Abstract
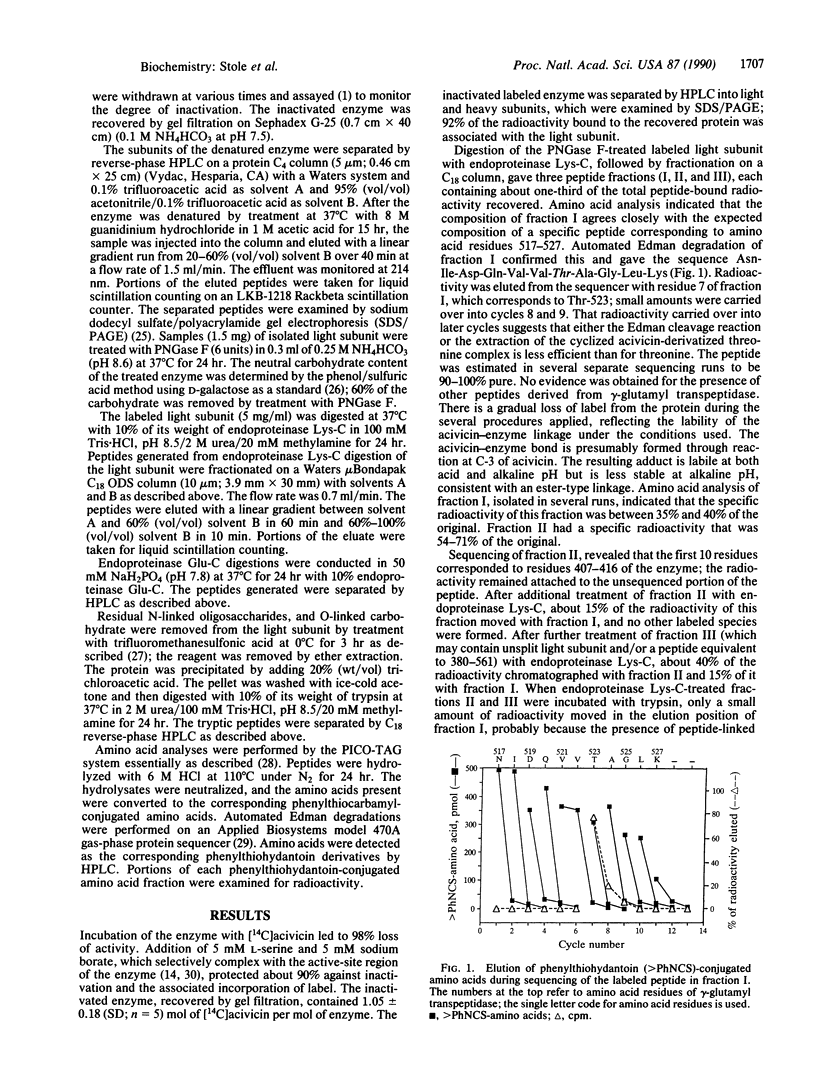
gamma-Glutamyl transpeptidase [(5-glutamyl)-peptide:amino-acid 5-glutamyltransferase, EC 2.3.2.2], an enzyme of major importance in glutathione metabolism, was inactivated by treating it with L-(alpha S,5S)-alpha-amino-3-chloro-4,5-dihydro-5-[3-14C]isoxazoleacetic acid. This selective reagent binds stoichiometrically to the enzyme; more than 90% of the label was bound to its light subunit. Enzymatic digestion of the light subunit gave a 14C-labeled peptide that corresponds to amino acid residues 517-527 of the enzyme and two incomplete digestion products that contain this labeled peptide moiety. The radioactivity associated with this peptide was released with threonine-523 during sequencing by the automated gas-phase Edman method. The light subunit contains 14 other threonine residues and a total of 19 serine residues; these were not labeled. Threonine-523 is situated in the enzyme in an environment that greatly increases its reactivity, indicating that other amino acid residues of the enzyme must also participate in the active-site chemistry of the enzyme.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen L., Meck R., Yunis A. The inhibition of gamma-glutamyl transpeptidase from human pancreatic carcinoma cells by (alpha S,5S)-alpha-amino-3-chloro-4,5-dihydro-5-isoxazoleacetic acid (AT-125; NSC-163501). Res Commun Chem Pathol Pharmacol. 1980 Jan;27(1):175–182. [PubMed] [Google Scholar]
- Allison R. D. gamma-Glutamyl transpeptidase: kinetics and mechanism. Methods Enzymol. 1985;113:419–437. doi: 10.1016/s0076-6879(85)13054-5. [DOI] [PubMed] [Google Scholar]
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Blow D. M., Birktoft J. J., Hartley B. S. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969 Jan 25;221(5178):337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- Capraro M. A., Hughey R. P. Processing of the propeptide form of rat renal gamma-glutamyltranspeptidase. FEBS Lett. 1983 Jun 27;157(1):139–143. doi: 10.1016/0014-5793(83)81132-6. [DOI] [PubMed] [Google Scholar]
- Coloma J., Pitot H. C. Characterization and sequence of a cDNA clone of gamma-glutamyltranspeptidase. Nucleic Acids Res. 1986 Feb 11;14(3):1393–1403. doi: 10.1093/nar/14.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAWID I. B., FRENCH T. C., BUCHANAN J. M. Azaserine-reactive sulfhydryl group of 2-formamido-N-ribosylacetamide 5'-phosphate: L-glutamine amido-ligase (adenosine diphosphate). II. Degradation of azaserine-C-14-labeled enzyme. J Biol Chem. 1963 Jun;238:2178–2185. [PubMed] [Google Scholar]
- Edge A. S., Faltynek C. R., Hof L., Reichert L. E., Jr, Weber P. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal Biochem. 1981 Nov 15;118(1):131–137. doi: 10.1016/0003-2697(81)90168-8. [DOI] [PubMed] [Google Scholar]
- Elce J. S. Active-site amino acid residues in gamma-glutamyltransferase and the nature of the gamma-glutamyl-enzyme bond. Biochem J. 1980 Feb 1;185(2):473–481. doi: 10.1042/bj1850473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frielle T., Curthoys N. P. Specific labelling of the hydrophobic domain of rat renal gamma-glutamyltransferase. Ciba Found Symp. 1983;95:73–91. doi: 10.1002/9780470720769.ch6. [DOI] [PubMed] [Google Scholar]
- Fushiki T., Iwami K., Yasumoto K., Iwai K. Evidence for an essential arginyl residue in bovine milk gamma-glutamyltransferase. J Biochem. 1983 Mar;93(3):795–800. doi: 10.1093/jb/93.3.795. [DOI] [PubMed] [Google Scholar]
- Gardell S. J., Tate S. S. Affinity labeling of gamma-glutamyl transpeptidase by glutamine antagonists. Effects of the gamma-glutamyl transferase and proteinase activities. FEBS Lett. 1980 Dec 29;122(2):171–174. doi: 10.1016/0014-5793(80)80430-3. [DOI] [PubMed] [Google Scholar]
- Griffith O. W., Meister A. Excretion of cysteine and gamma-glutamylcysteine moieties in human and experimental animal gamma-glutamyl transpeptidase deficiency. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3384–3387. doi: 10.1073/pnas.77.6.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman S. C. Glutaminase of Escherichia coli. I. Purification and general catalytic properties. J Biol Chem. 1968 Mar 10;243(5):853–863. [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Holcenberg J. S., Ericsson L., Roberts J. Amino acid sequence of the diazooxonorleucine binding site of Acinetobacter and Pseudomonas 7A glutaminase--asparaginase enzymes. Biochemistry. 1978 Feb 7;17(3):411–417. doi: 10.1021/bi00596a005. [DOI] [PubMed] [Google Scholar]
- Jekel P. A., Weijer W. J., Beintema J. J. Use of endoproteinase Lys-C from Lysobacter enzymogenes in protein sequence analysis. Anal Biochem. 1983 Oct 15;134(2):347–354. doi: 10.1016/0003-2697(83)90308-1. [DOI] [PubMed] [Google Scholar]
- Khedouri E., Anderson P. M., Meister A. Selective inactivation of the glutamine binding site of Escherichia coli carbamyl phosphate synthetase by 2-amino-4-oxo-5-chloropentanoic acid. Biochemistry. 1966 Nov;5(11):3552–3557. doi: 10.1021/bi00875a024. [DOI] [PubMed] [Google Scholar]
- Kuno T., Matsuda Y., Katunuma N. The conversion of the precursor form of gamma-glutamyltranspeptidase to its subunit form takes place in brush border membranes. Biochem Biophys Res Commun. 1983 Jul 29;114(2):889–895. doi: 10.1016/0006-291x(83)90864-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laperche Y., Bulle F., Aissani T., Chobert M. N., Aggerbeck M., Hanoune J., Guellaën G. Molecular cloning and nucleotide sequence of rat kidney gamma-glutamyl transpeptidase cDNA. Proc Natl Acad Sci U S A. 1986 Feb;83(4):937–941. doi: 10.1073/pnas.83.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C. W., Levitzki A., Koshland D. E., Jr The subunit structure and subunit interactions of cytidine triphosphate synthetase. J Biol Chem. 1970 Jan 10;245(1):80–87. [PubMed] [Google Scholar]
- Matsuda Y., Tsuji A., Katunuma N. Studies on the structure of gamma-glutamyltranspeptidase. I. Correlation between sialylation and isozymic forms. J Biochem. 1980 Apr;87(4):1243–1248. [PubMed] [Google Scholar]
- Meister A., Anderson M. E. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- Meister A., Tate S. S., Griffith O. W. Gamma-glutamyl transpeptidase. Methods Enzymol. 1981;77:237–253. doi: 10.1016/s0076-6879(81)77032-0. [DOI] [PubMed] [Google Scholar]
- Nash B., Tate S. S. Biosynthesis of rat renal gamma-glutamyl transpeptidase. Evidence for a common precursor of the two subunits. J Biol Chem. 1982 Jan 25;257(2):585–588. [PubMed] [Google Scholar]
- ORLOWSKI M., MEISTER A. GAMMA-GLUTAMYL-P-NITROANILIDE: A NEW CONVENIENT SUBSTRATE FOR DETERMINATION AND STUDY OF L- AND D-GAMMA-GLUTAMYLTRANSPEPTIDASE ACTIVITIES. Biochim Biophys Acta. 1963 Aug 6;73:679–681. doi: 10.1016/0006-3002(63)90348-2. [DOI] [PubMed] [Google Scholar]
- Peterson R. G., Richards F. F., Handschumacher R. E. Structure of peptide from active site region of Escherichia coli L-asparaginase. J Biol Chem. 1977 Mar 25;252(6):2072–2076. [PubMed] [Google Scholar]
- Pinkus L. M., Meister A. Identification of a reactive cysteine residue at the glutamine binding site of carbamyl phosphate synthetase. J Biol Chem. 1972 Oct 10;247(19):6119–6127. [PubMed] [Google Scholar]
- Sakamuro D., Yamazoe M., Matsuda Y., Kangawa K., Taniguchi N., Matsuo H., Yoshikawa H., Ogasawara N. The primary structure of human gamma-glutamyl transpeptidase. Gene. 1988 Dec 15;73(1):1–9. doi: 10.1016/0378-1119(88)90307-1. [DOI] [PubMed] [Google Scholar]
- Schasteen C. S., Curthoys N. P., Reed D. J. The binding mechanism of glutathione and the anti-tumor drug L-(alpha S, 5S)-alpha-amino-3-chloro-4,5-dihydro-5-isoxazoleacetic acid (AT-125;NSC-163501) to gamma-glutamyltransferase. Biochem Biophys Res Commun. 1983 Apr 29;112(2):564–570. doi: 10.1016/0006-291x(83)91501-2. [DOI] [PubMed] [Google Scholar]
- Szewczuk A., Connell G. E. The reaction of iodoacetamide with the active center of gamma-glutamyl transpeptidase. Biochim Biophys Acta. 1965 Aug 24;105(2):352–367. doi: 10.1016/s0926-6593(65)80159-x. [DOI] [PubMed] [Google Scholar]
- Tate S. S., Meister A. Serine-borate complex as a transition-state inhibitor of gamma-glutamyl transpeptidase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4806–4809. doi: 10.1073/pnas.75.10.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate S. S., Meister A. gamma-Glutamyl transpeptidase from kidney. Methods Enzymol. 1985;113:400–419. doi: 10.1016/s0076-6879(85)13053-3. [DOI] [PubMed] [Google Scholar]
- Tso J. Y., Bower S. G., Zalkin H. Mechanism of inactivation of glutamine amidotransferases by the antitumor drug L-(alpha S, 5S)-alpha-amino-3-chloro-4,5-dihydro-5-isoxazoleacetic acid (AT-125). J Biol Chem. 1980 Jul 25;255(14):6734–6738. [PubMed] [Google Scholar]
- Zalkin H. Glu-tRNAGln amidotransferase. Methods Enzymol. 1985;113:303–305. doi: 10.1016/s0076-6879(85)13043-0. [DOI] [PubMed] [Google Scholar]



