Abstract
Immunoglobulin heavy chain (IgH) alleles have ambivalent relationships: they feature both allelic exclusion, ensuring monoallelic expression of a single immunoglobulin (Ig) allele, and frequent inter-allelic class-switch recombination (CSR) reassembling genes from both alleles. The IgH locus 3′ regulatory region (3′RR) includes several transcriptional cis-enhancers promoting activation-induced cytidine deaminase (AID)-dependent somatic hypermutation (SHM) and CSR, and altogether behaves as a strong super-enhancer. It can also promote deregulated expression of translocated oncogenes during lymphomagenesis. Besides these rare, illegitimate and pathogenic interactions, we now show that under physiological conditions, the 3′RR super-enhancer supports not only legitimate cis-, but also trans-recruitment of AID, contributing to IgH inter-allelic proximity and enabling the super-enhancer on one allele to stimulate biallelic SHM and CSR. Such inter-allelic activating interactions define transvection, a phenomenon well-known in drosophila but rarely observed in mammalian cells, now appearing as a unique feature of the IgH 3′RR super-enhancer.
Keywords: super-enhancer, alleles, transvection, nuclear positioning, gene regulation
INTRODUCTION
Interactions and co-localization of non-homologous genes are favored by super-enhancers, i.e. major cis-regulatory elements controlling key cell identity genes [1–4]. In B-cells, the specific and frequent co-localization of immunoglobulin heavy chain (IgH) genes, Ig κ light chain genes (Igκ) and J-chain genes (IgJ) has been reported at all stages of B-cell differentiation including plasma cells where high expression of these loci often involves the same transcription factories and a nuclear organization that may optimize the nuclear export of their transcripts and further translation [1, 3, 4]. Functional interactions between those co-localized non-homologous genes occur at the level of transcription, with the 3′Eκ enhancer positively influencing IgH transcription [3], but such effects were not evaluated between alleles of each locus.
Stimulatory interactions between homologous paired alleles, defining transvection, are well known in drosophila [5], but rarely observed in mammals except in pathologic or artificial settings. For example, transvection was documented between an IgH-translocated CCND1 gene and its non-translocated counterpart [6]. Manipulating the mouse genome also yielded transvection from a mutant paternal and expressed Igfr2 allele which triggered expression of an otherwise silent maternal allele [7]. Inhibitory trans-regulation, supporting allelic exclusion, is by contrast frequent in mammalian physiology. Regarding Ig genes, ordered V(D)J recombination initiates allelic exclusion during early B cell development, when a functional rearrangement shuts off accessibility of the second allele, which correlates with modified epigenetic marks, interactions of IgH/Igκ loci, and changes in nuclear location [8, 9].
In mature activated B-cells, localization close to the nuclear periphery and co-localization with Igκ and IgJ genes preferentially mark the functionally rearranged IgH allele [3]. However, inter-allelic contacts are also repeatedly identified in 3C, 4C, and FISH-3D [3, 9, 10]. In parallel, it is noticeable that both transcription and AID-dependent IgH changes such as class switch recombination (CSR) and somatic hypermutation (SHM) usually affect both productive and non-productive alleles at similar levels [9, 11, 12].
In the IgH locus, the role of the 3′ regulatory region (3′RR) enhancers goes beyond transcription and also features recruitment of AID for CSR, SHM or deletional silencing of constant genes through locus suicide recombination [13–17]. The 3′RR super-enhancer features a high density of individual enhancers [15], eRNA transcription [13, 18], regulation by long non-coding RNA [4], active chromatin marks and the ability to promote higher-order chromosomal structures [19]. Its polymorphisms influence susceptibility to immune disorders, while it endows transgenes with B-cell specific expression and can also deregulate oncogenes [20–23]. As any cis-regulatory element, the 3′RR controls covalently linked genes within a legitimate territory bounded by insulators, and even the deregulation of oncogenes secondary to translocation falls into bona fide “cis” effects. It is by contrast unclear whether the gene co-localization observed in B-cells has functional implications in trans.
In this regard, while illegitimate off-target CSR-like junctions with non Ig genes remain exceptional during AID expression, legitimate junctions repairing CSR breaks are known to occur actively in both cis and trans [24–27]. Promiscuous trans-CSR junctions strongly suggest that IgH biallelic co-localization has functional implications at stages where proximity might secure legitimate synapsis between concomitantly generated biallelic DNA breaks. In order to explore whether these functional interactions precede trans-CSR, we thus searched for inter-allelic stimulatory interactions reminiscent to transvection, and witnessed pairing between homologous and legitimate AID target alleles. Since the 3′RR is a regulator of both long-range interactions and AID recruitment, we considered that inter-allelic proximity may yield legitimate effects in trans and in the absence of covalent linkage. Here we indeed demonstrate 3′RR-dependent IgH trans-interactions in B-cells with either additional copies of the 3′RR linked to transgenes or, by contrast, with homozygous or hemizygous loss of the 3′RR. The latter configuration also reveals the ability of the 3′RR super-enhancer to yield DNA accessibility and recruit AID not only in cis but also in trans.
RESULTS
3′RR-dependent interactions of IgH transgenes with of endogenous IgH genes
IgH-Igκ inter-chromosomic pairing has been shown to occur at all stages of B-cell differentiation (except pro-B), while IgH-IgH inter-allelic interactions were evaluated during V(D)J recombination in developing B-cells [10] and plasma cells [3]. We checked and validated that pairing of IgH alleles can also be detected by 3D FISH in mature resting and in vitro activated B-cells (Figure 1A).
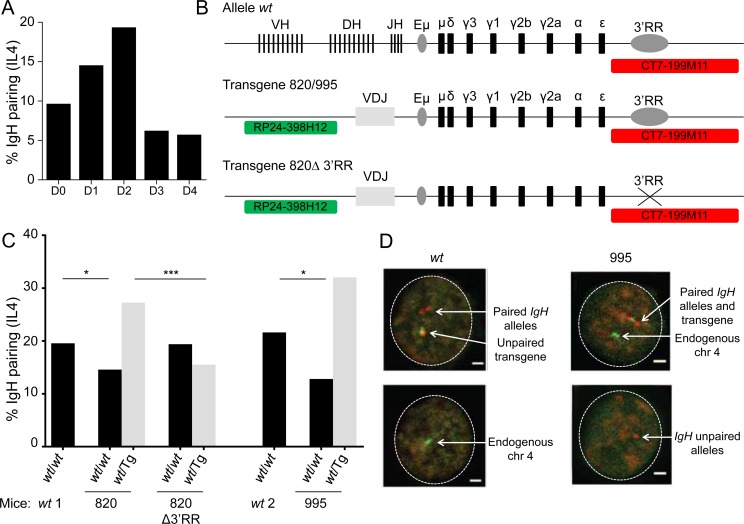
Figure 1. 3′RR dependent IgH pairing in wt and transgenic B-cells.
A. Kinetic of IgH alleles pairing in in vitro activated wt B-cells. Wt B splenocytes were stimulated 4 days in vitro with anti-CD40 + IL4 and IgH localization was studied by 3D FISH. IgH pairing (distance ≤ 1 µm) was quantified in resting (day 0) and activated B cells (D1, D2, D3, D4). Bars represent % of pairing from 2 experiments with at least 585 cells analyzed. B. Schematic representation of alleles and probes used for 3D FISH experiments (not scaled). Wt alleles contain all V(D)J genes, constant genes and 3′RR, transgenes include a rearranged VDJ gene, constant genes and 3′RR (820 and 995 transgenes) or not (820Δ transgene). Probes are specific for IgH (CT7-199M11, red) or for transgene integration site (RP24-398H12, green, chromosome 4). C. IgH pairing in in vitro activated B lymphocytes. Cells were analyzed at day 2 and IgH pairing (distance ≤ 1 µm) between wt and wt alleles (black) or between wt and transgenic alleles (grey) were quantified by 3D FISH. In a first set of experiments wt cells (wt1) were compared to 820 and 820Δ transgenic cells and in a separated experiment wt cells (wt2) were compared to 995 transgenic cells. Bars represent % of pairing from 2 experiments, with at least 50 cells per group. DNA probes used were BAC CT7-199M11 for 3´ IgH and BAC RP24-221C18 close to the insertion site of the IgH transgene on chromosome 7 or BAC RP24-398H12, which is located adjacent to the insertion site of the IgH transgene on chromosome 4. Fisher exact test for significance. D. Representative nuclei from 3D FISH experiments on in vitro activated B-cells (day 2). Examples of pairing between endogenous IgH alleles in wt cells and between transgene and IgH alleles in 995 transgenic cells are shown. IgH loci are in red, transgene and endogenous chromosome 4 are in green. Scale bar: 1µm.
To determine whether pairing of homologous IgH sequences depends upon the chromosomal context, we analyzed mice which carry an IgH transgene either as one copy on chromosome 7 (mouse line 820) or 3 copies on chromosome 4 (mouse line 995). This large IgH transgene included the IgH 3′RR (Figure 1B) [28]. In primary B-cells cultured in vitro (after 48h anti-CD40 + IL-4 stimulation of splenic cells from the 820 strain), we observed frequent pairing of the endogenous IgH locus with the IgH transgene. Pairing with the transgene reached a higher level (because there are two copies of the endogenous alleles with which it can pair) and competed with pairing of endogenous IgH alleles (Figure 1C and 1D). Similar results were obtained with cells from the second (995) transgenic mouse strain (Figure 1C and 1D). Together these experiments demonstrate that homologous IgH alleles or transgenes pair independent of chromosomal context.
The 3′ enhancer is required for pairing of an IgH transgene with the endogenous locus
We next examined splenic B cells from 820Δ mice, which bear the 820 transgene after cre-mediated deletion of the entire 3′RR [28]. Consistent with previous analyses, we observed no cell surface expression of Ig encoded by the rearranged transgene in activated splenic B cells under switching conditions (Figure S1A). Further, the 820Δ cells expressed 3-fold less transgenic germline transcripts compared to 820 cells (Figure S1B). Importantly by comparison to the parental 820 strain cells, pairing of the mutant transgene with endogenous IgH alleles was clearly reduced, while pairing between both endogenous IgH alleles was restored to normal levels (Figure 1C). Thus, the ability of an IgH transgene to pair with the homologous endogenous locus strongly depends on its inclusion of the 3´RR.
3′RR-dependent nuclear positioning of endogenous IgH loci
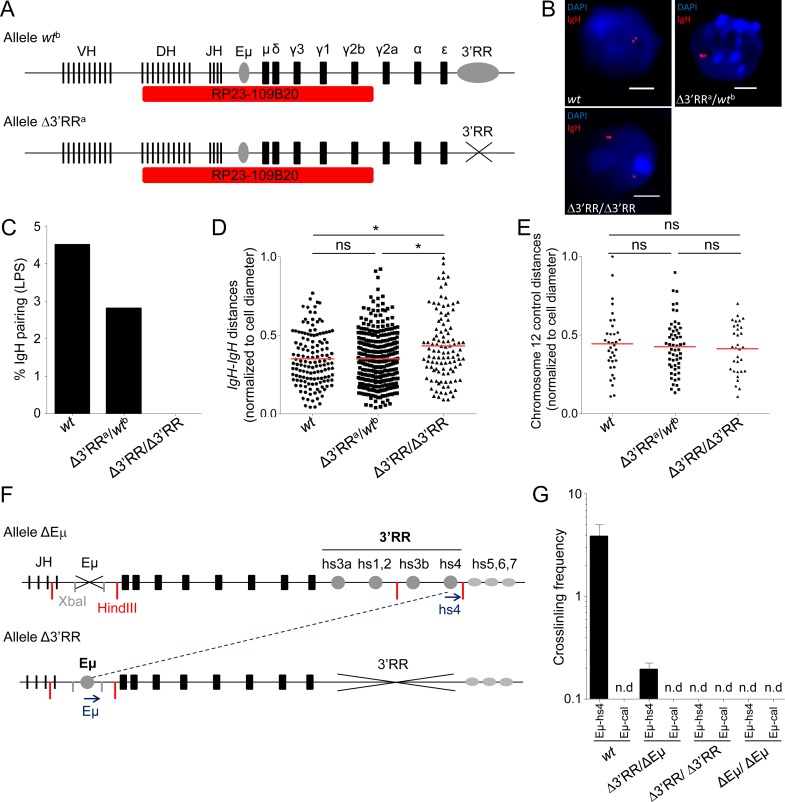
To evaluate whether inter-allelic pairing of endogenous IgH loci also relied on the 3′RR, inter-IgH distance was compared by 3D-FISH in wt, Δ3′RRa/wtb and homozygous Δ3′RRa/Δ3′RRa in vitro activated B-cells (72h with LPS) (Figure 2A and 2B). As for close inter-allelic interactions ≤ 1µm, none was detectable in Δ3′RR cells; they by contrast showed up in heterozygous cells, with a single 3′RR being sufficient to bring both IgH alleles into contact as in wt cells, albeit at a lower frequency (Figure 2C). In addition, the distribution of the IgH-IgH distance was clearly shifted toward increased distances for homozygous Δ3′RR B-cells compared to wt (p = 0.037) and Δ3′RRa/wtb (p = 0.024) (Figure 2D). A control experiment with a distant non-IgH probe from the same chromosome 12 showed no variation in the inter-chromosomal distance, demonstrating that effects of the 3′RR deletion do not extend beyond the IgH locus (Figure 2E).
Figure 2. 3′RR-dependent IgH inter-allelic interactions in wt, 3′RR hemizygous and 3′RR-deficient B cells.
A. Schematic representation of IgH alleles and probe (RP23-109B20, red) used for 3D FISH experiments (not scaled). Wtb allele includes the 3′RR, contrary to the Δ3′RRa allele. B. Representative nuclei from 3D FISH experiments on in vitro LPS activated B-cells (day 3). Examples of pairing observed in wt and Δ3′RRa/wtb cells, Δ3′RR/Δ3′RR failed to pair their IgH alleles. DAPI is in blue, IgH loci are in red. Scale bars: 4µm. C. IgH pairing in in vitro LPS activated B lymphocytes. Cells were analyzed at day 3 and IgH pairing (distance ≤ 1 µm) between alleles was quantified by 3D FISH. Bars represent % of pairing from at least 2 experiments, corresponding to 138 (wt), 107 (Δ3′RR) and 324 (Δ3′RRa/wtb) cells analyzed. D. Distribution of IgH inter-allelic distance in activated B-cells (with normalization according to cell diameter) from wt, Δ3′RR and Δ3′RRa/wtb cells are shown (from the same cells as in 2C). Mean is shown and KS test for significance. E. Distribution of inter-allelic distance (with normalization according to cell diameter) analyzed for a non-IgH chromosome 12 locus (probe RP23-436M24) in activated B-cells from wt (n = 35), Δ3′RR (n = 32) and Δ3′RR/wt (n = 56). (> 2 experiments with > 3 mice per group). Mean is shown and KS test for significance. F. Schematic representation of IgH ΔEµ and Δ3′RR alleles used for 3C experiments. Primers (blue arrows), XbaI (grey) and HindII (red) restriction sites are shown (not to scale). G. Relative inter-allelic interactions between IgH alleles determined by 3C experiments. Eµ-hs4 or Eµ-calreticulin (Eµ-cal) 3C crosslinking frequencies for wt, Δ3′RR/Δ3′RR, ΔEµ/ΔEµ and double heterozygous Δ3′RR/ΔEµ mice were determined from in vitro LPS activated B-cells (2 experiments with ≥ 3 mice per group). Mean is shown +/- s.e.m. (n.d., non-detected).
We also generated heterozygous Δ3′RR/ΔEµ mice lacking the 3′RR on one allele and Eµ on the other (Figure 2F), thereby suppressing known cis-interactions between these elements [19] in order to check whether inter-allelic interactions were still detectable in such conditions. 3C experiments in in vitro activated B-cells (72h with LPS) revealed persistent inter-allelic interactions, with Eµ from allele 1 contacting the 3′RR from allele 2. Parallel controls monitored the lack of any interaction with the unrelated calreticulin gene (as described in [19] (Figure 1G)).
Taken together these observations show a 3′RR-dependent inter-allelic interaction between IgH loci in mature B-cells, which might participate in the function of the super-enhancer.
Trans-activation by the wtb allele functional 3′RR during CSR
We explored if proximity and 3′RR trans-activation might support CSR on the second allele. Homozygous 3′RR deletions suppress CSR-accessibility of downstream switch (S) regions but have no effect on Sµ (and barely any on Sγ1) [17, 29]. This is partly rescued by heterozygosity, where trans-CSR is maintained and joins the accessible Sµ from the 3′RR-less allele to downstream S regions from the wt allele [24]. The IgHa locus (from SV129 strain) drives expression of IgMa, IgG1a, IgG2a, IgEa and IgAa (for which some allotype-specific antisera are available) instead of IgMb, IgG1b, IgG2c, IgEb and IgAb for the IgHb locus (from C57BL/6 strain). Whether using cis- or trans-CSR, heterozygous Δ3′RRa/wtb mice should thus overwhelmingly secrete switched Ig from the accessible IgHb constant genes, with the exception of IgG1 which is less affected by the 3′RR deletion in Δ3′RRa/Δ3′RRa homozygous mice [17]. We analyzed whether in heterozygous settings, the 3′RR-proficient IgH b allele can rescue some usage of the 3′RR-dependent C genes (that are not accessible) from the 3′RR-deleted IgH a allele.
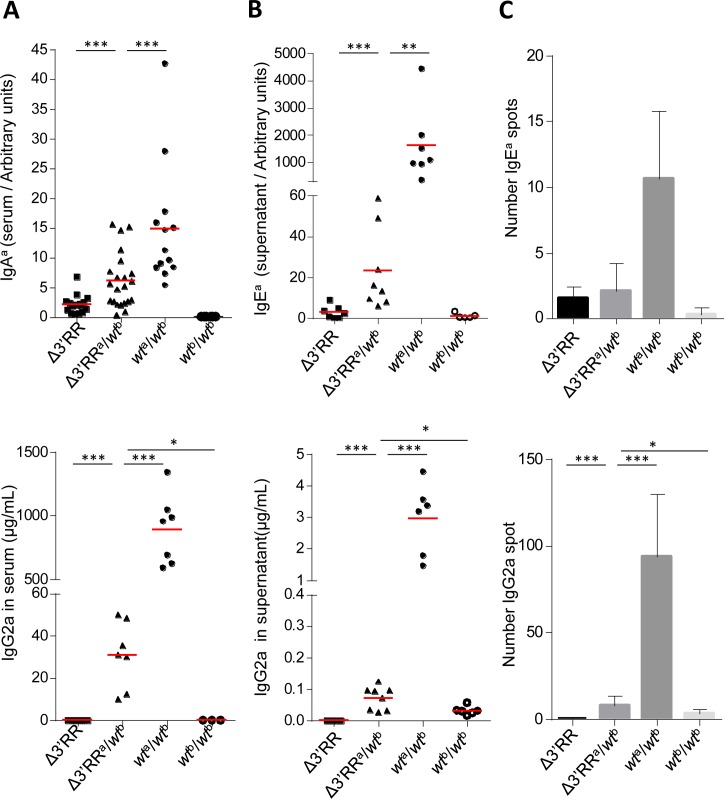
In sera, we observed that instead of following gene dosage, allotype-specific IgAa and IgG2a levels (measuring Cγa and Cγ2a CSR on the Δ3′RRa allele) significantly increased in heterozygous compared to homozygous Δ3′RRa mice (Figure 3A). The same was true for in vitro stimulated B-cells both by evaluating secretion of the IgEa and IgG2a allotypes in supernatants (Figure 3B) and by directly counting IgEa and IgG2a switched plasma cells by ELISpot (Figure 3C). Rescued CSR-accessibility of 3′RR-deleted IgHa C genes thus indicates inter-allelic 3′RR trans-activation.
Figure 3. The 3′RR partially rescues CSR in trans on a 3′RR-cis-deficient IgH allele.
A. In vivo IgHa allele CSR in Δ3′RRa/wtb mice: IgG2a (top) and IgAa (bottom) serum levels from homozygous Δ3′RR, Δ3′RRa/wtb, wta/b and wtb/b mice. Each point represents one mouse, mean is shown (≥ 3 independent experiments). Mann-Whitney test for significance. B. In vitro IgHa allele CSR in Δ3′RRa/wtb cells: IgG2a (top) and IgEa (bottom) levels in supernatant from Δ3′RR, Δ3′RRa/wtb, wta/b and wtb/b stimulated B-cells. Each point represents one stimulation from one mouse, mean is shown (≥ 3 independent experiments). Mann-Whitney test for significance. C. ELISpot specific for IgG2a (top) and IgEa (bottom) were performed on in vitro stimulated B-cells from Δ3′RR, Δ3′RRa/wtb, wta/b and wtb/b. Data shown are from 3 independent experiments with at least 2 mice. Mean +/- s.d. is shown. Mann-Whitney test for significance.
Trans-activation by the wtb allele functional 3′RR during SHM
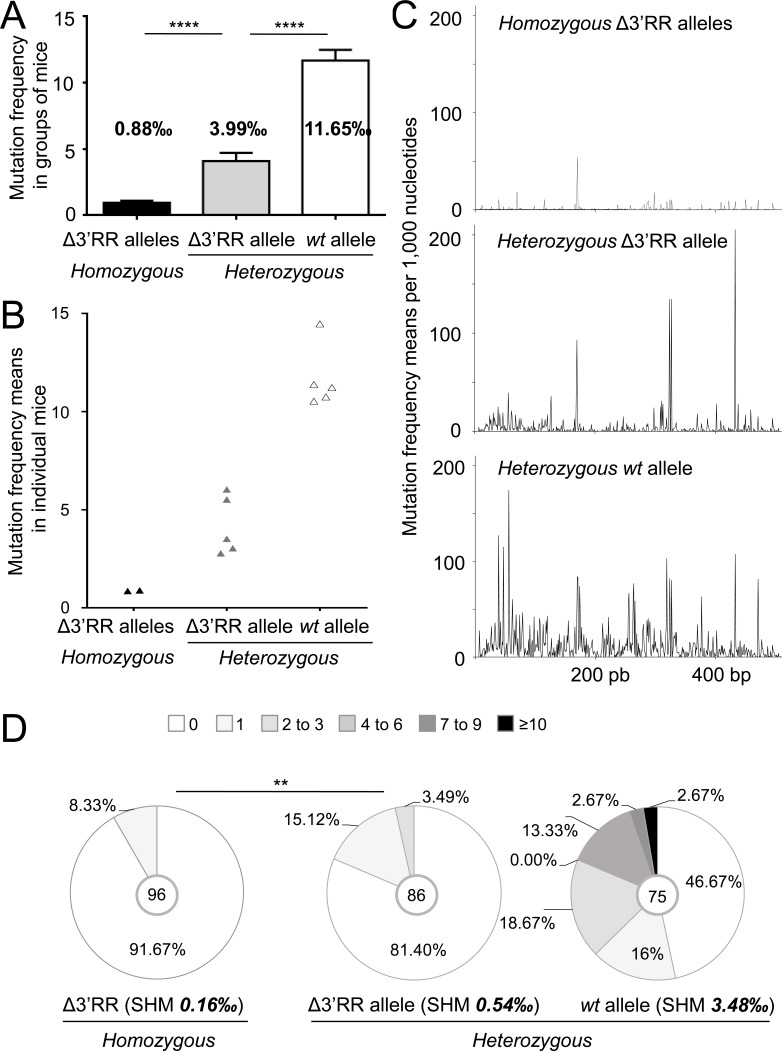
Potential trans-complementation of a SHM defect in Peyer's patches GC B-cells was checked downstream of rearranged JH4 segments (through either cloning/high-fidelity sequencing or high throughput next generation sequencing (NGS)). As previously documented [16], IgH SHM was deficient upon Δ3′RR homozygosity (0.89‰ bp in IgH vs 16.9‰ for Igκ) (Table 1, Figure 4A and 4B). In Δ3′RRa/wtb mice, SHM on the wtb allele reached 11.565‰ bp (Table 1, Figure 4A and 4B). Interestingly, the Δ3′RRa allele from heterozygous cells reached an intermediate 3.99‰ level, which although below the wtb level (p < 0.0001) is 4-fold higher than with a biallelic 3′RR defect (p < 0.0001) (Table 1, Figure 4A and 4B). Even though the higher error background level with NGS was less adapted for evaluating low-level SHM on the Δ3′RR allele, it confirmed cloning/sequencing data from a previous study [13] and additionally indicated on a high number of reads that SHM hotspots were superimposed in wtb and Δ3′RRa alleles in a heterozygous context providing trans-complementation (Figure 4C), Trans-complementation by 3′RRb rescues SHM with a typical AID pattern (increasing around RGYW/WRCY sequence targets) although this occurs at a lower level than in cis
Table 1. Rescued-SHM in Δ3′RRa/wtb mice.
| Genotypes | Δ3′RR | Δ3′RRa/wtb | |||
|---|---|---|---|---|---|
| Alleles | Igκ (control) | Allele a (Δ3′RR) | Allele a (Δ3′RR) | Allele b (wt) |
|
| Analyzed regions | 3′Jκ5 | 3′JH4 | 3′JH4 | 3′JH4 | |
| B220+/GL7+ | Total number of mutations | 515 | 663 | 2034 | 11,954 |
| Total length analyzed (base pairs) |
30,470 | 754,252 | 508,633 | 1,026,387 | |
| Mutation frequencies (per 1000 base pairs) |
16.90 | 0.89 | 3.99 | 11.65 | |
|
B220+/GL7+ /IgM+ |
Total number of mutations | 143 | 8 | 24 | 135 |
| Total length analyzed (base pairs) |
24,376 | 49,632 | 44,462 | 38,775 | |
| Mutation frequencies (per 1000 base pairs) |
5.87 | 0.16 | 0.54 | 3.48 | |
Mutation frequencies in 3′JH4 and 3′Jκ5 introns in B220+/GL7+ and B220+/GL7+/IgM+ cells from homozygous Δ3′RR and Δ3′RRa/wtb mice. Data were obtained from at least 2 experiments, ≥2 mice per group, with clonally related sequences counted only once.
Figure 4. The 3′RR partially rescues SHM in trans on a 3′RR-cis-deficient IgH allele.
A. Mutation frequency in 3′JH4 intron from Peyer's patches (B220+/GL7+) in Δ3′RR/Δ3′RR mice and in Δ3′RRa and wtb alleles from Δ3′RRa/wtb mice. Mean +/- s.d. is shown. Mann-Whitney test for significance. B. Mutation frequency as in (A), but with each point representing an individual mouse in order to evaluate the dispersion of the results when SHM is measured in groups of mice with homogeneous genotype. C. Graph of mutation frequencies along the sequenced JH4 flanking region in homozygous Δ3′RR mice (top) and in Δ3′RRa (middle) and wtb (bottom) alleles from Δ3′RRa/wtb mice. D. Mutation frequencies in 3′JH4 intron in Peyer's patches B220+/GL7+/IgM+ cells from homozygous Δ3′RR and Δ3′RRa/wtb heterozygous mice. Data were obtained from at least 3 mice per group and clonally related sequences were counted only once. Khi2 test for significance.
Some unsorted VDJa sequences might have been joined with 3′RRb through trans-CSR, then conventionally promoting SHM in cis. To exclude that, we also studied sorted unswitched IgM+/B220+/GL7+ cells: as expected SHM was lower but confirmed the above-described variations in 3′RR-less loci compared with either heterozygosity to homozygosity. Δ3′RR homozygous cells showed strongly decreased SHM (0.16‰ for IgH vs 5.87‰ for Igκ) (Table 1, Figure 4D). By contrast, in Δ3′RRa/wtb cells, SHM on the Δ3′RRa allele increased by 3-fold (0.54‰) (p < 0.01) (Table 1, Figure 4D), although lower than the 3.48‰ yielded in parallel on wtb alleles (Table 1, Figure 4D). Accordingly, the 3′RR-deficient allele showed less unmutated sequences when under heterozygosity, but more with 1-3 mutations, while no highly mutated sequence appeared in such IgM+ cells (Figure 4D).
Altogether, the 3′RR promotes inter-allelic interactions in both transgenic and endogenous IgH loci. In addition to its role in promoting interactions between IgH alleles, the 3′RR also supports AID recruitment on S (for CSR) and VH regions (for SHM), not only in cis but also in trans. We observed allele pairing in in vitro stimulated B-cells and measured CSR and SHM in trans showing 3′RR-dependent transvection in at least a fraction of activated B-cells. Even if 3′RR-dependent AID-initiated lesions occur less frequently in trans than in cis, they break the paradigm that the 3′RR is simply a cis-regulatory element and demonstrate a missing step that is important for generating the frequent interallelic CSR junctions (found in roughly 20% of B-cells). Functional transvection occurs during the window of time of AID activity and prior to any DNA recombination. Gene trans-activation occurs in yeast, drosophila and mammals and cis-regulatory elements can interact at long-distance [30]. IgH locus physiology is known to involve different types of intra-chromosomal loops that contribute to recombination [19, 31, 32]. In addition, we now demonstrate that the B-cell nucleus also supports IgH inter-allelic contacts allowing first transvection and eventually trans-CSR. It remains to be determined whether trans-interactions reflect bystander proximity after binding identical transcriptional factors, enzymes that may be rate-limiting and only concentrate in a few locations of the nucleus, or whether inter-allelic proximity is functionally important for optimal B cell maturation, by promoting use of the alternate trans-CSR pathway.
Non-coding RNAs might contribute to trans interactions as they were shown to modulate bridges and potential DNA loops in cis between distant IgH transcriptional regulatory elements, promoting AID accessibility [4].
Deleterious off-target interactions (which can promote translocations) affect Ig loci, and c-myc often co-localizes with the IgH transcription factory [33, 34]. AID is normally recruited at S regions by 3′RR-dependent chromatin marks [29] but c-myc proximity combined with a high frequency of off-target breaks can result in oncogenic translocations [35, 36]. An attractive hypothesis is that transvection between fragile IgH loci has evolved beside the major cis-CSR pathway in order to further promote legitimate and safe interactions between homologous alleles. Interestingly, this could then also help out-compete hazardous interactions with illegitimate partners. The 3′RR super-enhancer might thus favor transcription and remodeling of IgH loci (together with dangerous partners), and be an intrinsic caretaker of the B-cell genome.
EXPERIMENTAL PROCEDURES
Mice
Our research has been approved by the local ethics committee review board. Transgenic 995, 820 and 3´ enhancer deleted 820 (Δ820) mice were a kind gift of Dr. Wesley Dunnick (Figure 1B). 3′RR-mediated inter-allelic trans-interactions were studied using a 3′RR-deleted (Δ3′RR) locus (with altered CSR and SHM [16, 17]) either under homozygosity or with heterozygous Δ3′RRa / wild-type (wt)b alleles of different IgHa/b allotypes (Figure 2A). 3′RR-deficient (Δ3′RR) IgHa/a mice were bred with wt IgHb/b (C57BL/6) mice or enhancer Eµ-deficient mice (ΔEµ) mice [37] (Figure 2F). 8-10 weeks old littermates were used in all experiments. Allotypes known for several IgHa and IgHb constant genes can monitor the relative expression of both loci in heterozygous IgHa/b mice.
Heavy chain transgene chromosomal insertion site in lines 995 and 820
Genomic DNA from transgenic lines 995 and 820 was digested with MboI, diluted, and ligated to form circles, some of which include the 3´ end of the transgene and the adjacent chromosomal sequences from the host mouse (either C57BL/6 or SJL). PCR primers were selected for an orientation that would amplify a small portion of the 3´ end of the transgene and the adjacent chromosomal sequences in between. For line 820, the chromosomal sequences matched C57BL/6 chromosome 7 sequences (491 of 497 bases) at position 122 Mb (of 145 Mb total length).
Cell culture
Splenocytes were collected, red blood cells were lysed and CD43 depleted using CD43 microbeads (Miltenyi Biotec). B splenocytes were cultured 1, 2, 3, 4 or 5 days (for 3D-FISH) or 4 days (for ELISA and ELISpot) in RPMI containing 10% FCS with LPS (20µg/mL) or with anti-CD40 (5µg/ml) (RD systems) + IL4 (40ng/mL) (Peprotech) or LPS (20µg/mL) + IFNγ (2ng/mL) (RD systems).
3D-FISH
Interphase DNA FISH was performed as previously described [10] with minor adaptation. In vitro activated B-cells were dropped onto poly-L-lysine slides and fixed with 4% paraformaldehyde for 10 min at room temperature (RT). After washing with PBS, cells were permeabilized with pepsin 0.02% / HCl 0.1M for 15 min at RT, then washed and post-fixed with 1% paraformaldehyde for 5 min at RT, denatured for 5 min in 70% formamide, 2X SSC at 72°C and hybridized overnight at 37°C. The IgH (RP23-109B20) probe (encompassing an AID-targeted region from the DH cluster to Cγ2a) and RP23-436M24 was respectively labelled with dCTP-biotin (Invitrogen) or with dUTP-digoxigenin (Roche). Slides were washed in 1X SSC at 72°C for 5 min, incubated with steptavidin-Alexa Fluor 594 (1/200, BD biosciences) or with anti-digoxigenin-Alexa Fluor 488 (Abcam) 1h at RT, and mounted with vectashield containing DAPI (Vector labs). Images were acquired with an epifluorescence microscope (LEICA DMI6000B) or by confocal microscopy on a Leica SP5 AOBS system (Acousto-Optical Beam Splitter). Optical sections separated by 0.2 to 0.3µm were collected and stacks were de-convoluted and analyzed using Huygens and Volocity softwares, respectively. Separation of alleles was measured in 3D from the centre of mass of each signal. Volumetric pixel size was 0.064µm in xy and 0.2 µm in z-direction.
Chromosome conformation capture (3C)
107 cells were fixed with PBS 10% FCS and 1% formaldehyde for 10 min at RT, stopped with glycine (0.125M). Cells were lysed in Tris 10mM, NaCl 10mM, NP40 2%, supplemented with protease inhibitors, with 10 strokes of a dounce homogeneizer (Pestle A). Nuclei were resuspended in restriction buffer, permeabilized with SDS (0.1%) for 10min at 37°C and triton X-100 (1%) was added. DNA was restricted with HindIII (900U) overnight at 37°C. After HindIII inactivation DNA was ligated with T4 DNA ligase (NEB) overnight at 16°C in 8mL. The crosslink was reversed by proteinase K (500µg) overnight at 65°C and then 300µg of RNAse A was added and incubated 1h at 37°C. DNA was phenol/chloroform extracted and quantified with Qubit (Invitrogen).
The linear range of PCR amplification was determined by serial dilution of the control and the crosslinked chromatin templates. PCR was performed with Herculase Taq polymerase (Agilent) on 200ng DNA using Eµ primer: TTGACATTCTGGTCAAAACGGC and hs4: CAACCTGTGTCCCTAGAGGAGT or calreticulin primers F : CCCAAACCACCACTACCATTACA and R : GATGAACTGCCCTATCCTGAGTC (95°C 2min, 45 cycles 95°C 20sec, 56°C 20sec, 72°C 15sec and 72°C 3min). PCR products were quantified with Image J. Relative crosslinking frequencies were calculated as described [19]. BAC RP23-109B20, 199M11 and RP23-421H21 provided control templates.
Allotype-specific ELISA
ELISA for the presence of “a” allotype IgAa, IgG2a, and IgEa were performed on sera or supernatant from Δ3′RR homozygous, Δ3′RRa/wtb heterozygous, wta/b and wtb/b mice or in vitro stimulated B-cells. A pool of wta/b sera was used as standard. Plates were coated with monoclonal antibodies specific for IgAa (clone EC2, BD Pharmingen) or IgG2a (731926, Beckman coulter) overnight, samples were then incubated 2h at 37°C. After washing AP-conjugates goat anti mouse IgA (1040-04, Southern biotech) or IgG2a (1080-04, Southern biotech) were incubated 1h at 37°C. After washing and addition of AP substrate (Sigma), optic density was measured at 400 nm. For ELISA IgEa, black 96-well Immuno plates (Thermoscientific) were coated overnight with anti-IgE (732385, Beckman coulter). Samples were incubated 5h at 37°C and, after washing, with biotinylated anti-mouse IgEa (BLE408804, Ozyme) overnight at 37°C. After washing, streptavidin-AP (Sigma) was incubated 45 min at 37°C. After washing, AP activity was assayed using 4-methylumbelliferyl phosphate (Molecular probes) and fluorescence was read at 449 nm.
ELISpot
For evaluation of IgG2a or IgEa secretion, cells were seeded in duplicate at a density starting at 5 × 104/well, followed by 5-fold serial dilutions in culture medium on a 96-well MultiScreen HTS plate (Millipore) coated with 1.5 µg per well of anti-IgG2a or anti-IgEa. Cells were incubated 7 h at 37 °C and then removed by washing with PBS/Tween 0.01 %. Plates were then incubated 1 h at 37 °C with 1 µg/well of alkaline phosphatase-coupled anti-IgG2a or biotinylated-anti-IgEa. After washing with PBS/Tween 0.01 %, streptavidin-AP (Sigma) was incubated 45 min at 37°C (for IgEa). After washing, addition of 100 µL of BCIP/NBT alkaline phosphatase substrate (Millipore), new washing and drying, images were taken with NI-E microscope (Nikon) and analyzed for spots numbers with the Nis-Ar software (Nikon).
Flow cytometry and cell sorting
Cell suspensions from Peyer's patches were labeled with anti-B220-APC- (Clone RA3-6B2, Biolegend), GL7-PE- (BD), anti-IgM-FITC (Clone eB121-15F9, ebiosciences) -conjugated antibodies. Sorting of B220+/GL7+ or B220+/GL7+/IgM+ B-cells was performed on a FACS ARIA 3 (BD Biosciences). Cell sorting quality was validated when SHM frequency at the Igκ locus was superior to 5%.
Somatic hypermutation analysis
SHM analysis was performed as described [16] from sorted B220+/GL7+ or B220+/GL7+/IgM+ B-cells cells. For NGS experiment, PCR was performed using primers containing GS junior adaptors and sequences specific of V region (GCGAAGCTTARGCCTGGGRCTTCAGTGAAG) and JH4 intron (AGGCTCTGAGATCCCTAGACAG). Amplifications were performed with Phusion® High-Fidelity DNA Polymerase (New England Biolabs) according to the following program: DNA was denatured 30 s at 98°C and then submitted to 42 cycles consisting of 98°C for 10 s, 60°C for 30 s and 72°C for 30 s, and 1 cycle at 72°C for 10 min.
PCR products were first purified using NucleoSpin kit (Macherey-Nagel) and then using Ampure beads (Beckman Coulter). First a PCR emulsion (GS Junior+ emPCR Kit (Lib-A), Roche) was performed and finally PCR products were sequenced using the GS junior sequencing kit XL+ (Roche).
Statistical analysis
Statistical tests were performed using GraphPad Prism (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
SUPPLEMENTARY MATERIALS FIGURES AND REFERENCE
Acknowledgments
We thank Dr. Wesley Dunnick for providing IgH transgenic mice, and Dr. Max Cooper, Olivier Bernard, Michèle Goodhardt and Jeanne Cook-Moreau for critical reading of the manuscript. SLN and BL were supported by Association pour la Recherche sur le Cancer (ARC). We are grateful to M. Brousse, M. Faïsse, S. Desforges and B. Remerand for technical help, and to the Limoges University facilities for FACS sorting (C. Ouk) and sequencing (R. Moulinas). This work was supported by grants from ARC (PGA120150202338), Agence Nationale de la Recherche (ANR grant 16-CE15-0019-01), Institut National du Cancer (INCa grant #9363), Ligue Nationale contre le Cancer and Région Aquitaine-Limousin-Poitou-Charente.
Abbreviations
- IgH
Immunoglobulin heavy chain
- CSR
class-switch recombination
- 3′RR
3′ regulatory region
- AID
activation-induced cytidine deaminase
- SHM
somatic hypermutation
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no conflict of interest.
Authorship contributions
SLN, BL, CC, WD, SL and AG performed the experiments. EP and YD contributed essential reagents and mice. MC designed and supervised the study. SLN, BL, JS and MC analyzed the data and wrote the manuscript.
Editorial note
This paper has been accepted based in part on peer-review conducted by another journal and the authors’ response and revisions as well as expedited peer-review in Oncotarget.
REFERENCES
- 1.Chen J-L, Huisinga KL, Viering MM, Ou SA, C-tin Wu, Geyer PK. Enhancer action in trans is permitted throughout the Drosophila genome. Proc Natl Acad Sci USA. 2002;99:3723–8. doi: 10.1073/pnas.062447999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, Young RA. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–87. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park S-K, Xiang Y, Feng X, Garrard WT. Pronounced cohabitation of active immunoglobulin genes from three different chromosomes in transcription factories during maximal antibody synthesis. Genes Dev. 2014;28:1159–64. doi: 10.1101/gad.237479.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu Z-P, Economides AN, Bradner JE, Rabadan R, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–89. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis EB. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. American Naturalist. 1954;88:225–39. [Google Scholar]
- 6.Liu H, Huang J, Wang J, Jiang S, Bailey AS, Goldman DC, Welcker M, Bedell V, Slovak ML, Clurman B, Thayer M, Fleming WH, Epner E. Transvection mediated by the translocated cyclin D1 locus in mantle cell lymphoma. J Exp Med. 2008;205:1843–58. doi: 10.1084/jem.20072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herman H, Lu M, Anggraini M, Sikora A, Chang Y, Yoon BJ, Soloway PD. Trans allele methylation and paramutation-like effects in mice. Nat Genet. 2003;34:199–202. doi: 10.1038/ng1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewitt SL, Farmer D, Marszalek K, Cadera E, Liang H-E, Xu Y, Schlissel MS, Skok JA. Association between the Igk and Igh immunoglobulin loci mediated by the 3′ Igk enhancer induces “decontraction” of the Igh locus in pre-B cells. Nat Immunol. 2008;9:396–404. doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holwerda SJB, van de Werken HJG, Ribeiro de Almeida C, Bergen IM, de Bruijn MJW, Verstegen MJAM, Simonis M, Splinter E, Wijchers PJ, Hendriks RW, de Laat W. Allelic exclusion of the immunoglobulin heavy chain locus is independent of its nuclear localization in mature B cells. Nucleic Acids Res. 2013;41:6905–16. doi: 10.1093/nar/gkt491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, Farrar MA, Sleckman BP, Schatz DG, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–64. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delpy L, Sirac C, Le Morvan C, Cogné M. Transcription-dependent somatic hypermutation occurs at similar levels on functional and nonfunctional rearranged IgH alleles. J Immunol. 2004;173:1842–8. doi: 10.4049/jimmunol.173.3.1842. [DOI] [PubMed] [Google Scholar]
- 12.Yeap L-S, Hwang JK, Du Z, Meyers RM, Meng F-L, Jakubauskaitė A, Liu M, Mani V, Neuberg D, Kepler TB, Wang JH, Alt FW. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–37. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peron S, Laffleur B, Denis-Lagache N, Cook-Moreau J, Tinguely A, Delpy L, Denizot Y, Pinaud E, Cogne M. AID-Driven Deletion Causes Immunoglobulin Heavy Chain Locus Suicide Recombination in B Cells. Science. 2012;336:931–4. doi: 10.1126/science.1218692. [DOI] [PubMed] [Google Scholar]
- 14.Dunnick WA, Shi J, Fontaine C, Collins JT. Transgenes of the mouse immunoglobulin heavy chain locus, lacking distal elements in the 3′ regulatory region, are impaired for class switch recombination. PLoS ONE. 2013;8:e55842. doi: 10.1371/journal.pone.0055842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinaud E, Marquet M, Fiancette R, Péron S, Vincent-Fabert C, Denizot Y, Cogné M. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv Immunol. 2011;110:27–70. doi: 10.1016/B978-0-12-387663-8.00002-8. [DOI] [PubMed] [Google Scholar]
- 16.Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B, Pinaud E, Cogné M, Denizot Y. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210:1501–7. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogné N, Cogné M, Denizot Y. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1, 2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116:1895–8. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- 18.Meng F-L, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon K-R, Meyers RM, Amor C, Wasserman CR, Neuberg D, Casellas R, Nussenzweig MC, Bradner JE, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–48. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–22. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truffinet V, Pinaud E, Cogné N, Petit B, Guglielmi L, Cogné M, Denizot Y. The 3′ IgH locus control region is sufficient to deregulate a c-myc transgene and promote mature B cell malignancies with a predominant Burkitt-like phenotype. J Immunol. 2007;179:6033–42. doi: 10.4049/jimmunol.179.9.6033. [DOI] [PubMed] [Google Scholar]
- 21.Aupetit C, Drouet M, Pinaud E, Denizot Y, Aldigier JC, Bridoux F, Cogné M. Alleles of the alpha1 immunoglobulin gene 3′ enhancer control evolution of IgA nephropathy toward renal failure. Kidney Int. 2000;58:966–71. doi: 10.1046/j.1523-1755.2000.00253.x. [DOI] [PubMed] [Google Scholar]
- 22.Gostissa M, Yan CT, Bianco JM, Cogné M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature. 2009;462:803–7. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MGM, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, Reichert E, Kung AL, Rodig SJ, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–90. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dougier H-L, Reynaud S, Pinaud E, Carrion C, Delpy L, Cogné M. Interallelic class switch recombination can reverse allelic exclusion and allow trans-complementation of an IgH locus switching defect. Eur J Immunol. 2006;36:2181–91. doi: 10.1002/eji.200535529. [DOI] [PubMed] [Google Scholar]
- 25.Kingzette M, Spieker-Polet H, Yam PC, Zhai SK, Knight KL. Trans-chromosomal recombination within the Ig heavy chain switch region in B lymphocytes. Proc Natl Acad Sci USA. 1998;95:11840–5. doi: 10.1073/pnas.95.20.11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laffleur B, Bardet SM, Garot A, Brousse M, Baylet A, Cogné M. Immunoglobulin genes undergo legitimate repair in human B cells not only after cis- but also frequent trans-class switch recombination. Genes Immun. 2014;15:341–6. doi: 10.1038/gene.2014.25. [DOI] [PubMed] [Google Scholar]
- 27.Reynaud S, Delpy L, Fleury L, Dougier H-L, Sirac C, Cogné M. Interallelic class switch recombination contributes significantly to class switching in mouse B cells. J Immunol. 2005;174:6176–83. doi: 10.4049/jimmunol.174.10.6176. [DOI] [PubMed] [Google Scholar]
- 28.Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P, Papavasiliou FN. Switch recombination and somatic hypermutation are controlled by the heavy chain 3′ enhancer region. J Exp Med. 2009;206:2613–23. doi: 10.1084/jem.20091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saintamand A, Rouaud P, Saad F, Rios G, Cogné M, Denizot Y. Elucidation of IgH 3′ region regulatory role during class switch recombination via germline deletion. Nat Commun. 2015;6:7084. doi: 10.1038/ncomms8084. [DOI] [PubMed] [Google Scholar]
- 30.Kieffer-Kwon K-R, Tang Z, Mathe E, Qian J, Sung M-H, Li G, Resch W, Baek S, Pruett N, Grøntved L, Vian L, Nelson S, Zare H, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–20. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proudhon C, Hao B, Raviram R, Chaumeil J, Skok JA. Long-Range Regulation of V(D)J Recombination. Adv Immunol. 2015;128:123–82. doi: 10.1016/bs.ai.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha PP, Raviram R, Fu Y, Kim J, Luo VM, Aljoufi A, Swanzey E, Pasquarella A, Balestrini A, Miraldi ER, Bonneau R, Petrini J, Schotta G, et al. A Damage-Independent Role for 53BP1 that Impacts Break Order and Igh Architecture during Class Switch Recombination. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 34.Strongin DE, Groudine M, Politz JCR. Nucleolar tethering mediates pairing between the IgH and Myc loci. Nucleus. 2014;5:474–81. doi: 10.4161/nucl.36233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon K-R, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, Mathe E, Liang G, Cobell J, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocha PP, Micsinai M, Kim JR, Hewitt SL, Souza PP, Trimarchi T, Strino F, Parisi F, Kluger Y, Skok JA. Close proximity to Igh is a contributing factor to AID-mediated translocations. Mol Cell. 2012;47:873–85. doi: 10.1016/j.molcel.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marquet M, Garot A, Bender S, Carrion C, Rouaud P, Lecardeur S, Denizot Y, Cogné M, Pinaud E. The Eμ enhancer region influences H chain expression and B cell fate without impacting IgVH repertoire and immune response in vivo. J Immunol. 2014;193:1171–83. doi: 10.4049/jimmunol.1302868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.