Abstract
Purpose
TP53 and the TGFβ pathway are major mediators of pancreatic cancer metastasis. The mechanisms by which they cause hematogenous metastasis have not been fully explored.
Experimental Design
KPC (LSL-KRASG12D/+;LSL-Trp53R172H/+; Ptf1aCre/+) mice were generated and the frequency and morphology of organ-specific hematogenous metastases compared to that seen in KPTC and KTC littermates (Tgfbr2+/−). Key findings were validated in primary cells from each genotype and samples of human pancreatic cancer liver metastases.
Results
The frequency of hematogenous metastasis in KPTC mice was significantly lower than for KPC mice (41% vs 68%, p<0.05), largely due to a reduction in liver metastases. No differences were found between KPC and KPTC lung metastases whereas liver metastases in KPTC mice showed a profound extravasation deficiency characterized by sinusoidal growth and lack of desmoplastic stroma. Analogous findings were confirmed in liver samples from patients indicating their clinical relevance. Portal vein colonization as a direct mode of access to the liver was observed in both mice and humans. Secretome analyses of KPC cells revealed an abundance of secreted prometastatic mediators including Col6A1 and Lcn2 that promoted early steps of metastatic colonization. These mediators were overexpressed in primary tumors but not metastases suggesting the ability to colonize is in part developed within the primary site, a phenomenon we refer to as the “Cinderella effect”.
Conclusions
These findings establish a novel paradigm for understanding pancreatic cancer metastasis and the observed clinical latencies of liver versus lung metastases specifically.
Introduction
Metastasis, the growth of primary carcinoma cells in a distant organ, remains a major clinical problem for patients with pancreatic ductal adenocarcinoma (PDA). Due to the lack of effective screening methods to identify the disease at an earlier stage, metastasis is responsible for a large number of PDA deaths(1,2). Somatic alterations in a number of genes have been shown to play a role in PDA metastasis(3). In transgenic models, conditional deletion of p16/INK4A, Trp53 and/or SMAD4 result in aggressive PDA and varying degrees of metastasis(4-7). Given that these very same genes arise during pancreatic carcinogenesis(8) it appears that some aspects of metastatic propensity are present at the onset of invasion. This is supported by human studies in which the genetic status of the four most common driver genes, specifically TP53 and SMAD4 is highly correlated to subsequent metastatic burden(9,10).
Despite these established relationships, the mechanisms by which these genes specifically cause metastasis in secondary organs is unknown. Morton et al found that despite similar kinetics of PDA primary tumor formation, mutant Trp53R172H but not loss of Trp53 specifically promoted metastasis due to a loss of growth arrest(11). More recently, Whittle et al found that RUNX3 is a critical mediator of metastasis in the setting of mutant Trp53R172H due to its presumed interaction with and stabilization by the mutant Trp53 protein that contributes to a prometastatic gene expression program(12). Together, these findings suggest that Trp53 in association with other key genetic alterations that arise during pancreatic carcinogenesis sculpt the efficiency and dynamics of metastasis, a finding also seen in human PDA(10). However, the mechanisms by which PDA organ-specific metastasis occurs specifically has yet to be understood, a topic of tremendous significance for patients with this disease given the divergent sites of failure encountered in the clinical setting. For example, patients with lung-only metastases have significantly improved survival compared with those with liver-only or lung plus liver metastases (13). Indeed, durable long-term survival can be observed with removal of a solitary lung metastasis after surgery for primary PDA (14), which would never be attempted in the setting of a solitary liver metastasis. Thus, understanding the basis for these differences in disease biology can help inform patient management and the development of new therapeutics.
Methods
Mouse strains
The conditional LSL-KRASG12D mice (Jackson et al., 2001), 129S4-Trp53tm2Tyj, B6.129S6-Tgfbr2tm1Hlm and Ptf1atm1.1(cre)Cvw mice (Kawaguchi et al., 2002) were previously described. The LSL-KrasG12D knock-in strains 129S4-Trp53tm2Tyj and B6.129S6-Tgfbr2tm1Hlm were obtained from Jackson Laboratories. The Ptf1atm1.1(cre)Cvw mice were purchased from MMRRC (Mutant Mouse Regional Resource Centers). Mice were genotyped by PCR using primers specific to transgenic alleles (conditions available on request). This study was approved by the Animal Care and Use Committee.
Human Tissues
Postmortem primary and metastatic pancreatic cancer tissues from consenting patients were obtained following IRB approved protocols previously described in detail (15).
Antibodies
Primary antibodies raised against phospho-histone H3 (#9701) were purchased from Cell Signaling Technology; TGFBR1 (V-22, sc-398), TGFBR2 (L-21, sc-400), COL6A1 (H200, sc-20649), LCN2 (M-145, sc-50351) and E-cadherin (G-10, sc-8426) were purchased from Santa Cruz Biotechnology; rabbit polyclonal antibody raised against Trp53 (P53-CM5P) was purchased from Leica Biosystems.
Histology and Immunohistochemistry
Normal tissues, pancreata and metastatic cancer tissues of moribund mice or mice at specific age timepoints (4, 8, 12 weeks) were fixed with 10% buffered formalin solution overnight and embedded into paraffin by the institutional pathology core laboratory. Sections cut from each paraffin block were stained with hematoxylin and eosin following standard protocols and observed under a light microscope fitted with a micrometer for diagnoses and measurements. For immunolabeling, unstained 5-μm sections were cut from paraffin blocks and the slides deparaffinized by routine techniques followed by incubation in 1x sodium citrate buffer (diluted from 10 × heat-induced epitope retrieval buffer, Ventana-Bio Tek Solutions, Tucson, AZ) before steaming for 20 minutes at 80 °C. Slides were cooled 5 minutes and incubated for two hours to overnight with primary antibody. Immunolabeling was detected using the Ventana IVIEW Kit per kit instructions and sections were counterstained with hematoxylin.
Immunofluorescence
Cells grown on coverslips were fixed in 3.7% or 4.0% paraformaldehyde solution for 30 minutes at 37°C. All subsequent steps were carried out at room temperature. Coverslips were incubated with E-cadherin monoclonal antibodies (1:100 dilution; Santa Cruz) in PBS-1% BSA for 1 h and followed by incubation together with FITC-conjugated goat anti-mouse secondary antibody (Sigma). Coverslips were mounted in Prolong Gold Antifade reagent with DAPI (Molecular Probes), and images were captured with a Zeiss fluorescence microscopy system.
Cell proliferation assays
All cells were cultured in DMEM (GIBCO, Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mmol/L L-glutamine at 37 °C and 5% CO2. Cell proliferation assay was performed using cell counting Kit-8 (Dojindo Molecular Technologies, Inc., Gaithersburg, MD, USA) according to manufacturer protocols. Absorbance was measured at 490 nm using a microplate reader. Each experiment was carried out in triplicate.
Cell Migration & Invasion assays
Cell migration assay was performed using culture inserts (Cat. No., ECM 508, Millipore) and Invasion Assay was using insert (Cat. No., ECM 550, Millipore) according to manufacturer protocols. All cells were passaged 2-3 times, starved by incubating 24 hours prior to the assay. For cell migration assay, the starved cells were harvested in serum-free medium, counted cells and brought to a volume that gives 1.0 × 106 cells per mL. Add 300 μl of cells to each insert (Cat. No., ECM 508), 500 μl of 10% fetal bovine serum to the lower chamber and incubate the cells for 20 hours at 37 °C and 5% CO2. As for invasion assay, 300 μl of cells were added to insert (Cat. No., ECM 550) and incubated the cells for 72 hours at 37 °C and 5% CO2. Cells were treated with 5 ng/mL of Tgfb1 or vehicle control during incubating. On the day of assay, cells on the interior of the inserts were removed and the membranes were incubated with cell stain solution. Count cells by photographing the membrane through the microscope, then subsequently quantitate by dissolving stained cells in 10% acetic acid and detect on a standard microplate reader (560nm). Each experiment was carried out in triplicate.
Cell Apoptosis assay
Cell apoptosis assay was performed using NC 3000 system (ChemoMetec Inc., Davis, CA) according to manufacturer protocol. Seed 1 × 106 cells/well in a 6-well culture plate the day before the experiment. Cells were treated with 5 ng/mL of Tgfb1 or vehicle control during incubating. Cells were dissociated into single cell suspensions in PBS, counted, and incubated with Annexin V-CF488A and Hoechst 33342 (4 × 105 cells per sample) for 15 minutes at 37 °C. The cells were washed with Annexin V binding buffer twice and the cell pellet resuspended in 100 ml Annexin V binding buffer containing 10 μg/mL PI and then subject to Annexin V Assay. Data was derived from three independent experiments.
Cell Cycle assays
Cell cycle assay was performed using NC 3000 system (ChemoMetec Inc., Davis, CA) according to manufacturer protocol. Seed 3 × 105 cells/well in a 24-well culture plate the day before the experiment. Cells were treated with 5 ng/mL of Tgfb1 or vehicle control during incubating. Wash the cells with PBS, incubate the cells with lysis buffer containing 10 μg/mL DAPI for 5 minutes at 37 °C and then subject to 2-step cell cycle assay. Data was derived from three independent experiments.
Quantitative Secretome Analysis
KPC and KPTC cells were grown to confluence in tissue culture dishes, washed with serum-free media eight times and incubated in serum-free media for 24 hours. The conditioned media (CM) was harvested and centrifuged to eliminate any intact cells and the supernatants were then concentrated and desalted by centrifugation in Amicon Ultra-15 tubes (molecular weight cutoff 3000 Da; Millipore, Billerica, MA). Protein samples were quantified and analyzed by the Proteomics Core Facility of Johns Hopkins. The protein concentrations of KPC and KPTC CM samples were labeled with the iTRAQ reagent (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol, and then subjected to MS/MS analysis. Data analysis on the data files from the LC-MS/MS were performed with the Thermo Scientific™ Proteome Discoverer™ Software (Thermo Fisher Scientific, Inc., Waltham, MA). The identified proteins were further subjected to bioinformatic analysis. Proteomics data have been deposited in the Peptide Atlas Database with the accession number PASS00575.
Quantitative Real Time PCR
Total RNA was extracted from tissue samples using RNeasy mini Kit (Cat. No., 74104; Qiagen). RNA was treated with DNase I (Invitrogen) to digest remnant genomic DNA. cDNA was synthesized from 0.5 μg of total RNA by the SuperScriptTMIII Platinum® Two-Step qRT-PCR Kit (Invitrogen) according to the protocol recommended by the manufacturer. Real-time quantitative RT-PCR analysis was performed using an automated sequence detection instrument (7300 Real Time PCR System, Applied Biosystems, CA, USA) for the real-time monitoring of nucleic acid green dye fluorescence (SYBR®Green, Invitrogen Inc, CA, USA). Relative fold-changes of analyzed gene expression compared to the housekeeping gene β-actin were determined by calculation of the 2ΔΔCt. All analyses were performed in triplicate at least 2 times. Primer sequences will be provided upon request.
Western Blotting
Cell lysates were prepared by suspending cell pellets in RIPA buffer (20 mM Tris, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, pH 7.4) supplemented with protease inhibitor cocktail tablets (Roche diagnostics GmbH, Mannheim, Germany). Equal amounts of proteins were separated on SDS-polyacrylamide gel and transferred onto PVDF membranes (DuPont NEN, Boston, MA). Each membrane was hybridized with primary antibody followed by horseradish peroxidase (HRP)-linked IgG and visualized by the enhanced chemiluminescence (ECL) system (Amersham). Expression of β-actin or GAPDH were used as internal controls.
Col6a1 promoter reporter assay
A TP53 expression vector that included the R175H mutation was used which corresponds to the R172H mutation in our mouse models. GLuc/SEAP dual-reporter vector MPRM24422-LvPG04 (mCol6a1-pro) was used as a reporter that contains the Gaussia luciferase reporter gene (Gluc) driven by a mouse Col6a1 promoter. The secondary reporter, secreted Alkaline Phosphatase (SEAP), serves as an internal control. Expression vectors in various combinations were co-transfected with luciferase reporter vectors into cells using Effectene Transfection Reagent (QIAGEN). Twenty-four hours after transfection, the medium was exchanged to fresh medium and the cells treated with or without Tgfb1. Gluc and SEAP activities were measured 48 hrs after transfection using the Secrete-Pair™ Dual Luminescence Assay Kit (GeneCopoeia). Gluc and SEAP activity was determined with the Cytation 3 Cell Imaging Multi-Mode Reader (BioTek). Gluc activities were normalized by SEAP activities. Each transfection experiment was carried out in triplicate.
Stable shRNA knockdowns and Stable overexpressions
KPC cells grown to ~70% confluency in Dulbecco's modified medium containing 10%FBS were transfected with pRFP-C-RS vectors expressing 4 different 29-mer shRNA for mouse COL6A1 or mouse Lcn2 (OriGene, TF500404 and TR511895) as well as a Non-effective 29-mer scrambled shRNA cassette in pRFP-C-RS Vector (OriGene, TR30015) using Attractene reagent (Cat. No., 301005, Qiagen) following the Forward-fast transfection protocol. KPTC cells were transfected with pCMV6-AC-GFP vectors expressing mouse COL6A1 or Lcn2 (OriGene, MG223027 and MR226233). The Trp53 shRNA sh-1224, a kind gift of Dr. Scott Lowe (Memorial Sloan Kettering Cancer Center, NY), was used for Trp53 knock down experiments. 48 hours after transfection, cells were passaged into the appropriate selection medium and maintained until colonies appeared. Inhibition of gene expression by shRNA and over-expression of target gene were determined by Western blot analyses. Those colonies with the best knockdown efficiency or high expression levels of COL6A1 were used for next experiments.
Wound-healing assays
Cell wound-healing assay was performed using culture inserts (Cat. No., 80241, Ibidi) according to manufacturer protocols. All cells cultures were dissociated into single cell suspensions in completed DMEM medium, counted cells and brought to a volume that gives 6.0 × 105 cells per mL. Add 70 μl of cells to each well, fill the outer area with 200 μl of cell suspension. Cells are cultured in a confluent layer, and then a wound gap is created by removing the growth barrier. Images at time zero (t = 0 h) were captured to record the initial area of the wounds, and the recovery of the wounded monolayers due to cell migration toward the denuded area was evaluated at 16 and 24 h (t = Δ h). The images were captured using an inverted phase-contract microscope (EVOS FL Color Imaging Systems; Cat#, AMF 4300; 10 × objective) equipped with a motic digital camera. The area of wound was quantified by ImageJ (http://rsb.info.nih.gov/ij/download.html) using the polygon selection mode. The data was expressed as percentage of wound remaining: % of wound remaining = [Areat=Δh/Areat=0h]×100%, where, Areat=0h is the area of wound measured immediately after scratching, and Areat=Δh is the area of wound measured 16 or 24 h after scratching. Each experiment was carried out in triplicate.
Experimental Metastasis Assays
Tail veins, intracardiac as well as intrasplenic injections were performed to evaluate the tumor metastasis to organ sites. KPC and KPTC cell cultures were dissociated into single cell suspensions in PBS, counted, and injected into the tail veins, left cardiac ventricle or the splenic capsule of CD1nu/nu mice (5 × 105 cells per mouse). As for tail vein and intracardiac injection, after three weeks the animals were euthanized and their organs removed, rinsed in PBS and the number of gross (surface) lung nodules counted using a dissecting microscope. As for intrasplenic injection, mice were necropsied at 2 weeks post-injection and evaluation for hepatic metastasis was performed. These experiments were repeated and pooled metastasis counts averaged, with differences in average metastatic counts evaluated by Student's t-test, two tailed.
Results
TGFβ Influences Metastatic Patterns Conferred by Trp53
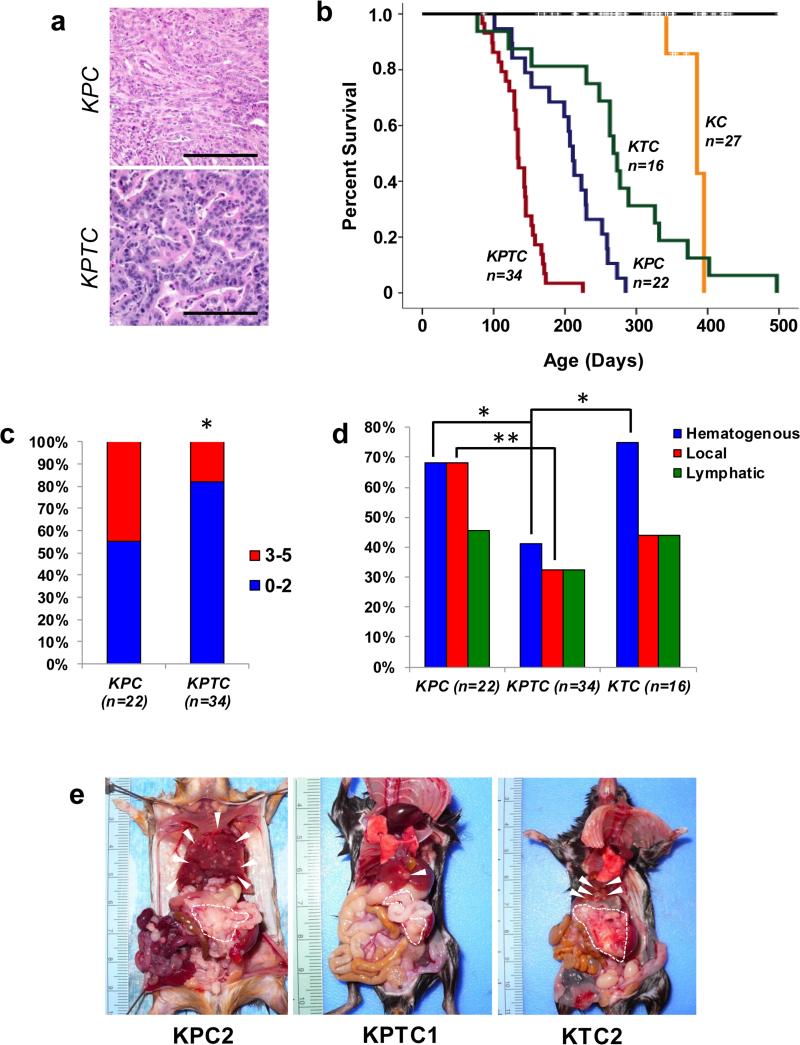
We used a well known murine model of PDA (LSL-KRASG12D/+;LSL-Trp53R172H/+; Ptf1aCre/+ mice, KPC) to characterize the organ-specific patterns of metastasis. In this model mice develop the full spectrum of pancreatic intraepithelial neoplasia (PanIN) that progresses at a high frequency to PDA(4). Histologic features of these PDAs recapitulate that of humans including the associated desmoplastic stromal and inflammatory reaction and frequent metastases to the liver, lung and peritoneum(4) (Fig 1A). We generated 27 KPC mice for study, 22 of which succumbed to PDA; at necropsy 17 of these 22 mice (77%) had gross or histologic evidence of metastasis to one or more organ sites consistent with the aggressive nature of this model.
Figure 1. Characterization of general patterns of metastases in murine models of PDA.
A, Representative poorly-differentiated PDA in KPC and well-differentiated PDA in KPTC mouse. Scale bars, 100 microns. B, Kaplan-Meier survival curves of KPTC, KPC, KTC, KC and control mouse cohorts. KPTC mice have a significantly shortened median survival compared to both KPC (p<0.0001) and KTC mice (p=<0.0001). Medians survivals were compared by pair-wise log-rank tests. C, Overall frequency of the gross or histologic metastases in KPC and KPTC mice. D, Frequency of hematogenous, peritoneal or abdominal lymph node metastases in KPC, KPTC and KTC mice. E, Gross findings in representative moribund KPC, KPTC or KTC mice with hematogenous and abdominal metastases. Dashed lines outline primary tumor, arrowheads indicate metastases. *, p<0.05; **, p<0.01.
TGFβ signaling figures prominently at several steps in the metastatic program and the p53 and TGFβ pathways are integrated at multiple levels(16-18). To determine the impact of TGFβ signaling on these patterns of metastasis, we generated LSL-KrasG12D/+;LSL-Trp53R172H/+; Tgfbr2flox/+; Ptf1aCre/+ (KPTC) mice (n=34) in which one allele of Tgfbr2 is conditionally deleted in pancreatic epithelial precursor cells (Supplemental Fig 1A). Haploinsufficiency for Tgfbr2 in mice produced a higher frequency of well-differentiated PDAs that invaded surrounding structures (19) (Fig 1A). However, KPTC mice exhibited a significantly shorter median survival than KPC mice (4.4 versus 7.0 months, p<0.0001) indicating that combined alterations of Trp53 and the TGFβ pathway cooperate to accelerate PanIN progression (Fig 1B and Supplemental Fig 1B) as previously described (19). In further support of this interpretation, LSL-KRASG12D/+; Tgfbr2flox/+; Ptf1aCre/+ (KTC) mice (n=16) generated during interbreeding exhibited a significantly prolonged latency to the development of PDA compared to KPC mice (8.9 months, p<0.0001) (Fig 1B). Thus, heterozygous mutation of both pathways together curtailed survival significantly more than either alone.
We next compared the frequency and pattern of metastasis in KPC and KPTC mice. KPTC mice had fewer overall gross or histologic metastases (77% vs 59%) and were less likely to have spread to more than two sites (p<0.05, Fig 1C). Specifically, KPTC mice showed a significant reduction in the frequency of hematogenous (41% vs 68%, p<0.05) and peritoneal metastases (32% vs 68%, p<0.01) (Fig 1D-E and Supplemental Table 1) although there was no difference in the frequency of abdominal lymph node metastases between the two genotypes (present in roughly one third of both cohorts) (Fig 1D). This decrease in metastatic disease burden is similar to that observed with perturbation further downstream in the TGFβ pathway (12). Collectively, these data indicate that the TGFβ pathway differentially influences the frequency and distribution of metastases to distinct sites. This influence is in part dependent on the co-existence of mutant Trp53, as KTC mice showed similar rates of hematogenous metastasis as KPC mice (Fig 1D and E).
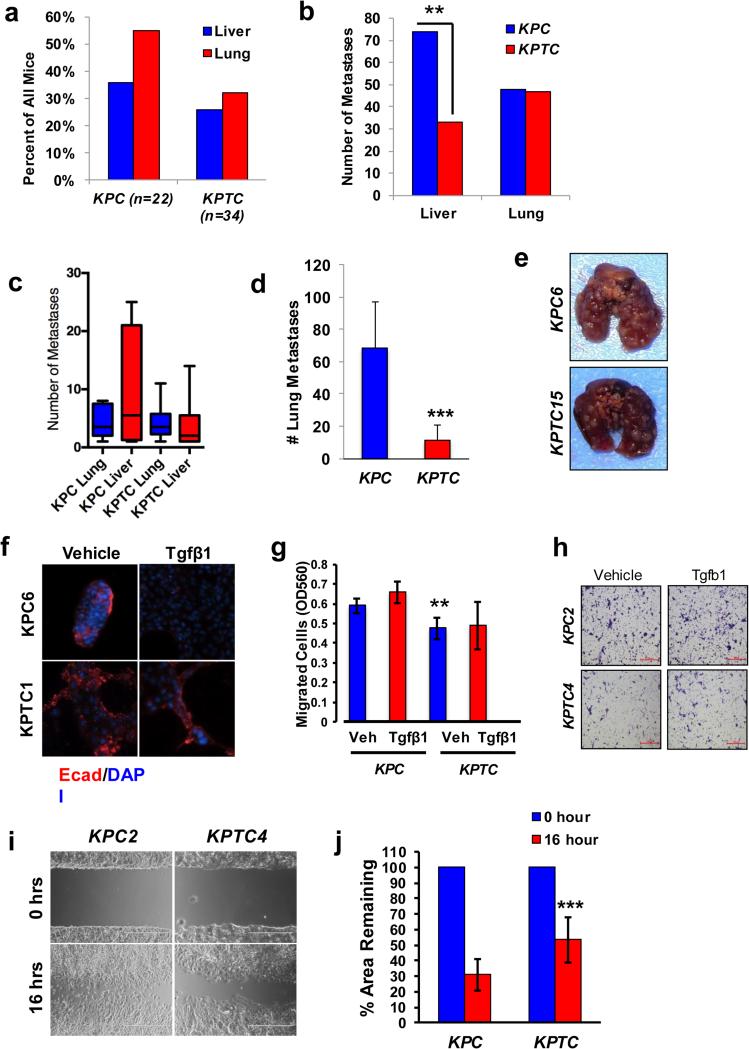
We next quantified the number of individual metastases in the liver and lung of all KPC and KPTC mice. To decrease the likelihood of scoring the same metastasis more than once we reviewed a single cross section of each lung and of each of the four lobes of the liver; a total of 202 hematogenous metastases (107 liver and 95 lung) were identified (Supplemental Table 2). KPC mice developed significantly more hematogenous metastases (15.3/animal; n=122 scored across 8 mice) than KPTC mice (8.9/animal; n=80 over 9 mice) and this difference was accounted for by a higher number of liver metastases in KPC mice (Fig 2A and B). The median number of liver metastases was 5.5 in KPC mice but only 2.0 in KPTC mice (Fig 2C). However, the median number of lung metastases was comparable between genotypes with hematogenous metastases (median 3.5 in both KPC and KPTC cohorts) These findings suggest that Tgfbr2 haploinsufficiency not only reduces the overall efficiency of hematogenous metastasis, it reduces the efficiency of metastasis to the liver to a greater extent than the lung.
Figure 2. Cell-autonomous Tgfβ signaling mediates the efficiency of hematogenous metastasis.
A, Frequency of liver and lung metastases in KPC (n=22) and KPTC (n=34) mice. B, Quantification of the number of metastases in the liver and lung based on histologic review in the cohort of KPC (n=8) and KPTC (n=9) mice with hematogenous metastases. C, The median and interquartile range of lung and liver metastases of mice with hematogenous metastases by genotype. D, Quantification of gross metastatic lung colonization at three weeks following tail vein injection of 1 × 105 primary KPC versus KPTC cells. E, Representative lungs in mice injected with KPC6 or KPTC15 primary cells. F, Immunoflourescence labeling for E-cadherin in representative KPC and KPTC primary cells treated with vehicle or TGFβ1. DAPI was used as a counterstain to visualize nuclei. G, Quantification of cell migration of KPC and KPTC cells. Data shown represent the mean ± standard deviation of three independent experiments per primary cell line. At least three cell lines per genotype were analyzed. H, Migration images in representative KPC and KPTC primary cells treated with vehicle or TGFβ1. I, Representative images of wound-healing in KPC2 and KPTC4 primary cells. J, Quantification of percentage of wound remaining in primary KPC versus KPTC cells. Data shown represent the mean ± standard deviation of at least three primary cell lines per genotype, with each line analyzed in triplicate per experiment. **, p<0.01; ***, p<0.001.
Disruption of TGFβ Signaling Abrogates Lung Metastasis
As KPTC mice succumbed earlier than KPC mice (Fig 1B) as a result of an increased primary tumor burden, we asked whether this explained the lower rates of hematogenous metastasis. If so, primary cells from KPTC and KPC carcinomas would be expected to behave similarly in experimental assays of metastatic potency. We therefore inoculated primary tumor cells prepared from individual KPC or KPTC mice into the bloodstream of CD1nu/nu mice and assessed the metastatic burden in the lungs three weeks later. KPTC cells generated lung metastases at a markedly reduced rate to KPC cells (Fig 2D and E, p<0.0001), mirroring our observations in KPTC engineered mice and indicating that the earlier time to death alone was not the cause of decreased metastasis.
Formation of a hematogenous metastasis requires acquisition of pro-metastatic traits, intravasation into and survival within the bloodstream, extravasation into the foreign organ and colonization by establishment of a metastatic niche (20). To examine abilities associated with earlier steps in this process, we treated primary cultured KPC and KPTC cells with vehicle or Tgfβ1 ligand. KPC cells underwent a profound loss of E-cadherin expression and transformed from tight epithelial clusters to loose aggregates in the presence of Tgfβ1, whereas no effect was seen with KPTC cells (Fig 2F), a finding also reported in LSL-KrasG12D/+;LSL-Trp53R172H/+;Dpc4flox/+;Ptf1aCre/+ (KPDC) cells haploinsufficient for Smad4 (12). KPC cells also showed significantly higher baseline rates of migration into Matrigel compared to KPTC cells (Fig 2G and H) and similar findings were observed in wound healing assays (Fig 2I and J). Together with our data from the tail vein assays, these findings suggest that KPTC cells have a reduced ability to form hematogenous metastases due to deficiencies at both proximal and distal steps of the metastatic cascade.
We next focused on the lung metastases in KPTC mice to identify the features that allowed their successful colonization. No overt morphologic differences among lung metastases between KPC or KPTC mice were apparent (Supplemental Fig 2A), nor were there differences in average cross sectional area (Supplemental Fig 2B) or pHH3 labeling indices (Supplemental Fig 2C). Immunolabeling for Tgfbr2 was consistently present across lung metastases from both genotypes indicating retention of the wild-type allele in KPTC mice (Supplemental Fig 2D). Interestingly, cell lines derived from KPTC primary tumors and metastases had no difference in Tgfbr2 mRNA expression suggesting that loss of the wild type Tgfbr2 allele was not required for metastasis (Supplemental Fig 2E), nor were there differences in Tgfbr1 mRNA expression (Supplemental Fig 2F). These data indicate that KPTC cells that successfully colonize the lung are essentially equivalent to KPC cells with respect to TGFβ signaling and that the overall number of KPTC cells equipped to accomplish each hurdle in the dissemination process must be vastly reduced.
TGFβ Cooperates with Trp53 to Colonize the Liver
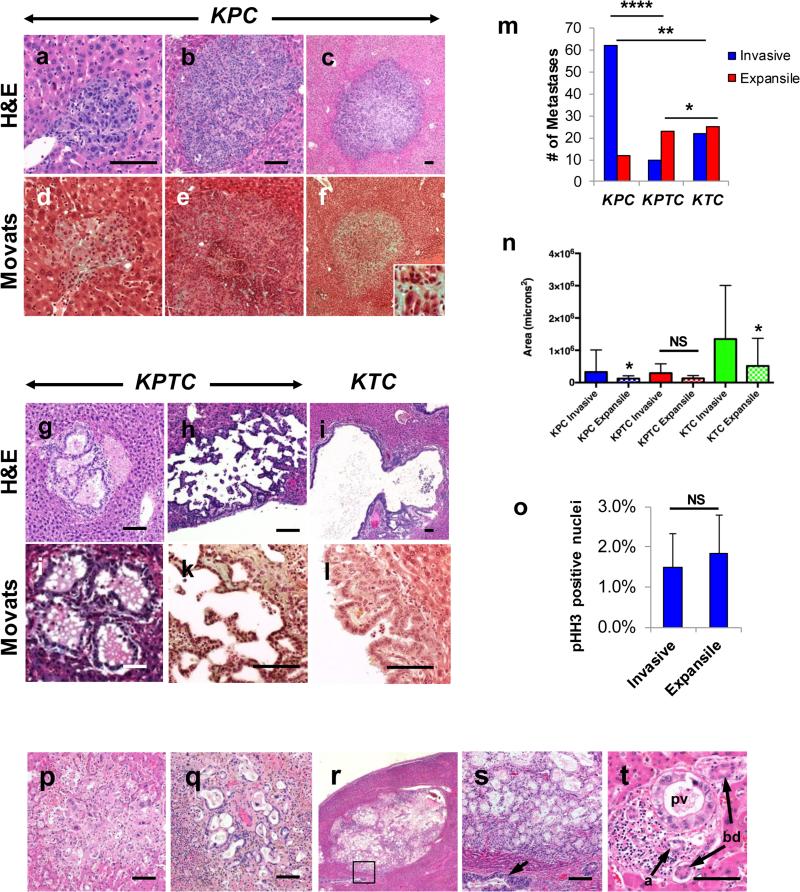
We next assessed the liver metastases in our model. Unlike lung metastases, liver metastases in KPC and KPTC mice showed striking dissimilarities. In KPC mice the metastases grew by invasive and destructive growth into the hepatic parenchyma (Fig 3A-C) with invasion recognized by the syncytial growth patterns of the neoplastic cells often with an associated myofibroblastic and inflammatory stromal response on routine and Movats pentachrome stains (Fig 3D-F). By contrast, in KPTC mice the metastases seen formed by expansile growth within the hepatic sinusoidal spaces with minimal to no stromal reaction (Fig 3G-H and 3J-K) and this finding was even more pronounced in metastases of KTC mice (Fig 3I and L). The frequency of invasive versus expansile morphology per genotype confirmed these striking differences in growth patterns (Fig 3M). To understand these features in greater detail, we measured the cross sectional area of each metastasis. In general invasive metastases were more than twice as large as expansive metastases regardless of genotype (3.2 × 105 μ2 versus 1.3 × 105 μ2, p<0.01, Fig 3N), although these differences were not consistently significant within a single genotype. Such a comparison is also an underestimate of cellularity as cystic metastases that occupy the same cross sectional volume as a solid invasive metastasis have fewer cells overall, both neoplastic and stromal. We also measured proliferative rates of the neoplastic epithelium in invasive versus expansile metastases and found no differences (Fig 3O). Thus, proliferative rates alone did not account for these divergent growth patterns.
Figure 3. Cell-autonomous Tgfβ signaling mediates the pattern of metastatic colonization of the liver.
Examples of invasive micrometastasis (A), invasive macrometastasis without a significant stromal response (B) and invasive macrometastasis with stromal reponse in KPC mice livers (C). D and F, Movats pentachrome stain of invasive liver metastasis in the same KPC mice. The inset in panel f highlights the collagen rich stroma. Representative expansile metastases in liver of KPTC (G and H) and KTC mice (I). Movats pentachrome stain of same metastases in liver in KPTC (J and K) and KTC mice (L). M, Frequency of invasive versus expansile metastases in KPC, KPTC and KTC mice. N, Quantification of the cross-sectional area of metastases in the liver of KPC, KPTC and KTC mice. O, Percent of pHH3 positive cells per 200x HPF in invasive versus expansile metastases in each genotype. P, Representative invasive liver metastasis in PDA patient. Q, Representative expansile liver metastasis in PDA patient. R, Large expansile liver metastasis forming cystic mass. S, Higher power view of metastasis shown in (R). Arrow indicates adjacent portal vein colonization. T, Representative portal vein colonization in area of otherwise normal liver in PDA patient. a, artery; bd, bile duct; pv, portal vein. Scale bars, 100 microns. *, p<0.05; **, p<0.01; ****, p<0.0001.
In some KPTC mice the PDA cells colonized large bile ducts (Supplemental Fig 3A) or portal veins (Supplemental Fig 3B) and these were the only sites of intrahepatic growth identified. Evidence that the neoplastic cells were located within bile ducts was supported by a Movats pentachrome stain that revealed a fibrotic cuff surrounding the enlarged bile duct adjacent to normal caliber hepatic arteries and portal vein (Supplemental Fig 3C); neoplastic cells within the portal veins were recognized by the presence of luminal red and white blood cells (Supplemental Fig 3B), a cuff of elastic fibers on Movats pentachrome stain (Supplemental Fig 3D) and adjacent normal caliber bile ducts. CD31 labeling further indicated that portal colonization led to replacement of the endothelial cells (not shown). Thus, not only does loss of TGFβ signaling reduce the efficiency of hepatic colonization, but it also alters the dynamics by which colonization itself occurs.
The TGFβ pathway transmits signals through a canonical pathway mediated by Smad4, and noncanonical pathways such as p38, JNK, Ras/Erk, or RhoA (21). To determine if the growth pattern observed in KPTC mice was also present in mice deficient for canonical signaling we examined the liver metastases in a set of previously generated KPDC mice that are haploinsufficient for Smad4 (12). KPDC mice showed the identical phenotype of expansile growth within sinusoidal spaces as KPTC mice (Supplemental Fig 3E and F). Moreover, KPDDC mice (which have complete loss of Smad4) had more severe cytologic atypia while maintaining expansile growth with limited to no stromal reaction (Supplemental Fig 3G), in some instances resulting in large cystic macrometastases (Supplemental Fig 3H). Finally, in two of two KC mice (which have wild type Trp53 and retain Smad4) that specifically had liver metastases at necropsy, both had an expansile morphology (Supplemental Fig 3I). These data suggested to us that expansile/sinusoidal growth indeed represents the initial phase of hepatic colonization and is mutant Trp53 independent, followed by cooperation of mutant Trp53 with TGFβ signaling to promote the second phase of hepatic colonization in part by the ability to induce a stromal response.
To determine the extent to which canonical signaling is affected in KPTC mice specifically we performed immunolabeling for phosphorylated Smad2/3, a marker of Smad4-dependent Tgfβ pathway activity. Positive pSmad2/3 labeling was present in both KPC and KPTC PDA tissues although labeling was heterogeneous and of lesser intensity in KPTC carcinomas than in KPC carcinomas (Supplemental Fig 3J). Western blotting also indicated a modest reduction of pSmad2/3 protein in KPTC primary cells compared to KPC primary cells whereas there was no difference in Smad4 content (Supplemental Fig 3K). We next determined the levels of Erk and Akt signaling in the same extracts to determine the extent to which Tgfbr2 haploinsufficiency affects noncanonical signaling (Supplemental Fig 3K). Phosphorylated Erk was detected in all four KPC cell lines whereas it was undetectable in two of three KPTC cell lines. Furthermore, no differences in pAkt levels were noted between the two genotypes although total Akt appeared to be more abundant in the KPTC cell lines, suggesting the relative proportion of pAkt was reduced in these tumors. Image quantification confirmed these observations with levels of pAKT approximately 25% less than that in KPC cells (Supplemental Fig 3L). Thus, both canonical and noncanonical signaling is reduced in KPTC cells, both of which may contribute to the reduction in overall metastatic efficiency.
We next wondered if expansile morphology represented a prolonged initial phase of colonization given the limited ability of KPTC, KTC, KPDC and KPDDC cells to generate a stromal response. The larger cross sectional areas of invasive compared with expansile metastases within a given genotype suggested this possibility as well (Fig 3N). If an expansile pattern of growth represented an early phase that was arrested in KPTC cells but KPC cells had been successfully transitioned through, then one would expect some invasive metastases to have a residual contiguous component of expansile morphology. Careful review of all invasive metastases in these cohorts indeed revealed a subset with such bimorphic features (Supplemental Fig 3M-N). Serial sectioning through several metastases confirmed these did not simply represent a “collision” between independent macrometastases with invasive or expansile morphology (Supplemental Fig 3O-P). There was no difference in the frequency of mixed morphology among invasive metastases in KPC and KPTC mice (23% versus 30%, p=NS), whereas in KTC mice the majority of metastases had a mixed morphology (68%, p<0.001 vs KPC mice) (Supplemental Fig 3Q).
A key metric of the significance of these findings is their presence in human samples. We selected 12 PDA patients with liver metastases at the time of autopsy (9) and genetic features that were concordant with our mouse models including activatingKRAS and TP53 missense mutations in all patients. A total of 25 micrometastases were identified in seven patients ranging from one to nine micrometastases per section analyzed. Of these micrometastases seven had invasive morphology (Fig 3P), seven had expansile morphology (Fig 3Q), and 11 had features of both expansile and invasive growth. In one patient the liver metastases reviewed formed macroscopic masses comprised solely of expansile morphology (Fig 3R-S). No relationship between these morphologies and the genetics of each PDA was discerned, although these numbers likely do not permit such an analysis. Moreover, for five of the 12 patients with intact Smad4 in their primary tumor, there was no heterogeneity observed for Smad4 labeling among different metastases within a single section consistent with our prior observations (10). Importantly nine of 12 patients (75%) had multifocal portal vein colonization that was often geographically distinct from macrometastases and in regions of otherwise normal liver, indicating this mode of hepatic access may be an underappreciated aspect of this disease (Fig 3T). En masse these observations indicate the divergent morphologic features and modes of hepatic colonization seen in our murine models are also common findings in PDA patients.
KPC Cells have a Different Secretome Compared to KPTC Cells
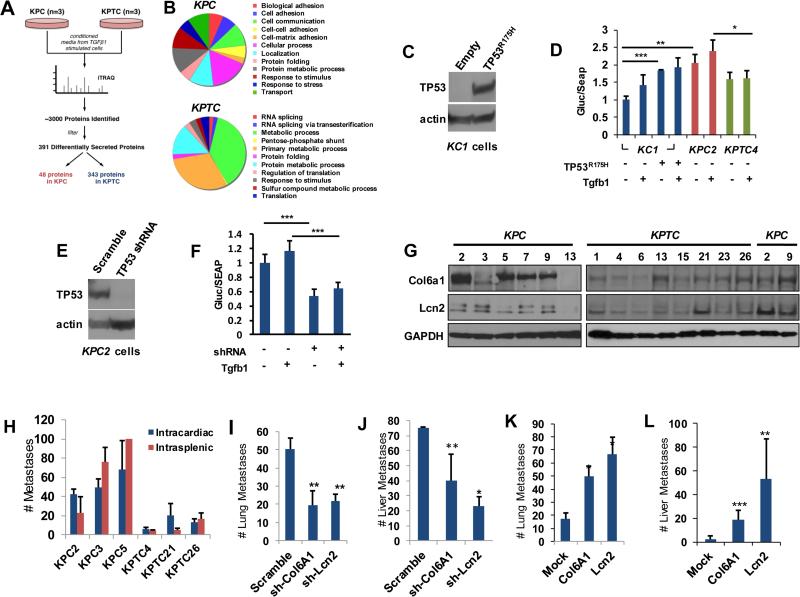
Given the ability to induce a stromal response was a defining characteristic of invasive versus expansile metastases we wondered if secreted proteins related to the extracellular matrix differ among KPC and KPTC cells. We therefore collected 24 hour conditioned media from Tgfβ stimulated KPC and KPTC cells and subjected them to iTRAQ-based quantitative proteomic analysis (Fig 4A). More than 3000 proteins were identified by this approach. Bioinformatic analyses to screen for the most differentially secreted proteins between KPTC and KPC cells resulted in a high-confidence list of 391 proteins with a false-discovery rate ≤0.001 and a p-value ≤0.001 (Supplemental Table 3).
Figure 4. Soluble mediators of metastasis are enriched in the secretome of KPC mice.
A, Schematic of mass-spectrophotometry experiment. B, PANTHER-based classification KPC or KPTC secretomes. C. Western blot illustrating endogenous expression of TP53-R175H in KC1 cells. D. Relative activity levels of the dual luminescence reporter vector driven by a Col6a1 promoter when transfected into KC1, KPC2 or KPTC4 cells with or without Tgfb1 ligand stimulation. Activity levels of KC1 cells after transfection with TP53-R175H are also shown. E. Western blot illustrating efficiency of knockdown of Trp53 in KPC2 cells. F. Relative reporter activity levels of KPC2 cells after knockdown of Trp53 with or without TGFb1 ligand stimulation. In both D and F Col6a1 promoter activities are presented as a ratio of Gluc activity to SEAP activity with each transfection experiment performed in triplicate. G, Western blots for Col6A1 and Lcn2 protein in KPC and KPTC primary cell lines. GAPDH is used as a loading control. H, Gross quantification of lung or liver metastases following intracardiac or intrasplenic injection of primary KPC versus KPTC cells. Data shown represent the mean ± standard deviation of at least three primary cell lines per genotype, with each line injected into at least four mice per experiment. I, Quantification of lung colonization by KPC2 cells stably transfected with scrambled, Col6A1 or Lcn2 shRNAs. J, Quantification of liver colonization following intrasplenic injection using KPC2 cells stably transfected with scrambled, Col6A1 or Lcn2 shRNAs. K, Quantification of lung colonization by KPTC4 cells stably transfected with mock, Col6A1 or Lcn2 expressing vector. L, Quantification of liver colonization following intrasplenic injection using KPTC4 cells stably transfected with mock, Col6A1 or Lcn2 vectors. For all experimental metastasis assays the data shown are the mean ± S.D of at least four animals per condition. Comparisons were made using a two-sided Student's T-Test. *, p<0.05; **, p<0.01; ***, p<0.001.
The secretomes of KPC and KPTC cells were dramatically different: 48 proteins were more abundant in the secretome of KPC cells and 343 proteins were more abundant in KPTC cells (Fig 4B). Overall, 13/40 proteins (33%) secreted by KPC cells were extracellular matrix related proteins, many of which have been mechanistically implicated in metastasis (12,22). Gene Ontology (GO) analysis of these 48 proteins confirmed a statistically significant overrepresentation of protein functions related to cell-matrix and cell-cell adhesion, among others (Fig 4B and Supplemental Table 3). By contrast, the same analysis of the 343 proteins enriched in the secretome of KPTC cells revealed a statistical overrepresentation of protein functions related to metabolism, RNA splicing and translation (Fig 4B and Supplemental Table 3).
To gauge the extent that a gain of function of the mutant Trp53 versus a reduction in Tgfβ signaling contribute to the observed differences in the two secretomes we assessed the ability of a transiently transfected TP53R175H mutant to activate Col6A1 expression, one representative protein enriched within the KPC secretome (12), in vitro by using a dual-luminescence promoter reporter assay. In 293T (not shown) and KC1 cells that have wild type TP53 or Trp53 alleles respectively, exogenous expression of TP53R175H significantly increased the reporter activity of the Col6a1 promoter and this increase was not significantly increased by the addition of TGFβ1 (Fig 4C,4D). This same Col6a1 reporter was activated in untreated KPC2 cells at levels similar to KC1 cells containing the Trp53R172H mutant also with no increase in reporter activity when TGFβ1 was added to the media. By contrast, activity levels in KPTC4 cells, containing the Trp53R172H mutant but haploinsufficient for Tgfbr2, were significantly lower than that of KPC2 cells and once again not affected by the addition of TGFβ1 (Fig 4D). These findings suggest that mutant Trp53 indeed influences Col6A1 expression through a gain of function mechanism, and this gain of function is in part dependent on TGFβ signaling as recently shown by Whittle et al (12). To further assess this possibility, we assessed Col6A1 reporter activity levels in KPC2 cells after short hairpin knock down of Trp53 (Fig 4E,4F). Consistent with this interpretation knockdown of Trp53 led to a significant reduction in reporter activity compared to a scrambled hairpin control (Fig 4F).
KPC Secreted Proteins Promote Hematogenous Metastasis
To determine if selected secreted proteins identified in the KPC secretome indeed influence metastatic ability we studied Col6A1 (collagen type IV A1) and Lcn2 (neutrophil-gelatinase associated lipocalin) for further evaluation and functional validation in our models (12,23-25). Immunoblotting for Col6A1 and Lcn2 in KPC cells confirmed the results of the secretome analysis, with KPC cells showing greater levels of expression for both proteins than primary KPTC cells (Fig 4G).
We first tested the ability of Col6A1 and Lcn2 to modulate metastases to the lung or liver in distinct experimental assays. Unmodified KPC cells showed a significantly greater ability than KPTC cells to colonize the lungs of CD1nu/nu mice following intracardiac injection; they also more readily colonized the liver following intrasplenic injection (Fig 4H). The majority of mice (seven of nine) that underwent intracardiac injection with KPC cells also developed pleural effusions in association with the lung metastases whereas this occurred only rarely (one of nine) in mice injected with KPTC cells. A similar pattern was noted for KPC cells injected via the spleen. In addition to liver colonization, the majority of mice (eight of nine) developed hemorrhagic ascites, tumor deposits in the pancreas and spleen, and metastases to the peritoneum, diaphragm and lymph nodes; in some mice the injected KPC cells diffusely replaced the liver parenchyma such that discrete nodules could no longer be resolved (not shown). These findings were never seen in mice injected with KPTC cells. The metastases formed by KPC and KPTC cells maintained the growth patterns seen in the original genetically engineered mice from which they were derived.
Stable short-hairpin knockdowns of Col6A1 and Lcn2 in highly metastatic KPC2 (Fig 4I and Supplemental Fig 4A) primary cells led to a significant reduction in lung colonization compared to mice injected with cells stably transfected with a scrambled shRNA. Knockdown of Col6A1 or Lcn2 in KPC2 cells also significantly reduced the number of liver metastases (Fig 4J) and impeded migration in wound healing assays (Supplemental Fig 4B and C). Conversely, stable overexpression of Col6A1 and Lcn2 in poorly metastatic KPTC4 cells (Supplemental Fig 4D) significantly increased their ability to form lung and liver metastases, together with the associated bloody ascites, peritoneal, diaphragm and lymph node metastases seen with baseline KPC cells (Fig 4K and L). Wound healing assays showed higher rates of migration as well (Supplemental Fig 4E and F). Collectively these data further indicate that Col6A1 and Lcn2 are soluble mediators of metastasis that are secreted from KPC cells with an intact Tgfβ pathway.
KPC Secreted Proteins Prime Cells for Colonization
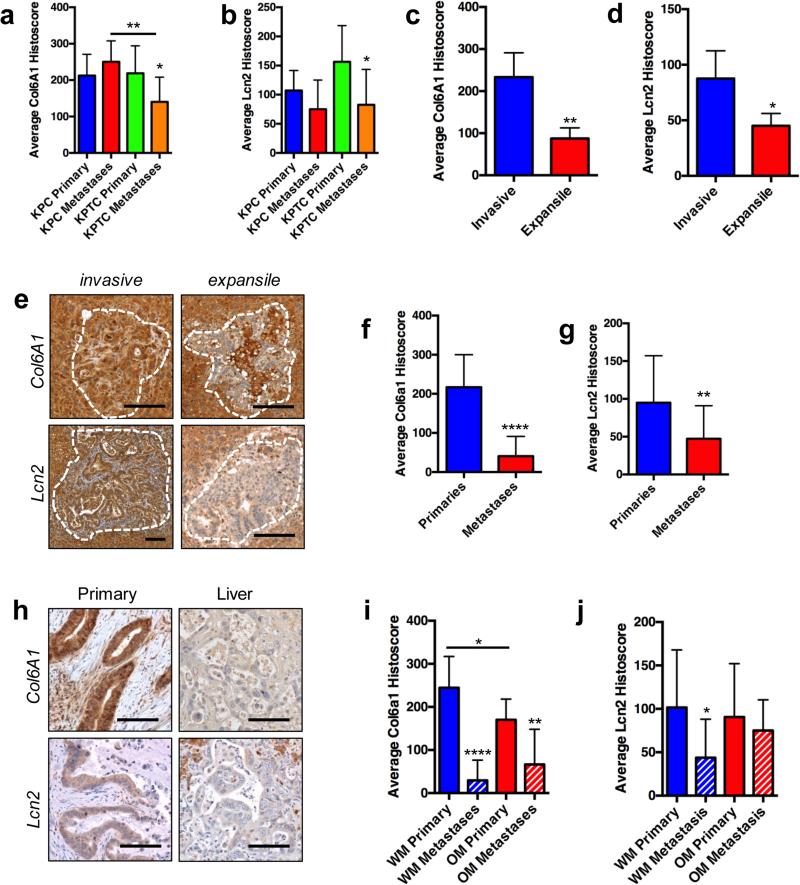
We next wondered at what point(s) in the metastatic cascade these proteins are overexpressed (26). Towards this end we performed immunolabeling for Col6A1 and Lcn2 in primary and metastatic tissues from KPC and KPTC mice. Both the intensity of labeling and the percent of positive labeling cells were considered, resulting in a “histoscore” for each marker per tissue section.
We found no difference in Col6A1 or Lcn2 histoscores between KPC and KPTC primary tissues, nor were there differences between primary and metastatic KPC tissues when all samples were analyzed. Two KPC tissues showed low expression of Col6A1 but did not have matched metastases for comparison. By contrast, KPTC metastases showed significantly lower Col6A1 and Lcn2 expression compared to KPTC primary tumors (Fig 5 A and B). KPTC metastases also showed lower Col6A1 expression compared to KPC metastases (Fig 5A). This intriguing observation suggests two features of KPC secreted proteins. First, that they are robustly activated within the primary site and second, that perturbation of TGFβ signaling attenuates the ability of the disseminated cells to maintain expression of these secreted proteins for efficient colonization in secondary sites. In agreement with this possibility invasive liver metastases had significantly higher histoscores for both Col6A1 and Lcn2 than expansile liver metastases (Fig 5C-E). By contrast, there were no differences in labeling for either protein in lung metastases from KPC or KPTC mice further underscoring the different thresholds required to colonize these two target organs.
Figure 5. Prometastatic protein expression is enabled within the primary site of mouse and human PDA.
A and B, Mean ± S.D. histoscores for Col6A1 and Lcn2 in primary and metastatic PDA tissues from KPC and KPTC mice. C and D, Mean ± S.D. histoscores for Col6A1 and Lcn2 in invasive versus expansile liver micrometastases. E, Representative Col6A1 or Lcn2 labeling of invasive and expansile liver metastases. Dashed lines outline the metastases. F and G, Mean ± S.D. histoscores for Col6A1 and Lcn2 in primary and metastatic PDA tissues from patients with PDA. H, Representative Col6A1 or Lcn2 labeling of matched human primary and liver metastases. I and J, Mean ± S.D. histoscores for Col6A1 and Lcn2 in primary and metastatic PDA tissues from patients categorized by having widely metastatic (WM) or oligometastatic (OM) PDA at autopsy. All distributions were compared by a two-sided Student's T-test. Scale bars, 100 microns. *, p<0.05; **, p<0.01; ****, p<0.0001.
To determine the extent to which this paradigm holds within human PDA we determined the expression of Col6A1 and Lcn2 in an extended set of primary and metastatic PDA tissues from 26 patients. This sample set included 19 primary PDAs and 23 metastases, 12 of which were matched sets from the same patient. Findings in human tissues were even more striking than that seen in mice with the average Col6A1 histoscore 216.7± 24.1 for primary carcinomas versus only 40.0±13.1 in metastases (Fig 5F, p<0.0001 and Fig 5H). Lcn2 labeling in human primary and metastatic PDAs showed a similar pattern albeit less robust pattern of higher expression within the primary carcinoma (94.9±13.0) accompanied by a relative loss of expression in secondary sites (47.2±10.3, p<0.01, Fig 5G and H) confirming that expression of these soluble mediators of metastasis are most highly activated within the stromal rich primary site compared to distant organs. These relationships were maintained when PDA samples were stratified based on the patients’ metastatic burden at autopsy (widespread metastasis to multiple organs, WM, versus oligometastatic, OM) (Fig 5I and J) and also when limited to matched primary and metastatic PDA tissues (not shown). There was no difference in the frequency or intensity of labeling among liver metastases for Col6A1 or Lcn2, although small sample sizes and the paucity of metastases from oligometastatic patients by nature in general likely confound this analysis. Collectively these findings support a model in which a mutant Trp53 gain of function cooperates with an intact TGFβ pathway in the primary site to upregulate soluble mediators of hematogenous metastases where they act in an autocrine manner to optimize cells for successful colonization should dissemination occur.
Discussion
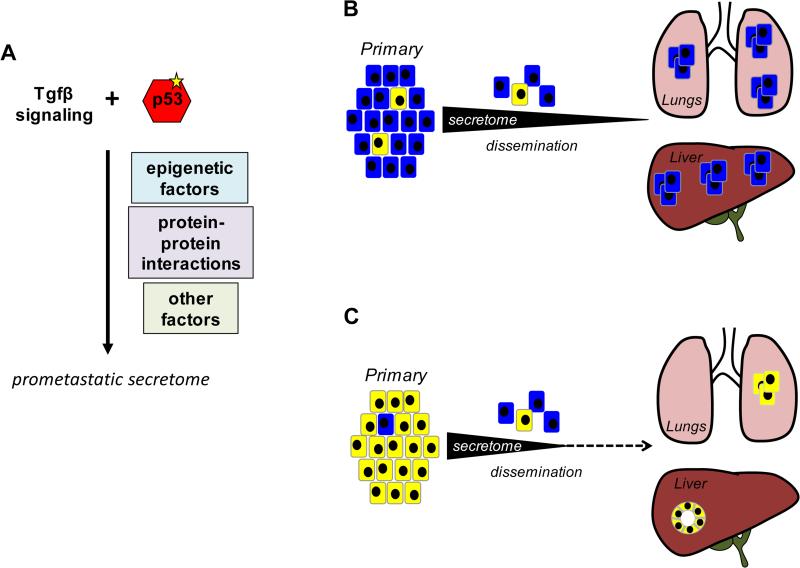
Pancreatic cancer metastasis is multifactorial in nature and influenced by both cell-autonomous and non-cell autonomous factors. (12,27-31). Pancreatic cancer metastasis is also a highly efficient phenomenon as evidenced by the frequency of patients diagnosed with stage IV disease and the high frequency of metastatic recurrence in patients who undergo potentially curative resection (1,32). We provide evidence of a potent cell-autonomous mechanism (Fig 6) based on mutant p53 and TGFβ signaling that promotes the efficiency of hematogenous metastasis. Novel insights included in this model are that the prometastatic features gained within the primary site are gradually lost upon exit from the TGFβ-rich microenvironment, a feature we refer to as the “Cinderella effect”. Explicitly, we use the term Cinderella to refer to the cancer cell itself that is metastatically enabled by the TGFβ rich microenvironment for a defined period of time after dissemination. Cells with high levels of soluble mediators maintain enough expression when they encounter the liver, permitting successful colonization of the liver and initiation of a stromal response. By contrast, those with relatively lower levels at the time of dissemination more quickly lose expression before reaching the liver resulting in vastly reduced colonization and an inability to mount a stromal response. Given strong evidence in humans that TP53 mutations are acquired during carcinogenesis (33,34) this buttresses the proposal that robust aspects of metastatic propensity are already established at the onset of invasion, a concept previously put forth based on descriptive data in humans (8). Thus, pancreatic cancer driver genes may be metastasis driver genes as well.
Figure 6. A Model for Organ-Specific Hematogenous Colonization by PDA.
A. A missense mutation in p53, denoted by the yellow star, confers a gain of function that cooperates with TGFβ signaling to produce a prometastatic secretome. The mechanism by which this cooperation occurs may be due to altered epigenetic regulation of specific genes, altered protein-protein interactions of the mutant p53 protein, or secondary effects on gene expression, among other possibilities. B. Mutant p53 together with TGFβ signaling leads to a high level of cellular fitness of the pancreatic cancer cells (blue cells) within the primary tumor. The high cellular fitness coupled with protein expression of prometastastic mediators supports efficient colonization of the liver and lung by disseminated cancer cells. C. Attenuation of TGFβ signaling decreases the prometastatic effects of mutant p53 and lowers the overall fitness of cancer cells (yellow cells). Upon dissemination they are unable to maintain expression of the secretome resulting in vastly reduced metastatic efficiency to hematogenous organs, an altered pattern of hepatic colonization, and a prolonged latency to development of clinically evident metastatic disease in general.
It is a curious fact that long-term survivors of pancreatic cancer most often fail in the lungs as an isolated metastasis (13,14). Long term survivors are also more likely to have both wild type TP53 and SMAD4 genes (10). Our findings clarify the mechanism for this phenomenon by revealing that the lungs have a lower threshold for permitting colonization than the liver. Thus, in pancreatic cancers predominated by cells with low overall fitness (i.e. more indolent biology) by chance and with enough time some will survive upon reaching the lungs and eventually undergo enough doublings to form a clinically detectable metastasis. By contrast, the liver appears a more fastidious and complex microenvironment to colonize, supporting the role of pre-metastatic niche formation by circulating factors (28). In patients with isolated pulmonary metastases, metastasectomy has been shown to be a safe procedure and doubles survival (14). Of interest, we have noted that in such patients who eventually died of their disease a complete gross and histologic review of their liver at autopsy failed to reveal unrecognized hepatic metastases (unpublished observation).
Our findings in mice and humans indicate that the formation of an extracellular matrix rich microenvironment is not required for formation of a macroscopic liver metastasis, and both stromal rich (invasive) and stromal poor (expansile/sinusoidal) metastases can be coexistent in the same liver. Moreover, colonization appears to occur via two different mechanisms, one in which invasion and extracellular matrix formation occurs rapidly upon seeding, and the second in which growth proceeds within the sinusoidal spaces leading to passive compression of the adjacent hepatocytes and formation of cystic masses. This finding is at odds with the definition of colonization as the ability to coopt the microevironment and invade the surrounding parenchyma (20). In some instances expansile colonization appeared to progress to invasion, although the possibility of selective clonal outgrowth within a polyclonal metastasis can also account for this observation (29). At the very least metastatic formation within the liver should not assume the presence of or requirement for a robust stromal microenvironment. However, we did note in both mice and humans that once an expansile metastasis reached a minimal size (macrometastasis) the frequency of a stromal component was quite high. Whether the characteristics of this stroma are identical irrespective of their colonization history remains to be seen.
While our data clearly show a relationship between mutant TP53 and the TGFβ pathway in PDA metastasis consistent with observational data in humans (10), many aspects of this relationship remain unknown. For example, whereas wild type TP53 largely exerts its effects through DNA binding, mutant TP53 proteins often lose DNA binding ability despite gains of oncogenic function (35,36). Thus, it remains to be determined if the prometastatic effects of mutant Trp53 in our model occur through direct versus indirect effects on gene expression, differential protein-protein interactions changes in post-translational modifications, or non-cell autonomous mechanisms (Figure 6), none of which may be mutually exclusive (12,18)(37). Different TP53 mutants may also have different gains of function (38) adding to the complexity of PDA metastasis. Future studies aimed at understanding the gains of function of different mutant TP53 proteins versus TP53 deletion, another common event in PDA (39), in modulating these mechanisms are expected to provide additional insights for eventual therapeutic exploitation.
Supplementary Material
Statement of Translational Relevance.
Metastasis, the growth of primary carcinoma cells in a distant organ, remains a major clinical problem for patients with pancreatic cancer. To date no studies have shown the mechanistic basis for organotropism to the liver or lungs. Here we show that TGFβ signaling modulates fitness of cancer cells at multiple levels of the metastatic cascade, colonization of the liver does not require an early stromal response, and portal vein colonization is a frequent and under-recognized mechanism of liver metastasis. Our findings also provide insight into why lung metastases are often the first and only site of recurrence in long term survivors and ultimately establish novel paradigms for understanding pancreatic cancer metastasis.
Acknowledgements
Supported by National Institutes of Health grants CA140599, CA179991, CA101955, CA62924, CA129537 and P30 CA008748.
Footnotes
None of the authors have any conflicts of interest to disclose.
References
- 1.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–72. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. American Association for Cancer Research. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Yachida S, Iacobuzio-Donahue CA. The pathology and genetics of metastatic pancreatic cancer. Arch Pathol Lab Med. 2009;133:413–22. doi: 10.5858/133.3.413. [DOI] [PubMed] [Google Scholar]
- 4.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 5.Bardeesy N, Aguirre AJ, Chu GC, Cheng K-H, Lopez LV, Hezel AF, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardeesy N, Cheng K-H, Berger JH, Chu GC, Pahler J, Olson P, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–43. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Yachida S, Iacobuzio-Donahue CA. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013 doi: 10.1038/onc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–13. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yachida S, White C, Naito Y, Zhong Y, Brosnan JA, Macgregor-Das AM, et al. Clinical Significance of the Genetic Landscape of Pancreatic Cancer and Implications for Identification of Potential Long Term Survivors. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:246–51. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell. 2015;161:1345–60. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz MHG, Wang H, Fleming JB, Sun CC, Hwang RF, Wolff RA, et al. Ann Surg Oncol. Vol. 16. Springer-Verlag; 2009. Long-term survival after multidisciplinary management of resected pancreatic adenocarcinoma. pp. 836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnaoutakis GJ, Rangachari D, Laheru DA, Iacobuzio-Donahue CA, Hruban RH, Herman JM, et al. Pulmonary resection for isolated pancreatic adenocarcinoma metastasis: an analysis of outcomes and survival. J Gastrointest Surg. 2011;15:1611–7. doi: 10.1007/s11605-011-1605-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Embuscado EE, Laheru D, Ricci F, Yun KJ, de Boom Witzel S, Seigel A, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–54. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113:301–14. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 17.Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, et al. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science. 2007;315:843. doi: 10.1126/science.1135961. [DOI] [PubMed] [Google Scholar]
- 18.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 19.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–60. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 21.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. Nature Publishing Group. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 22.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu K-H, Chang Y-H, Wu Y-S, Lee S-H, Liao P-C. Quantitative Secretome Analysis Reveals that COL6A1 is a Metastasis-Associated Protein Using Stacking Gel-Aided Purification Combined with iTRAQ Labeling. J Proteome Res [Internet] 2011;10:1110–25. doi: 10.1021/pr1008724. Available from: http://pubs.acs.org/doi/abs/10.1021/pr1008724. [DOI] [PubMed] [Google Scholar]
- 24.Candido S, Maestro R, Polesel J, Catania A, Maira F, Signorelli SS, et al. Oncotarget. Vol. 5. Impact Journals, LLC; 2014. Roles of neutrophil gelatinase-associated lipocalin (NGAL) in human cancer. pp. 1576–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volpe V, Raia Z, Sanguigno L, Somma D, Mastrovito P, Moscato F, et al. J Clin Endocrinol Metab. Vol. 98. Endocrine Society Chevy Chase; MD: 2013. NGAL controls the metastatic potential of anaplastic thyroid carcinoma cells. pp. 228–35. [DOI] [PubMed] [Google Scholar]
- 26.Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116:217–24. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- 27.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Nat Cell Biol. Vol. 17. Nature Publishing Group; 2015. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. pp. 816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddipati R, Stanger BZ. Pancreatic Cancer Metastases Harbor Evidence of Polyclonality. Cancer Discov. American Association for Cancer Research; 2015;5:1086–97. doi: 10.1158/2159-8290.CD-15-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu C-C, Simpson TR, et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer Cell [Internet] 2014;25:719–34. doi: 10.1016/j.ccr.2014.04.005. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1535610814001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–47. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter JM, Brennan MF, Tang LH, D'Angelica MI, Dematteo RP, Fong Y, et al. Survival after Resection of Pancreatic Adenocarcinoma: Results from a Single Institution over Three Decades. Ann Surg Oncol. 2012;19:169–75. doi: 10.1245/s10434-011-1900-3. [DOI] [PubMed] [Google Scholar]
- 33.DiGiuseppe JA, Hruban RH, Goodman SN, Polak M, van den Berg FM, Allison DC, et al. Overexpression of p53 protein in adenocarcinoma of the pancreas. Am J Clin Pathol. 1994;101:684–8. doi: 10.1093/ajcp/101.6.684. [DOI] [PubMed] [Google Scholar]
- 34.Boschman CR, Stryker S, Reddy JK, Rao MS. Expression of p53 protein in precursor lesions and adenocarcinoma of human pancreas. Am J Pathol. American Society for Investigative Pathology. 1994;145:1291–5. [PMC free article] [PubMed] [Google Scholar]
- 35.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 36.Garcia PB, Attardi LD. Illuminating p53 function in cancer with genetically engineered mouse models. Semin Cell Dev Biol. 2014;27:74–85. doi: 10.1016/j.semcdb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. Nature Research. 2014;14:359–70. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. Cell Death Differ. Vol. 20. Nature Publishing Group; 2013. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. pp. 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waddell N, Pajic M, Patch A-M, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.