Abstract
Noncoding RNAs (ncRNAs) have been demonstrated to closely associate with gene regulation and encompass the well-known microRNAs (miRNAs), as well as the most recently acknowledged long noncoding RNAs (lncRNAs). Current evidence indicates that lncRNAs can interact with miRNAs and these interactions play crucial roles in cancer metastasis, through regulating critical events especially the epithelial-mesenchymal transition (EMT). This review summarizes the types of lncRNA-miRNA crosstalk identified to-date and discusses their influence on the epithelial-mesenchymal plasticity and clinical metastatic implication.
Keywords: lncRNA, microRNA, epithelial-mesenchymal plasticity, cancer metastasis
INTRODUCTION
Cancer stems from the introduction of genetic mutations in normal cells to the biochemical changes of chromatin, signaling pathways and cell biological processes [1, 2]. The final manifestation of this disease usually comes to metastasis, responsible for more than 90% of cancer-associated mortality [3]. Many experimental and clinical studies have been tried to underlie the biology of this metastatic cascade. And there comes epithelial-mesenchymal plasticity, involving “changing faces” between epithelial cells and mesenchymal cells [4, 5]. Epithelial cells can undergo multiple biochemical changes to get a mesenchymal cell phenotype (EMT), and its reversible process, mesenchymal-epithelial transition (MET), can revert the mesenchymal cells back to epithelial cells [6, 7]. This bidirectional process contributes to the invasion-metastasis cascade from local invasion, then intravasation into nearby blood and lymphatic vessels, transition through the systems, extravasation into the parenchyma of distant tissues, finally colonization at particular distant sites [8, 9] (Figure 1). Given that emerging evidence has supported that de-differentiation of cancer cells through EMT with enhanced motility and dissemination, and re-differentiation through MET with colonization are critical in the course of multi-stage tumor progression, targeting EMT plasticity is thought to be a promising way to treat metastasis [10, 11]. Therefore, understanding the molecular mechanisms of governing EMT/MET in cancer metastasis cascade is vitally important.
Figure 1. schematic mode of the sites of EMT/MET in the metastasis of cancer.
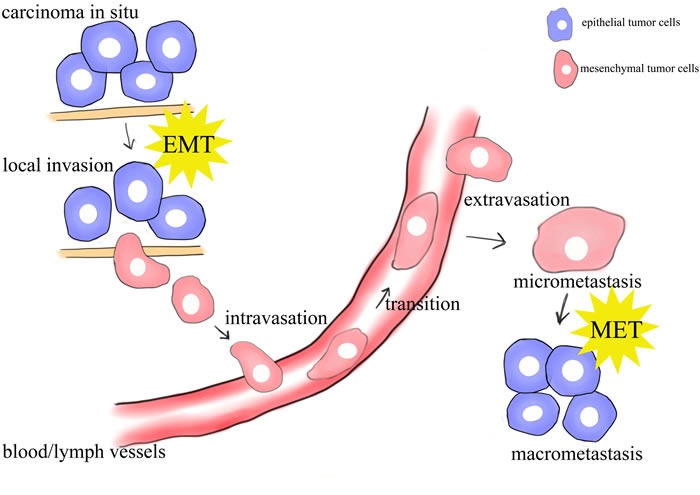
Epithelial cells undergo epigenetic changes and genetic alterations to become carcinoma in situ. Further alterations can induce local invasion of tumor cells, possibly through an EMT. The cells can intravasate into nearby blood and lymphatic vessels, be transported through the systems, and extravasate into the parenchyma of distant tissues. These cells may either be solitary or form a secondary tumor through a MET.
Noncoding RNAs (ncRNAs), though do not encode proteins, contain genetic information or have function in the biological process of cells. NcRNAs include structural RNAs such as rRNAs and tRNAs involved in mRNA translation, small nuclear RNAs (snRNAs) involved in splicing, and regulatory RNAs such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) [12]. All of them have important roles in regulating gene expression in development, physiology and pathology. Among these ncRNAs, the well-known miRNAs (~22nts), which are considered as central post-transcriptional gene regulators through their complementarity with the target mRNA sequences [13], and lncRNAs ( > 200nts), known as the “transcriptional noise”, which exhibit numerous functions in normal and abnormal tissues, are developing gradually [14, 15]. Recently, there is an interesting cross-regulation between lncRNAs and miRNAs, and the emerging evidence provides that this crosstalk has a great impact on the mechanisms of cancer metastasis [16]. In this review, we summarized miRNAs’ and lncRNAs’ control of EMT/MET, emphasized the influence of lncRNA-miRNA crosstalk in this multi-step process of human tumor progression, and harnessed this knowledge for translational medicine.
KEY REGULATORS OF EMT/MET
Key regulators of EMT were categorized into three groups, including EMT effectors, EMT core regulators, and EMT inducers [1]. EMT effectors usually are proteins, which demarcate the epithelial or mesenchymal identity of a cell such as E-cadherin, α-catenin, γ-catenin and vimentin or promote cell migration and invasion during EMT like fibronectin. Decreased E-cadherin, α-catenin, γ-catenin and increased vimentin, fibronectin are consistent markers during EMT. Among them, E-cadherin is regarded as the leading one [17]. EMT core regulators consist of transcription factors, including Snail1 and Snail2, Zeb1 and Zeb2, Twist1 and Twist2, and the newly discovered paired-related homeobox transcription factor 1 (Prrx1) [5, 17-20], which dynamically modulate EMT mainly by regulating the expression of E-cadherin [21, 22]. EMT inducers are many signaling pathways, including TGF-β, Wnt, Notch and growth factor receptor signaling cascades [23, 24]. Most notably, the TGF-β pathway appears to be a primary inducer of EMT [6, 25, 26]. Besides, tumor inflammation and hypoxia microenvironment also play fundamental roles in promoting EMT [27, 28].
Conversely, MET is characterized by decreased expression of mesenchymal markers, such as N-cadherin and vimentin, and concomitant increased expression of epithelial markers, such as E-cadherin and CK-19 [6]. Recent studies have pointed out that BMPs, as multifunctional cytokines of the TGF-β superfamily, have been involved in mediating MET programs and boosting metastatic outgrowth by antagonizing the activities of TGF-β [7].
Recently, EMT is reported to be regulated by post-transcription factors, such as miRNAs and lncRNAs, which exert their influence by regulating effectors, transcription factors and signaling pathways [29, 30]. The underlying molecular mechanisms of EMT/MET has refined our understanding of how this phenomenon may be affected by post-transcription factors like miRNAs and lncRNAs.
MICRORNAS’ CONTROL OF EPITHELIAL-MESENCHYMAL PLASTICITY
MiRNAs exert their influences, most of which are repressive, through targeting not only mRNAs, but also DNA and proteins [31]. Given that EMT can be regulated by post-transcription factors, a field of study has emerged requiring more efforts on the miRNAs control of EMT. Recently, a link between miRNAs and EMT has been extensively elucidated, and numerous miRNAs have been discovered to impact the process of EMT [32-35].
EMT-inhibiting miRNAs
MiR-200 family members, including miR-200a, b, c, miR-141 and miR-429, are the first discovered, and also the most widely studied EMT-related miRNAs [36-38]. Decreased expressions of them suppress E-cadherin and initiate EMT by targeting transcription factors ZEB1 and ZEB2. Conversely, ectopic expressions of these miRNAs in mesenchymal cells induce MET through downregulating ZEB1/2 levels, and thus increasing E-cadherin and decreasing N-cadherin [39-41]. Given the fact that miR-200 family members inhibit expression of ZEB by binding to highly conserved target sites in their 3’UTRs and ZEB factors in turn repress the genes of miR-200 family members by binding to highly conserved recognition sequences in their promoters, Emerging data have demonstrated that there is a double-negative feedback loop between them. This feedback loop controls EMT/MET process through balanced expression of miR-200 family and ZEB factors in cancer microenvironment [42]. Further studies find more evidence about the miR-200 family. miR-200c inhibits metastasis of breast cancer by downregulating high mobility group protein B1 (HMGB1), which enhances tumor cell motility and suppresses apoptosis [43]. The miR-200 family and the miR-183~96~182 cluster which are co-repressed in lung cancer inhibit EMT and metastasis by inducing forkhead box F2 (Foxf2), which correlates with ZEB1, represses E-cadherin and forms a double-negative feedback loop with the miR-200 family [44]. Though double-negative feedback loop between the miR-200 family and ZEB1/2 remains to be the most widely existed in many types of cancer [45, 46], it has also been applied in many other miRNAs such as miR-203 and miR-145. In breast cancer, Snail2 reduces expression of miR-203, while ectopic expression of miR-203 directly represses Snail2. The Snail2 and miR-203 regulatory loop is in concert with miR-200 and ZEB1/2, forming a feed-forward loop to regulate EMT and gene expression [47]. Similarly, the mutual control of miR-145 and ZEB2 contributes to prostate cancer progression and metastasis, wherein ZEB2 directly represses the transcription of miR-145, which in turn represses ZEB2 [48]. These reciprocal feedback loops may explain the reversibility of EMT and MET through an imbalanced expression of miRNAs and EMT transcription factors [49].
In addition to forming feedback loops, miRNAs can directly target transcription factors or signaling pathways of EMT. MiR-106b inhibits EMT and metastasis of endometrial cancer in vitro by directly downregulating Twist1 mRNA at the 3’UTR [50]. Reduced expression of miR-145 promotes lung cancer cell EMT and metastasis via targeting octamer-binding transcription factor 4 (Oct4) mediated Wnt/β-catenin signaling pathway [51]. Additionally, miRNAs also affect the integrity of the epithelial and mesenchymal architecture, thus regulating EMT. In gastric cancer cells, miR-30a directly targets the 3’UTR of vimentin, inhibits its protein level, thus decreasing EMT and cell invasion [52]. The expression of miR-30a is increased by overexpression of a putative tumor suppressor, Runt-related transcription factor 3 (RUNX3) [53]. MiR-506 suppresses EMT and metastasis of ovarian cancer through the direct downregulation of two mesenchymal marker proteins, vimentin and N-cadherin in vitro and in vivo [54]. Another EMT-inhibiting miRNA is miR-138, which suppresses EMT in head and neck squamous cell carcinoma cell lines via three distinct pathways: (a) by regulating the expression of vimentin, (b) by targeting ZEB2, (c) by the epigenetic regulator enhancer of zeste homolog (EZH2) [55, 56]. Except for the miRNAs mentioned above, many new scenes have taken on in this field. Stromal interaction molecule 1 (STIM1), an endoplasmic reticulum Ca2+ sensor, is regulated by a post-transcriptional regulatory mechanism mediated by a novel EMT-inhibiting miRNA named miR-185 in the metastasis cascade of colorectal cancer [57].
EMT-activating miRNAs
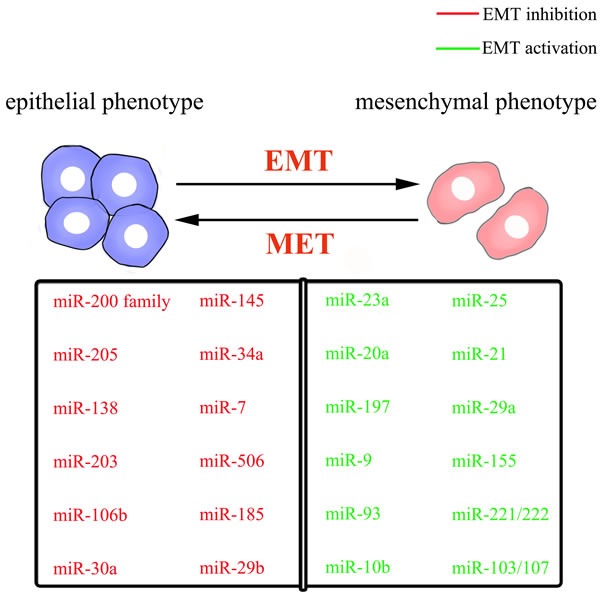
Compared with those miRNAs that suppress the EMT process, numbers of EMT-promoting miRNAs are relatively low. MiR-10b, the first reported metastasis-related miRNA, could trigger the EMT of laryngeal carcinoma by directly targeting E-cadherin mRNA [58, 59]. E-cadherin is also targeted by miR-9 and miR-23a. MiR-9, which is frequently overexpressed in esophageal squamous cell carcinoma, promotes metastasis by directly targeting E-cadherin and increasing β-catenin nuclear translocation, and subsequently inducing EMT [60]. Through TGF-β/Smad pathway [61], MiR-23a directly targets E-cadherin, promoting the mesenchymal phenotype with increased cell migration and invasion [62]. MiR-197 acts as an inducer of EMT in pancreatic cancer cells by indirectly targeting E-cadherin and regulating its membrane localization and trafficking via p120 catenin, an E-cadherin interaction protein [63]. Moreover, EMT-promoting miR-25 has been shown to be activated by the Wnt/β-catenin signaling pathway. Upregulation of miR-25 can inhibit the Rho GDP dissociation inhibitor alpha (RhoGDI1), enhancing expression of Snail and exerting its pro-metastatic function in hepatocellular carcinoma [64]. TGF-β1-induced miR-20a directly inhibits Smad7, thus enhancing the activity of β-catenin pathway and inducing EMT in gallbladder carcinoma [65]. Another refers to miR-93, which results in the attenuation of Smad-dependent TGF-β signaling and the activation of PI3K/Akt pathway by suppressing TGFBR2, promoting nasopharyngeal carcinoma cell uncontrolled metastasis and EMT-like process [66]. According to Lamouille's report, TGFBR2, also targeted by miR-302/miR-372 family and miR-106 family, plays a key role in regulating MET and maintaining the mesenchymal state [49]. In conclusion, these miRNAs profiles in the regulation of EMT/MET programs have developed gradually and become a tool to understand cancer metastasis (Figure 2).
Figure 2. schematic mode of epithelial-mesenchymal plasticity and selected miRNAs important for the epithelial or mesenchymal phenotype.
LNCRNAS’ CONTROL OF EPITHELIAL-MESENCHYMAL PLASTICITY
Although the functional roles of miRNAs in cancer metastasis are now well established, comparatively less is known about the regulatory roles of lncRNAs and their relevance to human diseases [67, 68]. Most of lncRNAs do not code for proteins, but interact with them [69, 70]. They are regulated like that of coding RNAs, subjecting to transcriptional regulation or even splicing, processing at the 5’ and 3’ ends, and exporting to the cytoplasm [71, 72]. Unlike microRNAs acting mainly as post-transcriptional repressors, Functional lncRNAs influence EMT in cancer metastasis through regulating gene expression at various levels, including chromatin modification, transcriptional and post-transcriptional processing [73, 74].
As the first disease-related ncRNA, MALAT-1, induced by TGF-β in cancer cells, promotes EMT by interacting with suppressor of zeste 12 (suz12), the subunit of polycomb repressive complex 2 (PRC2), then leading to E-cadherin downregulation [75, 76]. Suz12 functions as a histone H3 lysine 27 (H3K27) methyltransferase to bind E-cadherin promoter and suppress its expression in a PRC2-dependent way [77]. Besides chromatin modification, MALAT-1 also functions as transcription regulator through activating Wnt signaling pathway, which results in increasing of ZEB1, ZEB2, Snail2 and decreasing of E-cadherin in bladder cancer, or suppressing PI3K-AKT pathway and inhibiting EMT in breast cancer [78, 79]. Kan et al demonstrated that the expression of MALAT-1 can be enhanced by TADC-derived CCL5 in tumor microenvironment, subsequently increasing Snail expression [80]. Additionally, Shen et al found that silencing MALAT-1 may induce MET in the highly invasive subline of brain metastasis lung cancer cells, though the underlying mechanisms were not fully understood [81]. There are other lncRNAs functioning by chromatin modification, such as HOTAIR and H19. Both of them have been reported to interact with enhancer of zeste homolog 2 (EZH2), which also functions as a H3K27 methyltransferase when part of PRC2, to epigenetically inhibit genes responsible for suppressing cancer development, and increase metastasis [82-85].
LncRNAs perform their functions by regulating transcription through a broad spectrum of mechanisms. Linc00617 stimulates EMT via activating the transcription of Sox2 which promotes the oncogenic activity of breast cancer cells [86]. Wang demonstrated that lncRNA AOC4P inhibited EMT by binding vimentin and promoting its degradation in hepatocellular carcinoma [87]. Additionally, it was recently reported that some lncRNAs can be influenced by tumor microenvironment. An aberrant IL-6/STAT3/lncTCF7 signaling axis, in which IL-6 in tumor microenvironment induces lncTCF7 via activating STAT3, contributes to hapatocellular carcinoma cells aggressiveness through EMT induction [88]. Another example is a liver metastasis specific lncRNA, HULC, which can also be affected by liver micro-environment [89].
Furthermore, several lncRNAs can serve as antisense transcripts forming duplex with their corresponding mRNA counterparts to either induce or inhibit their translation, and thus influencing EMT. ZEB1-AS1 can induce EMT through positively regulating ZEB1 expression [90]. Ectopic overexpression of ZEB2NAT, which was founded in bladder cancer recently, prevents splicing of the ZEB2 5’-UTR, increases the levels of Zeb2 protein, and consequently downregulates E-cadherin mRNA and protein [91]. Further examples of antisense transcripts include 91H, ARNL (CDKN2B-AS1), and HNF1A-AS1 [92, 93].
In conclusion, Table 1 lists several selected lncRNAs with established roles in the EMT process. These representative lncRNAs are selected to illustrate the diverse mechanisms in EMT (Table 1).
Table 1. selected EMT-related lncRNAs showing diverse mechanisms of action.
| LncRNA | Mechanisms of action | References |
|---|---|---|
| MALAT-1 | Interacts with suz12 resulting in suppression of E-cadherin and activation of N-cadherin and fibronectin. Affected by tumor micro-environment. | Ji et al. [75] Fan et al. [76] Kan et al. [80] |
| HOTAIR | Reprograms chromatin state. | Qiu et al. [82] Wu et al. [83] Kim et al. [84] |
| BANCR | Histone de-acetylation suppresses BANCR to promote EMT. | Guo et al. [96] Sun et al. [97] |
| H19 | Chromatin modifier. | Matouk et al. [85] |
| Linc00617 | Stimulates EMT via activating the transcription of Sox2. | Li et al. [86] |
| LncRNA-HIT | Promotes TGF-β-induced EMT via downregulating the levels of E-cadherin. | Richards et al. [98] |
| AOC4P | Inhibits EMT via binding to vimentin and promoting its degradation. | Wang et al. [87] |
| HULC | Affected by tumor micro-environment. | Matouk et al. [89] |
| LncTCF7 | Affected by tumor micro-environment. | Hamada et al. [63] |
| ZEB1-AS1 | Upstream antisense RNA enhances ZEB1 expression. | Li et al. [90] |
| Zeb2NAT | Prevents splicing of the Zeb2 5’-UTR, increases Zeb2 and downregulates E-cadherin. | Zhuang et al. [91] |
| 91H | H19 antisense. Associated with H19 ICR methylation. | Deng et al. [92] Gao et al. [93] |
Abbreviations: AOC4P, amine oxidase, copper containing 4, pseudogene; BANCR, BRAF-activated non-protein coding RNA; EMT, epithelial-mesenchymal transition; HOTAIR, HOX transcript antisense RNA; HULC, highly up-regulated in liver cancer; lncRNA, long noncoding RNA; MALAT-1, metastasis-associated lung adenocarcinoma transcript 1; Sox2, SRY (sex determining region Y)-box 2; TGF-β, transforming growth factor beta; ZEB1/2, zinc-finger E-box-binding homeobox 1/2.
LNCRNA-MICRORNA INTERACTIONS’ CONTROL OF EPITHELIAL-MESENCHYMAL PLASTICITY
While the action of miRNAs and lncRNAs as controllers of EMT/MET in cancer metastasis has been discussed, a number of new findings over the past decade have begun to uncover the interactions between lncRNAs and miRNAs in this process [94, 95, 99]. In some cases, lncRNA stability will be reduced due to the interaction with specific miRNAs. In other cases, lncRNAs, also known as competing endogenous (ce)RNAs, can sequester miRNAs away from their target mRNAs by binding miRNAs, therefore antagonizing miRNAs [100]. LncRNAs can also compete with miRNAs by binding mRNAs. What's more, some lncRNAs can produce miRNAs, causing repression of target mRNAs. These studies suggested that interplay patterns between lncRNAs and miRNAs may have an impact on cancer development and progression, so it is necessary to further dig into these interactions in the biological process of cancer [95]. The lncRNA-miRNA interactions identified to-date are summarized in Table 2.
Table 2. EMT-related lncRNA-miRNA interactions identified to-date.
| LncRNA | MicroRNA | Mechanisms of interaction | Cancer types | References |
|---|---|---|---|---|
| MALAT-1 | miR-217 | miR-217-triggered MALAT-1 decay | lung cancer | Lu et al. [101] |
| miR-9 | miR-9-triggered MALAT-1 decay | osteosarcoma | Fang et al. [102] | |
| miR-1 | reciprocal negative control between MALAT-1 and miR-1 | breast cancer | Jin et al. [103] | |
| HOTAIR | miR-7 | HOTAIR downregulates miR-7 by inhibiting HoxD10 | breast cancer | Zhang et al. [114] |
| miR-568 | HOTAIR downregulates miR-568 by chromatin modification | breast cancer | Li et al. [115] | |
| lncRNA-ATB | miR-200s | lncRNA-ATB acts as sponge of miR-200s | hepatocellular and gastric cancer | Yuan et al. [104] Saito et al. [105] |
| H19 | let-7 | H19 acts as sponge of let-7 | pancreatic cancer | Ma et al. [106] |
| miR-138 miR-200a | H19 acts as sponge of miR-138 and miR-200a | colorectal cancer | Liang et al. [107] | |
| miR-141 | H19 acts as sponge of miR-141 | gastric cancer | Zhou et al. [108] | |
| miR-675 | H19 generates miR-675 | prostate cancer | Zhu et al. [112] | |
| lincRNA-ROR | miR-205 | lincRNA-ROR acts as sponge of miR-205 | breast cancer | Hou et al. [109] |
| ZFAS1 | miR-150 | ZFAS1 acts as sponge of miR-150 | hepatocellular carcinoma | Li et al. [110] |
| UCA1 | miR-145 | reciprocal negative control between UCA1 and miR-145 | bladder cancer | Xue et al. [111] |
Abbreviations: EMT, epithelial-mesenchymal transition; HOTAIR, HOX transcript antisense RNA; HoxD10, homeobox D10; lncRNA, long noncoding RNA; MALAT-1, metastasis-associated lung adenocarcinoma transcript 1; miRNA, microRNA; UCA1, the urothelial cancer associated 1.
miRNAs triggering lncRNAs to decay
MiR-217, a tumor suppressor, can inhibit MALAT-1 through the Ago2-mediated pathway, and thus inhibit EMT by suppressing EZH2-mediated H3K27me3, upregulating E-cadherin and downregulating N-cadherin and vimentin in cigarette smoke extract (CSE)-induced malignant transformation of HBE cells [101]. Similarly, the recruitment of miR-9 by 17β-Estradiol also causes decreased stability of MALAT-1 in osteosarcoma cell MG-63, and inhibits migration and invasion [102].
lncRNAs binding miRNAs to derepress mRNAs
There are lncRNAs which harbor similar miRNA target sequences, acting as miRNA sponges and hence sequestering miRNAs away from mRNAs, thereby derepressing mRNAs. For example, MALAT-1, which has complementary base pairing with miR-1, upregulates Slug expression through inhibiting miR-1 expression, and thus promotes EMT in triple-negative breast cancer [103]. Another was reported for lncRNA-ATB, which upregulated ZEB1 and ZEB2 by competitively binding the miR-200 family, and then induced EMT and invasion in both hepatocellular carcinoma and gastric cancer [104, 105]. Ma proposed that H19 promoted pancreatic ductal adenocarcinoma (PDAC) cell invasion and migration at least partially through antagonizing let-7 and then leading to derepression of its target high mobility group protein A2 (HMGA2) [106]. Additionally, other researches show that H19 also can act as ceRNAs for miR-138, miR-200a and miR-141 in different cancer types respectively. Liang demonstrated that H19 can antagonize functions of miR-138 and miR-200a and led to the derepression of their endogenous targets vimentin, ZEB1 and ZEB2, thus promoting EMT in colorectal cancer [107]. With regard to miR-141, Zhou was the first to demonstrate that H19 and miR-141 could compete with each other and affect their target genes in gastric cancer [108]. Other examples are involved in lincRNA-ROR, ZFAS1 and lncRNA-UCA1. These lncRNAs can act as ceRNAs and contribute to EMT and cancer metastasis [109-111].
lncRNAs generating miRNAs
LncRNAs are also processed to generate miRNAs. Zhu found that H19 could generate miR-675 which suppressed prostate cancer EMT and metastasis by downregulating transforming growth factor β induced protein (TGFBI), an extracellular matrix protein involved in cancer1metastasis [112]. This process during which H19 generates miR-675, is repressed by stress-response RNA binding protein HuR in a Drosha-dependent manner and dynamically regulated in vivo [113]. In addition, lncRNA HOTAIR, which is highly expressed in metastatic breast cancers, accelerates the EMT-dependent metastasis of breast cancer by inhibiting miR-7 through HoxD10 inhibition [114]. HOTAIR also transcriptionally inhibits the expression of miR-568 by directly targeting NFAT5 that promotes EMT in breast cancer [115]. However, further investigations are warranted to elucidate the relationship between lncRNAs and miRNAs for better therapeutic strategies.
SUMMARY AND FUTURE PERSPECTIVES
LncRNAs and miRNAs regulate gene expression involved in epithelial-mesenchymal plasticity on all levels. Through this sophisticated and multi-layered influence on protein expression patterns, these noncoding RNAs affect cancer metastasis and prognosis. Here, we have summarized the examples of a newly-developing mechanism - crosstalk between lncRNA and miRNA, and its influence on cancer metastasis cascade.
This crosstalk points out a novel way to understand the RNA networks. For example, the way miRNAs reduce lncRNA stability as described before is not entirely unexpected, since lncRNAs resemble mRNAs in many aspects. But whether or not miRNAs also regulate the transcription or splicing of lncRNAs requires further study [116, 117]. In addition, some lncRNAs have the ability of sequestering a handful of miRNAs as a ceRNA just like H19, and one miRNA can also control many genes, thus making this crosstalk more complicated [85]. Apart from the direct interactions mentioned before, there may exist some indirect actions. For example, lacking an mRNA specific target site, miRNA can transcriptionally repress mRNA through lowering the levels of a miRNA-related lncRNA. Another challenge to hamper the application of the crosstalk mechanism in clinic is which cellular conditions should exist for the network to work, since researches evidenced that the relative concentration of lncRNAs and miRNAs must be suitable for interaction [118, 119]. Moreover, EMT/MET, as a complex bidirectional process, seems difficult to target. Inhibiting EMT or blocking cancer cell invasion may be applicable in early stage carcinomas. But once cancer cells have disseminated from the primary site, inhibiting EMT may be counterproductive, since it is beneficial for MET [120]. So finding out the exact mechanism in the secondary site colonization is quite important.
In conclusion, lncRNA-miRNA crosstalk not only suggests the existence of a complex regulatory network in cancer, but also implies the possibility of cancer diagnosis and therapy using this panel of network. Though in its infancy, its ability to contribute to cancer metastasis is continually being validated.
Acknowledgments
This work was supported by National Natural Science Foundation of China grants (Nos. 81361120399, 81272961, 81372891, 81572650 and 81672672), by the Fundamental Research Funds of the Central Universities of China (2015), and by State Key Laboratory of Oral Diseases Special Funded Projects (SKLOD201512).
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest to disclose.
REFERENCES
- 1.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes & development. 2013;27:2192–206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Gupta GP, Massague J. Cancer metastasis: Building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Bu F, Royer C, Serres S, Larkin JR, Soto MS, Sibson NR, Salter V, Fritzsche F, Turnquist C, Koch S, Zak J, Zhong S, et al. ASPP2 controls epithelial plasticity and inhibits metastasis through β-catenin-dependent regulation of ZEB1. Nat Cell Biol. 2014;16:1092–104. doi: 10.1038/ncb3050. [DOI] [PubMed] [Google Scholar]
- 6.Morrison CD, Parvani JG, Schiemann WP. The relevance of the TGF-β Paradox to EMT-MET programs. Cancer Lett. 2013;341:30–40. doi: 10.1016/j.canlet.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer. 2012;12:425–36. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cursons J, Leuchowius KJ, Waltham M, Tomaskovic-Crook E, Foroutan M, Bracken CP, Redfern A, Crampin EJ, Street I, Davis MJ, Thompson EW. Stimulus-dependent differences in signalling regulate epithelial-mesenchymal plasticity and change the effects of drugs in breast cancer cell lines. Cell Commu Signal. 2015;13:26. doi: 10.1186/s12964-015-0106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turley EA, Veiseh M, Radisky DC, Bissell MJ. Mechanisms of disease: epithelial-mesenchymal transition--does cellular plasticity fuel neoplastic progression? Nat Clin Pract Oncol. 2008;5:280–90. doi: 10.1038/ncponc1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattick JS, Makunin IV, Non-coding RNA. Hum Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 13.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets foranticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liz J, Portela A, Solar M, Gómez A, Ling H, Michlewski G, Calin GA, Guil S, Esteller M. Regulation of pri-microRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55:138–47. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liz J, Esteller M. LncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–76. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Nantajit D, Lin D, Li JJ. The network of epithelial-mesenchymal transition: potential new targets for tumor resistance. J cancer Res Clin Oncol. 2015;141:1697–713. doi: 10.1007/s00432-014-1840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz-Lopez A, Diaz-Martin J, Moreno-Bueno G, Cuevas EP, Santos V, Olmeda D, Portillo F, Palacios J, Cano A. Zeb1 and Snail1 engage miR-200f transcriptional and epigenetic regulation during EMT. Int J Cancer. 2015;136:E62–73. doi: 10.1002/ijc.29177. [DOI] [PubMed] [Google Scholar]
- 19.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–94. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 20.Sun T, Zhao N, Zhao XL, Gu Q, Zhang SW, Che N, Wang XH, Du J, Liu YX, Sun BC. Expression and functional significance of Twist1 in hepatocellular carcinoma: its role in vasculogenic mimicry. Hepatology. 2010;51:545–56. doi: 10.1002/hep.23311. [DOI] [PubMed] [Google Scholar]
- 21.Ocaña OH, Córcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–24. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2012;22:699–701. doi: 10.1016/j.ccr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Said NA, Simpson KJ, Williams ED. Strategies and challenges for systematically mapping biologically significant molecular pathways regulating carcinoma epithelial-mesenchymal transition. Cells Tissues Organs. 2013;197:424–34. doi: 10.1159/000351717. [DOI] [PubMed] [Google Scholar]
- 24.Chang CC, Hsu WH, Wang CC, Chou CH, Kuo MY, Lin BR, Chen ST, Tai SK, Kuo ML, Yang MH. Connective tissue growth factor activates pluripotency genes and mesenchymal-epithelial transition in head and neck cancer cells. Cancer Res. 2013;73:4147–57. doi: 10.1158/0008-5472.CAN-12-4085. [DOI] [PubMed] [Google Scholar]
- 25.Tan EJ, Olsson AK, Moustakas A. Reprogramming during epithelial to mesenchymal transition under the control of TGFβ. Cell Adh Migr. 2015;9:233–46. doi: 10.4161/19336918.2014.983794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moustakas A, Heldin P. TGFβ and matrix-regulated epithelial to mesenchymal transition. Biochim Biophys Acta. 2014;1840:2621–34. doi: 10.1016/j.bbagen.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 27.O’Connell MP, Marchbank K, Webster MR, Valiga AA, Kaur A, Vultur A, Li L, Herlyn M, Villanueva J, Liu Q, Yin X, Widura S, Nelson J, et al. Hypoxia induces phenotypic plasticity and therapy resistance in melanoma via the tyrosine kinase receptors ROR1 and ROR2. Cancer Discov. 2013;3:1378–93. doi: 10.1158/2159-8290.CD-13-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faurobert E, Bouin AP, Albiges-Rizo C. Microenvironment, tumor cell plasticity, and cancer. Curr Opin Oncol. 2015;27:64–70. doi: 10.1097/CCO.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 29.Guo F, Kerrigan BCP, Yang D, Hu L, Shmulevich I, Sood AK, Xue F, Zhang W. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. J Hematol Oncol. 2014;7:19. doi: 10.1186/1756-8722-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Amato NC, Howe EN, Richer JK. MicroRNA regulation of epithelial plasticity in cancer. Cancer Lett. 2013;341:46–55. doi: 10.1016/j.canlet.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 31.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1845:255–65. doi: 10.1016/j.bbcan.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullock MD, Sayan AE, Packham GK, Mirnezami AH. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biol Cell. 2012;104:3–12. doi: 10.1111/boc.201100115. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31:653–62. doi: 10.1007/s10555-012-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Amato NC, Howe EN, Richer JK. MicroRNA regulation of epithelial plasticity in cancer. Cancer Lett. 2013;341:46–55. doi: 10.1016/j.canlet.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 35.Saitoh M. Epithelial-mesenchymal transition is regulated at post-transcriptional levels by transforming growth factor-beta signaling during tumor progression. Cancer Sci. 2015;106:481–8. doi: 10.1111/cas.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–9. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010;10:219–22. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344:166–73. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 41.Park DH, Jeon HS, Lee SY, Choi YY, Lee HW, Yoon S, Lee JC, Yoon YS, Kim DS, Na MJ, Kwon SJ, Kim DS, Kang J, et al. MicroRNA-146a inhibits epithelial mesenchymal transition in non-small cell lung cancer by targeting insulin receptor substrate 2. Int J Oncol. 2015;47:1545–53. doi: 10.3892/ijo.2015.3111. [DOI] [PubMed] [Google Scholar]
- 42.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–7. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang BP, Wang DS, Xing JW, Yang SH, Chu Q, Yu SY. miR-200c inhibits metastasis of breast cancer cells by targeting HMGB1. J Huazhong Univ Sci Technolog Med Sci. 2014;34:201–6. doi: 10.1007/s11596-014-1259-3. [DOI] [PubMed] [Google Scholar]
- 44.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, Tong P, Creighton CJ, Gibbons DL. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2015;23:71. doi: 10.1038/onc.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ, Huang X, Zhao J, Gu B, Zheng GX, Yang AG, Jia LT. MiR-200c suppresses TGF-beta signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int J Cancer. 2014;135:1356–68. doi: 10.1002/ijc.28782. [DOI] [PubMed] [Google Scholar]
- 46.Sundararajan V, Gengenbacher N, Stemmler MP, Kleemann JA, Brabletz T, Brabletz S. The ZEB1/miR-200c feedback loop regulates invasion via actin interacting proteins MYLK and TKS5. Oncotarget. 2015;6:27083–96. doi: 10.18632/oncotarget.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding X, Park SI, McCauley LK, Wang CY. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J Biol Chem. 2013;288:10241–53. doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renjie W, Haiqian L. MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015;356:568–78. doi: 10.1016/j.canlet.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–7. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong P, Kaneuchi M, Watari H, Sudo S, Sakuragi N. MicroRNA-106b modulates epithelial-mesenchymal transition by targeting TWIST1 in invasive endometrial cancer cell lines. Mol Carcinog. 2014;53:349–59. doi: 10.1002/mc.21983. [DOI] [PubMed] [Google Scholar]
- 51.Ling DJ, Chen ZS, Zhang YD, Liao QD, Feng JX, Zhang XY, Shi TS. MicroRNA-145 inhibits lung cancer cell metastasis. Mol Med Rep. 2015;11:3108–14. doi: 10.3892/mmr.2014.3036. [DOI] [PubMed] [Google Scholar]
- 52.Liu Z, Chen L, Zhang X, Xu X, Xing H, Zhang Y, Li W, Yu H, Zeng J, Jia J. RUNX3 regulates vimentin expression via miR-30a during epithelial-mesenchymal transition in gastric cancer cells. J Cell Mol Med. 2014;18:610–23. doi: 10.1111/jcmm.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li QL, Ito K, Sakakura C, Fukamachi H, Ki Inoue, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–24. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y, Hu L, Zheng H, Bagnoli M, Guo Y, Rupaimoole R, Rodriguez-Aguayo C, Lopez-Berestein G, Ji P, Chen K, Sood AK, Mezzanzanica D, Liu J, et al. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J Pathol. 2015;235:25–36. doi: 10.1002/path.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A, Dai Y, Zhou X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamasaki T, Seki N, Yamada Y, Yoshino H, Hidaka H, Chiyomaru T, Nohata N, Kinoshita T, Nakagawa M, Enokida H. Tumor suppressive microRNA138 contributes to cell migration and invasion through its targeting of vimentin in renal cell carcinoma. Int J Oncol. 2012;41:805–17. doi: 10.3892/ijo.2012.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z, Liu X, Feng B, Liu N, Wu Q, Han Y, Nie Y, Wu K, Shi Y, Fan D. STIM1, a direct target of microRNA-185, promotes tumor metastasis and is associated with poor prognosis in colorectal cancer. Oncogene. 2015;34:4808–20. doi: 10.1038/onc.2014.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen J, Zhang Y, Lai P, Fan X, Zhou X, Lin J, Li M, Ma W, et al. miR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. Cancer Res. 2014;74:3031–42. doi: 10.1158/0008-5472.CAN-13-2193. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Sun J, Wang B, Ren JC, Su W, Zhang T. MicroRNA-10b Triggers the Epithelial-Mesenchymal Transition (EMT) of Laryngeal Carcinoma Hep-2 Cells by Directly Targeting the E-cadherin. Appl Biochem Biotechnol. 2015;176:33–44. doi: 10.1007/s12010-015-1505-6. [DOI] [PubMed] [Google Scholar]
- 60.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L, Li J, Wang H, Qin Y, Zeng M, Guan XY, Li Y. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–80. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signaling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 62.Cao M, Seike M, Soeno C, Mizutani H, Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L, Gemma A. MiR-23a regulates TGF-beta-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol. 2012;41:869–75. doi: 10.3892/ijo.2012.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamada S, Satoh K, Miura S, Hirota M, Kanno A, Masamune A, Kikuta K, Kume K, Unno J, Egawa S, Motoi F, Unno M, Shimosegawa T. miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. J Cell Physiol. 2013;228:1255–63. doi: 10.1002/jcp.24280. [DOI] [PubMed] [Google Scholar]
- 64.Wang C, Wang X, Su Z, Fei H, Liu X, Pan Q. miR-25 promotes hepatocellular carcinoma cell growth, migration and invasion by inhibiting RhoGDI1. Oncotarget. 2015;6:36231–44. doi: 10.18632/oncotarget.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang Y, Liu C, Yang J, Liu G, Feng F, Tang J, Hu L, Li L, Jiang F, Chen C, Wang R, Yang Y, Jiang X, et al. MiR-20a triggers metastasis of gallbladder carcinoma. J Hepatol. 2013;59:518–27. doi: 10.1016/j.jhep.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 66.Lyu X, Fang W, Cai L, Zheng H, Ye Y, Zhang L, Li J, Peng H, Cho WC, Wang E, Marincola FM, Yao K, Cai H, et al. TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer. 2014;13:51. doi: 10.1186/1476-4598-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 70.Li W, Kang Y. A new Lnc in metastasis: long noncoding RNA mediates the prometastatic functions of TGF-β. Cancer Cell. 2014;25:557–9. doi: 10.1016/j.ccr.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 72.Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y, Chen H, Pan T, Jiang C, Zhao Z, Wang Z, Zhang J, Xu J, Li X. LncRNA ontology: inferring lncRNA functions based on chromatin states and expression patterns. Oncotarget. 2015;6:39793–805. doi: 10.18632/oncotarget.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin Z, Hu Y, Lai S, Xue M, Lin J, Qian Y, Zhuo W, Chen S, Si J, Wang L. Long Noncoding RNA: its partners and their roles in cancer. Neoplasma. 2015;62:846–54. doi: 10.4149/neo_2015_103. [DOI] [PubMed] [Google Scholar]
- 75.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 76.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–41. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 77.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–94. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 79.Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, Gao S, You Z, Zhan C, Liu F, Pang D. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015;8:4881–91. [PMC free article] [PubMed] [Google Scholar]
- 80.Kan JY, Wu DC, Yu FJ, Wu CY, Ho YW, Chiu YJ, Jian SF, Hung JY, Wang JY, Kuo PL. Chemokine (C-C Motif) Ligand 5 is Involved in Tumor-Associated Dendritic Cell-Mediated Colon Cancer Progression Through Non-Coding RNA MALAT-1. J Cell Physiol. 2015;230:1883–94. doi: 10.1002/jcp.24918. [DOI] [PubMed] [Google Scholar]
- 81.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–8. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 82.Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–8. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 83.Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 84.Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–30. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matouk IJ, Halle D, Raveh E, Gilon M, Sorin V, Hochberg A. The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decision. Oncotarget. 2015;7:3748–65. doi: 10.18632/oncotarget.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Zhu L, Xu L, Qin K, Liu C, Yu Y, Su D, Wu K, Sheng Y. Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol Carcinog. 2015;24:22338. doi: 10.1002/mc.22338. [DOI] [PubMed] [Google Scholar]
- 87.Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL, Hsieh TH, Shieh TM, Hsueh C, Chen TC. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget. 2015;6:23342–57. doi: 10.18632/oncotarget.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu J, Zhang J, Shen B, Yin K, Xu J, Gao W, Zhang L. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2015;34:116. doi: 10.1186/s13046-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly up-regulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–92. doi: 10.1097/meg.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- 90.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2015;15:223. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 91.Zhuang J, Lu Q, Shen B, Huang X, Shen L, Zheng X, Huang R, Yan J, Guo H. TGFβ1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, Li R, Ying H, Wang F, Liu X, Chen J, Wang S. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9:e103022. doi: 10.1371/journal.pone.0103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gao T, He B, Pan Y, Xu Y, Li R, Deng Q, Sun H, Wang S. Long non-coding RNA 91H contributes to the occurrence and progression of esophageal squamous cell carcinoma by inhibiting IGF2 expression. Mol Carcinog. 2015;54:359–67. doi: 10.1002/mc.22106. [DOI] [PubMed] [Google Scholar]
- 94.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semi Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2015;1859:169–76. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 96.Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869–75. doi: 10.3892/ol.2014.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, Chen JF, Zhang EB, De W, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y, Kong W, Dan S, Bui MM, Coppola D, Mao WM, Sellers TA, Cheng JQ. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290:6857–67. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–86. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 100.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479–90. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu L, Luo F, Liu Y, Liu X, Shi L, Lu X, Liu Q. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;289:276–85. doi: 10.1016/j.taap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 102.Fang D, Yang H, Lin J, Teng Y, Jiang Y, Chen J, Li Y. 17β-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochem Biophys Res Commun. 2015;457:500–6. doi: 10.1016/j.bbrc.2014.12.114. [DOI] [PubMed] [Google Scholar]
- 103.Jin C, Yan B, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour biology. 2015 doi: 10.1007/s13277-015-4605-6. [DOI] [PubMed] [Google Scholar]
- 104.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 105.Saito T, Kurashige J, Nambara S, Komatsu H, Hirata H, Ueda M, Sakimura S, Uchi R, Takano Y, Shinden Y, Iguchi T, Eguchi H, Ehata S, et al. A Long Non-coding RNA Activated by Transforming Growth Factor-β is an Independent Prognostic Marker of Gastric Cancer. Ann Surg Oncol. 2015;22:915–22. doi: 10.1245/s10434-015-4554-8. [DOI] [PubMed] [Google Scholar]
- 106.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 107.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–25. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou X, Ye F, Yin C, Zhuang Y, Yue G, Zhang G. The Interaction Between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cell Physiol Biochem. 2015;36:1440–52. doi: 10.1159/000430309. [DOI] [PubMed] [Google Scholar]
- 109.Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181–91. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 111.Xue M, Pang H, Li X, Li H, Pan J, Chen W. Long noncoding RNA UCA1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 2015;6:128–44. doi: 10.1111/cas.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–75. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 113.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Ighlr. Nat Cell Biol. 2012;14:659–65. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y, Dou J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–68. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 115.Li JT, Wang LF, Zhao YL, Yang T, Li W, Zhao J, Yu F, Wang L, Meng YL, Liu NN, Zhu XS, Gao CF, Jia LT, et al. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014;16:454. doi: 10.1186/s13058-014-0454-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.vanHeesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and poly-ribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–76. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–36. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


