Abstract
CD74 (invariant chain) plays a role in MHC class II antigen presentation. We assessed CD74 and MHCII expression in tumor cells, as well as CD8, CD4, and CD68 tumor infiltrating leucocyte (TIL) density by immunohistochemistry in a cohort of 492 breast cancer patients. CD74 expression was associated with poor prognostic markers including patient age, tumor grade, ER status, non-Luminal A subtypes, and with MHCII expression and higher TIL densities, particularly in the Basal-like subgroup. Univariate analysis showed a favorable prognostic effect of CD74 (Hazard ratio = 0.46, 95% CI = 0.26–0.89, p = 0.022) and for combined CD74/MHCII (Hazard ratio = 0.26, 95% CI = 0.17–0.81, p = 0.014) positive status for overall survival that was only manifested in the Basal-like subgroup. CD74 and MHCII expression is associated with patient survival in Basal-like breast cancer, and the association with TIL may reflect an effective intratumoral immune response.
Keywords: CD74, MHCII, CD4, CD8, CD68
INTRODUCTION
CD74 (invariant chain, Li) is expressed by breast tumor cells [1] as well as by several immune cell types. CD74 has dual roles as a component of the MHC class II antigen presentation pathway and as a cytokine receptor [2], as well as intracellular effects on activation of transcription [3, 4]. The MHCII is central to the process of presentation of peptides to CD4 T cells and consequently impacts many aspects of adaptive immunity including activation of effector CD8 T cells [5]. The macrophage inhibitory factor (MIF) is a prevalent cytokine that acts through CD74 to promote cell proliferation, migration and survival pathways in both immune and epithelial cell types [6–8].
CD74 expression in tumor cells might therefore be expected to mediate both pro and anti tumor effects attributable to its cytokine signaling and antigen presenting functions. Several previous studies based on mostly small cohorts have shown that CD74 is associated with ER negative and/or triple negative subgroups, but have been discordant with regard to the prognostic significance of CD74 [9, 10]. Given this dichotomy we set out to examine the relationship between CD74 and outcomes in breast cancer. We hypothesized that tumors that express CD74 along with MHCII, both key components of the antigen presenting machinery, represent those tumors most susceptible to a productive tumor infiltrating leukocyte (TIL) response and correspondingly good outcomes.
RESULTS
Cohort characteristics
A total of 492 patients with primary breast cancer diagnosed in the period 1988–1995 were studied. Follow-up outcomes data was available, with mean of 87 months (range 2 to 251 months). There were 195 breast specific deaths (mean time 55 months from diagnosis) and 297 survivors (mean time to last follow-up date 107 months). Primary therapy included surgical resection in all cases followed by adjuvant hormone, radiation, and chemotherapy in 373 (76%), 182 (37%), 102 (21%) cases respectively and 31 (6%) did not receive any form of systemic therapy. The clinical-pathological characteristics of the population are provided (Table 1).
Table 1. Clinical-pathological characteristics of the whole cohort.
| Parameter | Status | Cases | % |
|---|---|---|---|
| Age at diagnosis | ≤ 35 years | 13 | 3 |
| > 35years | 479 | 97 | |
| Tumor sizea | T1a/b | 4 | 1 |
| T1c | 124 | 25 | |
| T2 | 277 | 56 | |
| T3 | 57 | 12 | |
| Unknown | 30 | 6 | |
| Nodal status | Positive | 223 | 45 |
| Negative | 244 | 50 | |
| Unknown | 25 | 5 | |
| Tumor grade | 1 | 81 | 16 |
| 2 | 297 | 60 | |
| 3 | 111 | 23 | |
| Unknown | 3 | 1 | |
| ERb | Positive | 250 | 51 |
| Negative | 242 | 49 | |
| Unknown | 0 | 0 | |
| PRb | Positive | 250 | 51 |
| Negative | 242 | 49 | |
| Unknown | 0 | 0 | |
| Molecular subtypes | Luminal A | 197 | 40 |
| Luminal B | 52 | 11 | |
| Her2 | 72 | 15 | |
| TNNBc | 43 | 9 | |
| Basal-like | 73 | 15 | |
| Unclassified | 55 | 11 |
aTumor size: 0.1 cm < T1a/b < 1 cm; 1 cm ≤ T1c < 2 cm; 2 cm ≤ T2 < 5 cm; 5 cm ≤ T3.
bER negative defined as < 10 fmol/mg protein and PR negative as ≤ 15 fmol/mg protein (ligand binding assay).
cTNNB=Triple-negative-non-basal.
Expression of CD74 and association with clinical-pathological features
Expression of CD74 within tumor cells showed a predominantly cytoplasmic staining pattern with weak membrane staining visible in occasional cells, and was heterogeneous within positive staining tumors. CD74 expression was present in 139 (28%) cases (Figure 1). CD74 was associated with patient age (p = 0.001), tumor grade (p = 0.003), and ER status (p = 0.006) (Table 2). The frequency of CD74 expression between molecular intrinsic subtype classes was also significantly different, with high levels of CD74 present in a small proportion (21%) of Luminal A subtype tumors but significantly higher proportions (37%–38%) of Luminal B, Triple Negative Non-basal (TNNB), and Basal-like subtype tumors (p = 0.009, p = 0.022, p = 0.003 respectively) (Table 2).
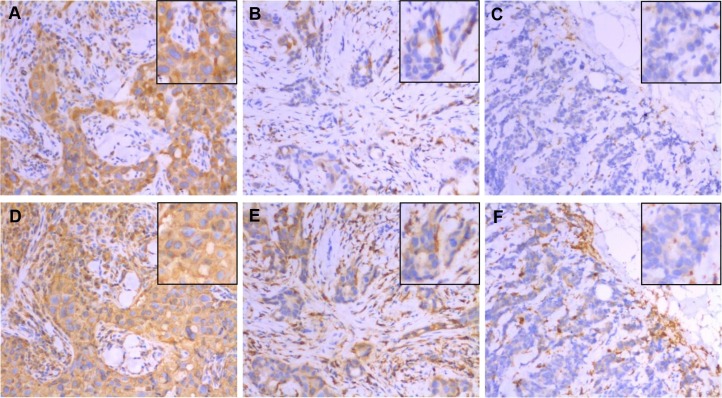
Figure 1. Expression of CD74 (top row) and MHCII (bottom row) in the same areas of three representative tumors as determined by immunohistochemistry.
These three cases scored as CD74 strong positive / MHCII strong positive (panels A and D), CD74 weak positive/MHCII weak positive (panels B and E), and CD74 negative/MHCII negative (panels C and F) for tumor cell expression. Main images taken at 200× original magnification with insets showing detail within the image. Note that TIL staining positive for CD74 and MHCII are present in stromal and epithelial compartments of all cases but were not included in scoring.
Table 2. Association between CD74, MHCII expression and clinical-pathological characteristics.
| Parameter | CD74 expression | MHCII expression | |||||
|---|---|---|---|---|---|---|---|
| Low (%) | High (%) | p-value | Low (%) | High (%) | p-value | ||
| Age at diagnosis (yrs) | ≤ 35 years | 4 (31%) | 9 (69%) | 0.001 | 4 (33%) | 8 (67%) | < 0.0001 |
| > 35 years | 349 (73%) | 130 (27%) | 352 (81%) | 81 (19%) | |||
| Tumour size (cm) | T1a/b | 4 (70%) | 0 (0%) | 0.555 | 2 (67%) | 1 (33%) | 0.195 |
| T1c | 87 (73%) | 37 (30%) | 65 (88%) | 9 (12%) | |||
| T2 | 201 (73%) | 76 (27%) | 223 (78%) | 63 (22%) | |||
| T3 | 38 (67%) | 19 (33%) | 41 (79%) | 11 (21%) | |||
| Nodal status | Positive | 178 (71%) | 72 (29%) | 0.616 | 185 (82%) | 41 (18%) | 0.537 |
| Negative | 179 (73%) | 65 (27%) | 154 (79%) | 40 (21%) | |||
| Tumor grade | 1 | 65 (80%) | 16 (20%) | 0.003 | 69 (93%) | 5 (7%) | < 0.0001 |
| 2 | 220 (74%) | 77 (26%) | 219 (82%) | 47 (18%) | |||
| 3 | 66 (59%) | 45 (41%) | 66 (64%) | 37 (36%) | |||
| ER status | Positive | 249 (76%) | 80 (24%) | 0.001 | 257 (87%) | 37 (13%) | < 0.0001 |
| Negative | 104 (64%) | 59 (36%) | 99 (66%) | 52 (34%) | |||
| PR status | Positive | 178 (74%) | 64 (26%) | 0.423 | 182 (84%) | 35 (16%) | 0.058 |
| Negative | 175 (70%) | 75 (30%) | 174 (76%) | 54 (24%) | |||
| Molecular subtypes | Luminal A | 156 (79%) | 41 (21%) | 156 (89%) | 20 (11%) | ||
| Luminal B | 32 (62%) | 20 (38%) | 0.009 | 37 (76%) | 12 (24%) | 0.020 | |
| Her2 | 51 (71%) | 21 (29%) | 59 (84%) | 11 (16%) | |||
| TNNBa | 27 (63%) | 16 (37%) | 0.022 | 22 (65%) | 12 (35%) | 0.0004 | |
| Basal-like | 45 (62%) | 28 (38%) | 0.003 | 39 (57%) | 29 (43%) | < 0.0001 | |
aTNNB = Triple Negative Non-basal.
Association of CD74 with outcomes
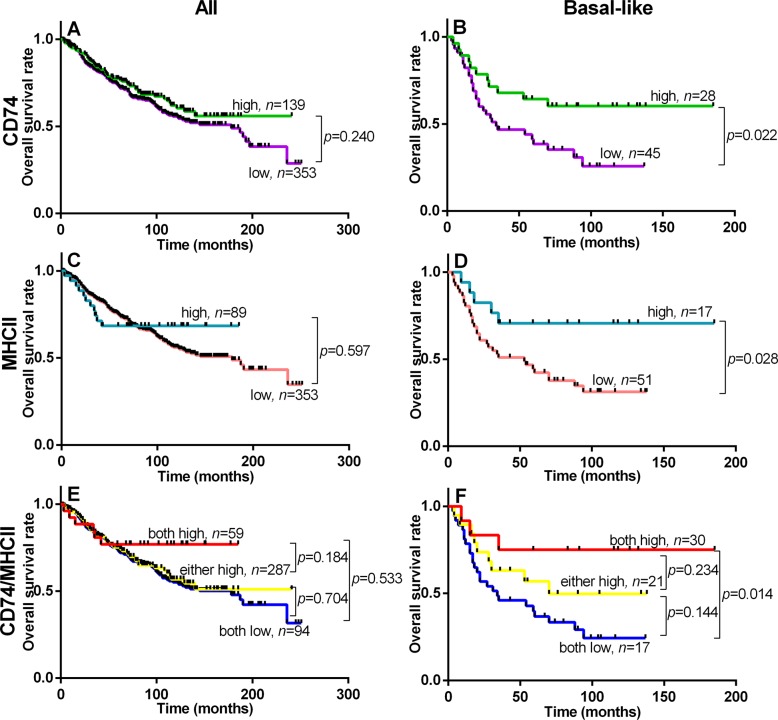
Univariate analysis of standard prognostic factors in the entire cohort confirmed patient age, high tumor grade, tumor size, nodal status, ER status and PR status as significant prognostic factors (Supplementary Table 1). Tumor subtype was also strongly prognostic with the rank order of good to poor overall survival subtypes as follows; Luminal A > Luminal B > Her2, TNNB, > Basal-like. CD74 was not prognostic for relapse free survival (RFS) or overall breast cancer specific survival (OS) in the overall cohort. However CD74 was associated with RFS and OS within the Basal-like subset (p = 0.018 and p = 0.022 respectively) (Figure 2, Table 3). CD74 was not prognostic in other subtypes (including Luminal A and Her2 subsets with larger or comparable subset sizes or Luminal B and TNNB subsets with relatively smaller case numbers). In multivariate analysis of CD74 with clinical prognostic factors within the Basal-like subset, only CD74 was independently prognostic and significant for both RFS and OS (Table 3). In addition, univariate analysis in the Basal-like subgroup showed that CD74/MHCII combined status was independently prognostic and significant for both RFS and OS (Table 4).
Figure 2. Overall Survival within entire cohort (All, left column) and Basal-like subgroup (Basal-like, right column) relative to status of CD74 (panels A and B), MHCII (panels C and D), and CD74/MHCII combined (panels E and F).
Kaplan Meier plots are shown with logrank test p values and n = total number of subjects within each curve.
Table 3. Relapse free survival and overall survival univariate log-rank and cox regression analysis for association of clinical parameters and CD74 or MHCII status in the Basal-like subgroup.
| A. Recurrence free survival | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| Parameter | Comparison | HR (95% CI) | p-value | HR (95% CI) | p-value |
| Age at diagnosis (yrs) | > 35 vs ≤ 35 | 0.71 (0.20–2.20) | 0.509 | 0.31 (0.09–1.11) | 0.072 |
| Tumour size (cm) | > 2 cm vs ≤ 2 cm | 2.71 (1.02–4.54) | 0.047 | 2.95 (0.82–10.67) | 0.100 |
| Nodal status | pos vs neg | 1.81 (0.90–3.37) | 0.101 | 2.33 (0.99–5.52) | 0.054 |
| Tumor grade | 2 vs 1 | 0.39 (0.15–1.41) | 0.186 | 0.43 (0.05–3.68) | 0.442 |
| 3 vs 1 | 1.31 (0.35–4.69) | 0.712 | 1.23 (0.50–3.00) | 0.652 | |
| MHCII expression | high vs low | 0.42 (0.23–1.02) | 0.058 | 0.61 (0.26–1.44) | 0.259 |
| CD74 expression | high vs low | 0.42 (0.23–0.86) | 0.018 | 0.25 (0.08–0.80) | 0.020 |
| B. Overall survival | Univariate | Multivariate | |||
| Parameter | Comparison | HR (95% CI) | p-value | HR (95% CI) | p-value |
| Age at diagnosis (yrs) | > 35 vs ≤ 35 | 0.78 (0.24–2.38) | 0.638 | 0.35 (0.10–1.20) | 0.095 |
| Tumour size (cm) | > 2 cm vs ≤ 2 cm | 1.60 (0.75–3.11) | 0.246 | 1.92 (0.63–5.86) | 0.251 |
| Nodal status | pos vs neg | 1.75 (0.90–3.16) | 0.107 | 1.82 (0.80–4.14) | 0.154 |
| Tumor grade | 2 vs 1 | 0.69 (0.23–2.19) | 0.552 | 0.39 (0.05–3.24) | 0.381 |
| 3 vs 1 | 1.05 (0.31–3.57) | 0.944 | 1.21 (0.51–2.91) | 0.665 | |
| MHCII expression | high vs low | 0.37 (0.23–0.91) | 0.028 | 0.82 (0.36–1.86) | 0.627 |
| CD74 expression | high vs low | 0.46 (0.26–0.89) | 0.022 | 0.28 (0.09–0.84) | 0.023 |
Table 4. Relapse free survival and overall survival univariate log-rank and cox regression analysis for association of clinical parameters and CD74/MHCII combined status in the Basal-like subgroup.
| A. Recurrence free survival | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| Parameter | Comparison | HR (95% CI) | p-value | HR (95% CI) | p-value |
| Age at diagnosis (yrs) | > 35 vs ≤ 35 | 0.79 (0.22–3.01) | 0.751 | 0.81 (0.15–4.34) | 0.809 |
| Tumour size (cm) | > 2 cm vs ≤ 2 cm | 2.88 (0.95–5.50) | 0.070 | 8.02 (1.01–63.78) | 0.049 |
| Nodal status | pos vs neg | 1.79 (0.78–3.92) | 0.155 | 2.66 (0.95–7.44) | 0.062 |
| Tumor grade | 2 vs 1 | 0.51 (0.12–2.82) | 0.501 | 2.72 (0.16–47.62) | 0.492 |
| 3 vs 1 | 1.07 (0.13–8.76) | 0.946 | 1.77 (0.63–4.97) | 0.278 | |
| CD74+MHCII expression | high vs low | 0.29 (0.17–0.90) | 0.031 | 0.29 (0.07–1.26) | 0.099 |
| B. Overall survival | Univariate | Multivariate | |||
| Parameter | Comparison | HR (95% CI) | p-value | HR (95% CI) | p-value |
| Age at diagnosis (yrs) | > 35 vs ≤ 35 | 0.77 (0.22–2.86) | 0.714 | 0.84 (0.17–4.18) | 0.829 |
| Tumour size (cm) | >2 cm vs ≤ 2 cm | 2.36 (0.88–4.72) | 0.100 | 3.73 (0.82–17.05) | 0.090 |
| Nodal status | pos vs neg | 1.59 (0.72–3.41) | 0.193 | 1.95 (0.75–5.07) | 0.172 |
| Tumor grade | 2 vs 1 | 0.56 (0.12–3.22) | 0.572 | 1.68 (0.14–20.55) | 0.686 |
| 3 vs 1 | 1.02 (0.14–7.59) | 0.983 | 1.75 (0.67–4.59) | 0.256 | |
| CD74+MHCII expression | high vs low | 0.26 (0.17–0.81) | 0.014 | 0.35 (0.11–1.18) | 0.091 |
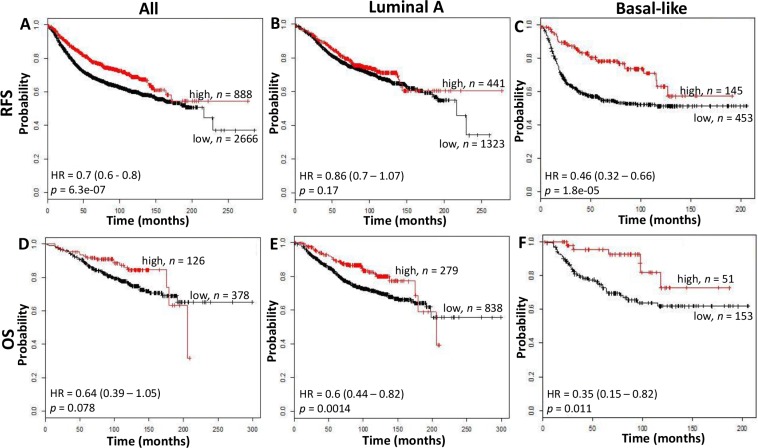
We conducted in-silico analysis of microarray gene expression data using an online survival analysis tool to validate the prognostic effect of CD74 in another cohort [9]. CD74 was prognostic for RFS but not OS within this overall cohort (RFS: p < 0.0001, OS: p = 0.078) and in the subset containing Basal-like subtype tumors (RFS: p < 0.0001; OS: p = 0.011), but not in Luminal A tumors (Figure 3) or other subtypes (data not shown).
Figure 3. The relation between CD74 and Relapse Free Survival (RFS, top row) and Overall Survival (OS, bottom row) was analyzed in a breast cancer cohort using the kmplotter tool.
RFS and OS were assessed in all cases (panels A and D), Luminal A subgroup (panels B and E), and Basal-like subgroup (panels C and F) using the Jetprobe set for CD74 (Affymetrix ID: 209619) and a cutpoint at the 75th expression quartile. Kaplan Meier plots are shown with hazard ratios (HR) and logrank test p values and n = total number of subjects within each curve.
Relation between CD74 and MHCII
We next examined the relation of CD74 with MHCII expression. Expression of MHCII within tumor cells showed a predominantly cytoplasmic staining pattern, but with membrane staining visible in some cells, and was relatively homogeneous within positive staining tumors. High expression of MHCII was present in 89 (20%) cases. CD74 expression was closely correlated with MHCII expression in the overall cohort (p < 0.0001) and also within all subtypes, and this association was significant in Luminal A (p < 0.0001), Luminal B (p = 0.0002), Her2 (p = 0.011) and Basal-like subsets (p = 0.005).
MHCII was also associated with patient age, tumor grade, and ER status (p < 0.0001) (Table 2). High levels of MHCII were present in 11% of Luminal A subtype tumors in comparison with significantly higher proportions of Luminal B (24%, p = 0.020), TNNB (35%, p = 0.0004) and Basal-like subtype tumors (43%, p < 0.0001).
Relation between CD74 and MHCII and intratumoral immune response
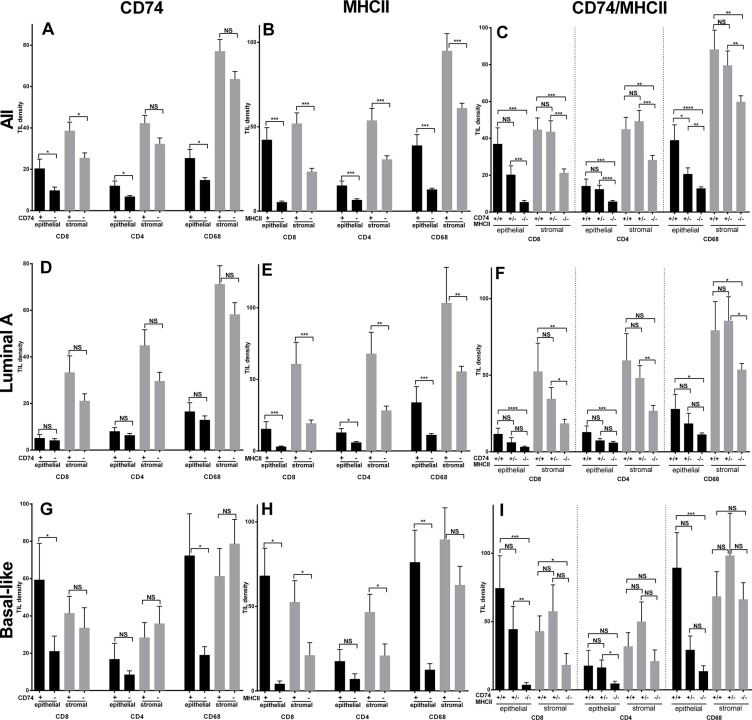
The intratumoral immune response was assessed by analysis of CD8, CD4, and CD68 infiltrates. In the entire cohort the TIL densities in intra-epithelial versus intra-stromal areas were lower but closely correlated and the mean (standard deviation) densities were as follows; CD8 –12 (33) vs 30 (48), CD4–8 (16) vs 36 (46), CD68–17 (27) vs 66 (59). High levels of CD74 were associated with higher mean densities of CD8, CD4, and CD68 TIL in the entire cohort within both epithelium and stroma, and this was significant for all three TIL types in epithelial areas but only for CD8 in stroma (Figure 4).
Figure 4. Tumor infiltrating leucocyte (TIL) cell densities in entire cohort (All, top row) and Luminal A subgroup (middle row) and Basal-like subgroup (bottom row) relative to positive and negative status of CD74 (panels A, D, G), MHCII (panels B, E, H), and CD74/MHCII combined (panels C, F, I).
Bars represent means +/– standard error. Black and grey bars represent intra-epithelial and intra-stromal densities of CD8, CD4, and CD68 positive TIL respectively. NS = no significance, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
Given the well documented differences between Luminal A and Basal-like type tumors with respect to genomic mutational signatures and intratumoral inflammation, and in this study with respect to CD74 and MHCII expression, we focused additional analysis on these two subgroups. Within the Luminal A subset, there were no significant differences in TIL densities between CD74 positive versus negative categories. However in the Basal-like subset of tumors, CD8 and CD68 were significantly higher in CD74 high tumors (p < 0.034 and p < 0.003) in the epithelial compartment. High levels of MHCII were also associated with higher CD8, CD4, and CD68 TIL and this was statistically significant for epithelium and stromal areas across tumor groups (i.e. all cases), and in both Luminal A, and Basal-like tumors. Further analysis of TIL infiltrates was conducted in relation to dual positive status for both CD74 and MHCII as compared to mixed status and double negative status. Tumors associated with combined CD74/MHCII positive status also showed much higher TIL levels in dual positive compared to mixed or dual negative status tumors across the entire cohort and within Luminal A and Basal-like subgroups (Figure 4). This TIL pattern was mirrored by RFS and OS curves of subgroups with combined positive, mixed, and dual negative status (Figure 2). Analysis of the relation between CD74 and MHCII expression and TIL densities within other subtypes showed comparable trends but only some weak positive associations with respect to individual TIL densities (data not shown).
DISCUSSION
We have shown that CD74 expression is associated with better prognosis in Basal-like subtype invasive breast cancer. This association correlates with higher levels of MHCII expression by tumor cells and with a dense TIL response.
CD74 is an important component in the functional presentation of MHCII restricted antigens, a key factor in anti-tumor immunity [11–13]. The MHCII complex is stabilized by CD74 in the endoplasmic reticulum allowing subsequent MHCII restricted peptide presentation at the cell surface. It has been shown that CD74 regulates the repertoire of antigens presented by MHCII [14–16] and that this leads to primary engagement of CD4 and then CD8 components of the adaptive immune response, leading to tumor rejection [17–19]. However CD74 is not required for the entire MHCII antigen presenting function and cells lacking CD74 show differences in antigen presentation of only some peptides [20]. The significance in terms of tumor recognition and rejection of this subset of peptides remains to be determined [11, 14]. CD74 can also perform a different function as the receptor for MIF [8]. MIF is a pleiotropic cytokine produced by many cell types that is associated with promotion of tumor growth and invasiveness [21] through effects on both breast epithelial and immune cells [1]. Finally, CD74 can be a target of regulated intramembrane proteolysis by the CatS protease resulting in release of the CD74 intracellular domain. This in turn leads to activation of transcription of chemokines such as CCL2 which is associated with poor prognosis in breast and other cancers [3, 4]. Therefore these different CD74 functions are contradictory with respect to effects on tumor outcome.
Our data here confirms the expected close association between CD74 (Li) and MHCII (HLADR) shown in previous studies [10, 22]. These studies, based on relatively small cohorts (n = 52 and n = 112 cases) also identified similar relationships between CD74 (and HLADR) and CD3 TIL [22], and high tumor grade. In outcomes analysis that included incorporation of the expression status of HLADM, a second MHCII processing chaperone molecule, co-expression of HLADR/CD74/HLADM conferred a better prognosis than tumors with partial or negative status for all three [10]. However this finding was in contrast to another more recent and larger study showing that associations with poor prognostic factors and in particular with the triple negative subset were observed [9]. This latter association has also been suggested by others [23, 24]. However the Tian study [9] included up to 20% of cases receiving neoadjuvant chemotherapy which may have influenced CD74 expression and complicated outcomes analysis and did not examine correlations with the immune response. Follow-up data was also limited to approximately 5 years. Our current study confirms previous observations that CD74 is closely associated with MHCII, TIL, and several poor prognostic factors. While there was no association with overall survival in the entire cohort, CD74 conferred better outcome in the Basal-like subtype category. Our results therefore suggest that the anti tumor effect predicted to be exerted by CD74's role in facilitating the immune response is dominant over the protumor effect involving MIF signal transduction, expected from laboratory studies on MIF [25–28].
The close association between combined CD74/MHCII expression on tumor cells and the presence of TIL across the entire cohort suggest that a functional relationship exists with antigen presentation on tumor cells. As might be expected, this relationship was present for TIL in general but was strongest for intra-epithelial rather than stromal TIL [29]. TIL assessment has recently emerged as a strong prognostic factor and recent recommendations have focused on scoring stromal TIL. However it should be noted that this is in the context of an ‘immunoscore’ based on H&E stained sections where distinction of intra-epithelial TIL is challenging and TIL subsets cannot be delineated. The role of CD74 in regulating the CCL2 cytokine may also be relevant to the association with TIL, especially macrophages [4, 30].
Dual CD74/MHCII positive status is only positively prognostic in the Basal-like tumor subgroup. One interpretation of our observation is that the functional implication of CD74/MHCII expression in terms of activation of immune responses is common to all subtypes but that tumor susceptibility to the immune system is peculiar to Basal-like tumor cells. The absence of a prognostic significance in the other triple negative non-basal subset may be attributable to the small subgroup size. It should also be noted that while subgroup assignment based only on immunohistochemistry can distinguish prognostic subgroups [31] it has limitations in terms of specificity [32, 33]. While the broad class of triple negative breast tumors is known to be associated with inflammatory infiltrates [34], the triple negative category is very heterogeneous and varies widely in clonal frequency [35]. Tumors of the Basal-like subtype show the most variation in clonal clusters at time of diagnosis, the highest mutation frequency amongst breast intrinsic subtypes, and have the lowest frequency of recurrent mutation in common genes [35, 36]. It might be speculated that these features promote recruitment and activation of multiple T cell clones and contribute to the high TIL density that is a feature of these tumors [37–39]. Both TIL density and immune gene expression signatures have been identified by several groups within Basal-like tumors to correlate with better outcomes [40, 41].
In conclusion, interpretation of our findings should be qualified by the relatively small Basal-like subsets in study and validation cohorts. However, overall these observations are consistent with the view that CD74 expression in tumor cells promotes the intratumoral immune response and is associated with a better prognosis.
MATERIALS AND METHODS
Case cohort
A cohort of 492 breast cancer cases was studied representing primary tumors collected by the Manitoba Breast Tumor Bank at time of diagnosis and initial surgical intervention. Age at diagnosis, tumor grade, size, nodal status, and outcomes in terms of relapses, and deaths were recorded [42]. All tumors were histologically classified and graded by one pathologist (PHW). The time of diagnosis and accrual by the bank (1988–1995) predated current biomarker assays. Therefore immunohistochemistry (IHC) was previously performed by the Bank using an auto-immunostainer (Discovery Staining Module, Ventana Medical Systems, AZ, USA) on TMA sections from the cohort for ER, PR, Ki67, CK5/6, EGFR and Her2 biomarkers. ER, PR, and Her2 were scored and positive status assigned according to ACP guidelines. Ki67, CK5/6 and EGFR were also scored and positive status assigned as > 14% (Ki67) or any positive tumor cell staining (CK5/6 and EGFR). On the basis of the IHC determined expression of these five biomarkers the cohort was classified by the Bank into five intrinsic molecular subtypes: Luminal A (ER+/Ki67–/Her–), Luminal B (ER+/Ki67+/Her–), Her2 (Her2+), Triple Negative Non-basal (TNNB, ER–/PR–/Her–/CK5/6–/EGFR–), and Basal-like (ER-/PR-/Her- and either CK5/6+ and/or EGFR+ [31, 43]. The Bank operates with approval of the University of Manitoba Biomedical Research Ethics Board and this research study was conducted under approval from the BC Cancer Agency Research Ethics Board. A report concerning the source of the biospecimens and data used according to the BRISQ guidelines [44] is provided in Supplementary Table 1. In addition, validation studies were performed on a second cohort using an online survival analysis tool based on gene expression and outcomes data from 3554 patients (available at www.kmplot.com) [45].
Tissue microarray (TMA) construction
Primary tumors were represented in tissue microarrays (TMAs) compiled by the Tumor Bank. To construct a TMA, all cases were initially selected from the database and then sections were re-reviewed to confirm and select areas for coring of corresponding blocks. Duplicate tissue cores (0.6 mm diameter) were taken from central cellular areas of each tumor with a tissue arrayer instrument (Beecher Instruments, Silver Spring, MD, USA).
Immunohistochemistry and TMA scoring
CD74, MHCII, CD8, CD4, CD68, was also performed on deparaffinized sections from TMAs using a Biocare Medical Intellipath FLX autostainer using reagents from Biocare (Concord, CA) unless otherwise noted. Slides were deparaffinized manually through xylene and graded alcohols then antigen retrieval performed in Biocare's decloaking chamber using Diva decloaking solution for 125°C for 30 seconds. Slides were loaded into the Intellipath FLX, subjected to non-specific blocking with Peroxidased-1 and background sniper then incubated with either CD74, MHCII, CD8, CD4 or CD68 (Supplementary Table 3) in Da Vinci Green diluents for 30 minutes at room temperature. The slides were then incubated with either Mach2 Mouse-(CD74, MHCII, CD8) or Rabbit-(CD4, CD68)-HRP polymer for 30 minutes at room temperature and then detected with IP DAB for 5 minutes followed by counterstaining with a 1:10 dilution of CAT hematoxylin, air drying and coverslipping with Ecomount.
IHC Scoring was performed in a blinded fashion by an experienced breast histopathologist (PHW). Immunostained TMA sections were initially assessed at low magnification to select the core with the highest density of positive cells. The two types of biomarker (CD74 and MHCII expressed by tumor epithelial cells and TIL markers indicating immune cell subsets) were assessed. Only CD74 staining within tumor cells was scored and this could be discriminated as a relatively distinct signal within individual cells and so was assessed by H score method whereby the expression is quantitated as the product of staining intensity (ranked from 0 to 3) and proportion of positive staining tumor cells (0 to 100%) to give an expression score range from 0 to 300; For analysis CD74 tumor cell expression was categorized into low or high expression levels based on the upper quartile (CD74 H scores ≥ 20; corresponding to 1+ intensity in ≥ 20% cells or 2+ intensity in ≥ 10% cells) to avoid any confusion with isolated infiltrating TIL within epithelial areas. Only MHCII staining within tumor cells was scored and this signal was relatively diffuse and so was assessed by assigning an expression score on a 4 point scale (0 to 3+) with 0 = absent, 1 = weak intensity/less than 10% cells, 2 = moderate intensity/10–50% cells, 3 = strong intensity/50–100% cells. As with CD74 expression, in order to avoid any confusion with TIL within epithelial areas, MHCII tumor cell expression status was categorized as low or high based on the upper quartile (MHCII scores (> 1). CD8, CD4, CD68 tumor infiltrating leucocytes (TIL) were assessed as described previously [46] by direct counting up to 20 cells or by estimation when in excess of this number (IHC score, range 0–100) within the selected core area. The area of the entire core occupied by tumor epithelium versus stroma was then assessed followed by estimation of the proportion of positive TIL that were intra-epithelial or intra-stromal (intra-epithelial localization, was defined as lymphocytes within tumor cell nests and/or adjacent to and in direct contact with tumor cells). Intra-epithelial and intra-stromal TIL density per core was then calculated for each type of TIL and for each case.
Statistical analysis
Associations of CD74 and MHCII with clinical-pathological features were evaluated using Fisher's exact test. Associations of CD74 with TILs were evaluated using t test. Survival was calculated using the Kaplan-Meier method and curves were compared with the log-rank test. Multivariate survival analyses were done using Cox regression analysis. All statistical tests were two-sided with significance established at p-values less than 0.05. Statistical analyses were performed using Graphpad Prism 6.0 (GraphPad, La Jolla, CA, USA) and SPSS statistics 17 (SPSS, Chicago, IL, USA).
SUPPLEMENTARY TABLES
ACKNOWLEDGMENTS AND FUNDING
This research was funded by the Canadian Cancer Society (Grant #702375). Clinical specimens were provided and/or processed by the Manitoba Breast Tumor Bank and the BC Cancer Agency Tumor Tissue Repository, both associated with the Canadian Tumor Repository Network (CTRNet).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Richard V, Kindt N, Saussez S. Macrophage migration inhibitory factor involvement in breast cancer (Review) Int J Oncol. 2015;47:1627–1633. doi: 10.3892/ijo.2015.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borghese F, Clanchy FI. CD74: an emerging opportunity as a therapeutic target in cancer and autoimmune disease. Expert Opin Ther Targets. 2011;15:237–251. doi: 10.1517/14728222.2011.550879. [DOI] [PubMed] [Google Scholar]
- 3.Becker-Herman S, Arie G, Medvedovsky H, Kerem A, Shachar I. CD74 is a member of the regulated intramembrane proteolysis-processed protein family. Mol Biol Cell. 2005;16:5061–5069. doi: 10.1091/mbc.E05-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson RDA, Magorrian SM, Williams R, Young A, Small DM, Scott CJ, Burden RE. CCL2 is transcriptionally controlled by the lysosomal protease cathepsin S in a CD74-dependent manner. Oncotarget. 2015;6:29725–29739. doi: 10.18632/oncotarget.5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang RF. The role of MHC class II-restricted tumor antigens and CD4+ T cells in antitumor immunity. Trends Immunol. 2001;22:269–276. doi: 10.1016/s1471-4906(01)01896-8. [DOI] [PubMed] [Google Scholar]
- 6.Bucala R, Shachar I. The integral role of CD74 in antigen presentation, MIF signal transduction, and B cell survival and homeostasis. Mini Rev Med Chem. 2014;14:1132–1138. doi: 10.2174/1389557515666150203144111. [DOI] [PubMed] [Google Scholar]
- 7.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian B, Zhang Y, Li N, Liu X, Dong J. CD74: a potential novel target for triple-negative breast cancer. Tumour Biol. 2012;33:2273–2277. doi: 10.1007/s13277-012-0489-x. [DOI] [PubMed] [Google Scholar]
- 10.Oldford SA, Robb JD, Codner D, Gadag V, Watson PH, Drover S. Tumor cell expression of HLA-DM associates with a Th1 profile and predicts improved survival in breast carcinoma patients. Int Immunol. 2006;18:1591–1602. doi: 10.1093/intimm/dxl092. [DOI] [PubMed] [Google Scholar]
- 11.Accolla RS, Lombardo L, Abdallah R, Raval G, Forlani G, Tosi G. Boosting the MHC Class II-Restricted Tumor Antigen Presentation to CD4+ T Helper Cells: A Critical Issue for Triggering Protective Immunity and Re-Orienting the Tumor Microenvironment Toward an Anti-Tumor State. Front Oncol. 2014;4:32. doi: 10.3389/fonc.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Accolla RS, Tosi G. Optimal MHC-II-restricted tumor antigen presentation to CD4+ T helper cells: the key issue for development of anti-tumor vaccines. J Transl Med. 2012;10:154. doi: 10.1186/1479-5876-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thibodeau J, Bourgeois-Daigneault MC, Lapointe R. Targeting the MHC Class II antigen presentation pathway in cancer immunotherapy. Oncoimmunology. 2012;1:908–916. doi: 10.4161/onci.21205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chornoguz O, Gapeev A, O'Neill MC, Ostrand-Rosenberg S. Major histocompatibility complex class II+ invariant chain negative breast cancer cells present unique peptides that activate tumor-specific T cells from breast cancer patients. Mol Cell Proteomics. 2012;11:1457–1467. doi: 10.1074/mcp.M112.019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuzaki J, Tsuji T, Luescher I, Old LJ, Shrikant P, Gnjatic S, Odunsi K. Nonclassical antigen-processing pathways are required for MHC class II-restricted direct tumor recognition by NY-ESO-1-specific CD4(+) T cells. Cancer Immunol Res. 2014;2:341–350. doi: 10.1158/2326-6066.CIR-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuzaki J, Tsuji T, Luescher IF, Shiku H, Mineno J, Okamoto S, Old LJ, Shrikant P, Gnjatic S, Odunsi K. Direct tumor recognition by a human CD4(+) T-cell subset potently mediates tumor growth inhibition and orchestrates anti-tumor immune responses. Sci Rep. 2015;5:14896. doi: 10.1038/srep14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortara L, Castellani P, Meazza R, Tosi G, De Lerma Barbaro A, Procopio FA, Comes A, Zardi L, Ferrini S, Accolla RS. CIITA-induced MHC class II expression in mammary adenocarcinoma leads to a Th1 polarization of the tumor microenvironment, tumor rejection, and specific antitumor memory. Clin Cancer Res. 2006;12:3435–3443. doi: 10.1158/1078-0432.CCR-06-0165. [DOI] [PubMed] [Google Scholar]
- 18.Ekkirala CR, Cappello P, Accolla RS, Giovarelli M, Romero I, Garrido C, Garcia-Lora AM, Novelli F. Class II transactivator-induced MHC class II expression in pancreatic cancer cells leads to tumor rejection and a specific antitumor memory response. Pancreas. 2014;43:1066–1072. doi: 10.1097/MPA.0000000000000160. [DOI] [PubMed] [Google Scholar]
- 19.Shi B, Vinyals A, Alia P, Broceno C, Chen F, Adrover M, Gelpi C, Price JE, Fabra A. Differential expression of MHC class II molecules in highly metastatic breast cancer cells is mediated by the regulation of the CIITA transcription Implication of CIITA in tumor and metastasis development. Int J Biochem Cell Biol. 2006;38:544–562. doi: 10.1016/j.biocel.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Thompson JA, Dissanayake SK, Ksander BR, Knutson KL, Disis ML, Ostrand-Rosenberg S. Tumor cells transduced with the MHC class II Transactivator and CD80 activate tumor-specific CD4+ T cells whether or not they are silenced for invariant chain. Cancer Res. 2006;66:1147–1154. doi: 10.1158/0008-5472.CAN-05-2289. [DOI] [PubMed] [Google Scholar]
- 21.Lue H, Thiele M, Franz J, Dahl E, Speckgens S, Leng L, Fingerle-Rowson G, Bucala R, Luscher B, Bernhagen J. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26:5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]
- 22.Oldford SA, Robb JD, Watson PH, Drover S. HLA-DRB alleles are differentially expressed by tumor cells in breast carcinoma. Int J Cancer. 2004;112:399–406. doi: 10.1002/ijc.20441. [DOI] [PubMed] [Google Scholar]
- 23.Metodieva G, Nogueira-de-Souza NC, Greenwood C, Al-Janabi K, Leng L, Bucala R, Metodiev MV. CD74-dependent deregulation of the tumor suppressor scribble in human epithelial and breast cancer cells. Neoplasia. 2013;15:660–668. doi: 10.1593/neo.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwood C, Metodieva G, Al-Janabi K, Lausen B, Alldridge L, Leng L, Bucala R, Fernandez N, Metodiev MV. Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. J Proteomics. 2012;75:3031–3040. doi: 10.1016/j.jprot.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 25.Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J Immunol. 2006;177:8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- 26.Conroy H, Mawhinney Donnelly SC. Inflammation and cancer: macrophage migration inhibitory factor (MIF)—the potential missing link. QJM. 2010;103:831–836. doi: 10.1093/qjmed/hcq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi S, Kim HR, Leng L, Kang I, Jorgensen WL, Cho CS, Bucala R, Kim WU. Role of macrophage migration inhibitory factor in the regulatory T cell response of tumor-bearing mice. J Immunol. 2012;189:3905–3913. doi: 10.4049/jimmunol.1102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Yan L, Kim JA. Modulating mammary tumor growth, metastasis and immunosuppression by siRNA-induced MIF reduction in tumor microenvironment. Cancer Gene Ther. 2015;22:463–474. doi: 10.1038/cgt.2015.42. [DOI] [PubMed] [Google Scholar]
- 29.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F, Perez EA, Thompson EA, Symmans WF, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steiner JL, Murphy EA. Importance of chemokine (CC-motif) ligand 2 in breast cancer. Int J Biol Marker. 2012;27:179–185. doi: 10.5301/JBM.2012.9345. [DOI] [PubMed] [Google Scholar]
- 31.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, Heikkila P, Heikkinen T, Nevanlinna H, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahreini F, Soltanian AR, Mehdipour P. A meta-analysis on concordance between immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) to detect HER2 gene overexpression in breast cancer. Breast cancer. 2015;22:615–625. doi: 10.1007/s12282-014-0528-0. [DOI] [PubMed] [Google Scholar]
- 33.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thurlimann B, Senn HJ, Panel M. -Tailoring therapies-improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann Oncol. 2015;2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto H, Koo SL, Dent R, Tan PH, Iqbal J. Role of inflammatory infiltrates in triple negative breast cancer. J Clin Pathol. 2015;68:506–510. doi: 10.1136/jclinpath-2015-202944. [DOI] [PubMed] [Google Scholar]
- 35.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran E, Rosenberg SA. T-cell therapy against cancer mutations. Oncotarget. 2014;5:4579–4580. doi: 10.18632/oncotarget.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, Wunderlich JR, Somerville RP, Rosenberg SA. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015 doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Disis ML, Stanton SE. Triple-negative breast cancer: immune modulation as the new treatment paradigm. Am Soc Clin Oncol Educ Book. 2015:e25–30. doi: 10.14694/EdBook_AM.2015.35.e25. [DOI] [PubMed] [Google Scholar]
- 40.Jezequel P, Loussouarn D, Guerin-Charbonnel C, Campion L, Vanier A, Gouraud W, Lasla H, Guette C, Valo I, Verriele V, Campone M. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015;17:43. doi: 10.1186/s13058-015-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West NR, Milne K, Truong PT, Macpherson N, Nelson BH, Watson PH. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13:R126. doi: 10.1186/bcr3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson PH, Snell L, Parisien M. The NCIC-Manitoba Breast Tumor Bank: a resource for applied cancer research. CMAJ. 1996;155:281–283. [PMC free article] [PubMed] [Google Scholar]
- 43.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 44.Moore HM, Kelly AB, Jewell SD, McShane LM, Clark DP, Greenspan R, Hayes DF, Hainaut P, Kim P, Mansfield E, Potapova O, Riegman P, Rubinstein Y, et al. Biospecimen reporting for improved study quality (BRISQ) J Proteome Res. 2011;10:3429–3438. doi: 10.1021/pr200021n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 46.West NR, Panet-Raymond V, Truong PT, Alexander C, Babinszky S, Milne K, Ross LA, Loken S, Watson PH. Intratumoral Immune Responses Can Distinguish New Primary and True Recurrence Types of Ipsilateral Breast Tumor Recurrences (IBTR). Breast Cancer. 2011;5:105–115. doi: 10.4137/BCBCR.S7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.