Abstract
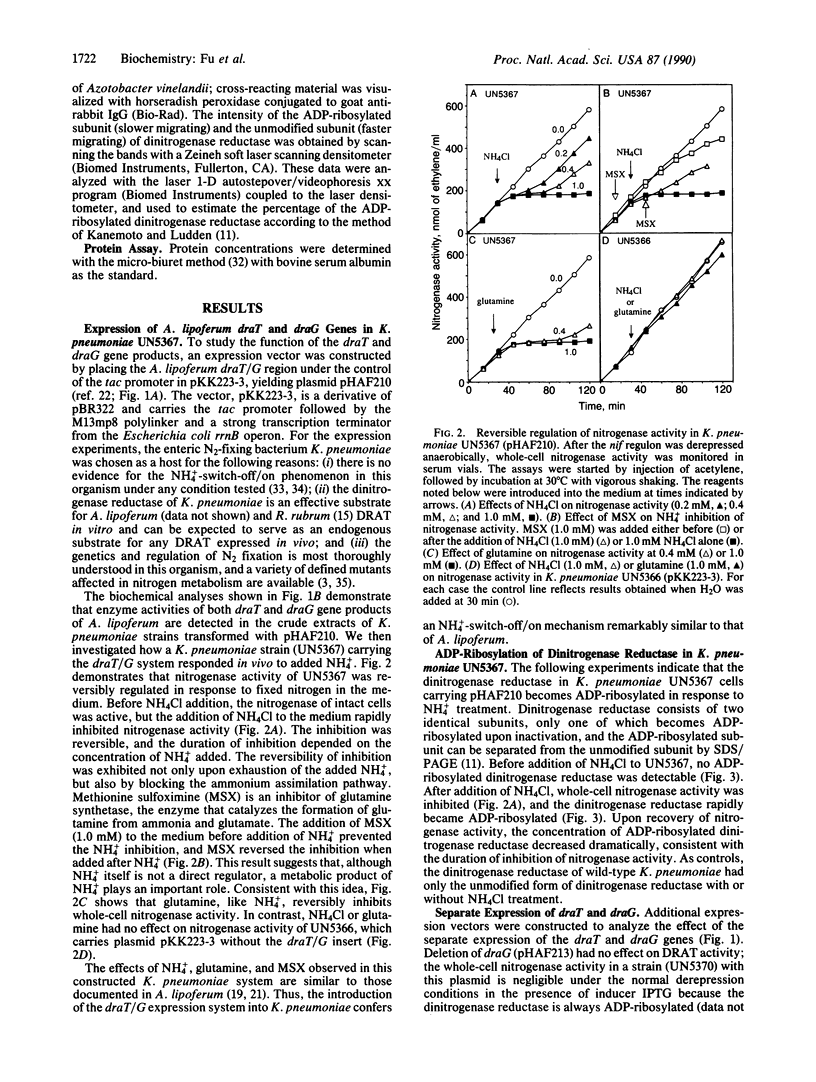
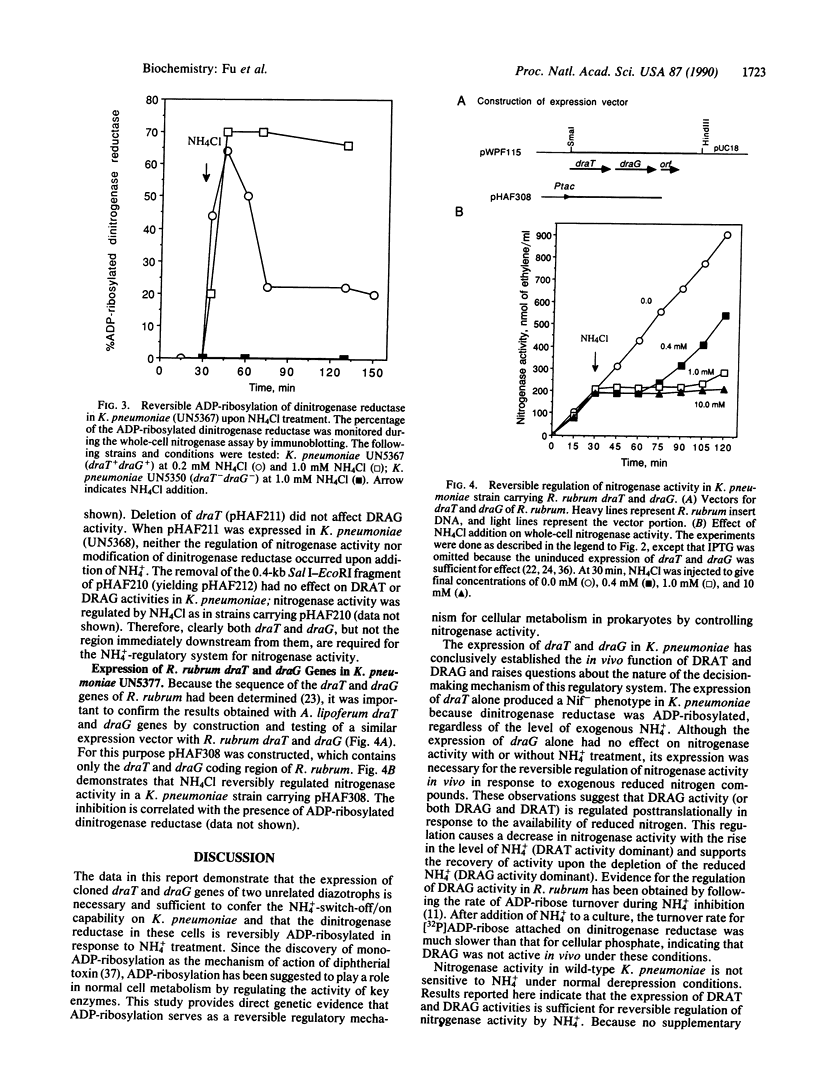
The primary product of biological nitrogen fixation, ammonia, reversibly regulates nitrogenase activity in a variety of diazotrophs by a process called "NH4(+)-switch-off/on." Strong correlative evidence from work in Azospirillum lipoferum and Rhodospirillum rubrum indicates that this regulation involves both the inactivation of dinitrogenase reductase by dinitrogenase reductase ADP-ribosyltransferase and the reactivation by dinitrogenase reductase activating glycohydrolase. The genes encoding these two enzymes, draT and draG, have been cloned from these two organisms, so that direct genetic evidence can be marshaled to test this model in vivo. The draT/G system has been transferred to and monitored in the enteric nitrogen-fixing bacterium Klebsiella pneumoniae, an organism normally devoid of such a regulatory mechanism. The expressed draT and draG genes allowed K. pneumoniae to respond to NH4Cl with a reversible regulation of nitrogenase activity that was correlated with the reversible ADP-ribosylation of dinitrogenase reductase in vivo. Thus, the expression of draT and draG genes in K. pneumoniae is necessary and sufficient to support NH4(+)-switch-off/on, and ADP-ribosylation serves as a reversible regulatory mechanism for controlling nitrogenase activity in prokaryotes.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Burris R. H. Nitrogen fixation--assay methods and techniques. Methods Enzymol. 1972;24:415–431. doi: 10.1016/0076-6879(72)24088-5. [DOI] [PubMed] [Google Scholar]
- Daesch G., Mortenson L. E. Sucrose catabolism in Clostridium pasteurianum and its relation to N2 fixation. J Bacteriol. 1968 Aug;96(2):346–351. doi: 10.1128/jb.96.2.346-351.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay R., Frey J., Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52(2-3):147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Saari L. L., Lowery R. G., Ludden P. W., Roberts G. P. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol Gen Genet. 1989 Aug;218(2):340–347. doi: 10.1007/BF00331287. [DOI] [PubMed] [Google Scholar]
- Fu H. A., Fitzmaurice W. P., Roberts G. P., Burris R. H. Cloning and expression of draTG genes from Azospirillum lipoferum. Gene. 1990 Jan 31;86(1):95–98. doi: 10.1016/0378-1119(90)90118-b. [DOI] [PubMed] [Google Scholar]
- Fu H. A., Hartmann A., Lowery R. G., Fitzmaurice W. P., Roberts G. P., Burris R. H. Posttranslational regulatory system for nitrogenase activity in Azospirillum spp. J Bacteriol. 1989 Sep;171(9):4679–4685. doi: 10.1128/jb.171.9.4679-4685.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. A., Wirt H. J., Burris R. H., Roberts G. P. Functional expression of a Rhodospirillum rubrum gene encoding dinitrogenase reductase ADP-ribosyltransferase in enteric bacteria. Gene. 1989 Dec 21;85(1):153–160. doi: 10.1016/0378-1119(89)90475-7. [DOI] [PubMed] [Google Scholar]
- GOA J. A micro biuret method for protein determination; determination of total protein in cerebrospinal fluid. Scand J Clin Lab Invest. 1953;5(3):218–222. doi: 10.3109/00365515309094189. [DOI] [PubMed] [Google Scholar]
- Gordon J. K., Shah V. K., Brill W. J. Feedback inhibition of nitrogenase. J Bacteriol. 1981 Dec;148(3):884–888. doi: 10.1128/jb.148.3.884-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Hartmann A., Fu H., Burris R. H. Regulation of nitrogenase activity by ammonium chloride in Azospirillum spp. J Bacteriol. 1986 Mar;165(3):864–870. doi: 10.1128/jb.165.3.864-870.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T., Nishizuka Y., Hayaishi O. Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J Biol Chem. 1968 Jun 25;243(12):3553–3555. [PubMed] [Google Scholar]
- Kamen M. D., Gest H. Evidence for a Nitrogenase System in the Photosynthetic Bacterium Rhodospirillum rubrum. Science. 1949 Jun 3;109(2840):560–560. doi: 10.1126/science.109.2840.560. [DOI] [PubMed] [Google Scholar]
- Kanemoto R. H., Ludden P. W. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J Bacteriol. 1984 May;158(2):713–720. doi: 10.1128/jb.158.2.713-720.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowery R. G., Chang C. L., Davis L. C., McKenna M. C., Stephens P. J., Ludden P. W. Substitution of histidine for arginine-101 of dinitrogenase reductase disrupts electron transfer to dinitrogenase. Biochemistry. 1989 Feb 7;28(3):1206–1212. doi: 10.1021/bi00429a038. [DOI] [PubMed] [Google Scholar]
- Lowery R. G., Ludden P. W. Purification and properties of dinitrogenase reductase ADP-ribosyltransferase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1988 Nov 15;263(32):16714–16719. [PubMed] [Google Scholar]
- Lowery R. G., Saari L. L., Ludden P. W. Reversible regulation of the nitrogenase iron protein from Rhodospirillum rubrum by ADP-ribosylation in vitro. J Bacteriol. 1986 May;166(2):513–518. doi: 10.1128/jb.166.2.513-518.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludden P. W., Burris R. H. Activating factor for the iron protein of nitrogenase from Rhodospirillum rubrum. Science. 1976 Oct 22;194(4263):424–426. doi: 10.1126/science.824729. [DOI] [PubMed] [Google Scholar]
- Ludden P. W., Okon Y., Burris R. H. The nitrogenase system of Spirillum lipoferum. Biochem J. 1978 Sep 1;173(3):1001–1003. doi: 10.1042/bj1731001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludden P. W., Roberts G. P. Regulation of nitrogenase activity by reversible ADP ribosylation. Curr Top Cell Regul. 1989;30:23–56. doi: 10.1016/b978-0-12-152830-0.50004-9. [DOI] [PubMed] [Google Scholar]
- MacNeil T., MacNeil D., Roberts G. P., Supiano M. A., Brill W. J. Fine-structure mapping and complementation analysis of nif (nitrogen fixation) genes in Klebsiella pneumoniae. J Bacteriol. 1978 Oct;136(1):253–266. doi: 10.1128/jb.136.1.253-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva-Gómez D., Roberts G. P., Klevickis S., Brill W. J. Electron transport to nitrogenase in Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 1980 May;77(5):2555–2558. doi: 10.1073/pnas.77.5.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope M. R., Murrell S. A., Ludden P. W. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenosine diphosphoribosylation of a specific arginine residue. Proc Natl Acad Sci U S A. 1985 May;82(10):3173–3177. doi: 10.1073/pnas.82.10.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari L. L., Pope M. R., Murrell S. A., Ludden P. W. Studies on the activating enzyme for iron protein of nitrogenase from Rhodospirillum rubrum. J Biol Chem. 1986 Apr 15;261(11):4973–4977. [PubMed] [Google Scholar]
- Saari L. L., Triplett E. W., Ludden P. W. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1984 Dec 25;259(24):15502–15508. [PubMed] [Google Scholar]
- Tubb R. S., Postgate J. R. Control of nitrogenase synthesis in Klebsiella pneumoniae. J Gen Microbiol. 1973 Nov;79(1):103–117. doi: 10.1099/00221287-79-1-103. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zumft W. G., Castillo F. Regulatory properties of the nitrogenase from Rhodopseudomonas palustris. Arch Microbiol. 1978 Apr 27;117(1):53–60. doi: 10.1007/BF00689351. [DOI] [PubMed] [Google Scholar]



