Abstract
One in six men will develop prostate cancer in his life time. Early detection and accurate diagnosis of the disease can improve cancer survival and reduce treatment costs. Recently, imaging of prostate cancer has greatly advanced since the introduction of multi-parametric magnetic resonance imaging (mp-MRI). Mp-MRI consists of T2-weighted sequences combined with functional sequences including dynamic contrast-enhanced MRI, diffusion-weighted MRI, and MR spectroscopy imaging. Due to the big data and variations in imaging sequences, detection can be affected by multiple factors such as observer variability and visibility and complexity of the lesions. In order to improve quantitative assessment of the disease, various computer-aided detection systems have been designed to help radiologists in their clinical practice. This review paper presents an overview of literatures on computer-aided detection of prostate cancer with mp-MRI, which include the technology and its applications. The aim of the survey is threefold: an introduction for those new to the field, an overview for those working in the field, and a reference for those searching for literature on a specific application.
Keywords: Prostate cancer, MR imaging, Image Quantification, Computer-aided Detection
INTRODUCTION
Prostate cancer (PCa) is currently the most common cancer in men and the second leading cause of cancer-related deaths among men in the United States [1]. In 2015, it is estimated that the number of estimated new cases and deaths will be 220,800 and 27,540, respectively, accounting for 26.0% of new cancer cases and 8.8% of cancer deaths for American men [1].
The prostate is subdivided into the base, mid-gland, and apex from superior to inferior. The prostate also has four anatomic zones: the transition zone (TZ), which contains 5% of the glandular tissue and accounts for around 25% of PCa; the central zone (CZ), which contains 20% of the glandular tissue and accounts for around 5% of PCa; the peripheral zone (PZ), which contains 70–80% of the glandular tissue and accounts for about 70% of PCa; and the non-glandular anterior fibromuscular stroma. Accurate localization of PCa within the TZ or the PZ is extremely important as TZ prostate cancer is associated with favorable pathologic features and better recurrence-free survival [2].
At present, the clinical standard for definitive diagnosis of prostate cancer is transrectal ultrasound (TRUS)-guided sextant or systematic biopsy. The PSA blood test and DRE results are considered to identify patients who need biopsy. The actual impact of MRI for prostate cancer management is through guided biopsies and improved cancer diagnosis and staging yield. In recent years, magnetic resonance imaging (MRI) targeted prostate biopsies have been showing better disease localization and more accurate sampling than conventional TRUS-guided biopsy in various studies [3–6]. MRI-based computer-assisted sophisticated imaging for individual patients would offer such a significant role in defining an optimal targeted biopsy and interventional approach. Several approaches have been explored to improve the accuracy of image-guided targeted prostate biopsy, including in-bore MRI-guided, cognitive fusion, and MRI/transrectal ultrasound fusion-guided biopsy [7].
MR imaging provides excellent soft-tissue contrast and has become an imaging modality of choice for localization of prostate tumors. Multiparametric MRI (mp-MRI) includes high-resolution T2-weighted (T2W) MRI, diffusion-weighted imaging (DWI), dynamic contrast-enhanced imaging (DCE-MR), and MR spectroscopy (MRS). The mp-MRI has proven to be an effective technique to localize high-risk prostate cancer [8, 9]. The combined use of anatomic and functional information provided by the multiparametric approach increases the accuracy of MR imaging in detecting and staging prostate cancer [8, 9]. It can also help guide biopsies to achieve a higher tumor detection rate and better reflect the true Gleason grade. The European Society of Urogenital Radiology (ESUR) in 2012 established the Prostate Imaging-Reporting and Data System (PI-RADS) scoring system for multiparametric MRI of the prostate [10]. The MR PI-RADS aims to enable consistent interpretation, communication and reporting of prostate mp-MRI findings [10, 11]. A joint steering committee formed by the American College of Radiology, ESUR, and the AdMeTech Foundation have recently announced an updated version of the proposals of PI-RADS Version 2 [12]. Prostate mp-MRI at 3 T had been recommended in PI-RADS Version 2. Generally, CAD systems are classified into two categories: computer-aided detection (CADe) and computer-aided diagnosis (CADx) systems. Currently, most CAD systems in prostate MRI focus on local suspicious lesions and discrimination between benign and malignant lesions; most of them are computer-aided diagnosis systems. As the combination of various MR images creates large amounts of data, supportive techniques or tools, such as computer-aided diagnosis (CADx), are needed in order to make a clinical decision in a fast, effective, and reliable way.
In the past 10 years, computer-aided techniques have developed rapidly. Automated computer-aided detection and diagnosis may help improve diagnostic accuracy of PCa, and reduce interpretation variation between and within observers [13, 14]. Prostate cancer diagnosis requires an experienced radiologist to read prostate MRI, and such expertise is not widely available. Addition of CADx may significantly improve the performance of less-experienced observers in prostate cancer diagnosis. When less-experienced observers used CADx, they reached similar performance as experienced observers [13]. In a more recent study, the use of CAD can also improve prostate mp-MRI study interpretation in experienced readers [15]. For cases in which radiologists are less confident, they can get higher performance by using the computer output. A recent study showed a pattern recognition system enables radiologists to have a lower variability in diagnosis, decreases false negative rates, and reduces the time to recognize and delineate structures in the prostate [16]. The benefit of CADx also includes guiding biopsy using cancer location information from MRI [14]. Therefore, along with rapid development of MR technique, CADx of prostate cancer has become an active field of research in the last five years.
This paper starts with the review of MR image acquisition technology and then focuses on a comprehensive review of the state-of-the-art image quantification methods. The part on validation and clinical applications is a reference of the literatures available in the clinical management of the disease. The paper closes with a discussion and future perspectives.
A PubMed electronic database search for the terms "computer-aided," "CAD," "prostate," and "MRI" was completed for articles about CAD of prostate cancer up to September 11, 2015.
MR IMAGE ACQUISITIONS
Contemporary MR imaging of the prostate combines anatomic images from high-resolution T1W and T2W sequences and functional information obtained from DWI, DCEI, and MRS. The combination of conventional anatomical and functional MRI is known as multiparametric MRI. The PI-RADS Prostate MR Guidelines published in 2012 suggest the use of T2W images plus 2 functional techniques [10]. The anatomy of the prostate gland is visualized with T2W images; DWI and MRS add specificity to lesion characterization, while DCE-MRI has a high sensitivity in cancer detection. In the PI-RADS ™ v2, the essential components of the mp-MRI prostate examination are T2W, DWI, and DCE [12]. For the PZ, DWI is the primary determining sequence. For the TZ, T2W is the primary determining sequence. In order to obtain high and stable accuracy, a combination of anatomical and functional imaging is necessary in clinical practice. Recent studies showed an increasing interest in developing CADx systems to detect and characterize prostate cancer on the basis of an mp-MR imaging approach [14, 15, 17, 18]. T2W MR images are frequently used in mp-MRI CADx systems. T2W plus DWI and DCE-MRI are also commonly used among the combinations.
T2WI and T2 mapping
The anatomy of the prostate gland is best visualized with T2W images. The acquisition of high-resolution T2W images of the prostate is the first and most important step in an mp-MR imaging protocol. In T2W images, the peripheral zone of the prostate has hyperintense signal, whereas the central and transition zones have low signal, allowing the zonal anatomy of the prostate to be clearly delineated (Figure 1). In T2W images (Figure 2), PCa in the peripheral zone is usually depicted as a low-signal area. However, the growth pattern and the aggressiveness of the tumor can alter its appearance. T2W MR imaging has been advocated as an accurate technique in the detection of PCa in the transition zone [19, 20]. The value of T2W MR images is also in predicting pathological stage and extra-capsular extension of PCa [21].
Figure 1.
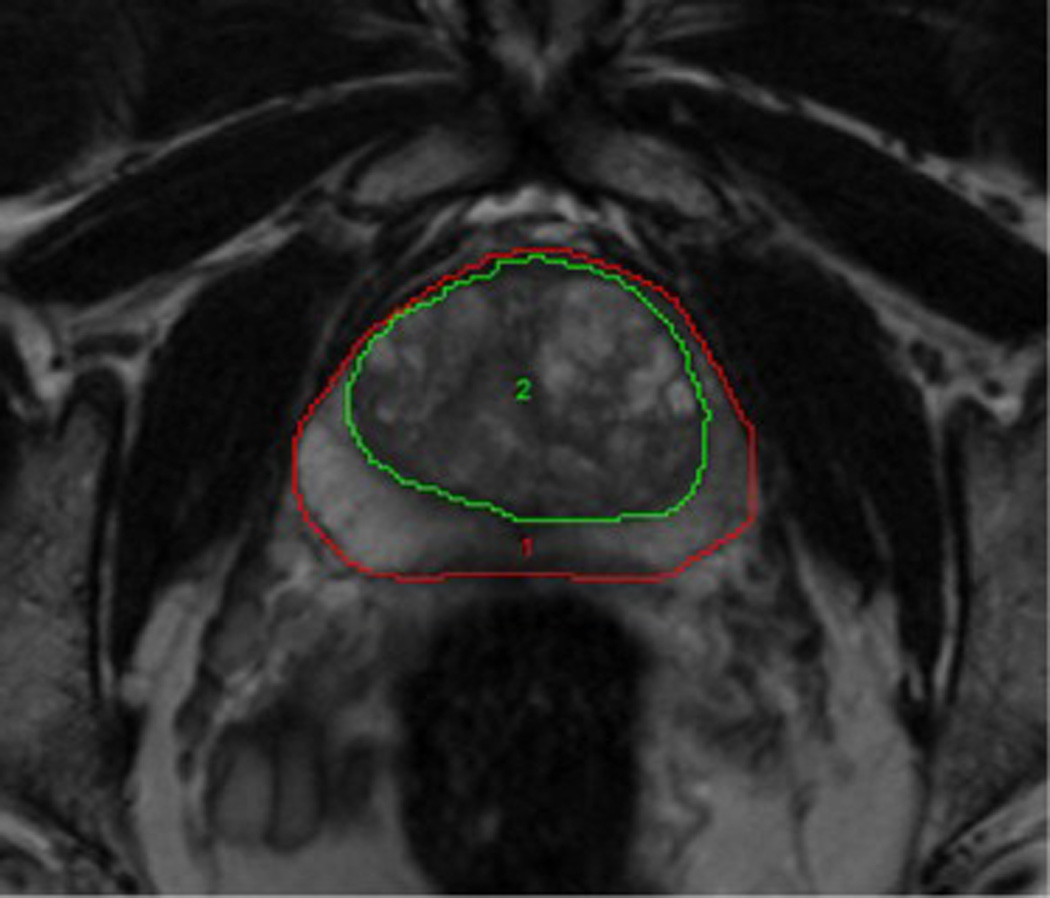
High-resolution T2-weighted MRI. T2-weighted MR images can differentiate the normal intermediate- to high-signal-intensity peripheral zone ( Region 1) from the low-signal-intensity central and transition zones (Region 2).
Figure 2.
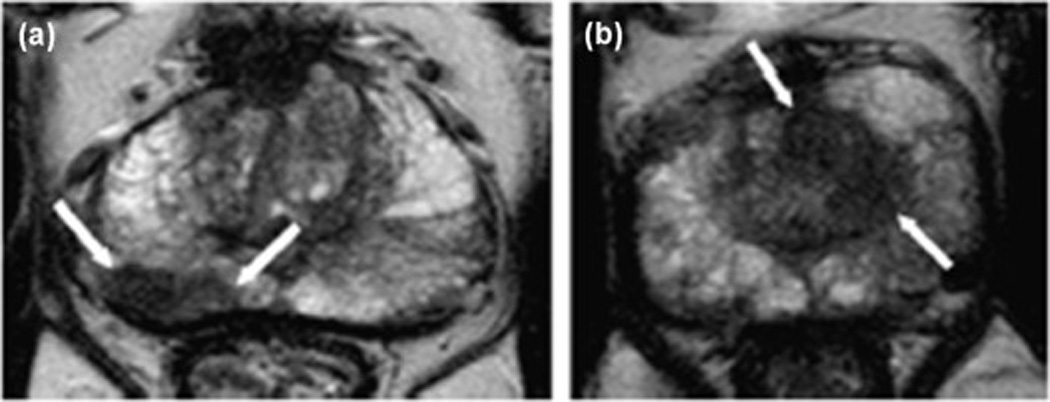
High-resolution T2-weighted MR images of prostate cancer. (A) There is a low–signal intensity lesion on the right peripheral zone (white arrows) at the mid-gland of the prostate. At prostatectomy, the lesion was classified as a Gleason grade 7 (4+3) prostate adenocarcinoma. (B) An ill-defined homogeneous low–signal intensity area at the left transition zone (white arrows) at mid-gland of the prostate in another patient. TRUS-guided biopsy showed a Gleason grade 8 (4 + 4) prostate adenocarcinoma on the corresponding position (Images from Neto JA, Parente DB: Multiparametric magnetic resonance imaging of the prostate. Magn Reson Imaging Clin N Am 2013, 21:409–426).
Because T2W MR images play an important role in both location and staging of PCa, T2W MRI is the basis and important sequence in CADx systems for PCa. In T2W MR images, the tumor region of interest (ROI) has more dark pixels than bright pixels, whereas the normal tissue ROI has more bright pixels than dark pixels. Different features, including fractal features, textural features and signal intensity can be used by CADx. Because prostate cancers at the central gland and peripheral zone usually have significantly different texture on T2W MR images [22], and because the use of mp-MRI may have challenges for detecting cancer at the transition zone [23], a CADx system that can analyze features based on the lesion’s location may be able to aid in the detection of suspicious lesions.
T2 maps offer quantitative T2 values. As the standard T2 mapping approach of performing multiple single spin-echo acquisitions with a range of TE settings requires excessive scan times, the T2 mapping is not include in most clinical applications. Recently, some new sequences can provide an effective approach to speed up T2 quantification [24, 25]. T2 values of histologically proven malignant tumor areas were significantly lower than the suspicious lesions but nonmalignant lesions or normal areas [26]. The use of quantitative T2 measurement improves the specificity and/or sensitivity of prostate cancer detection [27] and aggressiveness assessment [28, 29]. There is a potential benefit of incorporating quantitative T2 values into CADx systems.
Dynamic contrast-enhanced MRI (DCE-MRI)
DCE-MRI, which enables visualization of vascular permeability and perfusion, is an important tool in oncology to define tumor. DCE-MRI is sensitive to alterations in vascular permeability, extracellular space, and blood flow. The clinical application of DCE-MRI for prostate cancer is based on data showing that malignant lesions show earlier and faster enhancement and earlier contrast agent washout compared with healthy prostate tissues (Figure 3) [30].
Figure 3.
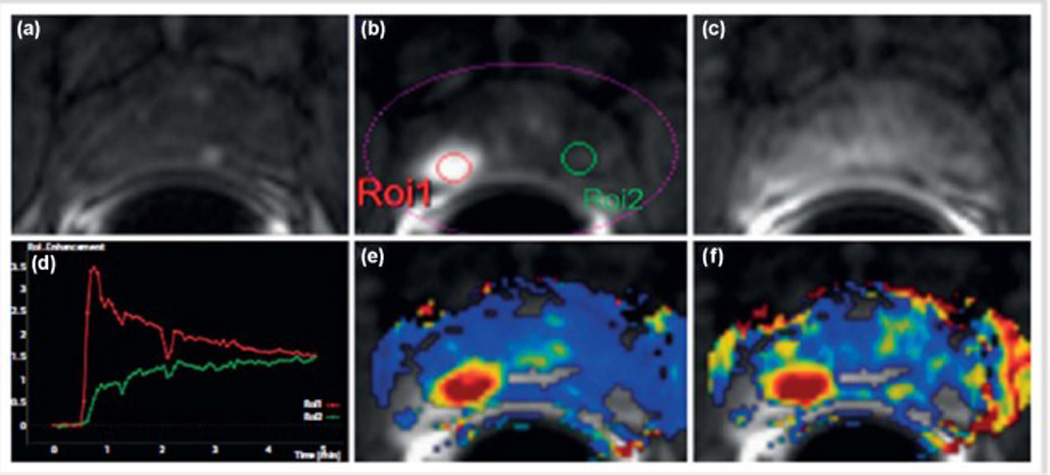
Dynamic contrast enhanced MRI (DCE-MRI) of the prostate. (a) Axial T1 GRE unenhanced image. After contrast agent administration, an area with early enhancement is seen on the right in the peripheral zone (b, ROI1) with significant washout in the late-phase image (c). The curve (red) with early enhancement is a typical finding in the case of prostate cancer, while healthy prostate tissue is characterized by a steady slow enhancement (green). High transport constants Ktrans (e) and kep (f) can confirm suspicion of prostate cancer. Prostate adenocarcinoma with a Gleason score of 4+5=9 was diagnosed after prostatectomy (Image from Durmus, T, Baur, A, Hamm, B: Multiparametric magnetic resonance imaging in the detection of prostate cancer. Aktuelle Urol 2014, 45:119–126).
The DCE-MRI data can be analyzed with various semiquantitative or quantitative models to extract parameters related to vascular permeability, extracellular space, blood flow, and water exchange [31]. Semi-quantitative DCE-MRI analysis: As semi-quantitative DCE-MRI data are only relative to the patient, the baseline intensity is highly variable depending on the patient and the MRI protocol. It is necessary to use indicators relating to signal amplitude. The most commonly used quantitative approach of analyzing DCE-MRI is two-compartment pharmacokinetic (PK) models that can be used to generate pharmacokinetic parameters such as Ktrans (transfer of gadolinium contrast from the vasculature to the tumor, representing forward vascular perfusion and permeability) and Kep (reverse transfer of contrast agent from the extracellular space back to the plasma, representing backward leakage) in order to quantify tumor enhancement and the contrast uptake and wash-out [32]. However, pharmacokinetic model implementation typically involves assuming some prior knowledge; and the arterial input function (AIF) estimation methodology can have significant effects on the parameters estimated by PK modeling [33]. The empirical approach based on phenomenological universalities (PUN) is able to reproduce experimental data from a DCE-MRI acquisition [34] [35].
Different CADx systems have been developed to analyze the DCE-MRI data. Vos et al. developed a CADx system capable of discriminating PCa from non-malignant disorders in the peripheral zone and achieved a diagnostic accuracy of 0.83 (0.75–0.92) [36]. They also developed an automated segmentation per patient calibration method to improve the diagnostic accuracy of CADx [37]. Puech et al. designed a prostate CADx software to provide a 5-level cancer suspicion score for suspicious foci detected in DCE-MRI and T1-weighted images [38, 39].
DCE-MRI usually has lower spatial resolution than other sequences, especially when DCE-MRI is performed rapidly in a short period of time. Limitations in the interpretation of DCE-MRI data include overlap in enhancement properties between benign and malignant regions in the transition zone. Benign prostatic hyperplasia and other benign inflammatory conditions within the transition zone also exhibit substantial hypervascularity [40]. Diagnostic models containing contrast enhancement parameters have reduced performance when applied across zones, so zone-specific models can improve classification of prostate cancer on multi-parametric MRI [41].
Diffusion-weighted MR imaging (DWI)
The diffusion properties of tissue are related to the amount of interstitial free water and permeability. In general, cancer tends to have more restricted diffusion than normal tissue, because of the higher cell densities and abundance of intra- and intercellular membranes in cancer [42]. Diffusion weighted MRI images can be used to detect prostate cancer from differences in the diffusion of water molecules of the normal and tumor tissues (Figure 4) [42]. The diffusion-weighted image is usually generated with different b values which can be used to calculate the apparent diffusion coefficient (ADC), and the ADC for each pixel of the image is displayed as ADC map. Diffusion of water molecules in tumor tissue is thought to reflect tissue architecture such as cell density and nucleus/cytoplasm ratio, and reductions in ADC values. For these reasons, ADC values have received the attention as a predictor of Gleason score in prostate cancer [43, 44]. Studies show that DWI findings may indicate tumor aggressiveness [27, 45, 46].
Figure 4.
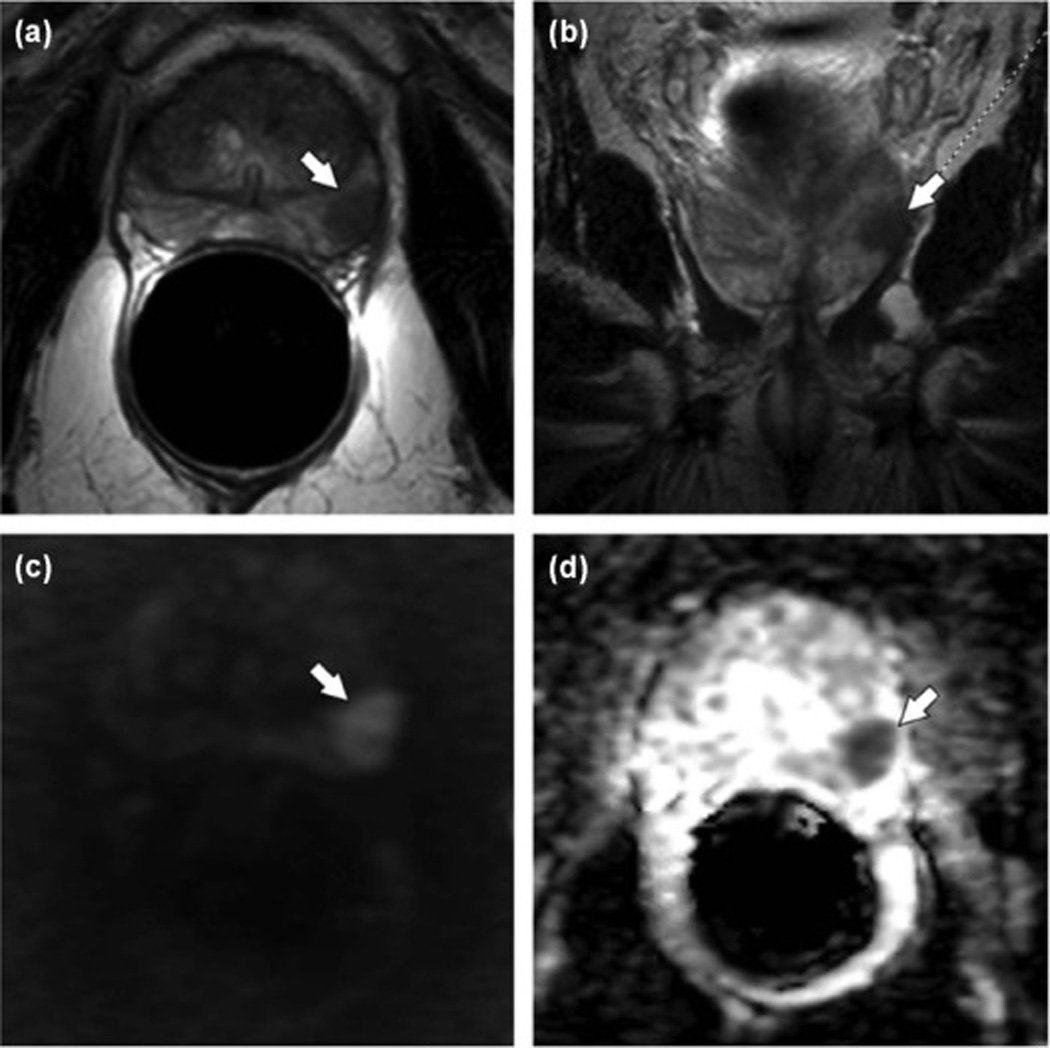
Multiparametric MRI (mp-MRI) of the prostate. Axial T2 TSE (A) and coronal T2 TSE (B) images show a well-defined T2 hypointense lesion in the peripheral zone (arrow) with corresponding high signal on DWI (C) and low signal on the ADC map (D). Biopsy of this region was positive for Gleason 4 + 3 prostate cancer (Images from Yacoub, JH, Oto, A, Miller, FH: MR Imaging of the Prostate. Radiologic Clinics of North America 2014, 52:811–837).
Technologic advances enable performance of diffusion-weighted imaging (DWI) at high b- or ultrahigh b-values (greater than 1,000 s/mm2). High b-value images can be obtained in one of two ways: either directly by acquiring a high b-value DWI sequence, or by calculating (synthesizing) the high b-value image by extrapolation from the acquired lower b-value data. Previous research has shown that high b-value DWI images allow for increased delineation between tumors and healthy tissue which makes the prostate cancer detection more robust [47, 48]. Whereas contrast in ADC maps does not significantly change with different b values, contrast ratios of DWI images are significantly higher at b-values of 1500 and 2000 s/mm2 in comparison to b values of 800 and 1000 s/mm2 [49].Wang et al. have reported that DWI images and ADC maps using b = 1500 s/mm2 should be considered more effective than those at b = 2000 s/mm2 or b = 1000 s/mm2 for detecting prostate cancer at 3 T MRI [50].
DWI images and ADC maps are the key component of the prostate mp-MRI exam. Several CADx systems adopting DWI images or ADC maps have been developed. DWI was mostly often combined with T2W in these CADx systems. Peng et al. demonstrated that the combination of 10th percentile ADC, average ADC, and T2-weighted skewness with CADx is promising in the differentiation of prostate cancer from normal tissue [27]. Niaf et al. presented a CADx system based on T2W, DWI and DCE for assisting cancer identification in the PZ [18]. Stember et al. develop a software system that identifies suspicious regions at the prostate transition zone (TZ) using signal and textural features on T2W and ADC maps, free of user input [51]. Kwak et al. recently designed a prostate CADx combined T2W and high b-value (b = 2000 s/mm2) DWI. They obtained an AUC of 0.89 [52].
MR Spectroscopy (MRS)
In MRS, the position of each metabolite peak in the output graph reflects the resonant frequencies or chemical shifts of its hydrogen protons, and the area of each peak reflects the relative concentration of that metabolite [53]. The dominant peaks observed in prostate MRS are from protons in citrate (2.60 ppm), creatine (3.04 ppm), and choline compounds (3.20 ppm) (Figure 5) [53].
Figure 5.
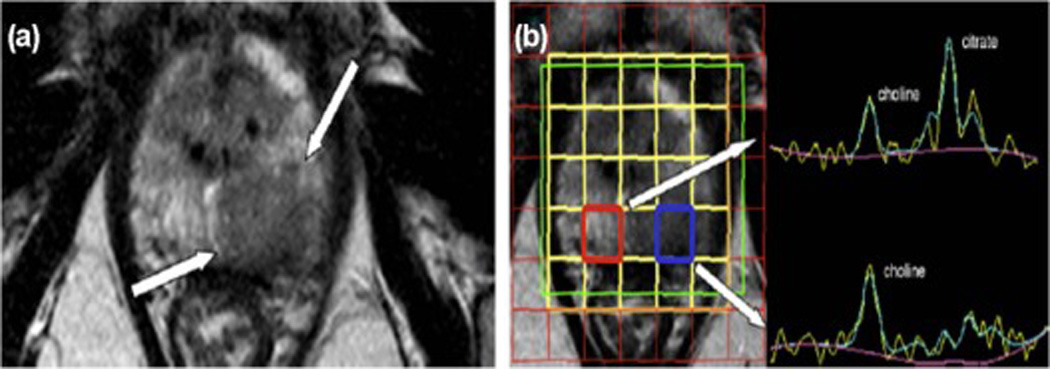
MR spectroscopy (MRS) of prostate cancer. (A) Axial T2-weighted MR images at the level of the prostate mid-gland to apex, shows a large hypointense lesion on the left peripheral zone. (B) A 3D MRS shows a normal spectrum on the right peripheral zone (red box) with normal choline plus creatine-to-citrate ratio of 0.48. In the voxel placed over the lesion on the left peripheral zone (blue box), the curve shows an increased choline peak and the citrate peak is markedly reduced. Random systematic biopsy showed a Gleason grade 9 (4 + 5) prostate adenocarcinoma on the left apex (Images from Neto JA, Parente DB: Multiparametric magnetic resonance imaging of the prostate. Magn Reson Imaging Clin N Am 2013, 21:409–426).
As a metabolic biomarker for PCa, MRS has not gained wide acceptance in routine clinical practice due to a variety of factors including the length and complexity of data acquisition, zonal anatomy, processing, and analysis. Visual interpretation of the spectra by a trained spectroscopist is time-consuming and requires accurate knowledge of prostate anatomy. Therefore, a method for automated analysis of prostate MRS data is necessary.
Over the last decade, with a view to assisting radiologists in interpretation and analysis of MRS data, several researchers have begun to develop CADx schemes for PCa identification from spectroscopy. Tiwari et al. developed an approach that integrated a manifold learning scheme (spectral clustering) with an unsupervised hierarchical clustering algorithm to identify spectra corresponding to cancer on prostate MRS [54]. The scheme successfully identified MRS cancer voxels with a sensitivity of 77.8%, a false positive rate of 28.92%, and a false negative rate of 20.88% [54]. They also presented a CADx scheme that integrated nonlinear dimensionality reduction (NLDR) with an unsupervised hierarchical clustering algorithm to automatically identify suspicious regions on the prostate using MRS [55]. They introduced the use of wavelet embedding to map MRS and T2-W texture features into a common space to identify the voxels that are affected by prostate cancer [56]. They recently presented a computerized decision support system called Semi Supervised Multi Kernel Graph Embedding (SeSMiK-GE) that may be developed into a powerful diagnostic and prognostic tool for distinguishing high and low grade PCa in vivo. Matulewicz et al. used an artificial neural network (ANN) model to automatically detect cancerous voxels from prostate MRS datasets and found that the additional information concerning the prostate’s zonal anatomy can improve the performance of the detection [57].
Other imaging methods
Although T2W, DWI, DCE-MRI and MRS are more commonly used in mp-MRI, some MRI methods, including diffusion tensor imaging (DTI), diffusion kurtosis imaging (DKI), and MR elastography (MRE), have been investigated for characterizing prostate cancer [58–61]. Other MRI methods, including proton density-weighted (PD-W) image [62] and T1 map[14], had also been added for feature calculation purposes in some CADx system.
DTI has been widely used in clinical applications, especially in neuro- and musculoskeletal imaging. Fractional anisotropy (FA) and apparent diffusion coefficient (ADC) values provided from DTI data reflect the degree of water diffusion restriction in different tissue. Pathological processes may cause change in normative FA values and disruption of fibers in tractography. The feasibility of performing DTI of the prostate had been demonstrated by some studies; and DTI tractography can successfully visualize fiber tracts around the prostate [58]. DTI tractography might be applicable to the estimation of structures of the prostate [59], the characterization of prostate cancer [60], and monitoring prostatic structural changes under radiotherapy [61].
The novel technique, diffusion kurtosis imaging, enables characterization of non-Gaussian water diffusion behavior. DK model may add value in PCa detection and diagnosis, and DKI potentially offers a new metric for assessment of PCa[63]. A recent study demonstrated no significant benefit of DKI for detection and grading of PCa as compared with standard ADC in the peripheral zone determined from b values of 0 and 800 s/mm [64].The mechanical properties of the tissue of interest are calculated from the wave fields and displayed as an image, commonly referred as an elastogram. In MR elastography (MRE), an external mechanical excitation is applied to the tissue of interest to induce tissue vibrations [65]. MRE has been shown to be of clinical value in MRI for its ability to detect tissue abnormalities in organs such as the liver [66], brain [67] and breast [68–70]. More recently, researchers have also focused on the development of MRE methods to detect prostate cancer [71–73]. The resulting wave fields are imaged using a motion-sensitized MRI pulse sequence. Elastograms may add another dimension to current mp-MRI techniques for diagnosing prostate cancer, and may further increase the sensitivity and specificity of such techniques.
T1 maps offer quantitative T1 values and can be produced by a variety of methods, such as multiple inversion or multiple repetition time acquisitions, typically requiring lengthy acquisition times. Another approach taken in the context of the prostate has been to employ spoiled gradient-echo (SPGR) sequences where it is possible to obtain T1 estimates in relatively short acquisition times by varying the RF flip angle [74]. The T1 mapping is not including in most CADx systems. Vos, et al. [14] had presented a fully automatic CADx by combining a histogram analysis on mpMR images including T1, pharmacokinetic, T2 and ADC maps.
MR lymphography has been used for the investigation of the lymphatic channels and lymph glands. Different imaging techniques, including nanoparticle-enhanced [75, 76] and non-contrast MR lymphography [77, 78], had been developed for detection of nodal metastases. MR lymphography is a noninvasive technique that is well suited for the examination of regional (intrapelvic) lymph node metastases in PCa.
MR IMAGE QUANTIFICATION METHODS
General framework
Development of computer-aided detection systems includes several aspects: image preprocessing, algorithm development, methodology for assessing CADx performance, validation using appropriate cases to measure performance and robustness, observer performance studies, assessing performance with a clinical trial, and ultimately commercialization. The development must confront several challenges. Computerized image procedure may cover different aspects of segmentation, registration, feature extraction, and classifiers. A computer algorithm should be developed based on the understanding of image reading by radiologists, such as how radiologists detect certain lesions, why they may miss some abnormalities, and how they can distinguish between benign and malignant lesions. It is important to develop CADx systems that extracts quantitative data in a more accurate and automated fashion.
Many different types of CADx systems are produced to locate/diagnose prostate cancer in MR imaging, including T2W, DWI, DCE-MRI and MRS. Considering the particularity of prostate cancer in anatomy, pathology and clinic, the core of a CADx system for the detection of prostate cancer is associated with its computerized algorithms. In general, the pipeline of the CADx system for prostate cancer is visualized schematically in Figure 6. In the initial stage, lesion candidates are selected within a likelihood map that is generated by a voxel classification of one or more images. Hereafter, the lesion candidates are segmented into a region of interest from which region-based features are extracted. Finally, the extracted information is fused by a classifier into malignancy likelihood. The following sections describe each step in detail.
Figure 6.
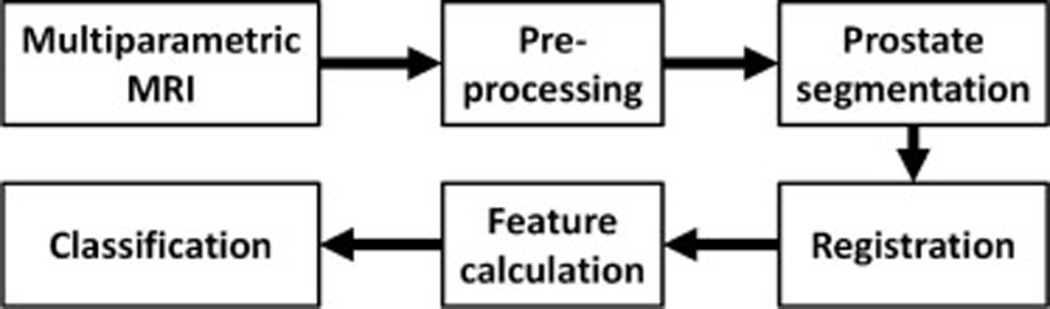
Flowchart for computer aided detection of prostate cancer in mp-MRI.
Preprocessing
The purpose of preprocessing is to normalize the MR data or to transform the MR data to a domain in which prostate lesions can be easily detected.
For T2W-MRI, the image intensities can vary, even using the same protocol and the same scanner. The quality of images depends on the acquisition conditions such as temperature, calibration adjustment, B0 intensity, coil position, and the receiver gain value. In addition, the intensity variation will increase when different scanners are used. This relationship must be taken into account for MR image analysis. Collewet et al. [79] proposed four schemes for the intensity normalization. The most used method is that intensities are proportionally normalized by defining the median+2* (inter-quartile range).
ADC maps calculated from DWI are useful for detecting prostate cancer with a relatively high specificity. However, it has lower resolution than T2W-MRI and is subject to magnetic susceptibility artifacts [17]. ADC represents a quantitative assessment of water diffusion. Lower ADC value is associated with higher rate of malignancy. Prostate cancer can be identified as a low signal region on ADC maps against a background of normal tissue with higher signal intensity [17].
Intensity inhomogeneity arises from the imperfections of the image acquisition, which can reduce the accuracy of segmentation, classification, and registration. The most intuitive method to correct intensity inhomogeneity is image smoothing or homomorphic filtering [80]. Vovk et al. [81] classify inhomogeneity correction methods into two categories, which are prospective and retrospective. Prospective methods aim at the calibration and improvement of image acquisition processes. Retrospective methods rely exclusively on the information of the acquired images or on a priori knowledge. Sled et al. [82] proposed a nonparametric non-uniform intensity normalization (N3) method for inhomogeneity correction, which is independent of pulse sequence. Tustison et al. [83] proposed a variant of N3 for bias field correction. Similar to the N3, the source code, testing, and technical documentation are publically available and the package is "N4ITK". This algorithm is available to the public through the Insight Toolkit of the National Institutes of Health.
Segmentation
The segmentation aims to reduce the burden of the classifier in the later stages. Therefore, the classifiers only focus on the prostate region obtained by segmentation methods. T2W imaging provides the best resolution and contrast to show the anatomy of the prostate and has a very high sensitivity for prostate cancer. Therefore, T2W-MRI is the most useful image sequence in determining the contours of prostate.
Extensive studies were developed to segment the prostate from MR images [62, 84–90]. It can be a challenging task to obtain accurate prostate volume in T2W-MRI. Firstly, the contrast between the prostate and the surrounding tissues can be low. Therefore, it may be difficult to accurately segment the boundary of the prostate. Secondly, the prostate shapes of different patients can be significantly different. Even for the same patient, the prostate motion at different patient positions can be large, which results in a shape difference on MR images. Thirdly, MR image appearance, quality, and the presence of artifacts can be affected by different scanners, which in turn can have a large influence on the performance of computerized algorithms. All these aspects need to be considered when developing a robust and accurate segmentation method for prostate MR images.
Contour and shape based methods [91–95]exploit edge and shape features to segment the prostate, which contains two categories. The first category is edge based segmentation methods. The edge detection operators are used to produce edges on MR images. The candidate edges are picked up and then connected to obtain the prostate boundary. Zwiggelaar et al [91] developed a semi-automatic method to segment the prostate in MRI data. Their method exploits the characteristics of the anatomical shape of the prostate when represented in a polar transform space. The edge detection and non-maximum suppression are used to track the boundary of the prostate.
The second category is deformable model based segmentation methods. Kass et al [96] proposed an active contour model and used the image gradient to evolve a curve. The internal spline force pushes the curve toward the salient image feature, while external force is responsible for putting the curve near the object. Chan and Vese [97] proposed a level set algorithm of the piecewise constant variant of the Mumford-Shah model [98] for segmentation.
Atlas based methods are also used to segment the prostate in MR images [99]. An atlas consists of original image data and its corresponding manual segmentation. The atlas can be used as a reference to segment the prostate of a new patient. Klein et al. [99] proposed an automatic method for segmenting the prostate in 3D MR images. Their method is based on non-rigid registration of a set of pre-labeled atlas images. The label images of the deformed atlas are fused to yield a segmentation of images from a new patient.
Besides the above methods, a global optimization algorithm called graph cut [100, 101] is becoming more and more popular due to its efficient global minimization. The segmentation problem can be formulated as a minimization of an energy minimization. Egger [102] proposed a graph-based approach to automatically segment prostate based on a spherical template. The minimal cost on the graph is optimized by a graph cut algorithm, which can get the segmentation of the prostate volume. Mahapatra and Buhmann [85] proposed a fully automatic method for prostate segmentation using random forests classifiers and graph cuts. The prostate probability map was generated based on a random forests classifier. The negative log-likelihood of the probability maps was used as the penalty cost in an energy function, which was minimized by graph cuts. Tian et al. [103] proposed a supervoxel-based segmentation method for the prostate. The prostate segmentation problem was considered as assigning labels to supervoxels. An energy function with both data and smoothness terms was used to model the labels, which was minimized using graph cuts. The segmentation results are shown in Figure 7. Other segmentation methods were also developed for the prostate [104, 105]. Ghose et al. [105] reviewed segmentation methods for the prostate in TRUS, MR and CT images. They studied the similarities and differences among the different methods, highlighted their advantages and disadvantages in order to assist in the choice of an appropriate segmentation methods. They also showed a comprehensive description of the existing methods in all TRUS, MR and CT images, and highlighted their key-points and features. They provided a strategy for choosing segmentation method for a given image modality.
Figure 7.
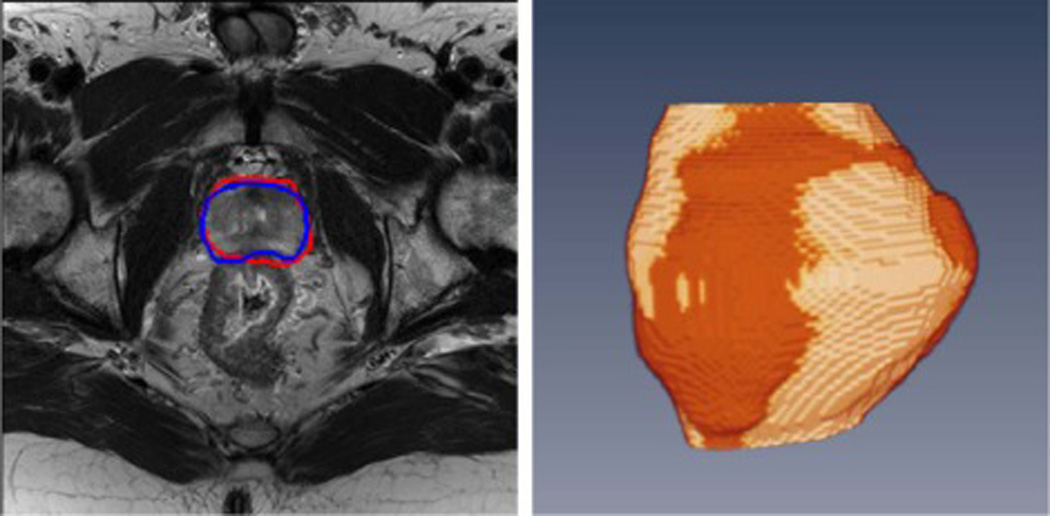
Prostate segmentation on MR images. Left: 2D MR image and segmentation results where the red curve represents the segmentation from a computer algorithm while the blue curve is the ground truth labeled by a radiologist. Right: 3D visualization after segmentation. The gold region is the prostate surface obtained by the computer algorithm while the red region is the ground truth.
A publicly available data set called MICCAI Challenge Prostate MR Image Segmentation (PROMISE12) [104] can be used to evaluate the performances of the new proposed methods. This data set contains 50 cases with ground truths for training, and 30 cases without ground truths for test, which are 3D T2w MR transverse images of the prostate. The MR images were obtained from multicenter, multivendor, and different acquisition protocols (i.e., with/without endorectal coil, differences in slice thickness).
Registration
Image registration is a process of aligning two or more images, which aims to find the optimal transformation that best aligns the structures of interest in the input images. Image registration is needed in order to integrate the features from different images of mp-MRI such as DCE-MRI and T2W MRI. The registration of images requires the selection of the feature space, a similarity measure, a transformation type, and a search strategy [106]. The DICOM header of MR images can provide coordination and orientation information that are useful for registering T2W, ADC, and Ktrans maps. T2W-MRI is considered as the reference. Other modalities can be registered to T2W-MRI by aligning the coordinates of their origins, which are obtained from the DICOM header. If necessary, resolution adjustment is also performed after the alignment.
Registration is also used to validate in vivo MR imaging using ex vivo histologic images [107, 108]. To obtain the reliable ground truth of the prostate cancer region, whole-mount histology is performed on ex vivo prostate. The pathologist labels the cancer region in the histology images. Based on the registration between the whole-mount histology and T2W MRI, the labelling of the cancer in histology can be mapped to T2W MRI for validation [107, 108]. Kalavagunta et al. [108] proposed a method to register MRI and histology using local affine transformations guided by internal structures. First, the histologic and MR images are first segmented, scaled, and translated. Second, the prostate capsule and internal structure masks are identified to constrain the pathology transformation. A transformation matrix is obtained by registering two images based on capsule and internal structure masks. Third, the pathology images are warped using a computed transformation matrix. Fourth, a transformation matrix is applied for each annotated cancer region. The warped cancer regions are superposed on registered pathology images. Last, the cancer regions in MRI can be obtained by mapping the cancer regions of pathologic images to MR images. In another study, Chappelow et al. [107] presented a new registration method that maximizes the combined mutual information shared by the intensity of the reference image and multiple representations of the floating images in multiple feature spaces. The method provides enhanced registration performance by combining the intensity information with transformed feature images from the images. These feature images are not as susceptible to intensity artifacts and provide additional similarity information regarding the reference image but not contained in the floating image. This method is particularly useful for registering MRI and histology.
Feature extraction
Feature extraction plays an important role in prostate MRI CADx systems. Classic features for medical images include intensity, shape, texture, and statistical features. For medical image classification, choosing the right features for a classifier is more important than choosing the classifier itself [62].
Litjens et al. classified the features into five types: intensity, pharmacokinetic, texture, blobness, and anatomical features [62]. For the intensity feature, a T2-estimate map is generated by using the MR signal equation, the proton density image and a reference tissue [88]. Anatomical features include the relative distance to the prostate boundary and the relative position feature. Both the relative distance and relative position features are calculated with respect to the prostate surface obtained by segmentation methods. For the pharmacokinetic feature, the traditional analysis is incorporated in their CADx system by using a curve fitting-technique to fit a bi-exponential curve to the time data, as presented in [109]. For the texture feature, a Gaussian texture bank was used to capture the textural distortions [22]. For the blobness feature, it was found that prostate cancer tends to appear as a blob-like lesion in DWI and DCE-MRI. The blobness-filter presented by Li et al. was chosen as a blobness measure [110]. Blobness is calculated on the ADC, tau and LateWash images, and on the Ktrans and Kep images as well [110].
Shah et al. [17] created an mp-MRI feature set for CADx systems (Figure 8). First, in order to reduce interpatient variability, normalized T2W maps were calculated from the transversal T2W intensities using the average fat signal adjacent to the prostate as a reference. Second, quantitative ADC maps were computed from the transversal DWI by fitting the dependence of the signal intensity in each pixel. Third, each dynamic curve was de-noised by using a wavelet filter for DCE-MRI. The pharmacokinetic parameters were extracted by using the generalized kinetic model (GKM) [111, 112]. Then, the GKM was fitted to the measured concentration time curves, using the linear least-squared method [112] to yield the volume transfer constant Ktrans, and the rate constant kep. Finally, the normalized T2W and ADC maps were resized to have a pixel resolution equal to the T1 and Ktrans and kep maps in order to form the final feature set for the CADx system.
Figure 8.
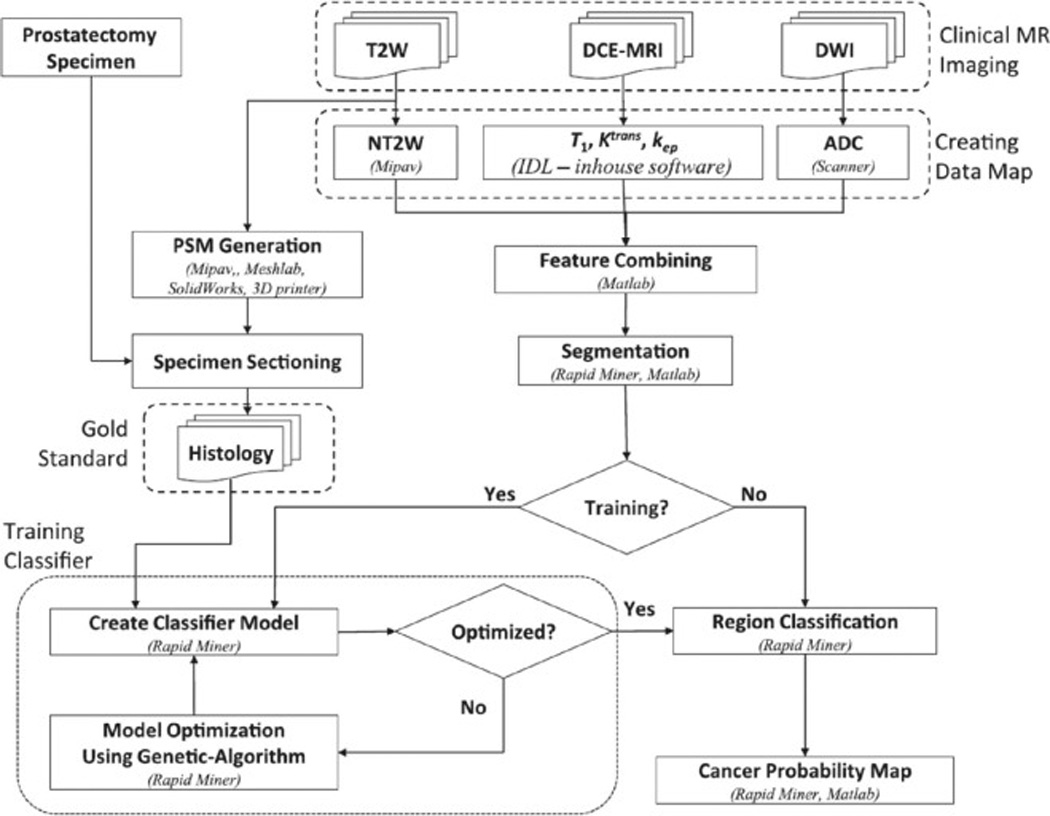
Flowchart for a CAD system based on a multiparametric MRI. The cancer probability map is the final outcome of the algorithm (Image from Shah V, Turkbey B, Mani H, Pang Y, Pohida T, Merino MJ, Pinto PA, Choyke PL, Bernardo M: Decision support system for localizing prostate cancer based on multiparametric magnetic resonance imaging. Med Phys 2012, 39:4093–4103).
Niaf et al. extracted about 140 kinds of features for a CADx system [18]. Most of these features were chosen based on their proven efficiency between cancer and non-cancer. Two categories of features were proposed: image features and functional features. For image features, there were three types: grey-level features, texture features, and gradient features. The image intensity values of T2, DCE and ADC maps were used as grey-level features. First order texture measurements were computed for each pixel over a local window, which includes mean, median, standard deviation and average deviation. Second order texture features were computed based on two neighboring pixels, which includes co-occurrence matrix. The Sobel and Kirsch filters and numerical gradient operators were used to compute gradient features (Figure 9).
Figure 9.

Image features for prostate cancer detection. (a) With prostate cancer superposed in green. (b) First order statistics (standard deviation). (c) Sobel-Kirsch feature. (d) second order statistics (contrast inverse moment). (e) Corresponding time-intensity curves for CaP (red) and benign (blue) regions are shown based on DCE-MRI data (Images from Viswanath S, Bloch BN, Rosen M, Chappelow J, Toth R, Rofsky N, Lenkinski R, Genega E, Kalyanpur A, Madabhushi A: Integrating Structural and Functional Imaging for Computer Assisted Detection of Prostate Cancer on Multi-Protocol 3 Tesla MRI. Proc Soc Photo Opt Instrum Eng 2009, 7260:72603I).
Radiomics is an emerging field for the quantification of tumor phenotypes by applying a large number of quantitative image features [113, 114]. Radiomics can provide complementary and interchangeable information to improve individualized treatment selection and monitoring. Since medical imaging technology is routinely used in clinical practice worldwide, radiomics may have a high clinical impact on future patient management. The workflow of radiomics consists of three steps [113]. The first step is the acquisition of standardized images for diagnostic or planning purposes. On the images, the tumor regions are extracted by an algorithm or an experienced radiologist. Second, quantitative imaging features are extracted from the tumor regions. These features involve tumor image intensity, texture, and shape and size of the tumor. Last, all the extracted features are analyzed and selected by a model. The most informative features are identified and incorporated into predictive models for treatment outcome. Radiomics, as a high dimensional mineable feature space, can be used for prostate cancer. Cameron et al. had constructs a comprehensive radiomics feature model to detect tumorous regions using mp-MRI [115]. New radiomics-driven texture feature models had been developed for the detection of prostate cancer and for the classification of prostate cancer Gleason scores by utilizing mp-MRI data [116–118].
Classification
Image classification involves training and testing with features extracted from image data and its corresponding labels [62]. A classifier is usually trained by using the labeled image data set and applied to unseen image data sets. Several classification techniques from the machine learning field have been developed for picking up discriminative features. Support vector machines (SVMs) and random forests could achieve good performance based on the positive and negative training samples [17, 119]. A pixel classification provides a likelihood between 0 and 1 for each pixel, with 0 indicating no suspicion of prostate cancer and 1 indicating high suspicion of cancer.
Litjens et al. [62] experimented with three different classifiers: a linear discriminate classifier, a Gentle Boost classifier [120], and a random forests classifier [119] with regression trees. Shah et al. used SVM to create a classifier model [17]. Because real data are not linearly separable, the SVM implementation was used to allow relaxed constraint for misclassified points. SVMs “kernel trick” was also implemented to enable operations to be performed in the input space rather than the potentially high-dimensional feature space [121].
Chan et al. [122] investigated the use of a statistical classifier for detecting prostate cancer by combining information from MR images. SVM is used to predict the tumor likelihood in the peripheral zone using the derived features. For SVM training, they randomly sampled 10% of the PZ data and retained all the tumor data to confine the training dataset to a manageable size for SVM training convergence. The radial basis function kernel was used for SVM. These works indicate that the SVM classifiers and random forests work well on the problem of classifying prostate tumors on mp-MRI.
VALIDATION
When developing a CADx system for prostate MRI, the accuracy of the “gold standard” is important. Histopathology, as the ground truth, usually includes findings from prostatectomy specimens or biopsy specimens. The validation of CADx systems is summarized in Table 2. In order to transfer the labels from pathology to MR images, MR images usually need to be registered with pathological sections of the prostate. An accurate registration of histologic and MR images serves as the bridge between in vivo anatomical information and ex vivo pathologic information, which is valuable in developing a CADx system.
Table 2.
Validation of CADx systems
| Reference | Ground truth on the histology |
Candidate on MR image | Image Registration |
|---|---|---|---|
| Chan et al. (2003)[122] |
Biopsy | MO | NA |
| Puech et al. (2007)[39] |
Needle biopsy or prostatectomy |
MO | NA |
| Tiwari et al. (2007) [54] |
Biopsy | Sextant location determined
by radiologist |
NA |
| Vos et al. (2008)[36] |
WMHS+ MO | MO | 3D rendering mode |
| Viswanath et al.(2008)[140] |
WMHS+ MO | MANTRA | Multimodal image registration |
| Viswanath et al.(2009)[129] |
WMHS | MANTRA | Multimodal image registration |
| Vos et al.(2009)[37] | WMHS | Not specified | Not specified |
| Liu et al.(2009)[141] |
WMHS+ MO | MO +ex vivo MRI | Manual |
| Tiwari et al.(2009)[55] |
WMHS+ sextant boundaries |
A joint review session of trial imagers and pathologists |
NA |
| Artan et al.(2010)[142] |
WMHS+ MO | Tumor location is transferred to the in vivo MRI from histologic images +ex vivo MRI |
NA |
| Vos et al.(2010)[143] |
WMHS+ MO | MO | Mutual information registration |
| Viswanath et al.(2011)[144] |
WMHS+ MO | Registration from
histologic images |
MACMI |
| Lopes et al.(2011)[145] |
WMHS+ drawn by urologists |
Drawn by urologists | manual correspondence |
| Liu and Yetik (2011)[26] |
WMHS+ MO | MO +ex vivo MRI | Manual registration |
| Sung et al.(2011)[146] |
Radical prostatectomy+ MO |
The radiologist matched the pathologic slices with corresponding MRI |
NA |
| Tiwari et al.(2012)[56] |
WMHS | MO +ex vivo MRI | Manual registration |
| Viswanath et al.(2012)[22] |
WMHS+MO | Registration from
histologic images |
Multimodal Elastic Registration |
| Vos et al.(2012)[14] | Needle biopsy | Combining the findings
with, histopathology of MR-guided samples by radiologist. |
NA |
| Niaf et al.(2012)[18] |
WMHS+MO | MO | Manual registration |
| Artan et al.(2012)[147] |
WMHS+MO | MO +ex vivo MRI | Manual registration |
| Shah et al.(2012)[17] |
WMHS+MO | Not specified | PSM |
| Matulewicz et al.(2013)[57] |
WMHS+MO | MO | Manual registration |
| Hambrock et al.(2013)[13] |
WMHS+MO | MO | Manual registration |
| Tiwari et al.(2013)[148] |
WMHS+MO | MO | Manual registration |
| Peng et al. (2013)[27] |
WMHS | MO | Manual registration |
| Ginsburg et al.(2014)[149] |
WMHS+MO | Registration from
histologic images |
Nonlinear registration |
| Stember et al.(2014)[51] |
Needle biopsy | Not specified | NA |
| Niaf et al.(2014)[150] |
Prostatectomy+MO | MO | Manual registration |
| Garcia Molina et al.(2014)[16] |
Prostatectomy+MO | MO | Manual registration |
| Litjens et al.(2014)[62] |
Needle biopsy | Not specified | NA |
| Kwak et al.(2015)[52] |
Needle biopsy | Determined by radiologists | NA |
| Zhao et al.(2015)[151] |
Biopsy | MO | NA |
MO: manual outlined regions of lesions
MANTRA: multi-Attribute, non-initializing, texture reconstruction based ASM
MACMI: multi-attribute, higher order mutual information based elastic registration scheme
PSM: patient-specific molds
WMHS: whole mount histological sections
NA: no registration was used
Whole-mount sections are generated from tissue slices and microscopic slices are stained with hematoxylin-eosin staining after being embedded in paraffin [111, 123]. Pathologists outline each lesion on the microscopic slices. Gleason scores of different regions may also be provided on the microscopic slices. For correlation between MR images and histopathologic images, the corresponding anatomical landmarks and cancerous regions are manually labeled by an expert. The urethra may serve as a guide for correlating the images. In order to improve the accuracy and efficiency of the correlation, some automatic methods have been developed [111, 124].
There are several challenges in establishing automatic correlation between in vivo MR images and histopathologic images. The orientation of the specimen and its sections may be different from that of in vivo MR imaging. There are mismatches between MR imaging and histopathology, which make it difficult to assess the true accuracy of MRI. Once the anatomic orientation in the body is lost, it may be difficult to section the prostate in the same plane as that of in vivo MR images. The specimen can be marked with separate colors on the left, right, and anterior aspects for anatomic orientation [111]. Using image processing, computer-aided design, and rapid prototyping technology, a customized mold has been used to process prostatectomy specimens for each patient [124]. The customized mold holds the prostate in the same position and the same shape as those of in vivo MR images and guides the cutting knife to obtain tissue blocks that correspond to the image slices.
The prostate is an easily deformable organ, hence, the gland deforms during and after prostatectomy. Additionally, prostate MRI is often performed by using an endorectal coil, which further deforms the gland. Specimen formalin fixation and paraffin embedding also induce variable tissue shrinkage. Deformable image registration provides a high degree of flexibility for registration of histologic images with in-vivo/ex-vivo MR images, and can assist in more accurate evaluation of MRI findings. Boundary landmarks and internal landmarks of the same prostate have been used in a deformable registration algorithm. Mazaheri et al. describe a semi-automatic method by using a free-form deformation (FFD) algorithm based on B-splines [125]. This method enabled successful registration of anatomical prostate MR images to pathologic slices. Jacobs et al.[126] proposed a method for the registration and warping of MR images to histologic sections. This method consists of a modified surface-based registration algorithm followed by an automated warping approach using nonlinear thin plate splines to compensate for the distortions between the datasets.
There are two general approaches to map ex vivo histological PCa extent to pre-operative MR images. The first method, perhaps the more intuitive approach, is to reconstruct the 3D histologic volume, and then register the 3D histologic volume with the 3D MR volume [127, 128]. The second approach is to register each 2D histology slice to its corresponding 2D MRI slice separately [107, 129]. In the first approach, one critical prerequisite was the accurate reconstruction of the histologic volume; while in the second approach, the prerequisite was to determine the histology-MRI slice correspondence. In some cases, the former prerequisite may not be achievable hence the only solution is to take the second approach. There is an increasing interest in the registration of 3D histopathology with prostate MR imaging. Three-dimensional reconstruction of prostate histology facilitates these registration-based evaluations by reintroducing 3D spatial information lost during histology processing [130, 131]. Patel et al. [132] presented a scheme for the registration of digitally reconstructed whole mount histology to pre-operative in vivo mp-MRI using spatially weighted mutual information. McGrath et al. [133] used reference landmarks that are visible in both data sets to assist 3D histopathology reconstruction and thus can provide important information on the deformation effects of fixation, and hence improved registration accuracy. Histostitcher, a software system designed to create a pseudo whole mount histology section from a stitching of four individual histology quadrant images, is another alternative for reconstructing pseudo whole-mount prostate images [134].
Registering pathologic information to mp-MRI is a challenging problem in developing a CADx system for mp-MRI (Figure 10). Chappelow et al. [135] described a method based on mutual information that registers T2W, DCE-MRI, and ADC. However, this method is based on 2D histology and requires considerable expertise to determine the correspondence between histologic and MR images. Orczyk et al. [136] described a method based on the present registration method and were the first to create a 3D counterpart within the same reference space between histology and both anatomical and functional sequences provided by prostate mp-MRI (Figure 11). The method enables a true, deformable transformation and achieves an accuracy of 1–2 mm. The registration of different MR images is critical considering prostate motion, especially related to rectal peristalsis. Orczyk et al. [136] used rigid registration to correct motion between difference sequences.
Figure 10.
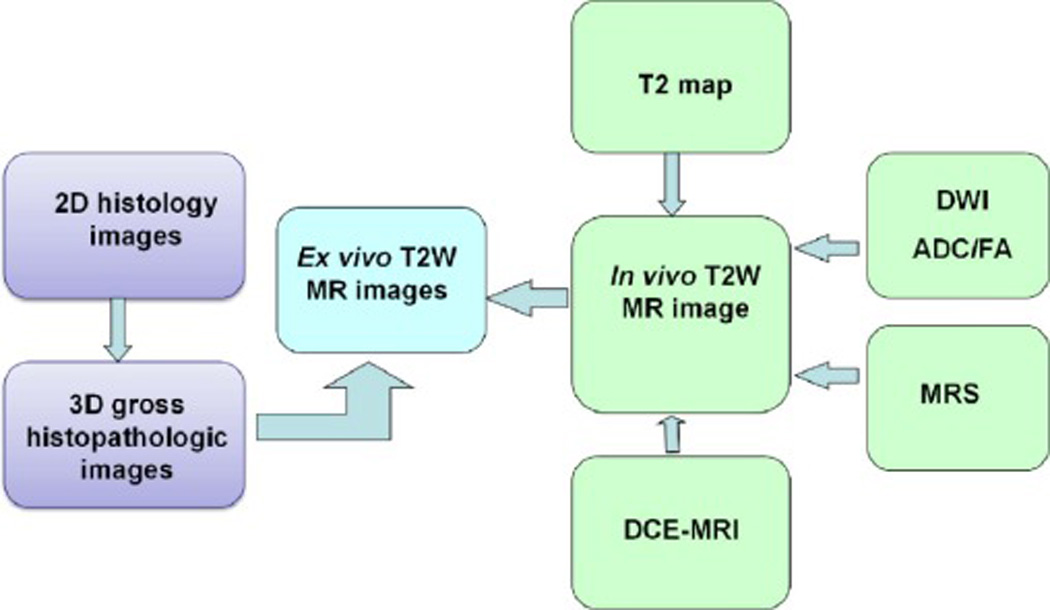
Registration between multiparametric MRI and histology.
Figure 11.
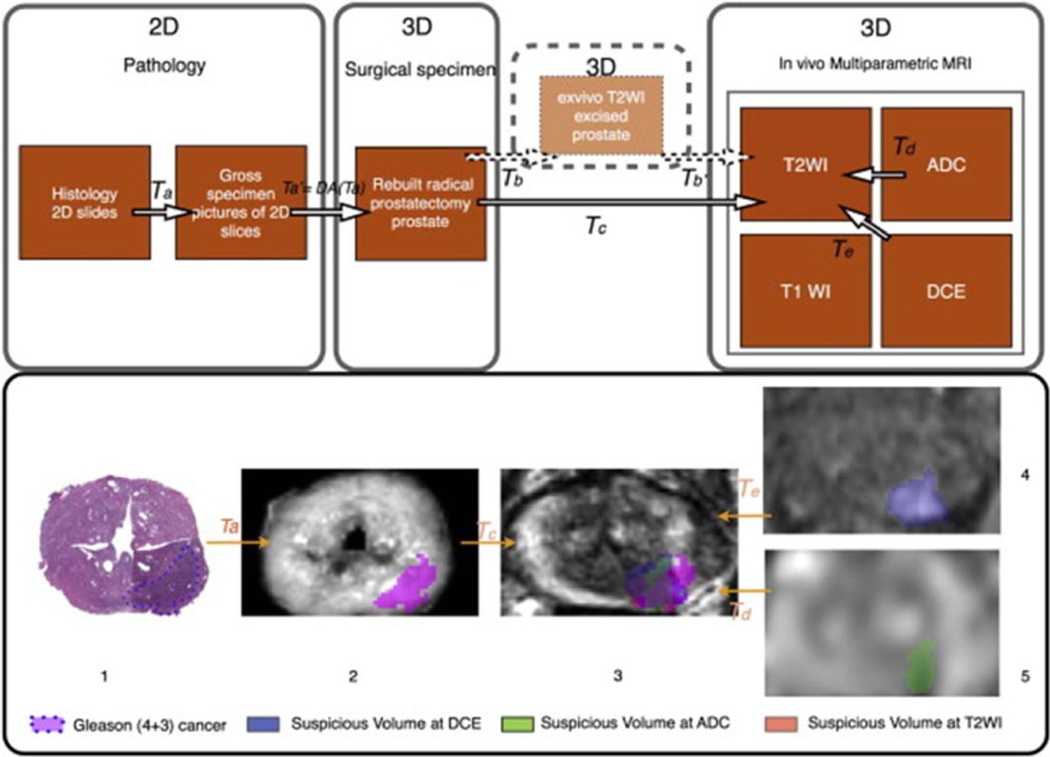
Registration between MRI and histology. Top: Workflow for pathology-mp-MRI registration in a surgical 3D space. Bottom: 3D deformable registration of virtual whole-mount histology (1), fresh specimen (2), T2 weighted MRI (3), perfusion (4), and diffusion (5) sequences (ADC) applied to prostate cancer (Image from Orczyk C, Rusinek H, Rosenkrantz AB, Mikheev A, Deng FM, Melamed J, Taneja SS: Preliminary experience with a novel method of three-dimensional co-registration of prostate cancer digital histology and in vivo multiparametric MRI. Clin Radiol 2013, 68:e652–658).
Although whole mount prostate histological analysis provides accurate label information for training a CADx system, whole mount histology is expensive and registering whole mount histologic slices with 3D mp-MRI is a challenging problem. Therefore, histologic interpretations from biopsy specimens are used to determine the ground truth in some studies [14, 62, 122, 137, 138]. In vivo biopsy can only label the pathology of the core inside the prostate. Radiologists must manually define lesion boundaries on mp-MRI retrospectively based on the biopsy results.
Meyer et al. [139] reviewed the registration methods of 3-D medical images and histopathology of the prostate. They examined the registration process and techniques for registering MRI or PET with whole-mounted prostatectomy specimens.
CLINICAL APPLICATIONS
Diagnosis
The functional MR imaging data, like DCE-MRI and MRS, are more complex and larger in amounts than anatomic MR imaging. There are clinical needs to develop fast, cost-effective, supportive techniques, such as computer-aided analysis tools, for easy and more reproducible diagnosis of prostate cancer. Researchers have focused on developing CADx methodology for automated prostate MRS classification and DCE-MRI analysis. Because all functional MR imaging techniques have their strengths and shortcomings, single technique cannot adequately detect and characterize PCa. The combination of anatomic (T2W) images and functional techniques has been shown to increase the accuracy of MR imaging for diagnosis of PCa. Table 1 compares the performance of the major published prostate CADx systems [13, 14, 16–18, 22, 26, 27, 36, 37, 39, 51, 52, 54–57, 62, 122, 129, 140–151]. Chan et al. were one of the first groups who implemented an mp-MRI CADx system for the diagnosis of prostate cancer [122]. In their approach they used line-scan diffusion, T2 and T2-weighted images to identify predefined areas of the peripheral zone of the prostate for the presence of prostate cancer. Viswanath et al. [129] present an mp-MRI CADx system for PCa detection by integrating functional and structural information obtained via DCE and T2W MRI. Liu et al. [141] present fuzzy MRF models for prostate cancer detection of multispectral MR prostate images. Tiwari et al. [55] investigated the use of MR spectroscopy in combination with T2W MRI to identify the voxels that are affected by prostate cancer. They also introduced the use of wavelet embedding to map MRS and T2-W texture features into a common space. In a study by Peng et al. [27], the combination of 10th percentile ADC, average ADC, and T2-weighted skewness with CADx yielded an AUC value of 0.95 in differentiating prostate cancer from normal tissue. The combination achieved higher accuracy than any MR parameter alone. In a more recent study by Litjens et al [62], they developed a fully automated computer-aided detection system which consists of two stages. The first (detection) stage consists of segmentation of the prostate on the transversal T2W MRI, extraction of voxel features from the image volumes, classification of the voxels and candidate selection. The second (diagnosis) stage consists of candidate segmentation, candidate feature extraction and candidate classification. The system was evaluated on a large consecutive cohort of 347 patients, and yielded an AUC value of 0.889.
TABLE 1.
Summary of representative studies in the literature.
| Reference | Modality | Validation | Region | Classifier | Data size |
Performance |
|---|---|---|---|---|---|---|
| Chan et al. (2003)[122] |
T2WI, ADC, T2 |
Biopsy | PZ | SVM, FLD | 15 | FLD,AUC=0.839; SVM, AUC=0.761 |
| Puech et al. (2007)[39] |
DCE | prostatectomy | PZ & TZ |
Software
titled "ProCAD" |
100 | PZ,Se/Sp=100/49%; TZ,Se/Sp=100/40% |
| Tiwari et al. (2007) [54] |
MRS | Biopsy | WP | Spectral clustering |
14 | Se=77.8%, FP=28.92%,
and FN=20.88% |
| Vos et al. (2008)[36] |
DCE | WMHS | PZ | SVM | 34 | AUC = 0.83 |
| Viswanath et al.(2008)[140] |
DCE | WMHS | WP | LLE
and Consensus Clustering |
6 | Se=60.72%, Sp= 83.24% |
| Viswanath et al.(2009)[129] |
T2WI, DCE |
WMHS | WP | Random forest | 6 | AUC = 0.815 |
| Vos et al.(2009)[37] |
DCE | WMHS | PZ | SVM | 38 | AUC = 0.80 |
| Liu et al.(2009)[141] |
T2W, T2,ADC, DCE |
WMHS | PZ | Fuzzy MRF Model |
11 | Se=89.58%, Sp= 87.50% |
| Tiwari et al.(2009)[55] |
MRS | prostatectomy | WP | NLDR | 18 | Se=89.33%,Sp=79.79% |
| Artan et al.(2010)[142] |
T2, ADC, DCE |
Biopsy | PZ | cost-sensitive CRF |
21 | AUC = 0.79 |
| Vos et al.(2010)[143] |
T2WI, DCE |
WMHS | PZ | SVM | 29 | AUC = 0.89 |
| Viswanath et al.(2011)[144] |
T2W, DWI, DCE |
WMHS | WP | EMPrAvISE | 12 | AUC=0.77 |
| Lopes et al.(2011)[145] |
T2WI | WMHS | WP | SVM,AdaBoost | 17 | SVM,Se/Sp=83/91%; AdaBoost,Se/Sp=85/93% |
| Liu and Yetik (2011)[26] |
T2W, DWI, DCE |
WMHS | WP | SVM | 20 | AUC = 0.89 |
| Sung et al.(2011)[146] |
DCE | prostatectomy | PZ & TZ |
SVM | 42 | PZ,Se/Sp=89/89%; TZ,Se/Sp=91/64% |
| Tiwari et al.(2012)[56] |
T2WI,MRS | WMHS | WP | Random forest | 36 | AUC = 0.89 |
| Viswanath et al.(2012)[22] |
T2WI | WMHS | PZ & CG |
QDA | 22 | CG,AUC=0.86; PZ, AUC=0.73 |
| Vos et al.(2012)[14] |
T1, T2, ADC, DCE |
Biopsy | WP | LDA | 200 | Se=0.74,at a FP level of 5 per patient. |
| Niaf et al.(2012)[18] |
T2W, DWI, DCE |
WMHS | PZ | SVM | 30 | AUC = 0.89 |
| Artan et al.(2012)[147] |
T2, ADC, T1-PC |
WMHS | WP | SVM | 15 | Se=76%,Sp=86% |
| Shah et al.(2012)[17] |
T2WI, ADC, DCE |
WMHS | PZ | SVM | 31 | f-measure=89% |
| Matulewicz et al.(2013)[57] |
MRS | WMHS | WP | ANN | 18 | AUC=0.968 |
| Hambrock et al.(2013)[13] |
T2WI, DWI, DCE |
prostatectomy | PZ & TZ |
in-house– developed CAD system |
34 | experienced observers, AUC=0.91 |
| Tiwari et al.(2013)[148] |
T2WI, MRS |
WMHS | WP | SeSMiK-GE | 29 | AUC = 0.89 |
| Peng et al. (2013)[27] |
T2WI, ADC, DCE |
prostatectomy | WP | LDA | 48 | AUC = 0.95 |
| Ginsburg et al.(2014)[149] |
T2WI, DWI, DCE |
WMHS | PZ and CG |
PCA-VIP | 108 | CG,AUC=0.85; PZ, AUC=0.79 |
| Stember et al.(2014)[51] |
T2WI, ADC |
Biopsy | TZ | Naive Bayes classifier |
18 | predicted TZ tumor in all test patients |
| Niaf et al.(2014)[150] |
T2WI, DWI, DCE |
prostatectomy | WP | P-SVM | 48 | AUC = 0.889 |
| Garcia Molina et al.(2014)[16] |
T2WI, ADC, DCE |
prostatectomy | PZ | incremental learning ensemble SVM |
12 | Se=84.4%,Sp=78.0% |
| Litjens et al.(2014)[62] |
T2WI, DWI, DCE, PDWI |
Biopsy | WP | Random Forest | 347 | AUC = 0.889 |
| Kwak et al.(2015)[52] |
T2WI, DWI |
Biopsy | PZ and TZ |
SVM | 244 | AUC of 0.89 |
| Zhao et al.(2015)[151] |
T2WI | biopsy/follow -up |
PZ and CG |
ANN | 71 | CG,AUC=0.821; PZ, AUC=0.849 |
Aggressiveness
Treatment choice for prostate cancer is based on initial PSA level, clinical stage of disease, and Gleason score, together with baseline urinary function, comorbidities, and patient age [152, 153]. Therefore, there is an urgent clinical need to detect high-grade cancers and to differentiate them from the indolent, slow-growing tumors. The Gleason system, using a rating system to determine the grade of prostate cancer, remains one of the widely used prognostic factors in prostate cancer. The higher grade tumors have a tendency to grow quickly and to spread faster than lower grade tumors.
DWI, DCE-MRI, and MRS are noninvasive assessment methods of PCa aggressiveness. The Gleason grading system is a fundamental indicator of the aggressive nature of prostate cancer. Studies found that ADC image features correlate with Gleason scores [27, 28, 46, 154–156]. A study by Yamamura et al. found a highly significant negative correlation between ADC-value and the Gleason score, while MRS did not show a significant correlation [157]. Recently, Zhang et al. found that transrectal ultrasound (TRUS)-guided, MRI-directed biopsies improved the prediction of PCa aggressiveness in comparison with 12-core TRUS-guided biopsies. DWI directed biopsies had a superior performance when compared with MRS directed biopsies in the peripheral zone [6]. Diffusion of water molecules in tumor tissue was thought to reflect tissue architecture such as cell density and nucleus/cytoplasm ratio, and reductions in ADC values in tumor tissue in fact correlates well with increases in cellular density [158–160]. For these reasons, ADC value has received more attention as a predictor of Gleason score in prostate cancer.
DCE-MRI is based on the permeability of blood vessels and extravasation of contrast agent into the surrounding tissue. Investigators have observed that quantitative parameters (Ktrans and Kep) and semi-quantitative parameters (wash-in and wash-out) derived from DCE-MRI have the potential to assess the aggressiveness of PCa. Oto et al. found a moderate correlation between kep and microvessel density of prostate cancer [154]. Peng et al. found Ktrans moderately correlate with Gleason scores [27].
In vivo MRS imaging has revealed a trend towards an increased (choline+ creatine)/citrate (CC/C) ratio with increased Gleason score [161, 162]. This relationship has also been demonstrated by ex vivo HR-MAS MRS [163]. However, other in vivo MRS imaging studies have found no correlation between metabolite ratios and aggressiveness [164, 165].
On T2W MRI, changes in signal intensity for prostate cancer detection have been associated with its aggressiveness [166]. In a large retrospective study with 220 patients [166], T2W MRI and MRS imaging scores based on a three-point scale for clinical prostate cancer aggressiveness were significantly correlated to biologic markers such as androgen receptor levels, which were associated with prostate cancer progression. In that study, the combination of biomarkers with T2W MRI and MRS imaging results can discriminate clinically unimportant prostate cancer. If mp-MRI can potentially aid in identifying low-grade disease in vivo, this might allow PCa patients to opt for active surveillance rather than immediately opting for aggressive therapy. Lee et al. demonstrated that the simple measurement of the diameter of suspicious tumor lesions on DWI could improve the prediction of insignificant prostate cancer in candidates for active surveillance therapy [167].
Although these MRI metrics are related to Gleason score, the power and threshold value of each metric are different and how to combine these anatomic and functional MRI information is still a problem. Developing a computerized decision support system may help in noninvasive assessment of PCa aggressiveness. Recently, a system called Semi-Supervised MultiKernel Graph Embedding (SeSMiK-GE), was developed to quantitatively combine T2WI and MRS data for distinguishing benign versus cancerous, and high- versus low-Gleason grade PCa regions in vivo [148].
Biopsy guidance
Transrectal ultrasound (TRUS)-guided sextant or systematic prostate biopsy is the clinical standard for definitive diagnosis of prostate cancer. The Gleason score derived from biopsy specimens is important for appropriate treatment selection. However, PCa is often heterogeneous and multicentric [168]. In addition, the biopsy, which samples a small portion of the prostate, might not represent the whole gland efficiently. Traditionally, it is believed that Gleason score in systematic random TRUS-guided biopsy tends to downgrade the surgical specimen, because a less differentiated pattern may not have been sampled in the biopsy [169, 170]. Systematic random TRUS-guided biopsies often require repeated biopsy procedures, which are associated with discomfort and potential morbidity [171]. In order to reduce the overtreatment and the number of biopsies, lesions must be accurately detected, characterized and targeted during biopsy. More effective imaging-guided targeted biopsy techniques are under investigation in order to improve the detection rate of prostate biopsies.
Optimization of prostate biopsy requires addressing the shortcomings of standard systematic TRUS-guided biopsy, including false-negative rates, incorrect risk stratification, detection of clinically insignificant disease and the need for repeat biopsy. MRI is an evolving noninvasive imaging modality that increases the accurate localization of prostate cancer at the time of biopsy, and thereby enhances clinical risk assessment and improves the ability to appropriately counsel patients regarding therapy.
Use of mp-MRI for targeted prostate biopsies has the potential to reduce the sampling error associated with conventional biopsy by providing better disease localization and sampling, and also has a potential role in avoiding biopsy and reducing over detection/overtreatment. MRI-compatible biopsy systems were developed for this purpose [172]. More accurate risk stratification through improved cancer sampling may impact therapeutic decision making. Optimal clinical application of MRI targeted biopsy remains under investigation.
There are three different manners in which an MRI-detected lesion can be targeted for biopsy: (1) Direct targeting within the magnet using MR-compatible devices, also called in-bore MRI guided biopsy; (2) Use of fusion software to allow an MRI–defined lesion to be identified on ultrasound during a TRUS-guided biopsy procedure (Figure 12); or (3) Cognitive targeting, in which the physician reviews the MRI data before the procedure and attempts to target the suspected area during the TRUS-guided biopsy using anatomic landmarks as reference [173]. An MRI-guided robotic prostate biopsy system, named APT-MRI robotic biopsy system, has been reported with an accuracy within 2 mm [174]. A real-time phase-only cross correlation (POCC) algorithm-based sequence has been used in transrectal 3T in-bore MR-guided prostate biopsies [175]. Fusion of pre-biopsy MR images onto interventional TRUS images might increase the overall biopsy accuracy [176, 177]. A novel method to identify the 2D axial MR slice from a pre-acquired MR prostate volume that closely corresponds to the 2D axial TRUS slice obtained during prostate biopsy has been reported by Mitra et al. [178].
Figure 12.
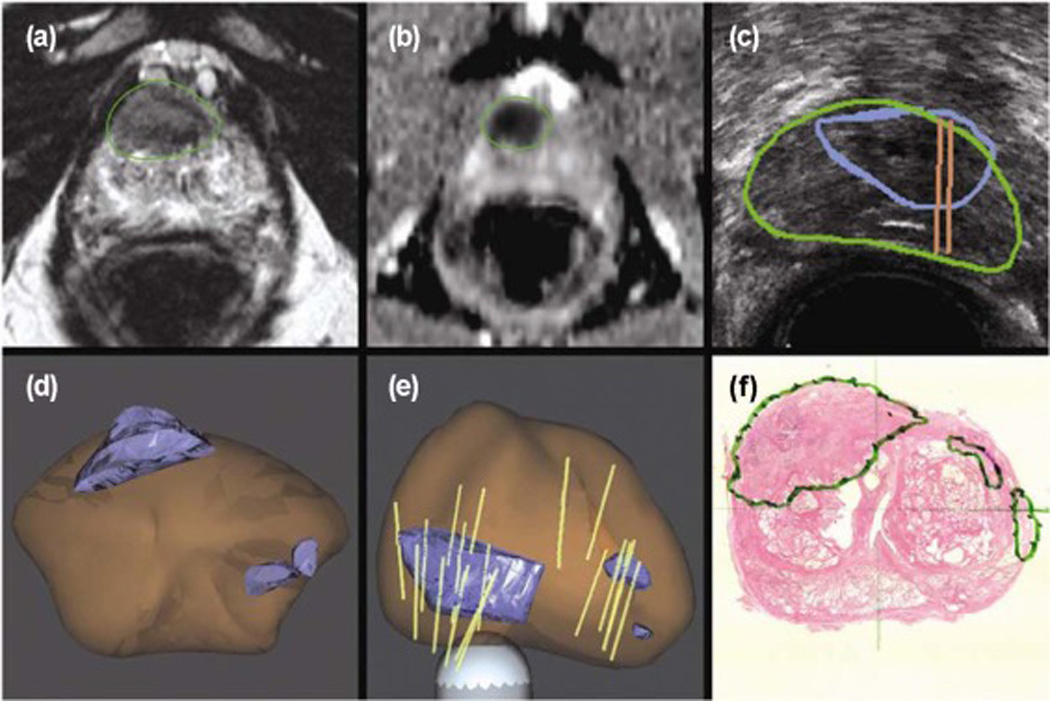
MRI and ultrasound fusion for targeted biopsy of the prostate. (A and B) Anterior lesion of the high suspicious lesion identified on mp-MRI. (C) Real-time ultrasound targeting the corresponding lesion. (D and E) 3D models demonstrate the target (blue), prostate (brown), and biopsy cores (tan cylinders). (F) Radical prostatectomy pathology confirmed a 2.3 cm Gleason 8 (4+4) cancer centered in the right anterior prostate (Images from Sonn, GA, Margolis, DJ, Marks, LS: Target detection: Magnetic resonance imaging-ultrasound fusion–guided prostate biopsy. Urologic Oncology: Seminars and Original Investigations 2014, 32:903–911).
Treatment planning and therapeutic response assessment
MRI-based techniques are used for computer-aided treatment procedures such as treatment planning of radiotherapy, MRI-guided radioactive seeds placement in prostate brachytherapy, and MRI-guided local ablation procedures [179–190].
The excellent soft-tissue contrast of MRI means that the technique is having an increasing role in contouring the gross tumor volume (GTV) and organs at risk (OAR) in radiation therapy treatment planning systems (TPS). MRI-planning scans from diagnostic MRI scanners are currently incorporated into the planning process by being registered to CT data. The soft-tissue data from the MRI provides target outline guidance and the CT provides a solid geometric and electron density map for accurate dose calculation on the TPS computer [191].
A number of minimally invasive, focal, organ-preserving methods have been used in recent years as further alternatives to the radical treatment of prostate cancer [170]. The focal therapy methods used to date for the prostate include cryotherapy, high-intensity focused ultrasound, laser-induced thermal ablation, and radioactive seed placement. Mp-MRI makes it possible to determine the exact location of tumor foci that are generally accessible for ablation or radioactive seed placement. Moreover, mp-MRI can also monitor treatment during and after minimally invasive therapy. A CADx system for the prostate may have potential value in helping clinicians to target tumor foci during treatment.
Mp-MRI can also be used as an imaging biomarker for monitoring therapeutic response, including radiotherapy of localized prostate cancer [191] and systemic therapy for metastatic disease. Successful treatment response to therapy is usually depicted by reductions in signal intensity accompanied by ADC increases [192–194]. There are clinical needs to develop mp-MRI-based CADx systems for monitoring therapeutic response of the prostate in the future.
DISCUSSION AND FUTURE DIRECTIONS
Unlike breast and lung cancer, prostate cancer CADx systems for MR images have not been widely used in daily clinical work for detection or diagnosis. The majority of the prostate CADx systems reported the AUC in the range from 0.80 to 0.89 [179], while one reported AUCs of 0.96 [46], which represented a high performance. However, most systems generated lesion candidates based on manually selected ROIs, which may be data set dependent, and employed a relatively small data set. Validation on a large-scale data set with several hundred patients is required. A prostate CADx system should be tested in multicenter trials to make the systems widely usable in clinical work.
One challenge of prostate CAD is related to mp-MRI protocols. Both 3T protocols and endorectal coils have the advantage of increasing the signal-to-noise ratio (SNR). At 3T without the use of an endorectal coil (ERC), image quality can be comparable with that obtained at 1.5 T with an endorectal coils [195]. Turkbey et al. found that dual-coil prostate MRI detected more cancer foci than non-endorectal coil MRI at 3T on T2W and DWI [196]. At 3 T MRI, DWI images and ADC maps using b = 1500 s/mm2 should be considered more effective than those at b = 2000 s/mm2 or b = 1000 s/mm2 for detecting prostate cancer [50]. Most members of the PI-RADS steering committee recommend 3 T for prostate MRI. There is no consensus among experts concerning the potential benefits of the use of endorectal coils [12]. The impact of the mp-MRI protocol on CADx systems should been considered and researched in the future. The combination of T2W, DWI, and DCE-MRI is the most commonly used set of parameters for the detection or diagnosis of prostate cancer. MRS with other parameters is also used in some research. The introduction of new imaging modalities or new modality combinations for mp-MRI may lead to better CADx systems. Combining CAD prediction and PIRADS into a combination score has the potential to improve diagnostic accuracy [197]. The MR PI-RADS system may provide a platform for CAD system development in the future.
The diagnostic value of these parameters for discrimination between benign and malignant tissue depend on the lesion’s location. The parameter values of PCa are in the range of those of nonmalignant diseases or conditions such as prostatitis, fibromuscular benign prostatic hyperplasia (BPH), post-biopsy hemorrhagic change, making for poor diagnostic value, especially in the transition zone. TZ and PZ cancer possess distinct quantitative imaging features on MRI. Computer-extracted parameters may be useful for cancer detection in the PZ, but are not suited in the TZ. In recent years, research focus has shifted from PZ prostate cancer to whole prostate cancer. There are more challenges in developing a CADx system for both PZ and TZ lesions than for PZ lesions only. Applications of anatomical segmentation from MRI as an additional input to ANN improve the accuracy of detecting cancerous voxels from MRSI [198]. A CAD system, utilizing two MRI sequences, such as T2-MRI and high-b-value (b = 2000 s/mm2) DWI, and texture features based on local binary patterns, is able to detect the discriminative texture features for cancer detection and localization, and the performance of the CADx system was not dependent on the specific regions of the prostate [52]. Future direction should also include whether zonal segmentation of the prostate is necessary when some new imaging sequences being used.
Ex vivo whole mount prostate histological analysis provides more accurate label information for training a CADx system. However, whole mount histology is expensive, and registering whole mount histological slices with 3D mp-MRI is a challenging problem. This is especially true during the preparation of the prostate histological data for training a CADx system. Pathologists must collect a large amount of training data from many patients, apply reliable biomarkers for each patient, prepare blocks, scan a large number of histological slices, and manually define lesion boundaries on histological slices. However, these are laborious and time-consuming procedures. Therefore, the histological image preparation procedures need to be performed by some automatic methods to improve efficacy. A software system has been designed to create a pseudo whole mount histology section [134]. A computer-aided system to automatically grade pathological images according to the Gleason grading system has also been investigated [199]. A scheme, including automatic diagnosis from histologic images, 3D histologic reconstruction and registration, should be developed for ground truth definition in the future.
Image quantification methods, such as accurate image registration for motion correction, compartment modeling for functional parameters estimation, feature extraction in high dimensional data, automatic image classification for differentiating cancer from normal tissue, and correlation analyses among radiological data and genomic information, will play key roles in the future development of intelligent CAD systems.
Radiomics, as a high dimensional extraction of large amounts of image features with high throughput from radiographic images, can provide valuable diagnostic, prognostic or predictive information. Cameron et al. had developed a quantitative radiomics feature model for performing prostate cancer detection using mp-MRI [115]. Khalvati et al. [118] present new texture feature models for radiomics-driven detection of prostate cancer utilizing mp-MRI data. Radiomics are emerging as a useful tool for prostate cancer detection. Further work is needed to build radiomics-based CAD systems for prostate cancer diagnosis, treatment planning, treatment prediction and treatment response evaluation.
The Gleason grade of PCa is the most widely used prognostic factor for prostate cancer. MR metrics on T2W, DWI, DCE-MRI and MRS imaging relate to microenvironment and microstructure. Therefore these MR metrics can predict the Gleason grade of the caner. Building a CAD system based on mp-MRI and Gleason score is feasible. It can play a significant role in prognostic prediction, guiding biopsy, identifying suitable patients under active surveillance and making a decision of appropriate treatment. CAD systems for prediction of Gleason score should be developed in the future.
As the anatomic information is important when analyzing functional data, T2W images are frequently used in mp-MRI CADx systems. T2W plus DWI and DCE-MRI are commonly used as the combinations. Chan et al. constructed a summary statistical map of the peripheral zone based on the utility of multichannel statistical classifiers by combining textural and anatomical features in PCa areas from T2W, DWI, proton density maps, and T2 maps [122]. Langer et al. included DCE-MRI and pharmacokinetic parameter maps as extra features to a CADx system for detecting prostate cancer at the peripheral zone [111]. They evaluated their system in predefined regions of interest, but on a per voxel basis. Vos et al. implemented a two-stage CADx system for prostate cancer using an initial blob detection approach combined with a candidate segmentation and classification using statistical region features [14]. Litjens et al. recently investigated a fully automated computer-aided detection system including a novel combination of segmentation, voxel classification, candidate extraction and classification }[62].
Promising preliminary results have been obtained with CADx systems that combine the analysis of statistical, structural, and functional MR imaging features and the use of an adapted classification scheme. Likelihood maps have been obtained by combining information from mp-MRI using mathematical descriptors. These studies showed the discrimination between benign and malignant tissues is feasible with good performances [62, 111].
CONCLUSION
We comprehensively reviewed mp-MRI based, computer aided technology for prostate cancer detection. Prostate CADx systems are a complicated composition of preprocessing, segmentation, registration, feature extraction, and classification modules. There are some challenges in accurate registration of MRI and histopathology, which is important for ground truth definition. Clinical applications of computer aided systems include localization, diagnosis, staging, aggressiveness assessment, guiding biopsy, treatment planning, and therapeutic response assessment. Although the performance of some CADx systems is good, there is no such a system that has been wildly used in clinic. It is likely that more improvements in quantitative image analysis and computer-aided methods would need to be made in order to meet the clinical needs in near future work.
Acknowledgments
This work was partially supported by NIH grants R01CA156775, R21CA176684, and P50CA128301. Z. Zhang was supported by the National Natural Science Foundation of China (81372274). The work was also partially supported by a Georgia Research Alliance Distinguished Scientists Award.
Abbreviation
- T2W
T2-weighted
- ADC
Apparent diffusion coefficient
- DCE
Dynamic contrast-enhanced
- MRS
Magnetic resonance Spectroscopy
- DWI
Diffusion-weighted imaging
- T1-PC
(principal component of T1-weighted dynamic series)
- T1
T1 mapping
- T2
T2 mapping
- SVM
Support vector machine
- P-SVM
Probabilistic SVM
- FLD
Fisher linear discriminant
- MRFs
Markov random fields
- NLDR
Nonlinear dimensionality reduction
- CRF
Conditional random fields
- EMPrAvISE
Enhanced Multi-Protocol Analysis via Intelligent Supervised Embedding
- QDA
Quadratic Discriminant Analysis
- ANN
Artificial neural network
- SeSMiK-GE
Semi Supervised Multi Kernel Graph Embedding
- LDA
Linear discriminant analysis
- PCA
Principal component analysis
- PCA-VIP
Variable importance on projection measure for PCA
- LLE
Locally linear embedding
- AUC
Area under a receiver operating characteristic curve
- Se
Sensitivity
- Sp
Specificity
- FP
False positive
- FN
False negative
- TZ
Transition zone
- PZ
Peripheral zone
- CG
Central gland
- WP
Whole prostate
- WMHS
Whole mount histological sections
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Lee JJ, Thomas IC, Nolley R, Ferrari M, Brooks JD, Leppert JT. Biologic differences between peripheral and transition zone prostate cancer. Prostate. 2015;75:183–190. doi: 10.1002/pros.22903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui MM, Rais-Bahrami S, Turkbey B, George AK, Rothwax J, Shakir N, Okoro C, Raskolnikov D, Parnes HL, Linehan WM, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. Jama. 2015;313:390–397. doi: 10.1001/jama.2014.17942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pokorny MR, de Rooij M, Duncan E, Schroder FH, Parkinson R, Barentsz JO, Thompson LC. Prospective study of diagnostic accuracy comparing prostate cancer detection by transrectal ultrasound-guided biopsy versus magnetic resonance (MR) imaging with subsequent MR-guided biopsy in men without previous prostate biopsies. Eur Urol. 2014;66:22–29. doi: 10.1016/j.eururo.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Schoots IG, Roobol MJ, Nieboer D, Bangma CH, Steyerberg EW, Hunink MG. Magnetic Resonance Imaging-targeted Biopsy May Enhance the Diagnostic Accuracy of Significant Prostate Cancer Detection Compared to Standard Transrectal Ultrasound-guided Biopsy: A Systematic Review and Meta-analysis. Eur Urol. 2014 doi: 10.1016/j.eururo.2014.11.037. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Xiu J, Dong Y, Wang M, Han X, Qin Y, Huang Z, Cai S, Yuan X, Liu Q. Magnetic resonance imagingdirected biopsy improves the prediction of prostate cancer aggressiveness compared with a 12core transrectal ultrasoundguided prostate biopsy. Mol Med Rep. 2014;9:1989–1997. doi: 10.3892/mmr.2014.1994. [DOI] [PubMed] [Google Scholar]
- 7.Brown AM, Elbuluk O, Mertan F, Sankineni S, Margolis DJ, Wood BJ, Pinto PA, Choyke PL, Turkbey B. Recent advances in image-guided targeted prostate biopsy. Abdom Imaging. 2015;40:1788–1799. doi: 10.1007/s00261-015-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puech P, Rouviere O, Renard-Penna R, Villers A, Devos P, Colombel M, Bitker MO, Leroy X, Mege-Lechevallier F, Comperat E, et al. Prostate cancer diagnosis: multiparametric MR-targeted biopsy with cognitive and transrectal US-MR fusion guidance versus systematic biopsy--prospective multicenter study. Radiology. 2013;268:461–469. doi: 10.1148/radiol.13121501. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence EM, Tang SY, Barrett T, Koo B, Goldman DA, Warren AY, Axell RG, Doble A, Gallagher FA, Gnanapragasam VJ, et al. Prostate cancer: performance characteristics of combined T(2)W and DW-MRI scoring in the setting of template transperineal re-biopsy using MR-TRUS fusion. Eur Radiol. 2014;24:1497–1505. doi: 10.1007/s00330-014-3159-0. [DOI] [PubMed] [Google Scholar]
- 10.Barentsz JO, Richenberg J, Clements R, Choyke P, Verma S, Villeirs G, Rouviere O, Logager V, Futterer JJ. ESUR prostate MR guidelines 2012. Eur Radiol. 2012;22:746–757. doi: 10.1007/s00330-011-2377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson L, Ahmed HU, Allen C, Barentsz JO, Carey B, Futterer JJ, Heijmink SW, Hoskin PJ, Kirkham A, Padhani AR, et al. Magnetic resonance imaging for the detection, localisation, and characterisation of prostate cancer: recommendations from a European consensus meeting. Eur Urol. 2011;59:477–494. doi: 10.1016/j.eururo.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, Margolis D, Schnall MD, Shtern F, Tempany CM, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol. 2015 doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hambrock T, Vos PC, Hulsbergen-van de Kaa CA, Barentsz JO, Huisman HJ. Prostate cancer: computer-aided diagnosis with multiparametric 3-T MR imaging--effect on observer performance. Radiology. 2013;266:521–530. doi: 10.1148/radiol.12111634. [DOI] [PubMed] [Google Scholar]
- 14.Vos PC, Barentsz JO, Karssemeijer N, Huisman HJ. Automatic computer-aided detection of prostate cancer based on multiparametric magnetic resonance image analysis. Phys Med Biol. 2012;57:1527–1542. doi: 10.1088/0031-9155/57/6/1527. [DOI] [PubMed] [Google Scholar]
- 15.Niaf E, Lartizien C, Bratan F, Roche L, Rabilloud M, Mege-Lechevallier F, Rouviere O. Prostate focal peripheral zone lesions: characterization at multiparametric MR imaging--influence of a computer-aided diagnosis system. Radiology. 2014;271:761–769. doi: 10.1148/radiol.14130448. [DOI] [PubMed] [Google Scholar]
- 16.Garcia Molina JF, Zheng L, Sertdemir M, Dinter DJ, Schonberg S, Radle M. Incremental learning with SVM for multimodal classification of prostatic adenocarcinoma. PLoS One. 2014;9:e93600. doi: 10.1371/journal.pone.0093600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah V, Turkbey B, Mani H, Pang Y, Pohida T, Merino MJ, Pinto PA, Choyke PL, Bernardo M. Decision support system for localizing prostate cancer based on multiparametric magnetic resonance imaging. Med Phys. 2012;39:4093–4103. doi: 10.1118/1.4722753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niaf E, Rouviere O, Mege-Lechevallier F, Bratan F, Lartizien C. Computer-aided diagnosis of prostate cancer in the peripheral zone using multiparametric MRI. Phys Med Biol. 2012;57:3833–3851. doi: 10.1088/0031-9155/57/12/3833. [DOI] [PubMed] [Google Scholar]
- 19.Akin O, Sala E, Moskowitz CS, Kuroiwa K, Ishill NM, Pucar D, Scardino PT, Hricak H. Transition zone prostate cancers: features, detection, localization, and staging at endorectal MR imaging. Radiology. 2006;239:784–792. doi: 10.1148/radiol.2392050949. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Sugimura K, Kaji Y, Kitamura Y, Fujii M, Hara I, Tachibana M. Conventional MRI capabilities in the diagnosis of prostate cancer in the transition zone. AJR Am J Roentgenol. 2006;186:729–742. doi: 10.2214/AJR.04.0775. [DOI] [PubMed] [Google Scholar]
- 21.Cornud F, Rouanne M, Beuvon F, Eiss D, Flam T, Liberatore M, Zerbib M, Delongchamps NB. Endorectal 3D T2-weighted 1mm-slice thickness MRI for prostate cancer staging at 1.5Tesla: should we reconsider the indirects signs of extracapsular extension according to the D’Amico tumor risk criteria? Eur J Radiol. 2012;81:e591–e597. doi: 10.1016/j.ejrad.2011.06.056. [DOI] [PubMed] [Google Scholar]
- 22.Viswanath SE, Bloch NB, Chappelow JC, Toth R, Rofsky NM, Genega EM, Lenkinski RE, Madabhushi A. Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 Tesla endorectal, in vivo T2-weighted MR imagery. J Magn Reson Imaging. 2012;36:213–224. doi: 10.1002/jmri.23618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeks CM, Hambrock T, Yakar D, Hulsbergen-van de Kaa CA, Feuth T, Witjes JA, Futterer JJ, Barentsz JO. Transition zone prostate cancer: detection and localization with 3-T multiparametric MR imaging. Radiology. 2013;266:207–217. doi: 10.1148/radiol.12120281. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Turkbey B, Senegas J, Remmele S, Xu S, Kruecker J, Bernardo M, Wood BJ, Pinto PA, Choyke PL. Accelerated T2 mapping for characterization of prostate cancer. Magn Reson Med. 2011;65:1400–1406. doi: 10.1002/mrm.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamauchi FI, Penzkofer T, Fedorov A, Fennessy FM, Chu R, Maier SE, Tempany CM, Mulkern RV, Panych LP. Prostate cancer discrimination in the peripheral zone with a reduced field-of-view T(2)-mapping MRI sequence. Magn Reson Imaging. 2015;33:525–530. doi: 10.1016/j.mri.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Yetik IS. Automated prostate cancer localization without the need for peripheral zone extraction using multiparametric MRI. Med Phys. 2011;38:2986–2994. doi: 10.1118/1.3589134. [DOI] [PubMed] [Google Scholar]
- 27.Peng Y, Jiang Y, Yang C, Brown JB, Antic T, Sethi I, Schmid-Tannwald C, Giger ML, Eggener SE, Oto A. Quantitative analysis of multiparametric prostate MR images: differentiation between prostate cancer and normal tissue and correlation with Gleason score--a computer-aided diagnosis development study. Radiology. 2013;267:787–796. doi: 10.1148/radiol.13121454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowak J, Malzahn U, Baur AD, Reichelt U, Franiel T, Hamm B, Durmus T. The value of ADC, T2 signal intensity, and a combination of both parameters to assess Gleason score and primary Gleason grades in patients with known prostate cancer. Acta Radiol. 2014 doi: 10.1177/0284185114561915. [DOI] [PubMed] [Google Scholar]
- 29.Hoang Dinh A, Souchon R, Melodelima C, Bratan F, Mege-Lechevallier F, Colombel M, Rouviere O. Characterization of prostate cancer using T2 mapping at 3T: a multi-scanner study. Diagn Interv Imaging. 2015;96:365–372. doi: 10.1016/j.diii.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Durmus T, Baur A, Hamm B. Multiparametric magnetic resonance imaging in the detection of prostate cancer. Aktuelle Urol. 2014;45:119–126. doi: 10.1055/s-0034-1371875. [DOI] [PubMed] [Google Scholar]
- 31.Ewing JR, Bagher-Ebadian H. Model selection in measures of vascular parameters using dynamic contrast-enhanced MRI: experimental and clinical applications. NMR Biomed. 2013;26:1028–1041. doi: 10.1002/nbm.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HB, Lee TY, Mayr NA, Parker GJ, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 33.Fedorov A, Fluckiger J, Ayers GD, Li X, Gupta SN, Tempany C, Mulkern R, Yankeelov TE, Fennessy FM. A comparison of two methods for estimating DCE-MRI parameters via individual and cohort based AIFs in prostate cancer: a step towards practical implementation. Magn Reson Imaging. 2014;32:321–329. doi: 10.1016/j.mri.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gliozzi AS, Mazzetti S, Delsanto PP, Regge D, Stasi M. Phenomenological universalities: a novel tool for the analysis of dynamic contrast enhancement in magnetic resonance imaging. Phys Med Biol. 2011;56:573–586. doi: 10.1088/0031-9155/56/3/004. [DOI] [PubMed] [Google Scholar]
- 35.Mazzetti S, Gliozzi AS, Bracco C, Russo F, Regge D, Stasi M. Comparison between PUN and Tofts models in the quantification of dynamic contrast-enhanced MR imaging. Phys Med Biol. 2012;57:8443–8453. doi: 10.1088/0031-9155/57/24/8443. [DOI] [PubMed] [Google Scholar]
- 36.Vos PC, Hambrock T, Hulsbergen-van de Kaa CA, Futterer JJ, Barentsz JO, Huisman HJ. Computerized analysis of prostate lesions in the peripheral zone using dynamic contrast enhanced MRI. Med Phys. 2008;35:888–899. doi: 10.1118/1.2836419. [DOI] [PubMed] [Google Scholar]
- 37.Vos PC, Hambrock T, Barenstz JO, Huisman HJ. Automated calibration for computerized analysis of prostate lesions using pharmacokinetic magnetic resonance images. Med Image Comput Comput Assist Interv. 2009;12:836–843. doi: 10.1007/978-3-642-04271-3_101. [DOI] [PubMed] [Google Scholar]
- 38.Puech P, Betrouni N, Makni N, Dewalle AS, Villers A, Lemaitre L. Computer-assisted diagnosis of prostate cancer using DCE-MRI data: design, implementation and preliminary results. Int J Comput Assist Radiol Surg. 2009;4:1–10. doi: 10.1007/s11548-008-0261-2. [DOI] [PubMed] [Google Scholar]
- 39.Puech P, Betrouni N, Viard R, Villers A, Leroy X, Lemaitre L. Prostate cancer computer-assisted diagnosis software using dynamic contrast-enhanced MRI. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:5567–5570. doi: 10.1109/IEMBS.2007.4353608. [DOI] [PubMed] [Google Scholar]
- 40.Padhani AR, Gapinski CJ, Macvicar DA, Parker GJ, Suckling J, Revell PB, Leach MO, Dearnaley DP, Husband JE. Dynamic contrast enhanced MRI of prostate cancer: correlation with morphology and tumour stage, histological grade and PSA. Clin Radiol. 2000;55:99–109. doi: 10.1053/crad.1999.0327. [DOI] [PubMed] [Google Scholar]
- 41.Dikaios N, Alkalbani J, Abd-Alazeez M, Sidhu HS, Kirkham A, Ahmed HU, Emberton M, Freeman A, Halligan S, Taylor S, et al. Zone-specific logistic regression models improve classification of prostate cancer on multi-parametric MRI. Eur Radiol. 2015;25:2727–2737. doi: 10.1007/s00330-015-3636-0. [DOI] [PubMed] [Google Scholar]
- 42.Joseph H, Yacoub M, Aytekin Oto, Frank H, Miller MR. Imaging of the Prostate. Radiol Clin N Am. 2014;52:811–837. doi: 10.1016/j.rcl.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Toivonen J, Merisaari H, Pesola M, Taimen P, Bostrom PJ, Pahikkala T, Aronen HJ, Jambor I. Mathematical models for diffusion-weighted imaging of prostate cancer using b values up to 2000 s/mm(2) : Correlation with Gleason score and repeatability of region of interest analysis. Magn Reson Med. 2015;74:1116–1124. doi: 10.1002/mrm.25482. [DOI] [PubMed] [Google Scholar]
- 44.Boesen L, Chabanova E, Logager V, Balslev I, Thomsen HS. Apparent diffusion coefficient ratio correlates significantly with prostate cancer gleason score at final pathology. J Magn Reson Imaging. 2015;42:446–453. doi: 10.1002/jmri.24801. [DOI] [PubMed] [Google Scholar]
- 45.Mazaheri Y, Shukla-Dave A, Hricak H, Fine SW, Zhang J, Inurrigarro G, Moskowitz CS, Ishill NM, Reuter VE, Touijer K, et al. Prostate cancer: identification with combined diffusion-weighted MR imaging and 3D 1H MR spectroscopic imaging--correlation with pathologic findings. Radiology. 2008;246:480–488. doi: 10.1148/radiol.2462070368. [DOI] [PubMed] [Google Scholar]
- 46.Peng Y, Jiang Y, Antic T, Giger ML, Eggener SE, Oto A. Validation of quantitative analysis of multiparametric prostate MR images for prostate cancer detection and aggressiveness assessment: a cross-imager study. Radiology. 2014;271:461–471. doi: 10.1148/radiol.14131320. [DOI] [PubMed] [Google Scholar]
- 47.Kitajima K, Takahashi S, Ueno Y, Yoshikawa T, Ohno Y, Obara M, Miyake H, Fujisawa M, Sugimura K. Clinical utility of apparent diffusion coefficient values obtained using high b-value when diagnosing prostate cancer using 3 tesla MRI: comparison between ultra-high b-value (2000 s/mm(2)) and standard high b-value (1000 s/mm(2)) J Magn Reson Imaging. 2012;36:198–205. doi: 10.1002/jmri.23627. [DOI] [PubMed] [Google Scholar]
- 48.Kim CK, Park BK, Kim B. High-b-value diffusion-weighted imaging at 3 T to detect prostate cancer: comparisons between b values of 1,000 and 2,000 s/mm2. AJR Am J Roentgenol. 2010;194:W33–W37. doi: 10.2214/AJR.09.3004. [DOI] [PubMed] [Google Scholar]
- 49.Wetter A, Nensa F, Lipponer C, Guberina N, Olbricht T, Schenck M, Schlosser TW, Gratz M, Lauenstein TC. High and ultra-high b-value diffusion-weighted imaging in prostate cancer: a quantitative analysis. Acta Radiol. 2015;56:1009–1015. doi: 10.1177/0284185114547900. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Qian Y, Liu B, Cao L, Fan Y, Zhang JJ, Yu Y. High-b-value diffusion-weighted MRI for the detection of prostate cancer at 3 T. Clin Radiol. 2014;69:1165–1170. doi: 10.1016/j.crad.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 51.Stember JN, Deng FM, Taneja SS, Rosenkrantz AB. Pilot study of a novel tool for input-free automated identification of transition zone prostate tumors using T2- and diffusion-weighted signal and textural features. J Magn Reson Imaging. 2014;40:301–305. doi: 10.1002/jmri.24375. [DOI] [PubMed] [Google Scholar]
- 52.Kwak JT, Xu S, Wood BJ, Turkbey B, Choyke PL, Pinto PA, Wang S, Summers RM. Automated prostate cancer detection using T2-weighted and high-b-value diffusion-weighted magnetic resonance imaging. Med Phys. 2015;42:2368–2378. doi: 10.1118/1.4918318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neto JA, Parente DB. Multiparametric magnetic resonance imaging of the prostate. Magn Reson Imaging Clin N Am. 2013;21:409–426. doi: 10.1016/j.mric.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 54.Tiwari P, Madabhushi A, Rosen M. A hierarchical unsupervised spectral clustering scheme for detection of prostate cancer from magnetic resonance spectroscopy (MRS) Med Image Comput Comput Assist Interv. 2007;10:278–286. doi: 10.1007/978-3-540-75759-7_34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tiwari P, Rosen M, Madabhushi A. A hierarchical spectral clustering and nonlinear dimensionality reduction scheme for detection of prostate cancer from magnetic resonance spectroscopy (MRS) Med Phys. 2009;36:3927–3939. doi: 10.1118/1.3180955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiwari P, Viswanath S, Kurhanewicz J, Sridhar A, Madabhushi A. Multimodal wavelet embedding representation for data combination (MaWERiC): integrating magnetic resonance imaging and spectroscopy for prostate cancer detection. NMR Biomed. 2012;25:607–619. doi: 10.1002/nbm.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matulewicz L, Jansen JF, Bokacheva L, Vargas HA, Akin O, Fine SW, Shukla-Dave A, Eastham JA, Hricak H, Koutcher JA, Zakian KL. Anatomic segmentation improves prostate cancer detection with artificial neural networks analysis of H magnetic resonance spectroscopic imaging. J Magn Reson Imaging. 2013 doi: 10.1002/jmri.24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panebianco V, Barchetti F, Sciarra A, Marcantonio A, Zini C, Salciccia S, Collettini F, Gentile V, Hamm B, Catalano C. In vivo 3D neuroanatomical evaluation of periprostatic nerve plexus with 3T-MR Diffusion Tensor Imaging. Eur J Radiol. 2013;82:1677–1682. doi: 10.1016/j.ejrad.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 59.Sinha S, Sinha U. In vivo diffusion tensor imaging of the human prostate. Magn Reson Med. 2004;52:530–537. doi: 10.1002/mrm.20190. [DOI] [PubMed] [Google Scholar]
- 60.Xu J, Humphrey PA, Kibel AS, Snyder AZ, Narra VR, Ackerman JJ, Song SK. Magnetic resonance diffusion characteristics of histologically defined prostate cancer in humans. Magn Reson Med. 2009;61:842–850. doi: 10.1002/mrm.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takayama Y, Kishimoto R, Hanaoka S, Nonaka H, Kandatsu S, Tsuji H, Tsujii H, Ikehira H, Obata T. ADC value and diffusion tensor imaging of prostate cancer: changes in carbon-ion radiotherapy. J Magn Reson Imaging. 2008;27:1331–1335. doi: 10.1002/jmri.21388. [DOI] [PubMed] [Google Scholar]
- 62.Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging. 2014;33:1083–1092. doi: 10.1109/TMI.2014.2303821. [DOI] [PubMed] [Google Scholar]
- 63.Suo S, Chen X, Wu L, Zhang X, Yao Q, Fan Y, Wang H, Xu J. Non-Gaussian water diffusion kurtosis imaging of prostate cancer. Magn Reson Imaging. 2014;32:421–427. doi: 10.1016/j.mri.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 64.Roethke MC, Kuder TA, Kuru TH, Fenchel M, Hadaschik BA, Laun FB, Schlemmer HP, Stieltjes B. Evaluation of Diffusion Kurtosis Imaging Versus Standard Diffusion Imaging for Detection and Grading of Peripheral Zone Prostate Cancer. Invest Radiol. 2015;50:483–489. doi: 10.1097/RLI.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 65.Suga M, Aga T, Minato K. Development of a magnetic resonance elastic microscope system. Conf Proc IEEE Eng Med Biol Soc. 2004;2:1025–1027. doi: 10.1109/IEMBS.2004.1403337. [DOI] [PubMed] [Google Scholar]
- 66.Venkatesh SK, Yin M, Glockner JF, Takahashi N, Araoz PA, Talwalkar JA, Ehman RL. MR elastography of liver tumors: preliminary results. AJR Am J Roentgenol. 2008;190:1534–1540. doi: 10.2214/AJR.07.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamhaber U, Klatt D, Papazoglou S, Hollmann M, Stadler J, Sack I, Bernarding J, Braun J. In vivo magnetic resonance elastography of human brain at 7 T and 1.5 T. J Magn Reson Imaging. 2010;32:577–583. doi: 10.1002/jmri.22294. [DOI] [PubMed] [Google Scholar]
- 68.Sinkus R, Tanter M, Xydeas T, Catheline S, Bercoff J, Fink M. Viscoelastic shear properties of in vivo breast lesions measured by MR elastography. Magn Reson Imaging. 2005;23:159–165. doi: 10.1016/j.mri.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 69.McKnight AL, Kugel JL, Rossman PJ, Manduca A, Hartmann LC, Ehman RL. MR elastography of breast cancer: preliminary results. AJR Am J Roentgenol. 2002;178:1411–1417. doi: 10.2214/ajr.178.6.1781411. [DOI] [PubMed] [Google Scholar]
- 70.Sinkus R, Lorenzen J, Schrader D, Lorenzen M, Dargatz M, Holz D. High-resolution tensor MR elastography for breast tumour detection. Phys Med Biol. 2000;45:1649–1664. doi: 10.1088/0031-9155/45/6/317. [DOI] [PubMed] [Google Scholar]
- 71.Arani A, Da Rosa M, Ramsay E, Plewes DB, Haider MA, Chopra R. Incorporating endorectal MR elastography into multi-parametric MRI for prostate cancer imaging: Initial feasibility in volunteers. J Magn Reson Imaging. 2013;38:1251–1260. doi: 10.1002/jmri.24028. [DOI] [PubMed] [Google Scholar]
- 72.Li S, Chen M, Wang W, Zhao W, Wang J, Zhao X, Zhou C. A feasibility study of MR elastography in the diagnosis of prostate cancer at 3.0T. Acta Radiol. 2011;52:354–358. doi: 10.1258/ar.2010.100276. [DOI] [PubMed] [Google Scholar]
- 73.Arani A, Plewes D, Krieger A, Chopra R. The feasibility of endorectal MR elastography for prostate cancer localization. Magn Reson Med. 2011;66:1649–1657. doi: 10.1002/mrm.22967. [DOI] [PubMed] [Google Scholar]
- 74.Fennessy FM, Fedorov A, Gupta SN, Schmidt EJ, Tempany CM, Mulkern RV. Practical considerations in T1 mapping of prostate for dynamic contrast enhancement pharmacokinetic analyses. Magn Reson Imaging. 2012;30:1224–1233. doi: 10.1016/j.mri.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fortuin AS, Smeenk RJ, Meijer HJ, Witjes AJ, Barentsz JO. Lymphotropic nanoparticle-enhanced MRI in prostate cancer: value and therapeutic potential. Curr Urol Rep. 2014;15:389. doi: 10.1007/s11934-013-0389-7. [DOI] [PubMed] [Google Scholar]
- 76.Thoeny HC, Triantafyllou M, Birkhaeuser FD, Froehlich JM, Tshering DW, Binser T, Fleischmann A, Vermathen P, Studer UE. Combined ultrasmall superparamagnetic particles of iron oxide-enhanced and diffusion-weighted magnetic resonance imaging reliably detect pelvic lymph node metastases in normal-sized nodes of bladder and prostate cancer patients. Eur Urol. 2009;55:761–769. doi: 10.1016/j.eururo.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 77.Derhy S, El Mouhadi S, Ruiz A, Azizi L, Menu Y, Arrive L. Non-contrast 3D MR lymphography of retroperitoneal lymphatic aneurysmal dilatation: a continuous spectrum of change from normal variants to cystic lymphangioma. Insights Imaging. 2013;4:753–758. doi: 10.1007/s13244-013-0290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arrive L, Derhy S, El Mouhadi S, Monnier-Cholley L, Menu Y, Becker C. Noncontrast Magnetic Resonance Lymphography. J Reconstr Microsurg. 2015 doi: 10.1055/s-0035-1549133. [DOI] [PubMed] [Google Scholar]
- 79.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magnetic Resonance Imaging. 2004;22:81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 80.Vovk U, Pernuš F, Likar B. MRI intensity inhomogeneity correction by combining intensity and spatial information. Physics in Medicine and Biology. 2004;49:4119. doi: 10.1088/0031-9155/49/17/020. [DOI] [PubMed] [Google Scholar]
- 81.Vovk U, Pernuš F, Likar B. A review of methods for correction of intensity inhomogeneity in MRI. Medical Imaging, IEEE Transactions on. 2007;26:405–421. doi: 10.1109/TMI.2006.891486. [DOI] [PubMed] [Google Scholar]
- 82.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 83.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qiu W, Yuan J, Ukwatta E, Sun Y, Rajchl M, Fenster A. Dual optimization based prostate zonal segmentation in 3D MR images. Medical image analysis. 2014;18:660–673. doi: 10.1016/j.media.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 85.Mahapatra D, Buhmann JM. Prostate MRI segmentation using learned semantic knowledge and graph cuts. Biomedical Engineering, IEEE Transactions on. 2014;61 doi: 10.1109/TBME.2013.2289306. [DOI] [PubMed] [Google Scholar]
- 86.Liao S, Gao Y, Oto A, Shen D. Representation learning: a unified deep learning framework for automatic prostate MR segmentation. Med Image Comput Comput Assist Interv. 2013;16:254–261. doi: 10.1007/978-3-642-40763-5_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Toth R, Madabhushi A. Multifeature Landmark-Free Active Appearance Models: Application to Prostate MRI Segmentation. IEEE Transactions on Medical Imaging. 2012;31:1638–1650. doi: 10.1109/TMI.2012.2201498. [DOI] [PubMed] [Google Scholar]
- 88.Litjens G, Debats O, van de Ven W, Karssemeijer N, Huisman H. A pattern recognition approach to zonal segmentation of the prostate on MRI. Med Image Comput Comput Assist Interv. 2012;15:413–420. doi: 10.1007/978-3-642-33418-4_51. [DOI] [PubMed] [Google Scholar]
- 89.Wu Qiu JY, Eranga Ukwatta, Yue Sun, Martin Rajchl, Aaron Fenster. Prostate Segmentation: An Efficient Convex Optimization Approach With Axial Symmetry Using 3-D TRUS and MR Images. IEEE Transactions on Medical Imaging. 2014;33:947–960. doi: 10.1109/TMI.2014.2300694. [DOI] [PubMed] [Google Scholar]
- 90.Yang M, Li X, Turkbey B, Choyke PL, Yan P. Prostate segmentation in MR images using discriminant boundary features. Biomedical Engineering, IEEE Transactions on. 2013;60:479–488. doi: 10.1109/TBME.2012.2228644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zwiggelaar R, Zhu Y, Williams S. In Pattern Recognition and Image Analysis. Springer; 2003. Semi-automatic segmentation of the prostate; pp. 1108–1116. [Google Scholar]
- 92.Flores-Tapia D, Thomas G, Venugopa N, McCurdy B, Pistorius S. Semi automatic MRI prostate segmentation based on wavelet multiscale products. In Engineering in Medicine and Biology Society, 2008 EMBS 2008 30th Annual International Conference of the IEEE; IEEE; 2008. pp. 3020–3023. [DOI] [PubMed] [Google Scholar]
- 93.Samiee M, Thomas G, Fazel-Rezai R. Semi-automatic prostate segmentation of MR images based on flow orientation. In Signal Processing and Information Technology, 2006 IEEE International Symposium on; IEEE; 2006. pp. 203–207. [Google Scholar]
- 94.Cootes TF, Hill A, Taylor CJ, Haslam J. In Information Processing in Medical Imaging. Springer; 1993. The use of active shape models for locating structures in medical images; pp. 33–47. [Google Scholar]
- 95.Zhu Y, Williams S, Zwiggelaar R. A hybrid ASM approach for sparse volumetric data segmentation. Pattern Recognition and Image Analysis. 2007;17:252–258. [Google Scholar]
- 96.Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. International journal of computer vision. 1988;1:321–331. [Google Scholar]
- 97.Chan TF, Vese LA. Active contours without edges. IEEE transactions on Image processing. 2001;10:266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 98.Mumford D, Shah J. Optimal approximations by piecewise smooth functions and associated variational problems. Communications on pure and applied mathematics. 1989;42:577–685. [Google Scholar]
- 99.Klein S, van der Heide UA, Lips IM, van Vulpen M, Staring M, Pluim JPW. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Medical Physics. 2008;35:1407–1417. doi: 10.1118/1.2842076. [DOI] [PubMed] [Google Scholar]
- 100.Boykov Y, Veksler O, Zabih R. Fast approximate energy minimization via graph cuts. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2001;23:1222–1239. [Google Scholar]
- 101.Boykov YY, Jolly M-P. Interactive graph cuts for optimal boundary & region segmentation of objects in ND images. In Computer Vision, 2001 ICCV 2001 Proceedings Eighth IEEE International Conference on; IEEE; 2001. pp. 105–112. [Google Scholar]
- 102.Egger J. PCG-Cut: Graph Driven Segmentation of the Prostate Central Gland. Plos One. 2013;8:6. doi: 10.1371/journal.pone.0076645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tian Z, Liu L, Fei B. A supervoxel-based segmentation for prostate MR images. In SPIE Medical Imaging. International Society for Optics and Photonics. 2015 doi: 10.1117/12.2082255. 941318-941318-941317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, Vincent G, Guillard G, Birbeck N, Zhang J. Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Medical image analysis. 2014;18:359–373. doi: 10.1016/j.media.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ghose S, Oliver A, Marti R, Llado X, Vilanova JC, Freixenet J, Mitra J, Sidibe D, Meriaudeau F. A survey of prostate segmentation methodologies in ultrasound, magnetic resonance and computed tomography images. Comput Methods Programs Biomed. 2012;108:262–287. doi: 10.1016/j.cmpb.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 106.Oliveira FP, Tavares JMR. Medical image registration: a review. Computer methods in biomechanics and biomedical engineering. 2014;17:73–93. doi: 10.1080/10255842.2012.670855. [DOI] [PubMed] [Google Scholar]
- 107.Chappelow J, Madabhushi A, Rosen M, Tomaszeweski J, Feldman M. Multimodal image registration of ex vivo 4 Tesla MRI with whole mount histology for prostate cancer detection. In Medical Imaging. International Society for Optics and Photonics. 2007 65121S-65121S-65112. [Google Scholar]
- 108.Kalavagunta C, Zhou X, Schmechel SC, Metzger GJ. Registration of in vivo prostate MRI and pseudo - whole mount histology using Local Affine Transformations guided by Internal Structures (LATIS) Journal of Magnetic Resonance Imaging. 2014 doi: 10.1002/jmri.24629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huisman HJ, Engelbrecht MR, Barentsz JO. Accurate estimation of pharmacokinetic contrast-enhanced dynamic MRI parameters of the prostate. Journal of Magnetic Resonance Imaging. 2001;13:607–614. doi: 10.1002/jmri.1085. [DOI] [PubMed] [Google Scholar]
- 110.Li Q, Sone S, Doi K. Selective enhancement filters for nodules, vessels, and airway walls in two-and three-dimensional CT scans. Medical physics. 2003;30:2040–2051. doi: 10.1118/1.1581411. [DOI] [PubMed] [Google Scholar]
- 111.Langer DL, van der Kwast TH, Evans AJ, Trachtenberg J, Wilson BC, Haider MA. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2009;30:327–334. doi: 10.1002/jmri.21824. [DOI] [PubMed] [Google Scholar]
- 112.Murase K. Efficient method for calculating kinetic parameters using T1-weighted dynamic contrast-enhanced magnetic resonance imaging. Magn Reson Med. 2004;51:858–862. doi: 10.1002/mrm.20022. [DOI] [PubMed] [Google Scholar]
- 113.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A. Radiomics: extracting more information from medical images using advanced feature analysis. European Journal of Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D. Radiomics: the process and the challenges. Magnetic resonance imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cameron A, Khalvati F, Haider M, Wong AMAPS. A Quantitative Radiomics Approach for Prostate Cancer Detection. IEEE Trans Biomed Eng. 2015 doi: 10.1109/TBME.2015.2485779. [DOI] [PubMed] [Google Scholar]
- 116.Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, Zheng J, Goldman D, Moskowitz C, Fine SW, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol. 2015;25:2840–2850. doi: 10.1007/s00330-015-3701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fehr D, Veeraraghavan H. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. 2015;112:E6265–E6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Khalvati F, Wong A, Haider MA. Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med Imaging. 2015;15:27. doi: 10.1186/s12880-015-0069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Breiman L. Random forests. Machine learning. 2001;45:5–32. [Google Scholar]
- 120.Friedman J, Hastie T, Tibshirani R. Additive logistic regression: a statistical view of boosting (with discussion and a rejoinder by the authors) The annals of statistics. 2000;28:337–407. [Google Scholar]
- 121.Baeza-Yates R, Ribeiro-Neto B. Modern information retrieval. New York: ACM press; 1999. [Google Scholar]
- 122.Chan I, Wells W, 3rd, Mulkern RV, Haker S, Zhang J, Zou KH, Maier SE, Tempany CM. Detection of prostate cancer by integration of line-scan diffusion, T2-mapping and T2-weighted magnetic resonance imaging; a multichannel statistical classifier. Med Phys. 2003;30:2390–2398. doi: 10.1118/1.1593633. [DOI] [PubMed] [Google Scholar]
- 123.Donati OF, Mazaheri Y, Afaq A, Vargas HA, Zheng J, Moskowitz CS, Hricak H, Akin O. Prostate cancer aggressiveness: assessment with whole-lesion histogram analysis of the apparent diffusion coefficient. Radiology. 2014;271:143–152. doi: 10.1148/radiol.13130973. [DOI] [PubMed] [Google Scholar]
- 124.Shah V, Pohida T, Turkbey B, Mani H, Merino M, Pinto PA, Choyke P, Bernardo M. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum. 2009;80:104301. doi: 10.1063/1.3242697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mazaheri Y, Bokacheva L, Kroon DJ, Akin O, Hricak H, Chamudot D, Fine S, Koutcher JA. Semiautomatic deformable registration of prostate MR images to pathological slices. J Magn Reson Imaging. 2010;32:1149–1157. doi: 10.1002/jmri.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jacobs MA, Windham JP, Soltanian-Zadeh H, Peck DJ, Knight RA. Registration and warping of magnetic resonance images to histological sections. Med Phys. 1999;26:1568–1578. doi: 10.1118/1.598671. [DOI] [PubMed] [Google Scholar]
- 127.Park H, Piert MR, Khan A, Shah R, Hussain H, Siddiqui J, Chenevert TL, Meyer CR. Registration methodology for histological sections and in vivo imaging of human prostate. Acad Radiol. 2008;15:1027–1039. doi: 10.1016/j.acra.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meyer CR, Moffat BA, Kuszpit KK, Bland PL, McKeever PE, Johnson TD, Chenevert TL, Rehemtulla A, Ross BD. A methodology for registration of a histological slide and in vivo MRI volume based on optimizing mutual information. Mol Imaging. 2006;5:16–23. [PMC free article] [PubMed] [Google Scholar]
- 129.Viswanath S, Bloch BN, Rosen M, Chappelow J, Toth R, Rofsky N, Lenkinski R, Genega E, Kalyanpur A, Madabhushi A. Integrating Structural and Functional Imaging for Computer Assisted Detection of Prostate Cancer on Multi-Protocol 3 Tesla MRI. Proc Soc Photo Opt Instrum Eng. 2009;7260:72603I. doi: 10.1117/12.811899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gibson E, Gaed M, Gomez JA, Moussa M, Pautler S, Chin JL, Crukley C, Bauman GS, Fenster A, Ward AD. 3D prostate histology image reconstruction: Quantifying the impact of tissue deformation and histology section location. J Pathol Inform. 2013;4:31. doi: 10.4103/2153-3539.120874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gibson E, Gaed M, Gomez JA, Moussa M, Romagnoli C, Pautler S, Chin JL, Crukley C, Bauman GS, Fenster A, Ward AD. 3D prostate histology reconstruction: an evaluation of image-based and fiducial-based algorithms. Med Phys. 2013;40:093501. doi: 10.1118/1.4816946. [DOI] [PubMed] [Google Scholar]
- 132.Patel P, Chappelow J, Tomaszewski J, Feldman MD, Rosen M, Shih N, Madabhushi A. Spatially weighted mutual information (SWMI) for registration of digitally reconstructed ex vivo whole mount histology and in vivo prostate MRI. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:6269–6272. doi: 10.1109/IEMBS.2011.6091547. [DOI] [PubMed] [Google Scholar]
- 133.McGrath DM, Vlad RM, Foltz WD, Brock KK. Technical note: fiducial markers for correlation of whole-specimen histopathology with MR imaging at 7 tesla. Med Phys. 2010;37:2321–2328. doi: 10.1118/1.3395575. [DOI] [PubMed] [Google Scholar]
- 134.Toth RJ, Shih N, Tomaszewski JE, Feldman MD, Kutter O, Yu DN, Paulus JC, Jr, Paladini G, Madabhushi A. Histostitcher: An informatics software platform for reconstructing whole-mount prostate histology using the extensible imaging platform framework. J Pathol Inform. 2014;5:8. doi: 10.4103/2153-3539.129441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chappelow J, Bloch BN, Rofsky N, Genega E, Lenkinski R, DeWolf W, Madabhushi A. Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information. Med Phys. 2011;38:2005–2018. doi: 10.1118/1.3560879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orczyk C, Rusinek H, Rosenkrantz AB, Mikheev A, Deng FM, Melamed J, Taneja SS. Preliminary experience with a novel method of three-dimensional co-registration of prostate cancer digital histology and in vivo multiparametric MRI. Clin Radiol. 2013;68:e652–e658. doi: 10.1016/j.crad.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lv D, Guo X, Wang X, Zhang J, Fang J. Computerized characterization of prostate cancer by fractal analysis in MR images. J Magn Reson Imaging. 2009;30:161–168. doi: 10.1002/jmri.21819. [DOI] [PubMed] [Google Scholar]
- 138.Rouviere O, Papillard M, Girouin N, Boutier R, Rabilloud M, Riche B, Mege-Lechevallier F, Colombel M, Gelet A. Is it possible to model the risk of malignancy of focal abnormalities found at prostate multiparametric MRI? Eur Radiol. 2012;22:1149–1157. doi: 10.1007/s00330-011-2343-8. [DOI] [PubMed] [Google Scholar]
- 139.Meyer C, Ma B, Kunju LP, Davenport M, Piert M. Challenges in accurate registration of 3-D medical imaging and histopathology in primary prostate cancer. European journal of nuclear medicine and molecular imaging. 2013;40:72–78. doi: 10.1007/s00259-013-2382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Viswanath S, Bloch BN, Genega E, Rofsky N, Lenkinski R, Chappelow J, Toth R, Madabhushi A. A comprehensive segmentation, registration, and cancer detection scheme on 3 Tesla in vivo prostate DCE-MRI. Med Image Comput Comput Assist Interv. 2008;11:662–669. doi: 10.1007/978-3-540-85988-8_79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu X, Langer DL, Haider MA, Yang Y, Wernick MN, Yetik IS. Prostate cancer segmentation with simultaneous estimation of Markov random field parameters and class. IEEE Trans Med Imaging. 2009;28:906–915. doi: 10.1109/TMI.2009.2012888. [DOI] [PubMed] [Google Scholar]
- 142.Artan Y, Haider MA, Langer DL, van der Kwast TH, Evans AJ, Yang Y, Wernick MN, Trachtenberg J, Yetik IS. Prostate cancer localization with multispectral MRI using cost-sensitive support vector machines and conditional random fields. IEEE Trans Image Process. 2010;19:2444–2455. doi: 10.1109/TIP.2010.2048612. [DOI] [PubMed] [Google Scholar]
- 143.Vos PC, Hambrock T, Barenstz JO, Huisman HJ. Computer-assisted analysis of peripheral zone prostate lesions using T2-weighted and dynamic contrast enhanced T1-weighted MRI. Phys Med Biol. 2010;55:1719–1734. doi: 10.1088/0031-9155/55/6/012. [DOI] [PubMed] [Google Scholar]
- 144.Viswanath S, Bloch BN, Chappelow J, Patel P, Rofsky N, Lenkinski R, Genega E, Madabhushi A. Enhanced Multi-Protocol Analysis via Intelligent Supervised Embedding (EMPrAvISE): Detecting Prostate Cancer on Multi-Parametric MRI. Proc SPIE Int Soc Opt Eng. 2011;7963 doi: 10.1117/12.878312. 79630u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lopes R, Ayache A, Makni N, Puech P, Villers A, Mordon S, Betrouni N. Prostate cancer characterization on MR images using fractal features. Med Phys. 2011;38:83–95. doi: 10.1118/1.3521470. [DOI] [PubMed] [Google Scholar]
- 146.Sung YS, Kwon HJ, Park BW, Cho G, Lee CK, Cho KS, Kim JK. Prostate cancer detection on dynamic contrast-enhanced MRI: computer-aided diagnosis versus single perfusion parameter maps. AJR Am J Roentgenol. 2011;197:1122–1129. doi: 10.2214/AJR.10.6062. [DOI] [PubMed] [Google Scholar]
- 147.Artan Y, Yetik IS. Prostate cancer localization using multiparametric MRI based on semi-supervised techniques with automated seed initialization. IEEE Trans Inf Technol Biomed. 2012;16:1313–1323. doi: 10.1109/TITB.2012.2201731. [DOI] [PubMed] [Google Scholar]
- 148.Tiwari P, Kurhanewicz J, Madabhushi A. Multi-kernel graph embedding for detection, Gleason grading of prostate cancer via MRI/MRS. Med Image Anal. 2013;17:219–235. doi: 10.1016/j.media.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ginsburg SB, Viswanath SE, Bloch BN, Rofsky NM, Genega EM, Lenkinski RE, Madabhushi A. Novel PCA-VIP scheme for ranking MRI protocols and identifying computer-extracted MRI measurements associated with central gland and peripheral zone prostate tumors. J Magn Reson Imaging. 2014 doi: 10.1002/jmri.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Niaf E, Flamary R, Rouviere O, Lartizien C, Canu S. Kernel-based learning from both qualitative and quantitative labels: application to prostate cancer diagnosis based on multiparametric MR imaging. IEEE Trans Image Process. 2014;23:979–991. doi: 10.1109/TIP.2013.2295759. [DOI] [PubMed] [Google Scholar]
- 151.Zhao K, Wang C, Hu J, Yang X, Wang H, Li F, Zhang X, Zhang J, Wang X. Prostate cancer identification: quantitative analysis of T2-weighted MR images based on a back propagation artificial neural network model. Sci China Life Sci. 2015;58:666–673. doi: 10.1007/s11427-015-4876-6. [DOI] [PubMed] [Google Scholar]
- 152.Saman DM, Lemieux AM, Nawal Lutfiyya M, Lipsky MS. A review of the current epidemiology and treatment options for prostate cancer. Dis Mon. 2014;60:150–154. doi: 10.1016/j.disamonth.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 153.Keyes M, Crook J, Morton G, Vigneault E, Usmani N, Morris WJ. Treatment options for localized prostate cancer. Can Fam Physician. 2013;59:1269–1274. [PMC free article] [PubMed] [Google Scholar]
- 154.Oto A, Yang C, Kayhan A, Tretiakova M, Antic T, Schmid-Tannwald C, Eggener S, Karczmar GS, Stadler WM. Diffusion-weighted and dynamic contrast-enhanced MRI of prostate cancer: correlation of quantitative MR parameters with Gleason score and tumor angiogenesis. AJR Am J Roentgenol. 2011;197:1382–1390. doi: 10.2214/AJR.11.6861. [DOI] [PubMed] [Google Scholar]
- 155.Anwar SS, Anwar Khan Z, Shoaib Hamid R, Haroon F, Sayani R, Beg M. Assessment of apparent diffusion coefficient values as predictor of aggressiveness in peripheral zone prostate cancer: comparison with Gleason score. 2014;2014:263417. doi: 10.1155/2014/263417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hambrock T, Somford DM, Huisman HJ, van Oort IM, Witjes JA, Hulsbergen-van de Kaa CA, Scheenen T, Barentsz JO. Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology. 2011;259:453–461. doi: 10.1148/radiol.11091409. [DOI] [PubMed] [Google Scholar]
- 157.Yamamura J, Salomon G, Buchert R, Hohenstein A, Graessner J, Huland H, Graefen M, Adam G, Wedegaetner U. MR Imaging of Prostate Cancer: Diffusion Weighted Imaging and (3D) Hydrogen 1 (H) MR Spectroscopy in Comparison with Histology. Radiol Res Pract. 2011;2011:616852. doi: 10.1155/2011/616852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gibbs P, Liney GP, Pickles MD, Zelhof B, Rodrigues G, Turnbull LW. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest Radiol. 2009;44:572–576. doi: 10.1097/RLI.0b013e3181b4c10e. [DOI] [PubMed] [Google Scholar]
- 159.Anderson AW, Xie J, Pizzonia J, Bronen RA, Spencer DD, Gore JC. Effects of cell volume fraction changes on apparent diffusion in human cells. Magn Reson Imaging. 2000;18:689–695. doi: 10.1016/s0730-725x(00)00147-8. [DOI] [PubMed] [Google Scholar]
- 160.Tanimoto K, Yoshikawa K, Obata T, Ikehira H, Shiraishi T, Watanabe K, Saga T, Mizoe J, Kamada T, Kato A, Miyazaki M. Role of glucose metabolism and cellularity for tumor malignancy evaluation using FDG-PET/CT and MRI. Nucl Med Commun. 2010;31:604–609. [PubMed] [Google Scholar]
- 161.Kobus T, Hambrock T, Hulsbergen-van de Kaa CA, Wright AJ, Barentsz JO, Heerschap A, Scheenen TW. In vivo assessment of prostate cancer aggressiveness using magnetic resonance spectroscopic imaging at 3 T with an endorectal coil. Eur Urol. 2011;60:1074–1080. doi: 10.1016/j.eururo.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 162.Zakian KL, Sircar K, Hricak H, Chen HN, Shukla-Dave A, Eberhardt S, Muruganandham M, Ebora L, Kattan MW, Reuter VE, et al. Correlation of proton MR spectroscopic imaging with gleason score based on step-section pathologic analysis after radical prostatectomy. Radiology. 2005;234:804–814. doi: 10.1148/radiol.2343040363. [DOI] [PubMed] [Google Scholar]
- 163.van Asten JJ, Cuijpers V, Hulsbergen-van de Kaa C, Soede-Huijbregts C, Witjes JA, Verhofstad A, Heerschap A. High resolution magic angle spinning NMR spectroscopy for metabolic assessment of cancer presence and Gleason score in human prostate needle biopsies. Magma. 2008;21:435–442. doi: 10.1007/s10334-008-0156-9. [DOI] [PubMed] [Google Scholar]
- 164.Garcia-Martin ML, Adrados M, Ortega MP, Fernandez Gonzalez I, Lopez-Larrubia P, Viano J, Garcia-Segura JM. Quantitative (1) H MR spectroscopic imaging of the prostate gland using LCModel and a dedicated basis-set: correlation with histologic findings. Magn Reson Med. 2011;65:329–339. doi: 10.1002/mrm.22631. [DOI] [PubMed] [Google Scholar]
- 165.Scheenen TW, Heijmink SW, Roell SA, Hulsbergen-Van de Kaa CA, Knipscheer BC, Witjes JA, Barentsz JO, Heerschap A. Three-dimensional proton MR spectroscopy of human prostate at 3 T without endorectal coil: feasibility. Radiology. 2007;245:507–516. doi: 10.1148/radiol.2451061444. [DOI] [PubMed] [Google Scholar]
- 166.Shukla-Dave A, Hricak H, Ishill NM, Moskowitz CS, Drobnjak M, Reuter VE, Zakian KL, Scardino PT, Cordon-Cardo C. Correlation of MR imaging and MR spectroscopic imaging findings with Ki-67, phospho-Akt, and androgen receptor expression in prostate cancer. Radiology. 2009;250:803–812. doi: 10.1148/radiol.2503080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lee DH, Koo KC, Lee SH, Rha KH, Choi YD, Hong SJ, Chung BH. Tumor lesion diameter on diffusion weighted magnetic resonance imaging could help predict insignificant prostate cancer in patients eligible for active surveillance: preliminary analysis. J Urol. 2013;190:1213–1217. doi: 10.1016/j.juro.2013.03.127. [DOI] [PubMed] [Google Scholar]
- 168.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 169.Cam K, Yucel S, Turkeri L, Akdas A. Accuracy of transrectal ultrasound guided prostate biopsy: histopathological correlation to matched prostatectomy specimens. Int J Urol. 2002;9:257–260. doi: 10.1046/j.1442-2042.2002.00456.x. [DOI] [PubMed] [Google Scholar]
- 170.Quann P, Jarrard DF, Huang W. Current prostate biopsy protocols cannot reliably identify patients for focal therapy: correlation of low-risk prostate cancer on biopsy with radical prostatectomy findings. Int J Clin Exp Pathol. 2010;3:401–407. [PMC free article] [PubMed] [Google Scholar]
- 171.Djavan B, Ravery V, Zlotta A, Dobronski P, Dobrovits M, Fakhari M, Seitz C, Susani M, Borkowski A, Boccon-Gibod L, et al. Prospective evaluation of prostate cancer detected on biopsies 1, 2, 3 and 4: when should we stop? J Urol. 2001;166:1679–1683. [PubMed] [Google Scholar]
- 172.Futterer JJ, Barentsz JO. MRI-guided and robotic-assisted prostate biopsy. Curr Opin Urol. 2012;22:316–319. doi: 10.1097/MOU.0b013e328354833c. [DOI] [PubMed] [Google Scholar]
- 173.Moore CM, Robertson NL, Arsanious N, Middleton T, Villers A, Klotz L, Taneja SS, Emberton M. Image-guided prostate biopsy using magnetic resonance imaging-derived targets: a systematic review. Eur Urol. 2013;63:125–140. doi: 10.1016/j.eururo.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 174.Xu H, Lasso A, Guion P, Krieger A, Kaushal A, Singh AK, Pinto PA, Coleman J, Grubb RL, 3rd, Lattouf JB, et al. Accuracy analysis in MRI-guided robotic prostate biopsy. Int J Comput Assist Radiol Surg. 2013;8:937–944. doi: 10.1007/s11548-013-0831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zamecnik P, Schouten MG, Krafft AJ, Maier F, Schlemmer HP, Barentsz JO, Bock M, Futterer JJ. Automated real-time needle-guide tracking for fast 3-T MR-guided transrectal prostate biopsy: a feasibility study. Radiology. 2014;273:879–886. doi: 10.1148/radiol.14132067. [DOI] [PubMed] [Google Scholar]
- 176.Fiard G, Hohn N, Descotes JL, Rambeaud JJ, Troccaz J, Long JA. Targeted MRI-guided prostate biopsies for the detection of prostate cancer: initial clinical experience with real-time 3-dimensional transrectal ultrasound guidance and magnetic resonance/transrectal ultrasound image fusion. Urology. 2013;81:1372–1378. doi: 10.1016/j.urology.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 177.Xu S, Kruecker J, Turkbey B, Glossop N, Singh AK, Choyke P, Pinto P, Wood BJ. Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies. Comput Aided Surg. 2008;13:255–264. doi: 10.1080/10929080802364645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mitra J, Ghose S, Sidibe D, Marti R, Oliver A, Llado X, Vilanova JC, Comet J, Meriaudeau F. Joint probability of shape and image similarities to retrieve 2D TRUS-MR slice correspondence for prostate biopsy. Conf Proc IEEE Eng Med Biol Soc. 2012;2012:5416–5419. doi: 10.1109/EMBC.2012.6347219. [DOI] [PubMed] [Google Scholar]
- 179.Wang S, Burtt K, Turkbey B, Choyke P, Summers RM. Computer Aided-Diagnosis of Prostate Cancer on Multiparametric MRI: A Technical Review of Current Research. Biomed Res Int. 2014;2014:789561. doi: 10.1155/2014/789561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fei BW, Lee ZH, Boll DT, Duerk JL, Lewin JS, Wilson DL. Image registration and fusion for interventional MRI guided thermal ablation of the prostate cancer. In: Ellis RE, Peters TM, editors. In Medical Image Computing and Computer-Assisted Intervention - Miccai 2003. Pt 2. Vol. 2879. 2003. pp. 364–372. Lecture Notes in Computer Science]. [Google Scholar]
- 181.Akbari H, Fei BW. 3D ultrasound image segmentation using wavelet support vector machines. Medical Physics. 2012;39:2972–2984. doi: 10.1118/1.4709607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fei BW, Lee ZH, Boll DT, Duerk JL, Sodee DB, Lewin JS, Wilson DL. Registration and fusion of SPECT, high-resolution MRI, and interventional MRI for thermal ablation of prostate cancer. Ieee Transactions on Nuclear Science. 2004;51:177–183. [Google Scholar]
- 183.Fei BW, Lee ZH, Duerk JL, Wilson DL. Image registration for interventional MRI guided procedures: Interpolation methods, similarity measurements, and applications to the prostate. In: Gee JC, Maintz JBA, Vannier MW, editors. In Biomedical Image Registration. Vol. 2717. 2003. pp. 321–329. Lecture Notes in Computer Science]. [Google Scholar]
- 184.Fei BW, Ng WS, Chauhan S, Kwoh CK. The safety issues of medical robotics. Reliability Engineering & System Safety. 2001;73:183–192. [Google Scholar]
- 185.Fei BW, Wang H, Meyers JD, Feyes DK, Oleinick NL, Duerk JL. High-field magnetic resonance imaging of the response of human prostate cancer to pc 4-based photodynamic therapy in an animal model. Lasers in Surgery and Medicine. 2007;39:723–730. doi: 10.1002/lsm.20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fei BW, Wang HS, Wu CY, Chiu SM. Choline PET for Monitoring Early Tumor Response to Photodynamic Therapy. Journal of Nuclear Medicine. 2010;51:130–138. doi: 10.2967/jnumed.109.067579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fei BW, Yang XF, Nye JA, Aarsvold JN, Raghunath N, Cervo M, Stark R, Meltzer CC, Votaw JR. MR/PET quantification tools: Registration, segmentation, classification, and MR-based attenuation correction. Medical Physics. 2012;39:6443–6454. doi: 10.1118/1.4754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang HS, Fei BW. Diffusion-Weighted MRI for Monitoring Tumor Response to Photodynamic Therapy. Journal of Magnetic Resonance Imaging. 2010;32:409–417. doi: 10.1002/jmri.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang HS, Fei BW. An MR image-guided, voxel-based partial volume correction method for PET images. Medical Physics. 2012;39:179–194. doi: 10.1118/1.3665704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang HS, Fei BW. Nonrigid point registration for 2D curves and 3D surfaces and its various applications. Physics in Medicine and Biology. 2013;58:4315–4330. doi: 10.1088/0031-9155/58/12/4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Song I, Kim CK, Park BK, Park W. Assessment of response to radiotherapy for prostate cancer: value of diffusion-weighted MRI at 3 T. AJR Am J Roentgenol. 2010;194:W477–W482. doi: 10.2214/AJR.09.3557. [DOI] [PubMed] [Google Scholar]
- 192.Padhani AR, Makris A, Gall P, Collins DJ, Tunariu N, de Bono JS. Therapy monitoring of skeletal metastases with whole-body diffusion MRI. J Magn Reson Imaging. 2014;39:1049–1078. doi: 10.1002/jmri.24548. [DOI] [PubMed] [Google Scholar]
- 193.Graham TJ, Box G, Tunariu N, Crespo M, Spinks TJ, Miranda S, Attard G, de Bono J, Eccles SA, Davies FE, Robinson SP. Preclinical evaluation of imaging biomarkers for prostate cancer bone metastasis and response to cabozantinib. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju033. dju033. [DOI] [PubMed] [Google Scholar]
- 194.Lee KC, Bradley DA, Hussain M, Meyer CR, Chenevert TL, Jacobson JA, Johnson TD, Galban CJ, Rehemtulla A, Pienta KJ, Ross BD. A feasibility study evaluating the functional diffusion map as a predictive imaging biomarker for detection of treatment response in a patient with metastatic prostate cancer to the bone. Neoplasia. 2007;9:1003–1011. doi: 10.1593/neo.07954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sosna J, Pedrosa I, Dewolf WC, Mahallati H, Lenkinski RE, Rofsky NM. MR imaging of the prostate at 3 Tesla: comparison of an external phased-array coil to imaging with an endorectal coil at 1.5 Tesla. Acad Radiol. 2004;11:857–862. doi: 10.1016/j.acra.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 196.Turkbey B, Merino MJ, Gallardo EC, Shah V, Aras O, Bernardo M, Mena E, Daar D, Rastinehad AR, Linehan WM, et al. Comparison of endorectal coil and nonendorectal coil T2W and diffusion-weighted MRI at 3 Tesla for localizing prostate cancer: correlation with whole-mount histopathology. J Magn Reson Imaging. 2014;39:1443–1448. doi: 10.1002/jmri.24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Litjens GJ, Barentsz JO, Karssemeijer N, Huisman HJ. Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI. Eur Radiol. 2015;25:3187–3199. doi: 10.1007/s00330-015-3743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Matulewicz L, Jansen JF, Bokacheva L, Vargas HA, Akin O, Fine SW, Shukla-Dave A, Eastham JA, Hricak H, Koutcher JA, Zakian KL. Anatomic segmentation improves prostate cancer detection with artificial neural networks analysis of 1H magnetic resonance spectroscopic imaging. J Magn Reson Imaging. 2014;40:1414–1421. doi: 10.1002/jmri.24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Huang PW, Lee CH. Automatic classification for pathological prostate images based on fractal analysis. IEEE Trans Med Imaging. 2009;28:1037–1050. doi: 10.1109/TMI.2009.2012704. [DOI] [PubMed] [Google Scholar]


