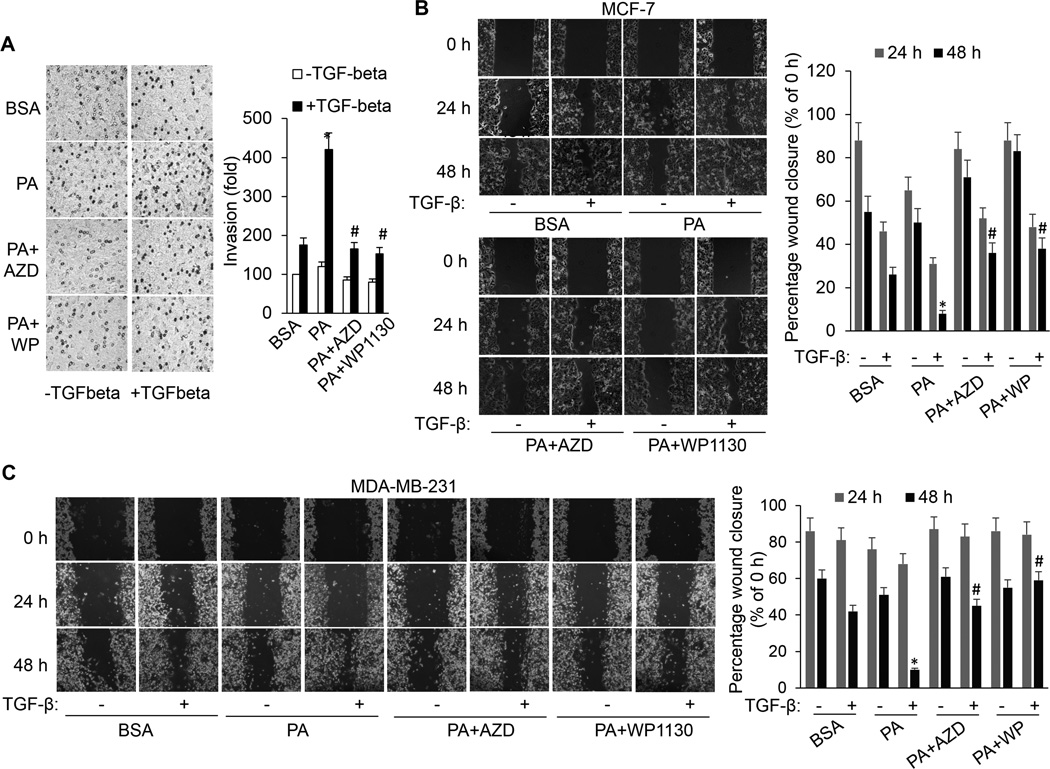
The capability of cancer cells to undergo invasion and migration allows them to change location within the tissues and is crucial for cancer progression and metastasis. Since TGF-β/SMAD signaling is the primary pathway regulating invasion in breast cancer (40), next, we determined whether FFA has an effect on TGF-β1–induced invasion and migration of breast cancer cells. PA treatment augments the TGF-β1–induced invasion and migration of MCF-7 and MDA-MB-231 cells (Fig. 3). To ascertain that the effect of FFA on the TGF-β1–induced invasion and migration is mainly dependent on ERK and USP9x, we inhibited ERK and USP9x using their pharmaceutical inhibitors. Inhibition of ERK and USP9x in MCF-7 and MDA-MB-231 cells significantly decreases the promoting effect of PA on the TGF-β1–induced invasive and migration ability of these cells (Fig. 3). Moreover, the scratch assays were also performed with one additional cell line, BT549 (Supplemental Fig. S3). Collectively, these results suggest that ERK and USP9x play critical roles in the regulation of TGF-β1–induced invasion and migration of breast cancer cells in response to high FFA.
Figure 3.
FFA promotes TGF-β-induced invasion and migration by activating ERK and USP9x. A, In vitro invasion assay performed on MCF-7 cells that were treated with BSA or PA in the presence or absence of AZD6244 or WP1130 with or without TGF-β1 for 20 hours. Each column (right panel) represents the mean (± SD) results of two independent experiments. *p<0.05 versus BSA; #p<0.05 versus PA. (B) MCF-7 and (C) MDA-MB-231 cells were scratched with a 20-µl pipette tip and then incubated with BSA, PA, PA plus AZD6244 or WP1130 in the absence or presence of TGF-β1. Migrating cells were photographed under a phase contrast microscope. The percentage of the wound closed was quantified from three independent replicates and is expressed as mean ± SD. *p<0.05 vs BSA/TGF-β; #p<0.05 vs PA/TGF-β.
SMAD4 T277 phosphorylation mediates FFA promotion of TGF-β-induced USP9x-SMAD4 interaction, nuclear SAMD4 retention and SMAD3-SMAD4 complex formation
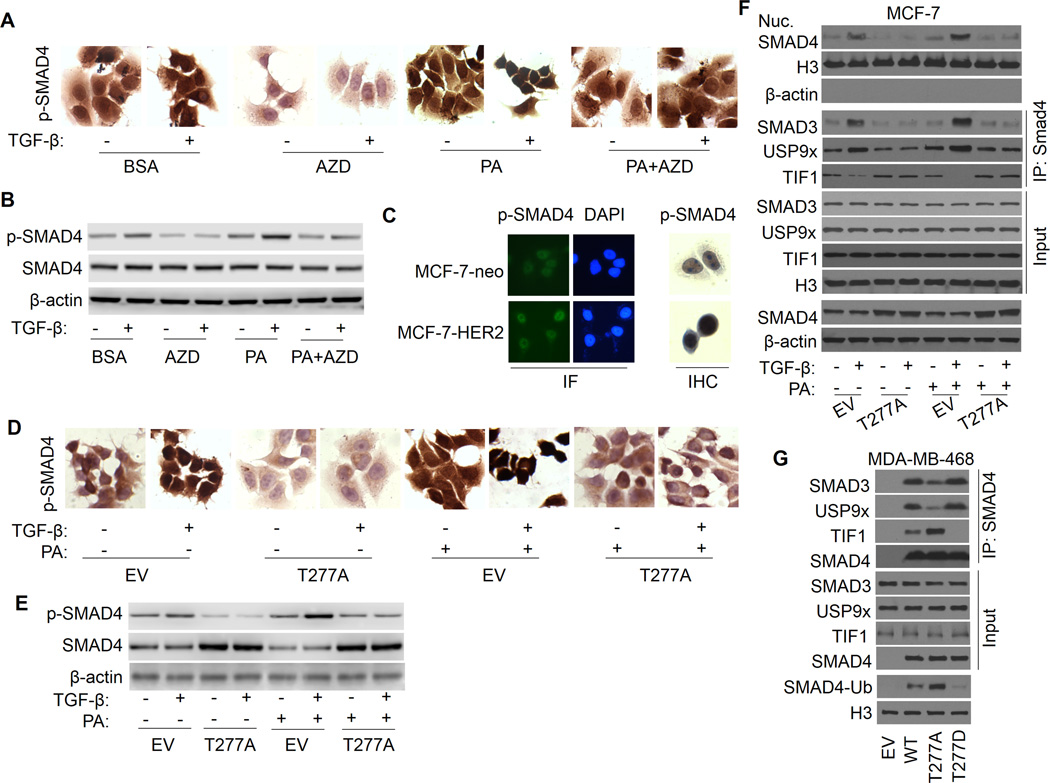
It has been shown that SMAD4 can be constitutively phosphorylated (41), nonetheless the phosphorylation sites, the upstream kinases related to these phosphorylations and the implication of the phosphorylations in homo sapiens have not yet been clarified. A study from Roelen et al. (42) suggests that MAP kinase can phosphorylate SMAD4 at threonine 276 (T276) in Sus scrofa kidney epithelial cells LLC-PK1, and that this phosphorylation is imperative for TGF-β-induced nuclear accumulation and transcriptional activity of SMAD4. Here, to determine the phosphorylation sites in human breast cancer cells that is responsible for PA/ERK-induced SMAD4 nuclear accumulation and its transcriptional activity, we analyzed SMAD4 T277, the orthologous residue in human. Cellular SMAD4 phosphorylation at T277 was measured by immunohistochemistry (IHC) and immunofluorescence (IF) staining with anti-T277 antibody. As indicated in Fig. 4A, PA treatment significantly increases TGF-β-induced SMAD4 phosphorylation at T277, and the phosphorylated SMAD4 gathers mainly in nuclear region. AZD6244 can significantly inhibit TGF-β-induced SMAD4 phosphorylation and abolishes PA promotion of TGF-β-induced SMAD4 phosphorylation. These results suggest that SMAD4 T277 is an important phosphorylation site that can be phosphorylated by ERK in human breast cancer cells and is associated with SMAD4 nuclear accumulation in response to FFA (Fig. 4A). SMAD4 T277 phosphorylation under different treatments was confirmed by Western blot (Fig. 4B and Supplementary Fig. S4B). Since HER-2 over-expressing MCF7 cells (MCF-7-HER2) have a significant increased ERK activation (43), we were interested to know whether this cell line has higher levels of SMAD4 phosphorylation at T277. As shown in Fig. 4C, MCF-7-HER2 cells have elevated levels of constitutive SMAD4 T277 phosphorylation and nuclear accumulation compared with its control MCF-7-neo cells.
Figure 4.
SMAD4 T277 phosphorylation is responsible for FFA promotion of TGF-β-induced USP9x-SMAD4 interaction, nuclear SAMD4 retention, SMAD3-SMAD4 complex formation. A, MCF-7 cells were treated with AZD6244, PA or PA plus AZD6244 in the presence or absence of TGF-β for 2 h and stained with IHC method using anti-phospho-SMAD4 (Thr277) antibody. B, Whole-cell extracts were prepared and subjected to Western analysis using anti-phospho-SMAD4, SMAD4 and actin antibodies. C, MCF-7-neo and MCF-7-HER2 cells were stained with IF and IHC method using anti-phospho-SMAD4 antibody. D, MCF-7 cells transfected with SMAD4-T277A or empty vector control (EV) were treated with BSA or PA in the presence or absence of TGF-β1 and stained with IHC method for anti-phospho-SMAD4 antibody. E, Whole-cell extracts were prepared and subjected to Western analysis using anti-phospho-SMAD4, SMAD4 and actin antibodies. F, MCF-7 cells were transfected with EV or SMAD4-T277A, and nuclear extract was prepared. SMAD4-SMAD3, SMAD4-USP9x, and SMAD4-TIF1γ interaction were determined by immunoprecipitation with SMAD4 antibody, followed by immunoblotting with SMAD3, USP9x, or TIF1γ antibody. G, MDA-MB-468 cells were transfected with EV, WT-SMAD4, SMAD4-T277A, or SMAD4-T277D, and nuclear SMAD4-SMAD3, SMAD4-USP9x, and SMAD4-TIF1γ interaction were determined by immunoprecipitation and immunoblotting as described above. SMAD4 monoubiquitination was also detected.
To further test this notion directly, MCF-7 cells were transfected with SMAD4 phosphorylation defect mutant T277A. As shown in Fig. 4D, 4E and Supplementary Fig. S4E, IHC staining and Western analysis demonstrates that T277A transfection significantly inhibits TGF-β-induced SMAD4 phosphorylation and abolishes PA promotion of TGF-β-induced SMAD4 phosphorylation, as well as diminishes SMAD4 nuclear retention. This result was also substantiated by Western analysis showing that T277A dramatically reverses TGF-β-induced SMAD4 nuclear accumulation and abolishes PA promotion of TGF-β-induced SMAD4 nuclear accumulation, accompanied by decreased USP9x-SMAD4 interaction and increased TIF1γ-SMAD4 binding (Fi. 4F and Supplementary Fig. S4F). These results suggest that T277 phosphorylation may regulate USP9x-SMAD4 interaction and SMAD4 ubiquitination status, influencing SMAD4 subcellular localization. Next, to further substantiate this possibility, SMAD4-null breast cancer cell line MDA-MB-468 was transfected with SMAD4 phosphorylation defect mutant T277A and phosphorylation mimic mutant T277D. As shown in Fig. 4G and Supplementary Fig. S4G, T277A undergoes less USP9x-SMAD4 interaction, more TIF1γ-SMAD4 binding and SMAD4 monoubiquitination than WT protein, while T277D shows lower level of SMAD4 monoubiquitination, implying that SMAD4 T277 phosphorylation facilitates USP9x-SMAD4 interaction and reduces the chance of SMAD4 to bind with TIF1γ, thus decreasing SMAD4 monoubiquitination, which is important for its nuclear localization, combination with SMAD3 and transcriptional activity.
USP9x selective binding to SMAD4 in competition with TIF1γ promotes nuclear SMAD4 retention, SMAD3-SMAD4 complex formation and target gene expression
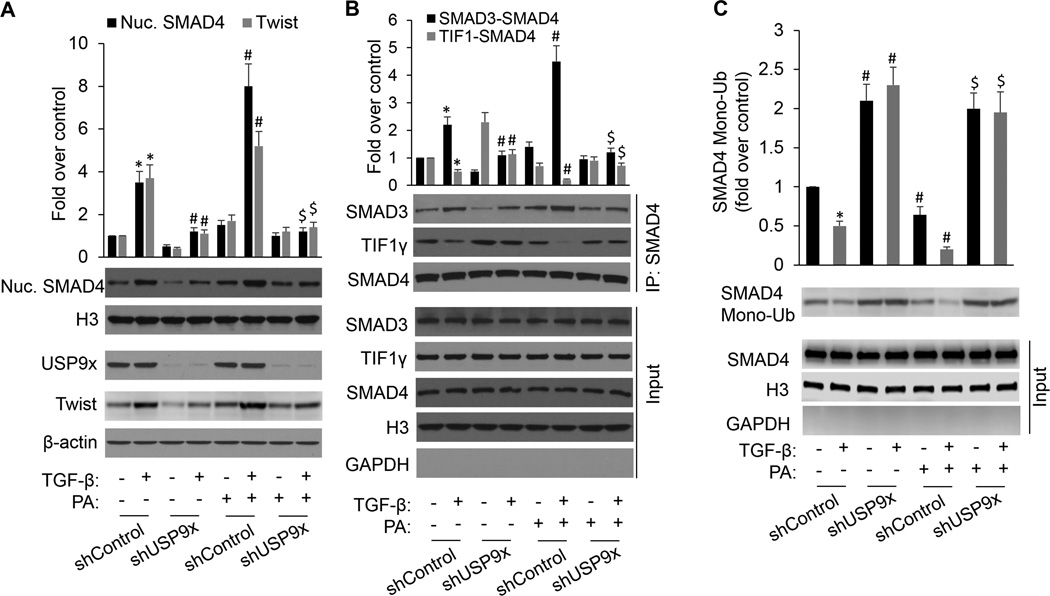
To further substantiate the physiological role of USP9x in cellular response to PA, we employed shRNA mediated knockdown of USP9x in MCF-7 cells. As with our USP9x knockdown studies, control shRNA was used as a control. Knockdown of USP9X was confirmed by western blot analysis (Fig. 5A). USP9x shRNA delivery significantly inhibits TGF-β-induced SMAD4 nuclear retention and eliminates PA promotion of TGF-β-induced SMAD4 nuclear retention. Accordingly, USP9x knockdown could block TGF-β-induced Twist expression and inhibited the promoting effect of PA on TGF-β-induced Twist expression. Intriguingly, USP9X-deficient cancer cells exhibit higher levels of TGF-β-induced TIF1γ-SMAD4 interaction and SMAD4 mono-ubiquitination, and lower levels of SMAD3-SMAD4 interaction under the conditions of either BSA or PA exposure relative to control cells (Fig. 5B, 5C and Supplementary Fig. S5). Taken together, these results demonstrate that USP9x selectively binds to SMAD4 in competition with TIF1γ and deubiquitinates SMAD4, promoting nuclear SMAD4 retention, SMAD3-SMAD4 complex formation and target gene expression. Our results, for the first time, directly show that SMAD4 posttranslational modifications including phosphorylation and ubiquitination regulate its nuclear-cytoplasmic shuttling.
Figure 5.
USP9x selective binding to SMAD4 in competition with TIF1γ facilitated nuclear SMAD4 retention, formation of the nuclear SMAD3-SMAD4 complex and target gene expression. A, MCF-7 cells stably expressing control shRNA or USP9x shRNA were incubated with BSA or PA in the presence or absence of TGF-β1 for 2 h. Nuclear SMAD4, total USP9x and Twist protein expression were analyzed by immunoblotting. B, Nuclear SMAD4-SMAD3 and SMAD4-TIF1γ interaction were determined by immunoprecipitation with SMAD4 antibody, followed by immunoblotting with SMAD3 or TIF1γ antibody. C, MCF-7 cells were transfected with WT-SMAD4, HA-ubiquitin, and the indicated shRNAs. The cells were treated with PA in the presence or absence of TGF-β1 for 2 h and nuclear extracts were made. The SMAD4 monoubiquitination (Mono-Ub) was detected by anti-SMAD4 immunoprecipitation and immunoblot with HA-ubiquitin. *p<0.01 vs shControl/-TGF-β/-PA; #p<0.01 vs shControl/−PA; $p<0.01 vs shControl/+PA.
ERK and USP9x is responsible for obesity-related breast cancer metastasis
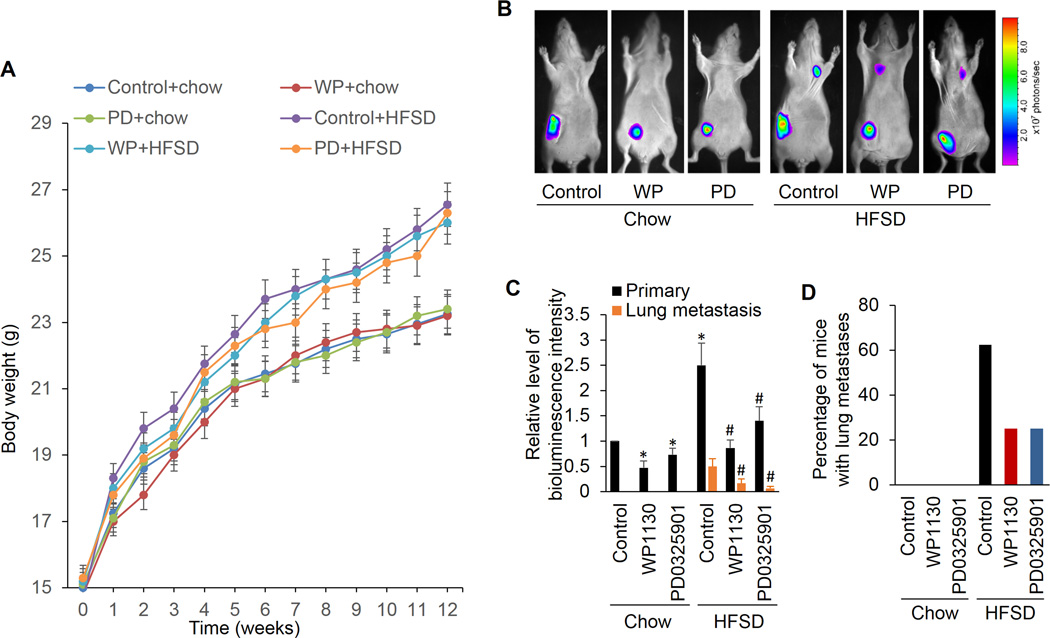
We next ascertained if the ERK and USP9x pathway observed in cell culture could modulate breast cancer metastasis in obese animal models. Plasma FFA levels are significantly increased in overweight/obese individuals (44) and hyperlipidemia in metabolic syndrome is characterized by an increase in FFA (45). To simulate such diet-induced metabolic abnormalities, a nude mouse model of diet-induced obesity was employed in the current study by feeding ovariectomized (OVX) athymic nude mice a high fat sucrose diet (HFSD) (46). As indicated in Figure 6A, the body weight is remarkably higher in mice fed the HFSD versus mice fed chow diet. The OVX athymic nude mice implanted with 4TO7 cells into the mammary fat pads do not produce spontaneous lung metastases when fed with chow diet (Fig. 6B). However, approximately 63% of the HFSD-induced obese mice produce lung metastases (Fig. 6B, 6C and 6D), suggesting that high fat diet and obesity are related to the metastatic phenotype. Importantly, the ability of the breast cancer cells to metastasize to the lung in nude mice and lung metastatic lesions were significantly repressed by inhibition of ERK and USP9x with administration of their inhibitors. These data suggest that ERK and USP9x are responsible for the metastasis formation in obese mice.
Figure 6.
The ERK and USP9x inhibitors inhibits the stimulative effect of obesity on metastases in vivo. A, Diet-induced obesity in ovariectomized (OVX) athymic nude mice that were fed with a high fat sucrose diet (HFSD). A standard chow-based diet (Chow) was used as the control diet (n=8/group). The mice were fed the diets for 12 weeks and the body weight of each mouse was measured every week. B, Tumor growth and metastasis in diet-induced obese OVX athymic nude mice versus the control animal group with the regular diet. After six weeks of feeding, luciferase-labeled 4TO7 tumor cells were implanted into the mammary fat pad on the right side of mice. One week after cell inoculations, PD0325901 was administered to mice (25 mg/kg dose) by gavage three times a week. Control mice were treated with vehicle by the same route. Tumor-bearing mice were administered with WP1130 suspension (40 mg/kg, i.p.) every other day. Six weeks after inoculation, mice were intraperitoneally injected with fluorescein substrate (1.5 mg/10 g). After anesthesia with saturated avertin (0.18 ml/10 g, i.p.), mice were placed in a Small Animal in Vivo Imaging System and images were acquired. C, The photon quantity in each mice group was analyzed (bottom), *p<0.05 vs Chow/Control; #p<0.05 vs HFSD/Control. D, Percentage of mice with lung metastases, as determined by lung bioluminescence.
ERK and SMAD4 phosphorylation levels correlate with twist expression and overweight/obesity-related metastasis in human breast cancer
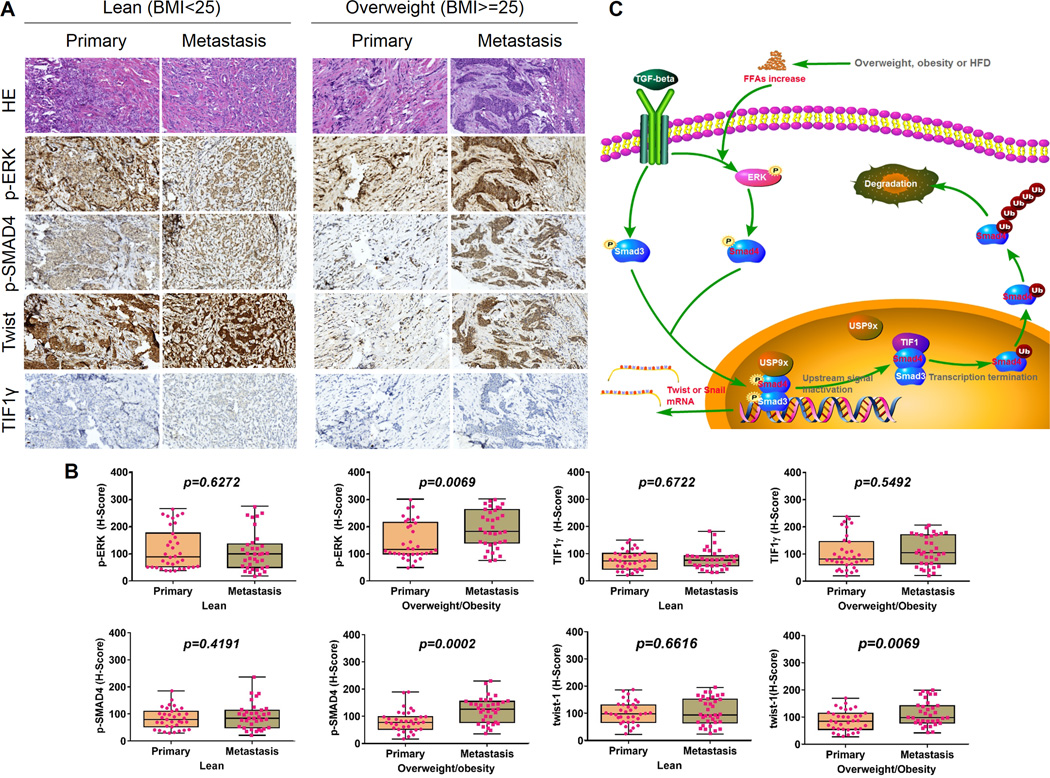
In an attempt to investigate the clinical relevance of ERK and SMAD4 pathway in cancer progression and metastasis, we examined the expression levels of p-ERK, p-SMAD4-T277, TIF1γ, and Twist in paired samples of breast primary cancers and matched metastases in lymph nodes from 35 lean (BMI 18.5–24.9) and 35 overweight (BMI 25.0–29.9) and obese patients (BMI>=30.0) via tissue microarray IHC staining. The levels of p-ERK, p-SMAD4 and Twist are significantly higher in the metastases than in primary cancers from overweight/obese but not lean patients (Fig. 7A and B). On the other hand, in this set of primary and metastatic tumors, expression level of TIF1γ is not related to metastatic status. This data are in corroboration with our findings in breast cancer cell lines and xenograft models and suggest that the induction of p-ERK, p-SMAD4 and Twist levels are important for metastasis, particularly in overweight/obese patients that usually have high serum FA levels. The levels of p-ERK and p-SMAD4 in the metastases but not primary cancers are significantly higher in overweight/obese than lean patients (Supplemental Fig. S6). Taken together, our results highlight the clinical relevance of ERK overactivation under the abnormal metabolic conditions which contributes to aberrant TGF-β signaling in breast cancer metastasis.
Figure 7.
Phospho-ERK and –SMAD4 expression show significant correlation with overweight/obesity-related metastasis in human breast cancer. A, The expression of phospho-ERK, phospho-SMAD4 and TIF1γ in representative cases of primary breast tumors and matched lymph node metastasis tissue specimens from 35 lean (BMI 18.5–24.9) and 35 overweight (BMI 25.0–29.9) or obese (BMI>=30.0) patients. Original magnification, ×200. B, Staining was analyzed by using a system based on the percentage of positively stained cells and the staining intensity. Integrated optical density of all the positive staining in each image was determined, and its ratio to total area of each photograph was calculated as density. C, The identified signaling pathway in this study indicating that FFA promotes metastasis via the TGF-β1 pathway. TGF-β transiently activates the ERK and subsequently marks SMAD4 for activation via Thr277 phosphorylation, which facilitates SMAD4-USP9x interaction, SMAD4 nuclear retention, SMAD3-SMAD4 complex, and stimulates TGF-β/SMAD3–mediated transcriptional activity of Twist and Snail. USP9x competitively inhibited TIF1γ from binding and monoubiquitinating SMAD4. In the presence of high FFA, FFA further facilitates the described pathway by sustaining TGF-β-induced ERK activation, SMAD4 phosphorylation, SMAD4 nuclear retention and SMAD3/SMAD4 formation, thus over-activating this system and promoting TGF-β–dependent cancer progression.
Discussion
In the present study, we reveal that the USP9x-SMAD4 interaction represents a pivotal mechanism for regulating TGF-β signaling and BC cell invasion and metastasis. TGF-β activates ERK and subsequently marks SAMD4 for phosphorylation at Thr277. In this study for the first time we showed, that in human breast cancer cells, phosphorylation of SMAD4 at Thr277 facilitates USP9x-SMAD4 interaction. Such interaction, contributes to low monoubiquitination status of SMAD4, SMAD4 nuclear retention, SMAD3-SMAD4 complex formation and implement their transcription function. Under normal physiological conditions, induction of signaling by TGF-β is not sustained for very long and the signaling cascade is terminated upon the restoration of ERK activity, leading to the reinstatement of SAMD4 Thr277 phosphorylation. Consequence of such event is USP9x-SMAD4 disassociation and then SMAD4-TIF1γ interaction, which marks SMAD4 for mono-ubiquitination, resulting in SMAD4 nuclear export. In both humans and rodents, consuming HFD or being overweight/obese is usually accompanied with high levels of plasma FFA. In this study for the first time we showed that high FFA intensifies TGF-β-induced ERK activation and SAMD4 phosphorylation, USP9x-SMAD4 interaction, and nuclear accumulation, thus promoting TGF-β–dependent cancer development and progression. On the other hand inhibition of ERK and USP9x with pharmaceutical or genetic approach in breast cancer cells abolishes FFA promotion of TGF-β–induced SMAD4-USP9x interaction, SMAD4 nuclear retention, SMAD3/SMAD4 complex formation, and target gene expression, resulting in decrease in cancer cell invasion and metastasis (Figure 7C).
In vivo studies further substantiate that inhibition of ERK and USP9x suppressed obesity-induced metastasis. These findings are highlighted by the significant clinical observation that phospho-ERK levels positively correlated with activated TGF-β signaling and metastasis in overweight/obese breast cancer patients. These data explain not only how TGF-β signaling pathway functions under the physiological and pathological conditions, but also provides a novel mechanistic explanation of why overweight/obesity/high FFA increases the risk of developing distant metastatic recurrence and is associated with poor prognosis of BC. In addition, we provide the evidence that ERK and USP9x specific inhibitors can be used as therapeutic targets for treatment of obesity-related breast cancer.
SMAD4 plays a crucial role in all TGF-β signaling pathways and has been identified as a tumor suppressor. The role of SMAD4-mediated TGF-β signaling in tumor progression and metastasis is contentious. Here, we demonstrate that SMAD4 nuclear-cytoplasm shuttling is dependent on its posttranslational modifications including phosphorylation and monoubiquitination which play counterbalancing roles for maintaining a delicate and an accurate TGF-β physiological signaling. We also found that FFA/ERK-induced SMAD4 T277 phosphorylation is indispensable in SMAD4-USP9x interaction and its deubiquitination as well as subsequent nuclear accumulation, which is required for TGF-β–induced breast cancer invasion and metastasis under the high FFA/obesity conditions. These findings significantly extend our understanding of molecular mechanisms underlying SMAD4-mediated TGF-β signaling in breast cancer progression, which might facilitate the development of effective therapies targeting TGF-β signaling for the treatment of human tumors.
The normal cellular protein ubiquitination levels are maintained by the ubiquitin ligases and deubiquitinating enzymes (DUB), two enzyme families with opposite activities (47). The X-linked deubiquitinase USP9X belongs to the family of DUB enzymes. It controls various cellular functions e.g., mediating cell survival (48), regulating cell adhesion molecules, apoptosis, cell polarity, chromosome segregation, NOTCH, mTOR, and TGF-β signaling (39,49–51), by deubiquitinating and stabilizing its substrates. A decade ago, in a study in Xenopus Dupont et al. (52) identified USP9x as the SMAD4 deubiquitinase and described its counteracting protein, SMAD4 ubiquitin ligase TIF1γ. They also revealed that TIF1γ antagonizes TGF-β signals through binding to SMAD4 and enhancing its ubiquitylation and that USP9x is a TGF-β pathway component required for SMAD4 activity. In addition, studies in the fly wing suggested the epistatic relationship between USP9x and TIF1γ in the competition for SMAD4 (39). Consistent with these studies, we, for the first time, observed that in human breast cancer cells, USP9x selectively binds to SMAD4 in competition with TIF1γ. In these cells USP9x facilitates nuclear SMAD4 deubiquitination and retention, which in turn promots target gene transcription. Therefore, the counterbalancing activities of USP9x and TIF1γ fine tune SMAD4 function in response to TGF-β signals. More importantly, depletion of USP9X hinders motility of MDA-MB-231 metastatic breast cancer cells (53), which may be an indication for the role of this protein in tumor cells invasion and metastasis. Clinically, USP9x overexpression correlates with poor prognosis in human non-small cell lung cancer, multiple myeloma and esophageal squamous cell carcinoma (54,55). Moreover, tumors with low USP9x expression are particularly sensitive to some of the conventional therapeutic agents (48). Accordingly, here we demonstrate that the inhibition of USP9x in BC cells abolishes FFA promotion of TGF-β–induced SMAD4 nuclear retention, SMAD3/SMAD4 complex formation, and target gene expression, resulting in suppression of cancer cell invasion and metastasis. Furthermore, inhibition of USP9x in vivo suppressed obesity-induced breast cancer metastasis.
In conclusion, our results indicate that TGF-β transiently activates ERK and subsequently marks SMAD4 for activation via Thr277 phosphorylation, which facilitates SMAD4-USP9x interaction, SMAD4 nuclear retention, SMAD3-SMAD4 complex, and promotes TGF-β-mediated target gene transcription. USP9x competitively inhibits TIF1γ from binding and monoubiquitinating SMAD4, thus maintains SMAD4 nuclear retention and stabilizes the SMAD3/SMAD4 complex in the nucleus. High FFA further intensifies TGF-β-induced ERK activation, SMAD4 phosphorylation, SMAD4 nuclear retention and SMAD3/SMAD4 formation, thus over-activating this system and promoting TGF-β–dependent cancer progression (Figure 7C). Our findings suggest that FFA overactivation of ERK stimulates cancer cell invasion and metastasis, whereas inhibition of ERK or USP9x hinders obesity/high FFA-associated breast cancer metastasis. These findings shed light on mechanisms of cancer progression and invasion and provide strategies for therapeutic intervention of cancer progression.
Supplementary Material
Acknowledgments
Financial support: This work was supported in part by NIH-NIMHD U54MD007598, NIH/NCI 1U54CA14393; U56 CA101599-01;Department-of-Defense Breast Cancer Research Program grant BC043180, NIH/NCATS CTSI UL1TR000124 (to J.V. Vadgama) and Accelerating Excellence in Translational Science Pilot Grants G0812D05, NIH/NCI SC1CA200517 (to Y. Wu).
Footnotes
Disclosure of Potential Conflicts of Interest The authors declare no competing financial interests.
REFERENCES
- 1.World Health Organization. http://www.who.int/mediacentre/factsheets/fs297/en/index.html%5d.
- 2.MacLennan M, Ma DW. Role of dietary fatty acids in mammary gland development and breast cancer. Breast cancer research : BCR. 2010;12(5):211. doi: 10.1186/bcr2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stebbing J, Sharma A, North B, Athersuch TJ, Zebrowski A, Pchejetski D, et al. A metabolic phenotyping approach to understanding relationships between metabolic syndrome and breast tumour responses to chemotherapy. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2012;23(4):860–866. doi: 10.1093/annonc/mdr347. [DOI] [PubMed] [Google Scholar]
- 4.Sarkissyan M, Wu Y, Vadgama JV. Obesity is associated with breast cancer in African-American women but not Hispanic women in South Los Angeles. Cancer. 2011;117(16):3814–3823. doi: 10.1002/cncr.25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demark-Wahnefried W, Platz EA, Ligibel JA, Blair CK, Courneya KS, Meyerhardt JA, et al. The role of obesity in cancer survival and recurrence. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21(8):1244–1259. doi: 10.1158/1055-9965.EPI-12-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ligibel J. Obesity and breast cancer. Oncology. 2011;25(11):994–1000. [PubMed] [Google Scholar]
- 7.Kushi L, Giovannucci E. Dietary fat and cancer. The American journal of medicine. 2002;113(Suppl 9B):63S–70S. doi: 10.1016/s0002-9343(01)00994-9. [DOI] [PubMed] [Google Scholar]
- 8.Freedman LS, Clifford C, Messina M. Analysis of dietary fat, calories, body weight, and the development of mammary tumors in rats and mice: a review. Cancer research. 1990;50(18):5710–5719. [PubMed] [Google Scholar]
- 9.Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. Journal of the National Cancer Institute. 1981;66(6):1191–1308. [PubMed] [Google Scholar]
- 10.Kinlaw WB, Baures PW, Lupien LE, Davis WL, Kuemmerle NB. Fatty Acids and Breast Cancer: Make them on Site or have them Delivered. Journal of cellular physiology. 2016 doi: 10.1002/jcp.25332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boden G. Obesity and free fatty acids. Endocrinology and metabolism clinics of North America. 2008;37(3):635–646. viii–ix. doi: 10.1016/j.ecl.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv W, Yang T. Identification of possible biomarkers for breast cancer from free fatty acid profiles determined by GC-MS and multivariate statistical analysis. Clinical biochemistry. 2012;45(1–2):127–133. doi: 10.1016/j.clinbiochem.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Saadatian-Elahi M, Norat T, Goudable J, Riboli E. Biomarkers of dietary fatty acid intake and the risk of breast cancer: a meta-analysis. International journal of cancer Journal international du cancer. 2004;111(4):584–591. doi: 10.1002/ijc.20284. [DOI] [PubMed] [Google Scholar]
- 14.Shannon J, King IB, Moshofsky R, Lampe JW, Gao DL, Ray RM, et al. Erythrocyte fatty acids and breast cancer risk: a case-control study in Shanghai, China. The American journal of clinical nutrition. 2007;85(4):1090–1097. doi: 10.1093/ajcn/85.4.1090. [DOI] [PubMed] [Google Scholar]
- 15.Menendez JA, Decker JP, Lupu R. In support of fatty acid synthase (FAS) as a metabolic oncogene: extracellular acidosis acts in an epigenetic fashion activating FAS gene expression in cancer cells. Journal of cellular biochemistry. 2005;94(1):1–4. doi: 10.1002/jcb.20310. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura I, Kimijima I, Zhang GJ, Onogi H, Endo Y, Suzuki S, et al. Fatty acid synthase expression in Japanese breast carcinoma patients. International journal of molecular medicine. 1999;4(4):381–387. doi: 10.3892/ijmm.4.4.381. [DOI] [PubMed] [Google Scholar]
- 17.Louie SM, Roberts LS, Mulvihill MM, Luo K, Nomura DK. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochimica et biophysica acta. 2013;1831(10):1566–1572. doi: 10.1016/j.bbalip.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer research. 2011;71(9):3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 19.Olsen AM, Eisenberg BL, Kuemmerle NB, Flanagan AJ, Morganelli PM, Lombardo PS, et al. Fatty acid synthesis is a therapeutic target in human liposarcoma. International journal of oncology. 2010;36(5):1309–1314. doi: 10.3892/ijo_00000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts AB. Molecular and cell biology of TGF-beta. Mineral and electrolyte metabolism. 1998;24(2–3):111–119. doi: 10.1159/000057358. [DOI] [PubMed] [Google Scholar]
- 21.Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 22.Roberts AB, Russo A, Felici A, Flanders KC. Smad3: a key player in pathogenetic mechanisms dependent on TGF-beta. Annals of the New York Academy of Sciences. 2003;995:1–10. doi: 10.1111/j.1749-6632.2003.tb03205.x. [DOI] [PubMed] [Google Scholar]
- 23.Wels J, Kaplan RN, Rafii S, Lyden D. Migratory neighbors and distant invaders: tumor-associated niche cells. Genes & development. 2008;22(5):559–574. doi: 10.1101/gad.1636908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massague J. TGF-beta signal transduction. Annual review of biochemistry. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 25.ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends in biochemical sciences. 2004;29(5):265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annual review of cell and developmental biology. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Zhou H, Wu K, Lee S, Li R, Liu X. PTEN phosphorylation and nuclear export mediate free fatty acid-induced oxidative stress. Antioxidants & redox signaling. 2014;20(9):1382–1395. doi: 10.1089/ars.2013.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. The Journal of clinical investigation. 2014;124(2):564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Song P, Xu J, Zhang M, Zou MH. Activation of protein phosphatase 2A by palmitate inhibits AMP-activated protein kinase. J Biol Chem. 2007;282(13):9777–9788. doi: 10.1074/jbc.M608310200. [DOI] [PubMed] [Google Scholar]
- 30.Wu Y, Shang X, Sarkissyan M, Slamon D, Vadgama JV. FOXO1A is a target for HER2-overexpressing breast tumors. Cancer Res. 2010;70(13):5475–5485. doi: 10.1158/0008-5472.CAN-10-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Sarkissyan M, McGhee E, Lee S, Vadgama JV. Combined inhibition of glycolysis and AMPK induces synergistic breast cancer cell killing. Breast Cancer Res Treat. 2015;151(3):529–539. doi: 10.1007/s10549-015-3386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer cell. 2011;20(4):427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyng MB, Laenkholm AV, Pallisgaard N, Ditzel HJ. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC cancer. 2008;8:20. doi: 10.1186/1471-2407-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristensen CA, Hamberg LM, Hunter GJ, Roberge S, Kierstead D, Wolf GL, et al. Changes in vascularization of human breast cancer xenografts responding to antiestrogen therapy. Neoplasia. 1999;1(6):518–525. doi: 10.1038/sj.neo.7900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SH, Hu LL, Gonzalez-Navajas J, Seo GS, Shen C, Brick J, et al. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nature medicine. 2010;16(6):665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L. Regulation of Smad activities. Biochimica et biophysica acta. 2006;1759(11–12):503–513. doi: 10.1016/j.bbaexp.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nardozzi JD, Lott K, Cingolani G. Phosphorylation meets nuclear import: a review. Cell communication and signaling : CCS. 2010;8:32. doi: 10.1186/1478-811X-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hough C, Radu M, Dore JJ. Tgf-beta induced Erk phosphorylation of smad linker region regulates smad signaling. PloS one. 2012;7(8):e42513. doi: 10.1371/journal.pone.0042513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 40.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, et al. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16(17):5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roelen BA, Cohen OS, Raychowdhury MK, Chadee DN, Zhang Y, Kyriakis JM, et al. Phosphorylation of threonine 276 in Smad4 is involved in transforming growth factor-beta-induced nuclear accumulation. Am J Physiol Cell Physiol. 2003;285(4):C823–C830. doi: 10.1152/ajpcell.00053.2003. [DOI] [PubMed] [Google Scholar]
- 43.Siddiqa A, Long LM, Li L, Marciniak RA, Kazhdan I. Expression of HER-2 in MCF-7 breast cancer cells modulates anti-apoptotic proteins Survivin and Bcl-2 via the extracellular signal-related kinase (ERK) and phosphoinositide-3 kinase (PI3K) signalling pathways. BMC cancer. 2008;8:129. doi: 10.1186/1471-2407-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni Y, Zhao L, Yu H, Ma X, Bao Y, Rajani C, et al. Circulating Unsaturated Fatty Acids Delineate the Metabolic Status of Obese Individuals. EBio Medicine. 2015;2(10):1513–1522. doi: 10.1016/j.ebiom.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocrine reviews. 2002;23(2):201–229. doi: 10.1210/edrv.23.2.0461. [DOI] [PubMed] [Google Scholar]
- 46.Schech A, Yu S, Goloubeva O, McLenithan J, Sabnis G. A nude mouse model of obesity to study the mechanisms of resistance to aromatase inhibitors. Endocrine-related cancer. 2015;22(4):645–656. doi: 10.1530/ERC-15-0168. [DOI] [PubMed] [Google Scholar]
- 47.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 48.Harris DR, Mims A, Bunz F. Genetic disruption of USP9X sensitizes colorectal cancer cells to 5-fluorouracil. Cancer Biol Ther. 2012;13(13):1319–1324. doi: 10.4161/cbt.21792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagai H, Noguchi T, Homma K, Katagiri K, Takeda K, Matsuzawa A, et al. Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol Cell. 2009;36(5):805–818. doi: 10.1016/j.molcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 50.Theard D, Labarrade F, Partisani M, Milanini J, Sakagami H, Fon EA, et al. USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J. 2010;29(9):1499–1509. doi: 10.1038/emboj.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agrawal P, Chen YT, Schilling B, Gibson BW, Hughes RE. Ubiquitin-specific peptidase 9, X-linked (USP9X) modulates activity of mammalian target of rapamycin (mTOR) J Biol Chem. 2012;287(25):21164–21175. doi: 10.1074/jbc.M111.328021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, et al. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121(1):87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 53.Xie Y, Avello M, Schirle M, McWhinnie E, Feng Y, Bric-Furlong E, et al. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 2013;288(5):2976–2985. doi: 10.1074/jbc.M112.430066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Liu Y, Yang B, Cao H, Yang CX, Ouyang W, et al. Elevated expression of USP9X correlates with poor prognosis in human non-small cell lung cancer. J Thorac Dis. 2015;7(4):672–679. doi: 10.3978/j.issn.2072-1439.2015.04.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng J, Hu Q, Liu W, He X, Cui L, Chen X, et al. USP9X expression correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Diagn Pathol. 2013;8:177. doi: 10.1186/1746-1596-8-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.