Abstract
Thiopurines are widely used as anticancer and immunosuppressive agents. However, life-threatening myelotoxicity has been noticed and largely explained by genetic variations, including NUDT15 polymorphisms (e.g., rs116855232). In this study, we conduct a meta-analysis to investigate the impact of rs116855232 on thiopurines-induced myelotoxicity susceptibility (1752 patients from 7 independent cohorts), as well as on thiopurines intolerance dose (2745 patients from 13 cohorts). Variant allele of rs116855232 contributes 7.86-fold (P < 0.00001, 95% CI: 6.13–10.08) higher risk to develop leucopenia with high specificity (91.74%) and sensitivity (43.19%), and lower thiopurines intolerance dose (P < 0.00001). Through bioinformatics prediction, amino acid changes induced by genetic variants are considered to reduce the stability, and break an α helix of NUDT15, which is part of the thiopurine binding pocket. Additionally, we conduct an expression quantitative trait loci (eQTL) analysis for NUDT15, and find a promoter-located eQTL signal (rs554405994), which may act as a potential marker to predict thiopurines-induced myelotoxicity. In conclusion, genetic polymorphisms in NUDT15 are strongly associated with adverse drug reaction (ADR) of thiopurines, although more evidences are needed to determine values of all functional NUDT15 polymorphisms for clinical regimen, rs116855232 should be considered as a highly credible pharmacogenetic indicator for thiopurines using espcially is Asians.
Keywords: meta-analysis, NUDT15, thiopurines-induced myelotoxicity, intolerance dose
INTRODUCTION
As immunosuppressive and anticancer pro-drug, thiopurines (e.g., azathioprine [AZA], mercaptopurine [6-MP]), alone or in combination with other agents, remain a gold standard medical therapy for the maintenance of disease remission in patients with acute lymphoblastic leukemia (ALL), inflammatory bowel diseases (IBD), and so on [1, 2]. AZA and 6-MP can convert to active metabolites 6-thioguanine nucleotides (6-TGNs) via multiple sequential anabolic reactions [3, 4], which (e.g., deoxythioguanosine triphosphate) can incorporate into double-stranded DNA to trigger futile mismatch repair and lead multiple types of cells (e.g., T lymphocytes) to apoptosis and subsequent resolution of inflammation [5–8]. Meanwhile, 6-MP and its metabolites can be methylated by thiopurine methyltransferase (TPMT) and interrupt intervening DNA synthesis [9, 10]. Therefore, TPMT activity has been noticed to be negatively related to 6-TGN content in plasma [11].
Large interindividual variations in dose responses and ADR susceptibility has been noticed due to the narrow therapeutic index of thiopurine drugs, with common dose-dependent toxicities, including myelotoxicity and hepatotoxicity [8, 12]. One well reported and accepted explanation is that patients with TPMT deficiency induced by genetic variants can increase sensitivity to myelotoxicity effects of thiopurine drugs [13, 14]. Actually, several single nucleotide polymorphisms (SNPs) in TPMT gene (e.g., rs1142345) have been labeled as pharmacogenetic markers, and strongly recommended to be genotyped for clinic usage of thiopurines [15–18]. However, racial diversity of TPMT SNPs in terms of variant allele frequencies limits their prediction values. For example, rs1142345, which is the most common TPMT SNP (also indicated as TPMT*3C), has allele frequency of 4% in Caucasians, but only 1.3% in East Asians. Paradoxically, thiopurines-induced leukopenia is more common in Asians, and quite a few patients with wild-type TPMT are intolerant to full dose of thiopurine drugs [19, 20], suggesting the existence of other underlying race specific genetic polymorphisms in thiopurine response. Recently, two independent studies have identified a variant in NUDT15 gene (i.e., rs116855232, inducing p.Arg139Cys) to be associated with intolerance to thiopurines or thiopurines-induced ADR in patients with ALL and IBD, respectively [2, 12]. Such association has been replicated by multiple independent studies [14, 21–28], and expanded to several other NUDT15 SNPs, including rs147390019 (inducing p.Arg139His) [24]. Large genetic population studies (e.g., ExAC project) demonstrate that variant allele of rs116855232 of NUDT15 is most common in East Asians (10.4%) and Hispanics (7.1%), rare in Europeans (0.46%), but barely detected in Africans, while rs147390019 is mostly in Hispanic (1.75%) [29], contributing to ancestry-related differences in thiopurine drugs tolerance [12, 19, 30].
NUDT15 is deemed to dephosphorylate the thiopurine active metabolites TGTP and TdGTP, preventing their incorporation into DNA and negatively affecting the cytotoxic effects of thiopurines [2, 3, 14, 21–24, 28, 31–33]. Crystal structure of NUDT15 has been characterized, making it possible to estimate the impact of Arg139Cys and Arg139His on NUDT15 activity, and subsequent cell sensitivity to thiopurine treatment. Indeed, in vitro pharmacological analyses and cellular drug response examinations have been done and determined the NUDT15 deficiency induced by not only genetic variants, but also the expression level of NUDT15 [24], highlighting the importance of NUDT15 SNPs genotyping for clinic use of thiopurine drugs.
In this study, we aim to conduct a systematic review and meta-analysis to investigate the association of NUDT15 SNPs with clinic thiopurine response on the basis of existing researches, and examine the impact of these common variants on NUDT15 structure through bioinformatics analyses. Finally, eQTL analyses are proceeded to search more pharmacogenetic markers for thiopurine induced ADR in NUDT15 gene, in order to increase the prediction sensitivity.
RESULTS
Meta-analyses
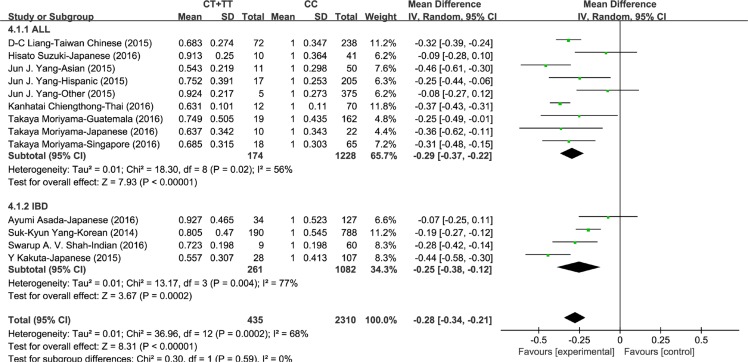
Through literature searching (see Methods), 20 independent cohort studies that demonstrated in 11 articles met the inclusion criteria for meta-analysis (Figure 1). Characteristics of these studies were summarized in Table 1. We conducted meta-analyses on association of rs116855232 with thiopurines-induced myelotoxicity susceptibility, as well as thiopurines intolerance dose. First, 7 studies were included with a total of 602 cases (patients with thiopurines-induced myelotoxicity) and 1150 controls (patients without myelotoxicity) for myelotoxicity susceptibility analysis. Fixed effect model was used since no heterogeneity was observed in the allele model (P = 0.68, and I2 = 0%). Compared to C allele, variant T allele significantly exhibited a 7.86-fold (OR = 7.86, 95% CI: [6.13, 10.08]) increased risk to develop thiopurines-induced myelotoxicity in both IBD and ALL (P < 0.00001, Figure 2). Totally, the presence of rs116855232 variant allele had a sensitivity of 43.19% (260/602) and specificity of 91.74% (1055/1150) for all myelotoxicity events, while the specificity reached 84.59% (1323/1564) for early myelotoxicity events (Supplementary Tables 1 and 2). Additionally, Consistent association was also observed in dominant model (P < 0.00001, OR = 9.48, 95% CI: [7.20, 12.47]), and recessive model (P < 0.00001, OR = 18.10, 95% CI: [6.34, 51.68]). Secondly, 13 studies assessed the association between rs116855232 and thiopurines intolerance dose with a sample size of 2745. Random model was employed in dosage maintenance meta-analysis since the high heterogeneity among studies. Compared to CC carriers (as reference group), T allele carriers (CT and TT genotypes) required 28% (P < 0.00001, 95% CI: [–0.34, –0.21]) lower mean daily thiopurines dose. Because thiopurine dosage used in ALL patients is significantly higher than that in IBD patients, we separated the patients into two groups in terms of disease types, and found similar risk of thiopurines-induced myelotoxicity and thiopurine maintenance dosage reduction rate for T allele (Figure 3).
Figure 1. Flow chart of included studies for the meta-analysis.
Table 1. Principle characteristics of the studies included in the Meta-Analysis for SNPs at NUDT15 rs116855232 locus.
| Year | Author [*] | Ethnicity | Sample sizea | Genotype counts (case) | Genotype counts (control) | AZA dose (mg/m2) (mean ± SD) (normalized dose)b | Diseases | Type of study | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | TT | CT | CC | TT | CT | CC | TT | CT | CC | |||||
| 2014 | Suk-Kyun Yang [2] | Korean | 346 | 632 | 14 | 133 | 199 | 0 | 43 | 589 | 2.335 ± 0.485 (0.522 ± 0.108) | 3.697 ± 2.145 (0.827 ± 0.480) | 4.472 ± 2.436 (1 ± 0.545) | IBD | myelotoxicity susceptibility and intolerance dose |
| 2015 | Y Kakuta [22] | Japanese | 34 | 101 | 5 | 10 | 19 | 0 | 13 | 88 | NA | 1.613 ± 0.891 (0.557 ± 0.307) | 2.915 ± 1.203 (1 ± 0.413) | IBD | myelotoxicity susceptibility and intolerance dose |
| 2016 | Ayumi Asada [21] | Japanese | 45 | 116 | 2 | 18 | 25 | 0 | 14 | 102 | 2.12 (0.872) | 2.26 ± 1.130 (0.930 ± 0.465) | 2.43 ± 1.270 (1 ± 0.523) | IBD | myelotoxicity susceptibility and intolerance dose |
| 2016 | X. Zhu [27] | Chinese Han | 65 | 188 | 4 | 36 | 25 | 0 | 17 | 171 | NA | NA | NA | IBD | myelotoxicity susceptibility |
| 2016 | Swarup A. V. Shah [25] | Indian | 6 | 63 | 1 | 5 | 0 | 0 | 3 | 60 | 2.066 ± 0.566 (0.723 ± 0.198) | 2.858 ± 0.566 (1 ± 0.198) | IBD | intolerance dose | |
| 2015 | Jun J. Yang [12] | East Asian | 61 | NA | 1 | 10 | 50 | NA | NA | NA | 10.125 (0.169) | 35.55 ± 11.25 (0.594 ± 0.188) | 59.85 ± 17.85 (1 ± 0.298) | ALL | intolerance dose |
| Hispanic | 222 | NA | 1 | 16 | 205 | NA | NA | NA | 2.175 (0.033) | 52.425 ± 13.4 (0.796 ± 0.355) | 65.85 ± 16.65 (1 ± 0.253 ) | ||||
| Other | 380 | NA | 0 | 5 | 375 | NA | NA | NA | NA | 59.475 ± 13.95 (0.924 ± 0.217) | 64.35 ± 17.55 (1 ± 0.273) | ||||
| 2015 | Yoichi Tanaka [14] | Japanese | 38 | 54 | 5 | 13 | 20 | 1 | 5 | 48 | NA | NA | NA | ALL | myelotoxicity susceptibility |
| 2015 | D-C Liang [23] | Taiwan Chinese | 310 | NA | 2 | 70 | 238 | NA | NA | NA | 18.8 ± 7.4 (0.213 ± 0.084) | 61.4 ± 23.4 (0.696 ± 0.265) | 88.2 ± 30.6 (1 ± 0.347) | ALL | intolerance dose |
| 2016 | Kanhatai Chiengthong [28] | Thai | 28 | 54 | 1 | 9 | 18 | 1 | 1 | 52 | 54.608 ± 8.719 (0.631 ± 0.101) | 86.542 ± 9.525 (1 ± 0.110) | ALL | myelotoxicity susceptibility and intolerance dose | |
| 2016 | Takaya Moriyama [24] | Guatemala | 181 | NA | 1 | 18 | 162 | NA | NA | NA | 8.944 (0.128) | 54.954 ± 34.516 (0.789 ± 0.496) | 69.638 ± 30.261 (1 ± 0.435) | ALL | intolerance dose |
| Singaporean | 83 | NA | 1 | 17 | 65 | NA | NA | NA | 5.522 (0.06) | 65.894 ± 25.765 (0.721 ± 0.282) | 91.354 ± 27.674 (1 ± 0.303) | ||||
| Japanese | 32 | NA | 1 | 9 | 22 | NA | NA | NA | 5.013 (0.05) | 69.950 ± 28.912 (0.702 ± 0.290) | 99.674 ± 34.231 (1 ± 0.343) | ||||
| 2016 | Hisato Suzuki [26] | Japanese | 46 | 5 | 0 | 10 | 36 | 0 | 0 | 5 | NA | 59.946 ± 16.405 (0.913 ± 0.250) | 65.647 ± 23.887 (1 ± 0.364) | ALL | myelotoxicity susceptibility and intolerance dose |
aCase and Control indicates patients with or without thiopurines-induced myelotoxicity, respectively;
b6-MP dose was converted to AZA equivalent dose using a conversion factor of 2.08, Meeh-Rubner formula was used to unify the units into mg/m2, AZA dose of each genotype was also normalized against CC (wildtype) ;
*numbers in the brackets represent the references in the manuscript
NA: not available.
Figure 2. Forest plot of association of rs116855232 with thiopurines-induced leukopenia in allele model.
Figure 3. Forest plot of thiopurines intolerance dose associated with rs116855232 (T carriers compared to CC carriers).
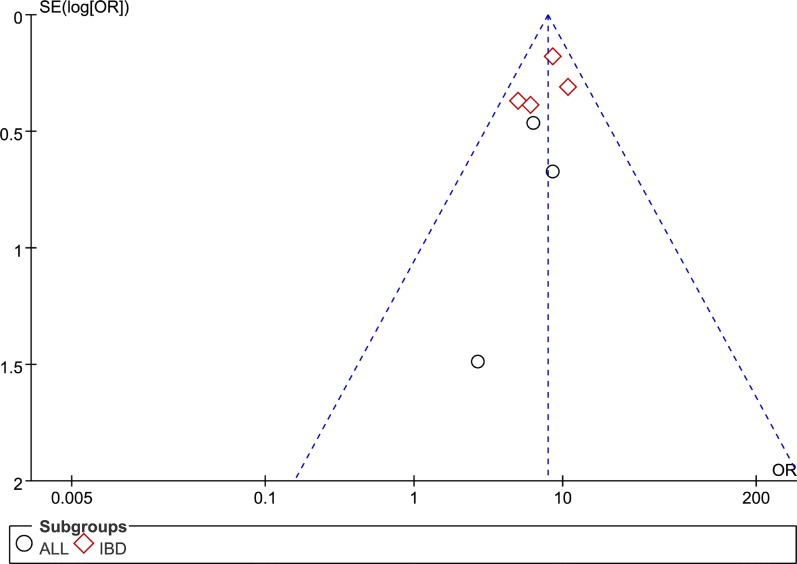
For both meta-analyses, we used Begg's test and Egger's test to measure the publication bias for all model, no evidence of obvious asymmetry was observed, such as Figure 4. Sensitivity analyses were also carried out by removing each study one at a time, the ORs remain stable, suggesting that the conclusion of rs116855232 impact on thiopurines-induced myelotoxicity susceptibility and thiopurines intolerance dose were robust and reliable.
Figure 4. Funnel plot of publication bias test for association between rs116855232 polymorphism and thiopurines-induced myelotoxicity susceptibility.
Crystal structure and protein stability prediction
Besides rs116855232, two additional functional SNPs (i.e., rs186364861 [inducing Val18Ile], and rs147390019 [inducing Arg139His]) were also reported recently. To investigate the effect of the Val18Ile, Arg139His and Arg139Cys mutants on the protein function of NUDT15, we firstly constructed their mutant models through mutating selected residue of the crystal structure, and optimize the mutant region structure by using loop refinement. Through bioinformatics prediction, we noticed that Arg139His and Arg139Cys were located at the second α-helix, may perturb the α-helix loop and the base of the substrate binding pocket (Figure 5A, 5B, 5C). Additionally, Arg139Cys could lead to the formation of a disulfide bond with the adjacent cysteine residue (Cys140) that may further reduce the enzyme activity of NUDT15 (Figure 5B). However, Val18Ile mutant at the first β-sheet has less effect on the protein structure. Subsequently, mutant energy calculation showed similar results (data not shown). For protein stability estimation, mutant energy of Arg139His and Arg139Cys variants are greater than 0.5 kcal/mol, largely decrease the protein stability compared to wildtype and Val18Ile mutant, indicating the possible role of these genetic variants on NUDT15 function (Figure 5D).
Figure 5. Impact of rs116855232 on protein structure and stability structures are illustrated for.
(A) wildtype, (B) Arg139Cys and (C) Arg139His for NUDT15, and subsequent protein stability has also been analyzed (D).
Cis-eQTL and epigenome regulation analyses for NUDT15
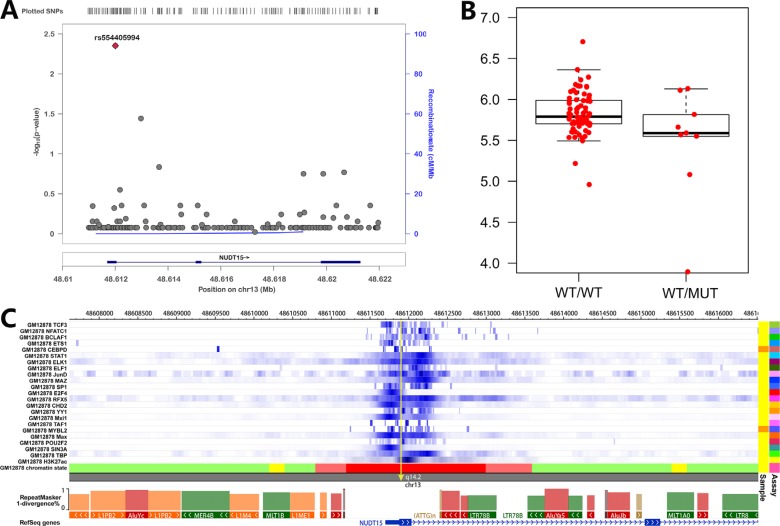
Although ~90% of early thiopurines-induced myelotoxicity can be predicted by rs116855232, sensitivity of such SNP for late myelotoxicity is still low, indicating more variants may be involved in such ADR. As down-regulation of NUDT15 in cell lines can also sensitize leukemia cells to thiopurines [24], we thus conducted an eQTLs analyses for NUDT15 to search additional potential pharmacogenetic markers in this gene. Expression level and SNP genotypes of LCLs were retrieved from the public resource (see Methods), and submitted for association analyses. Only one SNP (rs554405994) achieved statistical significance (P = 0.004) (Figure 6A), and LCLs from Asians with variant allele are related to lower NUDT15 expression (Figure 6B). rs554405994 locates in the promoter region of NUDT15, with a high GC content and DNase I sensitivity. Multiple strong transcription factors and H3K27Ac binding signal can be detected around this rs554405994-located region according to the epigenomic information from public resource (i.e., Epigenome Browser [38], Figure 6C), indicating variant allele of rs554405994 may be associated with NUDT15 expression through altering the binding affinity of transcription factors with gene promoter. Interestingly, rs554405994 also induces a GlyVal insertion between position 18 and 19 of NUDT15, and causes a slight reduction in NUDT15 activity according to the previous report [24]. Therefore, rs554405994 can impact thiopurine metabolism through both down-regulating gene expression and reducing enzyme activity, thus may be considered as a new causal variant for thiopurines-induced myelotoxicity.
Figure 6. eQTL and epigenetic analysis for NUDT15 SNPs.
(A) eQTL has been screened around NUDT15 locus and illustrated with online tool (i.e., LocusZoom); (B) association of NUDT15 expression with the top eQTL signal (rs554405994) was shown with boxplot; (C) Epigenomic signals at NUDT15 locus was illustrated by using online tool (i.e., Epigenome Browser).
DISCUSSION
Ethnic diversity in genetics is an important factor for inherited predisposition to disease susceptibility, as well as drug treatment outcomes [39, 40]. For instance, SNPs at ARID5B locus are associated with ALL susceptibility with varied odds radio among ethnicities [41, 42], while missense SNP in CDKN2A exhibits significance only in Caucasians [43]. 6-MP is commonly used in ALL chemotherapy and can induce severe ADR events in some patients (mainly myelotoxicity), which can be largely explained by TPMT variants in Caucasians and blacks but not Asians due to the low variant allele frequency [15, 29]. Recently, missense SNP (i.e., rs116855232) in NUDT15 has been linked to thiopurines-induced myelotoxicity in ALL as well as IBD by genome-wide association studies [2, 12]. Such association has been replicated in multiple independent follow-up studies by considering either myelotoxicity event or intolerance dose, and also exhibited ethnic specific mainly because the risk allele frequency is high in Asians and Hispanics (e.g., ~10% in Asians), but rare in Caucasians (0.2%) and not detected in blacks [12]. Therefore, rs116855232 of NUDT15 testing is of greater diagnostic value than TPMT genotyping for prospective risk assessment of thiopurines-induced myelotoxicity in Asian population.
In this study, we conducted a meta-analysis to estimate the impact of rs116855232 in NUDT15 on thiopurines-induced myelotoxicity as well as thiopurine intolerant. With allele model, T allele carriers have ~8-fold higher risk compared to C allele carriers to develop leucopenia no matter in ALL or IBD. However, patients with CT genotype have a significant lower myelotoxicity rate (229 out of 322, 71.10%) than TT genotype (31 out of 33, 93.94%), thus lower risk to develop leucopenia compared to CC genotype in additive model, with OR = 7.60 (95% CI: 5.77–10.03) and 18.10 (95% CI: 6.34–51.68), respectively. Moreover, the ADR rate varied largely in patients with CT genotype range from 43.48% to 100% (median = 72.22%), probably due to the starting dosage of thiopurines. Actually the incidence of thiopurines-induced myelotoxicity will increase with the higher standard dose (e.g., 35% vs. ~25% IBD patients suffered ADR in Suk-Kyun's study compared with others), but the T allele frequency are similar within patients with ADR among studies. However, statistical analysis can't be done due to the limited number of studies and complicated study design. For meta-analysis for thiopurine maintenance dose, high heterogeneity was observed in both IBD and ALL studies, possibly induced by drug response variations in patients with CT genotype as described above. However, we didn't use additive model to estimate the contribution of TT genotype to thiopurine intolerant compared to CC genotype, because 11 out of 13 studies have no more than one patient with TT genotype of rs116855232. Overall, our results indicate the important diagnostic value of rs116855232 in NUDT15, especially patients with TT genotype. Moreover, Suk-Kyun Yang et al demonstrated the significant higher diagnostic value of rs116855232 as indicator for early (OR = 35.13) than late leukopenia (OR = 5.29), with accordingly sensitivity of 89.40% (59/66) and 31.43% (88/280), respectively [2], suggesting not only the strong effect of rs116855232 on thiopurines-induced myelotoxicity, but also some other factors may be involved in late myelotoxicity events. Interestingly, additional SNPs in NUDT15 described by Takaya et al are related to final maintenance thiopurines dosage, including rs147390019 (only common in Hispanic [1.7%]) and rs186364861 (only common in Asian [1.6%]) [24]. Compound heterozygote of these SNPs illustrated similar effect on thiopurines dosage adjustment as homozygous variant of rs116855232, indicating more attention should be paid for all functional NUDT15 SNPs. However, meta-analysis is not applicable due to the lack of independent studies for these SNPs. In addition, interaction of TPMT and NUDT15 variants have also been considered in some studies, heterozygous of functional SNPs of both genes show significant lower intolerant thiopurine dose than mono-heterozygous SNPs, but higher than homozygous variants in either SNPs [24]. However, more independent studies are needed for validation because the sample size of patients with either compound heterozygous of NUDT15 variants or heterozygous genotype in both TMPT and NUDT15 is small.
According to the previous report, NUDT15 is a nudix hydrolase which can degrade dGTP and dGDP in vitro, suggesting that it may reduce the active metabolites of thiopurine in vivo [44]. Indeed, loss-of-function of NUDT15 enzymatic activity induced by amino acid change of Arg139Cys (induced by rs116855232 risk allele), can largely explain thiopurines-induced myelotoxicity and thiopurines maintain dose. Other clinical relevant genetic variants were also conducted to estimate their impact on NUDT15 function. Interestingly, rs147390019 introduces amino acid change at the same position as rs116855232, resulting in Arg139His. As the crystal structure of wildtype NUDT15 and the importance of the position Arg139 in thiopurine has been characterized [45, 46], we further figured out that both Arg139Cys and Arg139His greatly impact on the crystal structure of NUDT15 by breaking α-helix of the active domain and reducing the stability of mutated NDUT15. However, it can't explain the effect of Val18Ile (induced by rs186364861) and Val18_Val19insGlyVal (induced by rs554405994) since little change has been seen in NUDT15 structure with these altered amino acids (data not shown). Due to the possibility of prediction error, experimental crystallization and structure analysis for mutated NUDT15 is needed to determine the impact of these SNPs. Additionally, Takaya et al established stable cell lines and observed significant increase of TGTP/TGMP ratio as well as DNA-TG content in NUDT15 knockdown cells [24], raising a possibility that eQTL for NUDT15 may also be related to clinical ADR events. We thus conducted an eQTL analysis and screened out the top eQTL signal (i.e., rs554405994), which is predicted to be located at the promoter region with multiple binding sites of regulatory factors. Interestingly, this SNP has been described above as an inframe indel, inducing two amino acids (GlyVal) insertion between position 18 and 19. Therefore, the risk allele of rs554405994 is related to lower expression level and decreased enzyme activity of NUDT15 [24], thus could be considered as another pharmacogenetic marker for thiopurine response. However, further clinical confirmation studies are needed to determine the improved sensitivity for thiopurines-induced myelotoxicity after considering rs554405994, especially for the late ADR.
In conclusion, our meta-analysis demonstrates the strong significant association of the SNP (rs116855232) at NUDT15 with thiopurines-induced myelotoxicity susceptibility and thiopurines intolerance dose in either ALL or IBD. Although rs116855232 has already been labeled as pharmacogenomics marker in PharmGKB (www.pharmgkb.org), we considered the recommended level could be upgrade from 1B to 1A for thiopurines usage. Besides, more independent studies are needed to estimate the impact other functional SNPs of NUDT15 to guide the clinical usage of thiopurines.
MATERIALS AND METHODS
Literature and study acquisition
Systematically literature searching was carried out independently by two investigators from PubMed, Google Scholar and the Chinese National Knowledge Infrastructure (CNKI) date to July 27, 2016 according to following search terms: “rs116855232”, or “NUDT15”, or “thiopurine drugs” and “polymorphisms” and “NUDT15”, or “thiopurines” and “NUDT15”, or “polymorphism” and “NUDT15”. All papers were restricted to English (N = 30). Initially, checking of the titles as well as the abstracts was conducted to remove the duplicated articles along with papers that did not meet our subject. Then, we read through every remaining studies and retained valuable papers which meet the following criteria: (1) thiopurine drugs therapy based toxicity studies; (2) association of SNPs at NUDT15 with thiopurine drugs intolerance or susceptibility to toxicity was evaluated; (3) information of patient number was provided, including patients with or without ADR, respectively; (4) provided the genotype counts or sufficient data to impute the genotypes, (5) data without overlap (N = 11). When multiple publications reported on the same or overlapping data, only the publication with the most updated or detailed data was included. The literature screening flow presented in Figure 1. Neither Ethical approval nor patient consent was needed, because all the information was acquired from published studies.
Data extraction and verification
Using strict inclusion and exclusion criteria, detail information was extracted from each publications, including first author, ethnicity, sample size, and etc. 6-MP dose was converted to AZA equivalent dose using a conversion factor of 2.08 [2], and Meeh-Rubner formula was used to unify the units into mg/m2. Corresponding authors were contacted with if datasets were not accessible or incomplete for the required data. For accuracy, all the information was double checked and reviewed by another investigator. Detail information about the included papers was listed in Table 1 and Supplementary Tables 3, 4 and 5.
Meta-analyses
By using Review Manager 5.3 software [34], we intended to analyze the association of rs116855232 polymorphism with thiopurine induced leukopenia susceptibility, or thiopurines intolerance dose, with allele model (variant T allele vs. wildtype C allele), dominant model (TT+TC vs.CC), and recessive model (TT vs. TC+CC). Carriers of rs116855232 CC was defined as a reference group in SNP genetic models, while CT or TT was defined as “rs116855232 T carriers”. To remove any heterogeneity caused by pharmacodynamic differences in thiopurine drugs sensitivity among the study populations, the thiopurine drugs maintenance dose (mean ± SD) in each genotype (including CT, TT, and T carriers, respectively) was normalized against the reference group [35]. Heterogeneity among those studies were evaluated by the Q statistic and the I2 statistic, of which Q approximately follows a χ2 distribution with k-1 (k indicates the number of studies) degrees of freedom. P value was used to detect the significance level of heterogeneity. I2 = (Q-(k-1))/Q*100%, ranging from 0–100%. I2 was considered as a critical value, when I2 < 50% and P > 0.1, fixed-effect model was used to calculate summary odds ratios (OR) and 95% confidence interval (95% CI), while the random-effect model should be employed under the circumstances of I2 > 50% and P < 0.1 because of high heterogeneity.
Crystal structure prediction and protein stability estimation
The initial three dimensional geometric coordinates of the X-ray crystal structure of NUDT15 (PDB code: 5BON) were downloaded from the Protein Databank (PDB). The Val18Ile, Arg139His and Arg139Cys mutant models were constructed using the Build Mutants protocol of Discovery Studio 3.5 (Accelrys Inc., USA). Disulfide bridges of Arg139Cys mutant model was selected manually. Loop refinement protocol, which uses a looper algorithm to optimize the structure of a selected non-terminal loop region of a protein structure, was performed to evaluate the protein structure change of the mutants. Mutation energy (stability) was calculated using pH-dependent mode to investigate the effect of single-point mutations on protein stability.
Cis-eQTL analyses
Expression level of NUDT15 gene was obtained from public RNA-seq data resource of Lymphoblastoid cell lines for CHB/JPT (GSE11582) [36], and genotypes of SNPs (Chr13: 48582000-48622000, hg19 human genome version) around NUDT15 were obtained from the 1000 genome project website (http://browser.1000genomes.org/). SNPs with variant allele detected in less than three individuals were excluded. Genotype-expression association was assessed through a linear regression model for the available individuals (N = 447). Regional plots were constructed by plotting the negative logarithm of the P value for each SNP in a 11.2-kb window at the NUDT15 locus using LocusZoom [37].
SUPPLEMENTARY TABLES
ACKNOWLEDGMENTS AND FUNDING
We thank Dr. Jun Yang, Dr. Takaya Moriyama, and Dr. Wenjian Yang from St. Jude Childrens’ Research Hospital to provide us the detail information of their study. This study was supported by the National Natural Science Foundation of China (No. 81400120, No. 81522028, No. 81673452), and National Key Research Development Program (No. 2016YFC0905000), and Heng Xu are supported by the grant from “the Recruitment Program of Global Young Experts” (known as “the Thousand Young Talents Plan”).
Abbreviations
- AZA
azathioprine
- 6-MP
6-mercaptopurine
- IBD
inflammatory bowel diseases
- ALL
acute lymphoblastic leukemia
- SNP
single nucleotide polymorphism
- 95% CI
95% confidence intervals
- OR
odds ratios
- eQTL
expression quantitative trait loci
- CNKI
Chinese National Knowledge Infrastructure.
Footnotes
CONFLICTS OF INTEREST
None.
REFERENCES
- 1.Timmer A, Patton PH, Chande N, McDonald JW, MacDonald JK. Azathioprine and 6-mercaptopurine for maintenance of remission in ulcerative colitis. The Cochrane database of systematic reviews. 2016:CD000478. doi: 10.1002/14651858.CD000478.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang SK, Hong M, Baek J, Choi H, Zhao W, Jung Y, Haritunians T, Ye BD, Kim KJ, Park SH, Park SK, Yang DH, Dubinsky M, et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nature genetics. 2014;46:1017–1020. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts RL, Barclay ML. Update on thiopurine pharmacogenetics in inflammatory bowel disease. Pharmacogenomics. 2015;16:891–903. doi: 10.2217/pgs.15.29. [DOI] [PubMed] [Google Scholar]
- 4.Haglund S, Taipalensuu J, Peterson C, Almer S. IMPDH activity in thiopurine-treated patients with inflammatory bowel disease - relation to TPMT activity and metabolite concentrations. British journal of clinical pharmacology. 2008;65:69–77. doi: 10.1111/j.1365-2125.2007.02985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong FC, Leung AW, Kwok JS, Chan MH, Li CK, Yuen YP. NUDT15 variant and thiopurine-induced leukopenia in Hong Kong. Hong Kong medical journal. 2016;22:185–187. doi: 10.12809/hkmj154783. [DOI] [PubMed] [Google Scholar]
- 6.Karran P, Attard N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nature reviews Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 7.Hedeland RL, Hvidt K, Nersting J, Rosthoj S, Dalhoff K, Lausen B, Schmiegelow K. DNA incorporation of 6-thioguanine nucleotides during maintenance therapy of childhood acute lymphoblastic leukaemia and non-Hodgkin lymphoma. Cancer chemotherapy and pharmacology. 2010;66:485–491. doi: 10.1007/s00280-009-1184-5. [DOI] [PubMed] [Google Scholar]
- 8.Fotoohi AK, Coulthard SA, Albertioni F. Thiopurines: factors influencing toxicity and response. Biochemical pharmacology. 2010;79:1211–1220. doi: 10.1016/j.bcp.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Kroplin T, Iven H. Methylation of 6-mercaptopurine and 6-thioguanine by thiopurine S-methyltransferase. A comparison of activity in red blood cell samples of 199 blood donors. European journal of clinical pharmacology. 2000;56:343–345. doi: 10.1007/s002280000137. [DOI] [PubMed] [Google Scholar]
- 10.Kroplin T, Fischer C, Iven H. Inhibition of thiopurine S-methyltransferase activity by impurities in commercially available substrates: a factor for differing results of TPMT measurements. European journal of clinical pharmacology. 1999;55:285–291. doi: 10.1007/s002280050630. [DOI] [PubMed] [Google Scholar]
- 11.Lennard L, Lilleyman JS, Van Loon J, Weinshilboum RM. Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet. 1990;336:225–229. doi: 10.1016/0140-6736(90)91745-v. [DOI] [PubMed] [Google Scholar]
- 12.Yang JJ, Landier W, Yang W, Liu C, Hageman L, Cheng C, Pei D, Chen Y, Crews KR, Kornegay N, Wong FL, Evans WE, Pui CH, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. Journal of clinical oncology. 2015;33:1235–1242. doi: 10.1200/JCO.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gearry RB, Barclay ML, Burt MJ, Collett JA, Chapman BA. Thiopurine drug adverse effects in a population of New Zealand patients with inflammatory bowel disease. Pharmacoepidemiology and drug safety. 2004;13:563–567. doi: 10.1002/pds.926. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka Y, Kato M, Hasegawa D, Urayama KY, Nakadate H, Kondoh K, Nakamura K, Koh K, Komiyama T, Manabe A. Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. British journal of haematology. 2015;171:109–115. doi: 10.1111/bjh.13518. [DOI] [PubMed] [Google Scholar]
- 15.Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nature reviews Cancer. 2006;6:117–129. doi: 10.1038/nrc1800. [DOI] [PubMed] [Google Scholar]
- 16.Krynetski EY, Evans WE. Genetic polymorphism of thiopurine S-methyltransferase: molecular mechanisms and clinical importance. Pharmacology. 2000;61:136–146. doi: 10.1159/000028394. [DOI] [PubMed] [Google Scholar]
- 17.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526:343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, Krynetski EY, Pui CH, Evans WE. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. Journal of the National Cancer Institute. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 19.Bhatia S, Landier W, Hageman L, Kim H, Chen Y, Crews KR, Evans WE, Bostrom B, Casillas J, Dickens DS, Maloney KW, Neglia JP, Ravindranath Y, et al. 6MP adherence in a multiracial cohort of children with acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2014;124:2345–2353. doi: 10.1182/blood-2014-01-552166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takatsu N, Matsui T, Murakami Y, Ishihara H, Hisabe T, Nagahama T, Maki S, Beppu T, Takaki Y, Hirai F, Yao K. Adverse reactions to azathioprine cannot be predicted by thiopurine S-methyltransferase genotype in Japanese patients with inflammatory bowel disease. Journal of gastroenterology and hepatology. 2009;24:1258–1264. doi: 10.1111/j.1440-1746.2009.05917.x. [DOI] [PubMed] [Google Scholar]
- 21.Asada A, Nishida A, Shioya M, Imaeda H, Inatomi O, Bamba S, Kito K, Sugimoto M, Andoh A. NUDT15 R139C-related thiopurine leukocytopenia is mediated by 6-thioguanine nucleotide-independent mechanism in Japanese patients with inflammatory bowel disease. Journal of gastroenterology. 2016;51:22–29. doi: 10.1007/s00535-015-1142-4. [DOI] [PubMed] [Google Scholar]
- 22.Kakuta Y, Naito T, Onodera M, Kuroha M, Kimura T, Shiga H, Endo K, Negoro K, Kinouchi Y, Shimosegawa T. NUDT15 R139C causes thiopurine-induced early severe hair loss and leukopenia in Japanese patients with IBD. The pharmacogenomics journal. 2016;16:280–285. doi: 10.1038/tpj.2015.43. [DOI] [PubMed] [Google Scholar]
- 23.Liang DC, Yang CP, Liu HC, Jaing TH, Chen SH, Hung IJ, Yeh TC, Lin TH, Lai CL, Lai CY, Shih LY. NUDT15 gene polymorphism related to mercaptopurine intolerance in Taiwan Chinese children with acute lymphoblastic leukemia. The pharmacogenomics journal. 2015 doi: 10.1038/tpj.2015.75. [DOI] [PubMed] [Google Scholar]
- 24.Moriyama T, Nishii R, Perez-Andreu V, Yang W, Klussmann FA, Zhao X, Lin TN, Hoshitsuki K, Nersting J, Kihira K, Hofmann U, Komada Y, Kato M, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nature genetics. 2016;48:367–373. doi: 10.1038/ng.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah SAV, Paradkar M, Desai D, Ashavaid TF. Nudt15C415t Variant as a Predictor For Thiopurine Induced Toxicity in Indian Patients. Journal of gastroenterology and hepatology. 2016 doi: 10.1111/jgh.13494. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H, Fukushima H, Suzuki R, Hosaka S, Yamaki Y, Kobayashi C, Sakai A, Imagawa K, Iwabuchi A, Yoshimi A, Nakao T, Kato K, Tsuchida M, et al. Genotyping NUDT15 can predict the dose reduction of 6-MP for children with acute lymphoblastic leukemia especially at a preschool age. Journal of human genetics. 2016;61:797–801. doi: 10.1038/jhg.2016.55. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Wang XD, Chao K, Zhi M, Zheng H, Ruan HL, Xin S, Ding N, Hu PJ, Huang M, Gao X. NUDT15 polymorphisms are better than thiopurine S-methyltransferase as predictor of risk for thiopurine-induced leukopenia in Chinese patients with Crohn's disease. Alimentary Pharmacology & Therapeutics. 2016;44:967–975. doi: 10.1111/apt.13796. [DOI] [PubMed] [Google Scholar]
- 28.Chiengthong K, Ittiwut C, Muensri S, Sophonphan J, Sosothikul D, Seksan P, Suppipat K, Suphapeetiporn K, Shotelersuk V. NUDT15 c.415C > T increases risk of 6-mercaptopurine induced myelosuppression during maintenance therapy in children with acute lymphoblastic leukemia. Haematologica. 2016;101:e24–26. doi: 10.3324/haematol.2015.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kham SK, Soh CK, Liu TC, Chan YH, Ariffin H, Tan PL, Yeoh AE. Thiopurine S-methyltransferase activity in three major Asian populations: a population-based study in Singapore. European journal of clinical pharmacology. 2008;64:373–379. doi: 10.1007/s00228-007-0426-x. [DOI] [PubMed] [Google Scholar]
- 31.Sandborn W, Sutherland L, Pearson D, May G, Modigliani R, Prantera C. Azathioprine or 6-mercaptopurine for inducing remission of Crohn's disease. The Cochrane database of systematic reviews. 2000:CD000545. doi: 10.1002/14651858.CD000545. [DOI] [PubMed] [Google Scholar]
- 32.Park SK, Hong M, Ye BD, Kim KJ, Park SH, Yang DH, Hwang SW, Kwak MS, Lee HS, Song K, Yang SK. Influences of XDH genotype by gene-gene interactions with SUCLA2 for thiopurine-induced leukopenia in Korean patients with Crohn's disease. Scandinavian journal of gastroenterology. 2016;51:684–691. doi: 10.3109/00365521.2015.1133698. [DOI] [PubMed] [Google Scholar]
- 33.Sato M, Harada M, Oishi H, Wada-Hiraike O, Hirata T, Nagasaka K, Koga K, Fujii T, Osuga Y. Vaginal Stenosis After Gonadotropin-Releasing Hormone Agonist Therapy During Treatment for Acute Lymphoblastic Leukemia. Journal of lower genital tract disease. 2016;20:e11–13. doi: 10.1097/LGT.0000000000000175. [DOI] [PubMed] [Google Scholar]
- 34.Tian ZQ, Su XF, Lin ZY, Wu MC, Wei LX, He J. Meta-analysis of laparoscopic versus open liver resection for colorectal liver metastases. Oncotarget. 2016;7:84544–84555. doi: 10.18632/oncotarget.13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindh JD, Holm L, Andersson ML, Rane A. Influence of CYP2C9 genotype on warfarin dose requirements—a systematic review and meta-analysis. European journal of clinical pharmacology. 2009;65:365–375. doi: 10.1007/s00228-008-0584-5. [DOI] [PubMed] [Google Scholar]
- 36.Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, Rivas M, Dermitzakis ET, Cahir-McFarland E, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS genetics. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, Kuhn RM, Zhu J, Smirnov I, et al. The Human Epigenome Browser at Washington University. Nature methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JJ, Cheng C, Devidas M, Cao X, Fan Y, Campana D, Yang W, Neale G, Cox NJ, Scheet P, Borowitz MJ, Winick NJ, Martin PL, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nature genetics. 2011;43:237–241. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, Robinson GW, Huang J, Lim JY, Zhang H, Bass JK, Broniscer A, Chintagumpala M, Bartels U, Gururangan S, Hassall T, Fisher M, Cohn R, et al. Common variants in ACYP2 influence susceptibility to cisplatin-induced hearing loss. Nature genetics. 2015;47:263–266. doi: 10.1038/ng.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, Pei D, Scheet P, Burchard EG, Eng C, Huntsman S, Torgerson DG, Dean M, et al. Novel susceptibility variants at 10p12.31–12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. Journal of the National Cancer Institute. 2013;105:733–742. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu H, Cheng C, Devidas M, Pei D, Fan Y, Yang W, Neale G, Scheet P, Burchard EG, Torgerson DG, Eng C, Dean M, Antillon F, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. Journal of clinical oncology. 2012;30:751–757. doi: 10.1200/JCO.2011.38.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H, Zhang H, Yang W, Yadav R, Morrison AC, Qian M, Devidas M, Liu Y, Perez-Andreu V, Zhao X, Gastier-Foster JM, Lupo PJ, Neale G, et al. Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nature communications. 2015;6:7553. doi: 10.1038/ncomms8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hori M, Satou K, Harashima H, Kamiya H. Suppression of mutagenesis by 8-hydroxy-2′-deoxyguanosine 5′-triphosphate (7,8-dihydro-8-oxo-2′-deoxyguanosine 5′-triphosphate) by human MTH1, MTH2, and NUDT5. Free radical biology & medicine. 2010;48:1197–1201. doi: 10.1016/j.freeradbiomed.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Valerie NC, Hagenkort A, Page BD, Masuyer G, Rehling D, Carter M, Bevc L, Herr P, Homan E, Sheppard NG, Stenmark P, Jemth AS, Helleday T. NUDT15 Hydrolyzes 6-Thio-DeoxyGTP to Mediate the Anticancer Efficacy of 6-Thioguanine. Cancer research. 2016;76:5501–5511. doi: 10.1158/0008-5472.CAN-16-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter M, Jemth AS, Hagenkort A, Page BD, Gustafsson R, Griese JJ, Gad H, Valerie NC, Desroses M, Bostrom J, Warpman Berglund U, Helleday T, Stenmark P. Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2. Nature communications. 2015;6:7871. doi: 10.1038/ncomms8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.