Abstract
The functionality of many microsome-associated proteins which exhibit altered abundance in response to iron limitation in Aspergillus fumigatus is unknown. Here, we generate and characterize eight gene deletion strains, and of most significance reveal that MirC (AFUA_2G05730) contributes to the maintenance of intracellular siderophore [ferricrocin (FC)] levels, augments conidiation, confers protection against oxidative stress, exhibits an intracellular localization and contributes to fungal virulence in the Galleria mellonella animal model system. FC levels were unaffected following deletion of all other genes encoding microsome-associated proteins. MirC does not appear to play a role in either siderophore export from, or uptake into, A. fumigatus. Label-free quantitative proteomic analysis unexpectedly revealed increased abundance of siderophore biosynthetic enzymes. In addition, increased expression of hapX (7.2 and 13.8-fold at 48 and 72 h, respectively; p < 0.001) was observed in ΔmirC compared to wild-type under iron-replete conditions by qRT-PCR. This was complemented by significantly elevated extracellular triacetylfusarinine C (TAFC; p < 0.01) and fusarinine C (FSC; p < 0.05) siderophore secretion. We conclude that MirC plays an important role in FC biosynthesis and contributes to the maintenance of iron homeostasis in A. fumigatus.
Keywords: siderophore, fungal proteomics, NRPS, membrane proteins, oxidative stress
Introduction
Many iron-requiring fungi utilize high affinity iron acquisition strategies to procure essential iron. However, an ability to exist in two oxidative states; ferric (Fe3+) and ferrous (Fe2+), means that iron can drive the generation of deleterious reactive oxygen species (Halliwell and Gutteridge, 1984). Paradoxically, iron is also required as a co-factor for antioxidant enzymes, such as peroxidases and catalases, in the form of heme. Thus, processes of oxidative stress tolerance and iron regulation are tightly interwoven to maintain cellular homeostasis (Brandon et al., 2015). In fungi, careful management of the utilization of high affinity iron uptake mechanisms is an important facet of iron metabolism. Iron sensitive transcription factors, SreA and HapX are core players underlying the global “omic” shift observed when Aspergillus species face iron starvation (Oberegger et al., 2001; Schrettl et al., 2008, 2010). SreA is a GATA-type transcription factor that represses high affinity iron acquisition pathways during iron sufficiency. Conversely, HapX is a bZip transcription factor that represses iron-consuming pathways during iron starvation. One of the most notable responses to iron starvation in fungi is the production of siderophores. Siderophores are low molecular mass compounds capable of solubilising ferric iron. They are secreted in the desferri-form and following external iron chelation are taken back up for intracellular storage or direct use in metabolism (Haas, 2014).
Aspergillus fumigatus, a filamentous fungus which produces ubiquitous airborne conidia, is the predominant causative agent of invasive aspergillosis (IA); an infection mainly affecting immune-deficient hosts with a notoriously high mortality/morbidity rate (Dagenais and Keller, 2009). A. fumigatus produces four siderophores; two intracellular siderophores and two extracellular siderophores. Intracellular siderophores include ferricrocin (FC) that governs iron storage and distribution in hyphae and hydroxyferricrocin (HFC) that does so in conidia. In contrast, extracellular siderophores, fusarinine C (FSC) and its acetylated derivative, triacetylfusarinine C (TAFC) are secreted from hyphae for iron uptake (Schrettl et al., 2007). The siderophore biosynthetic pathway begins with the hydroxylation of ornithine by L-ornithine N5-oxygenase, SidA to generate N5-hydroxy-L-ornithine (Schrettl et al., 2004). Following this step, the pathway splits for the production of extracellular and intracellular siderophores. The ergosterol biosynthetic intermediate, mevalonate is directed into extracellular siderophore biosynthesis following enzymatic modification with mevalonyl-CoA ligase, SidI and mevalonyl-CoA hydratase, SidH to generate anhydromevalonyl (Yasmin et al., 2012). This process occurs in peroxisomes where transacylase SidF then mediates the addition of anhydromevalonyl to N5-hydroxy-L-ornithine (Gründlinger et al., 2013b). This N5-anhydromevalonyl-N5-hydroxy-L-ornithine moiety serves as a substrate for non-ribosomal peptide synthetase (NRPS), SidD which directs the generation of FSC consisting of 3 N5-anhydromevalonyl-N5-hydroxy-L-ornithine groups linked by ester bonds. TAFC is then derived from SidG-mediated N2 acetylation of FSC (Schrettl et al., 2007). During intracellular siderophore biosynthesis, the N5-hydroxy-L-ornithine is instead acetylated by SidL and another unknown transacylase to generate N5-acetyl-N5-hydroxy-L-ornithine (Blatzer et al., 2011). This moiety then serves as the substrate for NRPS, SidC along with glycine and serine to generate FC. FC is then hydroxylated by an unknown hydroxylase to generate HFC (Schrettl et al., 2007). A. fumigatus secretes desferri-FSC/TAFC (FSC−Fe/TAFC−Fe) through an unknown mechanism. FSC−Fe/TAFC−Fe chelate Fe3+ and the resulting siderophore:iron complexes are taken back up by siderophore-iron transporters (SITs) (Haas et al., 2003; Philpott and Protchenko, 2008). FSC+Fe/TAFC+Fe are then cytosolically hydrolysed by esterases and the released iron is used by metabolic machinery, stored in vacuoles or ferri-FC (FC+Fe) (Kragl et al., 2007; Gründlinger et al., 2013a).
The ability to synthesize and utilize siderophores constitutes an important part of the pathogen weaponry at the interface with the host (Cassat and Skaar, 2013). Importantly, using a SidA deletion mutant, siderophore mediated iron acquisition was shown to be essential for virulence of A. fumigatus in mice specifically indicating siderophore production as a plausible therapeutic target for this potentially devastating infection (Schrettl et al., 2004; Hissen et al., 2005). Targeting siderophore-mediated iron acquisition has been proposed as a therapeutic strategy against microbial infection via “Trojan Horse” drug delivery or interference with siderophore biosynthesis (Miethke and Marahiel, 2007; Lamb, 2015). Moreover, siderophore transporters represent one of the few protein families unique to fungi and thus, represent promising antifungal targets.
SITs are a fungal-specific protein family within the major facilitator superfamily (MFS) (Pao et al., 1998; Saier et al., 1999). Interestingly, SITs are also conserved in fungi incapable of producing siderophores (Philpott and Protchenko, 2008). A Saccharomyces cerevisiae strain deficient in high affinity iron acquisition was used to determine the substrate specificity of SITs from Aspergillus nidulans. Using this approach, MirA was found to be specific for enterobactin and MirB for TAFC+Fe (Haas et al., 2003). The A. fumigatus genome encodes seven putative SITs (Haas et al., 2008), which include five transporters induced under iron limitation (Moloney et al., 2016); Sit1, AFUA_3G03670, MirA/D, MirB, and MirC. Though MirA and MirB have been implicated in siderophore transport in A. nidulans, a role for MirC has yet to be demonstrated (Haas et al., 2003).
Significant recent advances including extensive transcriptional and proteomic analysis, has yielded insight into the regulation and functional remodeling undertaken during iron acquisition in A. fumigatus. However, the location and mechanisms underlying the membrane transport of siderophores remain to be fully elucidated in this important pathogen. Thus, we set out to functionally explore and characterize eight putative membrane proteins including MirC to further elucidate their role in iron metabolism in A. fumigatus.
Methods
Fungal strains
Strains used in this study are shown in Table S1.
General molecular techniques
PCR reactions for generation of DNA manipulation constructs were performed using the AccuTaq LA kit (Sigma-Aldrich). For general cloning procedures the bacterial strain Escherichia coli DH5α was used which was cultivated in LB (1% (w/v) Bacto-tryptone, 0.5% (w/v) yeast extract, [1% (w/v) NaCl, pH 7.5] medium. Genomic DNA was extracted using the Zymogen Fungal DNA Extraction Kit (Zymo Research Corporation, USA).
Deletion and complementation of A. fumigatus iron-responsive genes
The bipartite marker technique (Nielsen et al., 2006) was used to generate gene deletion strains in either A. fumigatus ATCC46645 or Afs77. For each deletion, either A. fumigatus strain was co-transformed with two DNA constructs, each containing an incomplete fragment of a pyrithiamine resistance gene (ptrA) (Kubodera et al., 2002) fused to 1 kb of up and downstream sequences which flanked the regions to be deleted. Fragments shared a 557 bp overlap within ptrA, serving as a potential recombination site during transformation. Two rounds of PCR generated each fragment. First, each flanking region was amplified from ATCC46645 genomic DNA using the primers ending with P1 and P2 for flanking region A, and P3 and P4 for flanking region B (Table S2). Subsequent to gel-purification, the fragments were digested with their corresponding restriction enzymes. The ptrA selection marker was released from plasmid pSK275 (a kind gift from Professor Sven Krappmann, Erlangen, Germany) by digestion with corresponding restriction enzymes, and ligated with the two flanking regions A and B described above. Two overlapping fragments were amplified from the ligation product using primers P5 and PTRoptrA2 for fragment C and primers P6 and PTRoptrA1 for fragment D. Subsequently ATCC46645 or Afs77 was transformed simultaneously with the overlapping fragments C and D. To complement mirC, a PCR fragment containing the mirC locus, including the promoter, was amplified using primers mirC-comp-F-XhoI and mirC-comp-R-SacI with ATCC46645 gDNA as a template. This PCR product was inserted into the pCR® 2.1-TOPO® TA vector. This vector containing the mirC fragment was digested with XhoI and SacI and inserted into the XhoI and SacI sites in the pAN7-1 plasmid, which contains the hph selection marker. The final vector (mirC-hph) was linearized with XhoI and transformed into A. fumigatus ΔmirC. Gene disruption at the correct locus was confirmed by Southern hybridization (wildtype: 1.79 kb, ΔmirC: 5.49 kb). gDNA was digested with MfeI and a 3′ Digoxigenin (DIG) probe was generated using the primers mirC-P3 and mirC-P6. A single copy ectopic integration in the complemented mutant strain (ΔmirCC1) as well as a complemented strain containing multiple integrations (ΔmirCC2) was verified by Southern hybridization. Disruption of genes AFUA_7G04730 (sit2), AFUA_6G09980 (ncr1), AFUA_1G01690 (impA), AFUA_5G10510 (impB), AFUA_3G03670 (impC), AFUA_5G07970 (impD), and AFUA_6G06620 (impE) were also confirmed by Southern hybridization.
A. fumigatus ATCC46645 mirC::GFP fusion and mirCGFP/RFP strain
A fragment containing mirC plus 1,454 bp upstream, to ensure the native promoter was included, was generated by PCR using primers mirC-GFP-F-KpnI and mirC-GFP-R-XmaI with ATCC46645 gDNA as a template. The PCR product was digested with KpnI and XmaI to insert into the KpnI and XmaI sites in the eGFP vector pUCGH which contains the hygromycin gene (hph) as the selection marker (Langfelder et al., 2001). The mirC::GFP pUCGH construct was transformed into competent E. coli OneShot Mach1-T1 (Invitrogen) and plated on LB agar containing ampicillin (50 μg/ml). Overnight growth at 37°C resulted in colonies and plasmid DNA was prepared using Qiaprep Spin Miniprep kit (Qiagen). The final construct was linearised using KpnI. A. fumigatus ΔmirC was transformed with this linearised mirC::GFP pUCGH construct. Single copy ectopic integration of the mirC::GFP (mirCGFP) was verified by Southern hybridization. The mirCGFP strain was then transformed with the pME3857 plasmid (a kind gift from Dr. Özgür Bayram) which is a mRFP::Histone2A vector with a phleoR marker (Bayram et al., 2012), thereby generating mirCGFP/RFP.
A. fumigatus growth in iron-deplete conditions
Glassware used for culturing in iron-deplete conditions was treated with 1 mM EDTA followed by HCl to remove trace iron. Minimal media (MM), contained 1% (w/v) glucose, 7 mM KCl, 2 mM MgSO4.7H2O, 11 mM KH2PO4, and trace elements as previously described (Pontecorvo et al., 1953). Following autoclaving, filter sterilized L-glutamine was added to 20 mM final concentration. For iron-replete cultures, FeSO4 (10 μM) was included. A. fumigatus ATCC46645 and Afs77 cultures at 106 conidia/ml were grown at 200 rpm, 37°C for up to 72 h. Plate assays were performed on MM agar with 5 × 103 conidia inoculum per spot. FeSO4 was included at 10 μM and 10 mM to represent iron-replete and –excess, respectively. Iron starvation was enhanced with the addition of iron chelator bathophenanthroline disulfonate (BPS; 400 μM), unless otherwise stated. Hydrogen peroxide was included at 1 and 2 mM to represent oxidative stress-inducing conditions. Plates were incubated at 37°C and colony diameters measured at 48 h. Statistical analysis was carried out using Student's t-test where mutant strains were compared to the wild-type (WT) strain.
Siderophore detection
Following preliminary analysis for siderophore production using the SideroTec assay (http://www.emergenbio.com, Ireland), culture supernatants from iron-deplete and iron-replete cultures of A. fumigatus were analyzed by RP-HPLC for the detection of extracellular siderophores at 254 nm. Supernatants were brought to 1.5 mM FeSO4 and ferrated siderophores were detected by absorbance at 440 nm (λmax of ferrated siderophores). Peaks associated with FSC+Fe and TAFC+Fe were collected and identity confirmed via LC-MS/MS. The analysis of intracellular siderophores was adapted from Szigeti et al. (2014) and Winkelströter et al. (2015). Briefly, mycelia from 72 h cultures were harvested and lyophilized in triplicate. Mycelia (50 mg) from each of the strains was added to 1,000 μl of deionized H2O and homogenized by bead beating for 10 min using tungsten beads. Lysates were centrifuged for 10,000 g for 10 min and supernatants (200 μl) removed and ferrated to a final concentration 1.5 mM FeSO4.
FSC+Fe and TAFC+Fe uptake experiments
WT, ΔmirC, and ΔmirCC2 strains were grown in iron-deplete MM for 21 h at 37°C (n = 3 each, 50 ml). Mycelia were removed using miracloth and washed with 100 ml of iron-deplete media followed by the removal of excess moisture using sterile tissue. The mycelia were wet-weighed under sterile conditions and an equal amount of mycelia was placed in each flask containing fresh iron-deplete media (50 ml). FSC+Fe (Final concentration 3.8 μM) or TAFC+Fe (Final concentration 25 μM) were spiked into each of the cultures and 1 ml aliquots of the supernatants were taken every hour for 3 h. Supernatants were brought to 1.5 mM FeSO4 and FSC+Fe or TAFC+Fe uptake was analyzed by RP-HPLC.
RP-HPLC analysis
RP-HPLC analysis was carried out using an Agilent Series 1200 HPLC System with a diode array detector (DAD) and separation across a water:acetonitrile gradient with 0.1% (v/v) TFA. For analysis and quantification of siderophores, a gradient condition of 5–55% acetonitrile over 22 min at 2 ml/min was used on a C18 column (Agilent Zorbax Eclipse XDB-C18 Semi- Preparative; 5 μm particle size; 9.4 × 250 mm) with DAD detection at 254 and 440 nm. Peaks associated with extracellular siderophores were collected and identity confirmed via LC-MS/MS.
Siderophore and protein mass spectrometry
LC-MS/MS analysis of RP-HPLC purified siderophores was carried out using an Agilent 6340 Ion Trap LC-MS System (Agilent Technologies, Santa Clara, CA). RP-HPLC purified siderophores were applied to a Zorbax SB-C18 HPLC-Chip with a 40 nl enrichment column and a 75 μm × 43 mm (5 μm particle and 300 Å pore size) analytical column (Collins et al., 2013; Owens et al., 2014; Moloney et al., 2016). For label-free quantitative (LFQ) comparative proteomics, A. fumigatus WT, ΔmirC, or ΔmirC2 were cultured for 72 h in either iron-deplete or iron-replete conditions (n = 4 for iron-deplete samples, n = 3 for iron-replete samples). Mycelial lysates were prepared in lysis buffer (100 mM Tris-HCL, 50 mM NaCl, 20 mM EDTA, 10% (v/v) glycerol, 1 mM PMSF, 1 μg/μl pepstatin A, pH 7.5) with grinding, sonication and clarified using centrifugation. The protein lysates were then precipitated using TCA/acetone and resuspended in 6 M urea, 2M thiourea, 0.1 M Tris-HCL (pH 8). After treatment with DTT and IAA, for reduction and alkylation respectively, sequencing grade trypsin combined with ProteaseMax surfactant was added (Collins et al., 2013). Digested samples were desalted prior to analysis using Zip Tips with C18 resin (Millipore). The desalted samples were analyzed via a Thermo Fisher Q-Exactive mass spectrometer coupled to a Dionex RSLC nano. LC gradients ran from 14 to 35% B (A: 0.1% (v/v) formic acid, B: 80% (v/v) acetonitrile, 0.1% (v/v) formic acid) over 2 h, and data was collected using a Top 15 method for MS/MS scans (O'Keeffe et al., 2014).
Data analysis for proteomic profiling
LFQ comparative proteomic data analysis was carried out using MaxQuant (version 1.3.0.5), as previously described (O'Keeffe et al., 2014), with LFQ algorithm engaged for protein ratio determination. Database searching was performed using Andromeda, against a protein database consisting of A. fumigatus Af293 and A1163 strains. Protein and peptide FDRs were set to 1%, with a reverse database search used. Perseus (version 1.4.1.3) was used for the quantitative analysis with a minimum of 2 peptides per protein accepted for identification. Results were further filtered to include only proteins identified from a minimum of 3 replicates from either sample set. Qualitative results were generated based on unique protein detection in either condition. This was dependant on the detection of a protein in 3 biological samples from one condition, and absence of detection in all replicates of the second condition. Functional enrichment analysis of proteins was performed using the application FungiFun2 (https://elbe.hki-jena.de/fungifun/fungifun.php) with Gene Ontology (GO) and Functional Categories (FunCat) derived ontologies (Priebe et al., 2015).
Culture conditions for microscopy
Conidia of WT, mirCGFP, and ΔmirCGFPRFP (4 × 105 per well in a 6 well plate) strains were inoculated into 4 ml of either iron-replete MM or iron-deplete MM containing BPS (400 μM), with each glass well containing a coverslip. Plates were incubated statically at 37°C for 48 h. Mycelia were visualized by both differential interference microscopy and fluorescence microscopy using an Olympus Fluo View 1,000 Laser Scanning microscope, as per Dolan et al. (2017).
RNA extraction and mRNA isolation
RNA was isolated from mycelia, ground to a fine powder in liquid N2 using the RNeasy™ Plant Mini kit (Qiagen), according to the manufacturer's instructions.
Reverse transcription quantitative PCR (RT-qPCR)
RNA was DNase treated using a DNase kit supplied by Sigma-Aldrich. cDNA synthesis was performed using qScript™ cDNA SuperMix (Quanta Biosciences) following the kit instructions. The constitutively expressed gene A. fumigatus calmodulin (calm) (Burns et al., 2005) was used as a reference gene. RT-qPCR was performed on the Light-Cycler® 480 Real-Time PCR System using the Light-Cycler® Sybr Green 1 Master Mix (Roche) as described previously (O'Hanlon et al., 2011). Three biological replicates for each treatment and three technical replicates for each biological replicate were used for the qRT-PCR analysis.
Galleria mellonella infection experiments
G. mellonella larvae (n = 30) were inoculated into the hind pro-leg with a 20 μl inoculum volume of 5 × 106 conidia. Mortality, defined by lack of movement in response to stimulation and discolouration (melanization) rates were recorded at 24 h intervals up to 96 h after injection. Kaplan-Meier survival curves were analyzed using the Log-Rank (Mantel-Cox) test for significance.
Results
Selection of genes to further study the ability of A. fumigatus to survive in iron-deplete conditions
Proteins with differential abundance under iron starvation were identified in microsomal preparations of A. fumigatus (Moloney et al., 2016). Target proteins with elevated abundance under iron-deplete conditions, in particular putative transporters, were selected for further characterisation using gene knockout strategies (Table 1). Proteins without previous assignation were designated Imp (Iron-modulated protein). Successful gene deletion of transporters was confirmed using Southern hybridization (Figures S1A,B, S2). mirC is predicted to encode a siderophore transporter of 611 amino acid residues with 12 membrane spanning domains. Residues 65–253 and 411–506 align to that of a functional family termed “siderophore iron transporter 1” which includes TAFC+Fe transporter, MirB using CATH (http://www.cathdb.info) (Haas et al., 2003; Raymond-Bouchard et al., 2012; Sillitoe et al., 2015). Absence of mirC expression in A. fumigatus ΔmirC, and restoration of expression in ΔmirCC2 and mirCGFP and was confirmed by RT-PCR (Figure S2H).
Table 1.
Targets for gene knockouts in A. fumigatus.
| Accession | Protein name | Description | p-value | Log2 (Fold Increase) | Strain |
|---|---|---|---|---|---|
| AFUA_1G01690 | ImpA | Heme binding activity | 0.000927 | 17.1 | Afs77 Ku |
| AFUA_5G10510 | ImpB | Role in transmembrane transport | 4.6E-06 | 9.4 | Afs77 Ku |
| AFUA_3G03670 | ImpC | ABC multidrug transporter | 0.000257 | 2.9 | ATCC46645 |
| AFUA_2G05730 | MirC | Putative siderophore transporter | 3.44E-06 | 1.9 | ATCC46645 |
| AFUA_6G09980 | Ncr1 | Putative sphingolipid transporter | 0.00631 | 1.0 | Afs77 Ku |
| AFUA_7G04730 | Sit2 | Putative siderophore transporter | NA | Unique | Afs77 Ku |
| AFUA_5G07970 | ImpD | Bilirubin transmembrane transporter activity | NA | Unique | Afs77 Ku |
| AFUA_6G06620 | ImpE | Role in ER to Golgi vesicle-mediated transport | NA | Unique | Afs77 Ku |
Proteins with significantly increased abundance in the A. fumigatus microsome in response to iron starvation (p < 0.05), as identified by LFQ comparative quantitative mass spectrometry with Progenesis LC-MS analysis (Moloney et al., 2016).
Growth of ΔmirC is reduced during iron limitation
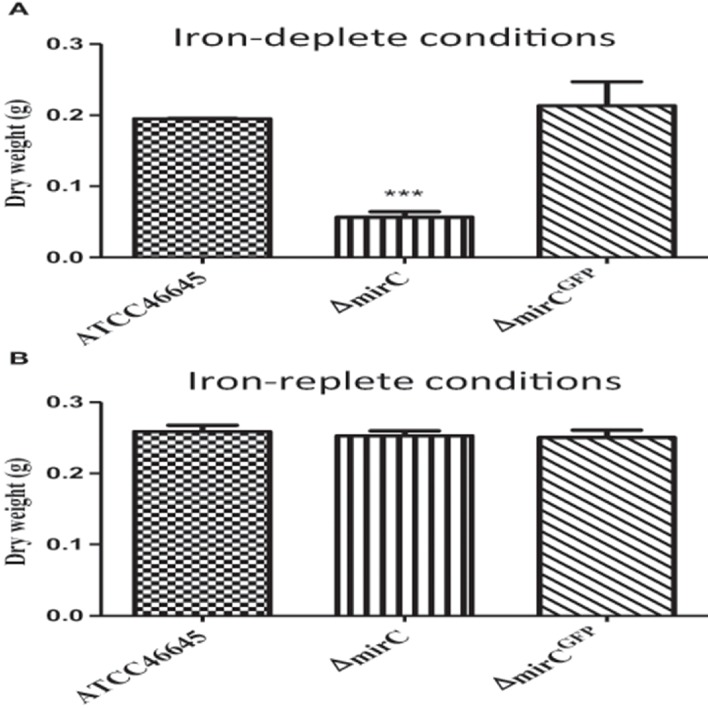
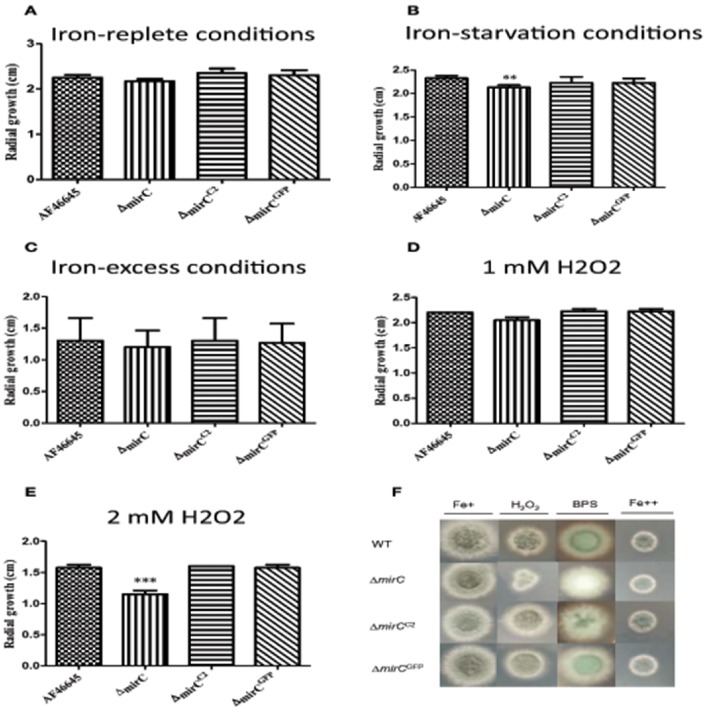
The dry weight of ΔmirC was significantly (p < 0.001) decreased at 72 h compared to WT, while there was no significant difference between mirCGFP and WT under iron-deplete conditions (Figure 1). During iron-replete growth, no difference between the dry weights of the three strains was observed (Figure 1). To confirm that ΔmirC growth was impeded under iron limitation, assays were carried out on MM plates containing the iron chelator BPS (400 μM). ΔmirC exhibited reduced conidiation and significantly decreased radial growth (p < 0.01) compared to the WT and complemented strains (ΔmirCC2 and ΔmirCGFP) under iron starvation while no difference was observed under iron-replete or iron-excess conditions (Figure 2). There was no significant difference in the dry weights under iron-deplete conditions or radial growth under iron-replete, iron starvation or iron-excess conditions of Δsit2 (AFUA_7G04730), Δncr1 (AFUA_6G09980), ΔimpA (AFUA_1G01690), ΔimpB (AFUA_5G10510), ΔimpC (AFUA_3G03670), ΔimpD (AFUA_5G07970), and ΔimpE (AFUA_6G06620) compared with the relevant WT strains (Figures S3, S4).
Figure 1.
A. fumigatus ΔmirC is sensitive to iron-deplete conditions. Dry weight of A. fumigatus WT, ΔmirC and ΔmirCGFP grown in (A) iron-deplete MM or (B) iron-replete MM for 72 h at 37°C. The dry weight of ΔmirC was significantly decreased (p < 0.001) compared to the WT while there was no statistical difference between the WT and ΔmirCGFP. There was no difference in dry weight between the strains in iron-replete MM. Statistical analyses were carried out using t-tests. (***p < 0.001).
Figure 2.
Phenotypic analysis of A. fumigatus ATCC46645, ΔmirC, ΔmirCC2, and ΔmirCGFP strains. The radial growth was measured in A. fumigatus WT and deletion strains grown on MM plates under (A) iron sufficiency, (B) iron starvation (400 μM BPS), (C) iron excess (10 mM FeSO4), (D) 1 mM hydrogen peroxide, and (E) 2 mM hydrogen peroxide after 48 h at 37°C. Radial growth of ΔmirC was significantly decreased in response to iron starvation (p < 0.01) and oxidative stress at 2 mM hydrogen peroxide (p < 0.001). ΔmirC also displayed reduced conidiation compared to the WT and complemented strains when exposed to BPS (400 μM). (F) A. fumigatus WT, ΔmirC, ΔmirCC2, and ΔmirCGFP grown on MM plates containing iron-replete (Fe+), H2O2 (2mM), BPS (400 μM), and iron-excess (Fe++) for 48 h at 37°C. ΔmirC was significantly more sensitive to H2O2 compared to the WT and complemented strains. ΔmirC also has decreased conidiation compared to the WT and complemented strain when exposed to BPS (400 μM). **p < 0.01; ***p < 0.001.
A. fumigatus ΔmirC is more sensitive to hydrogen peroxide
Growth of ΔmirC in 2 mM hydrogen peroxide was significantly reduced compared to the WT (p < 0.001), while there was no difference between the WT and the complemented strains (ΔmirCC2 and ΔmirCGFP) (Figure 2). There was no significant difference in the radial growth of Δsit2, Δncr1, ΔimpA, ΔimpB, ΔimpC, ΔimpD, and ΔimpE compared with the relevant WT strains under different concentrations of hydrogen peroxide (Figures S3D,E, S4D,E).
Differential extracellular siderophore levels in ΔimpC but not ΔmirC under iron limitation
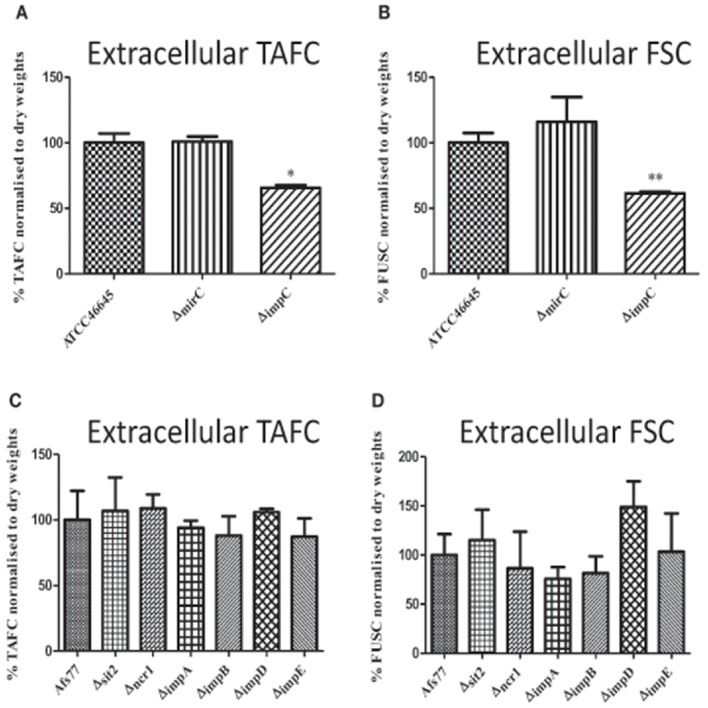
Extracellular siderophore levels from WT, ΔmirC, ΔimpC, and ΔimpD grown under iron limitation for 72 h were analyzed by RP-HPLC. Immediately prior to analysis, supernatants were brought to 1.5 mM FeSO4. Siderophore levels were normalized to biomass and the identity of peaks associated with FSC+Fe and TAFC+Fe was confirmed via LC-MS/MS (Figure S5). There was no significant difference in the level of extracellular siderophores produced by ΔmirC at 72 h (Figures 3A,B). Interestingly, ΔimpC showed a significant decrease in levels of TAFC+Fe (p < 0.05) and FSC+Fe (p < 0.01) (Figures 3A,B). Finally, there was no significant difference in the levels of TAFC+Fe or FSC+Fe secreted by Δncr1, Δsit2, ΔimpA, ΔimpB, ΔimpD, ΔimpE compared to the WT strain, under iron-deplete conditions (Figures 3C,D). There was no significant difference in the uptake of TAFC+Fe and FSC+Fe between WT, ΔmirC, and ΔmirCC2 at the measured time-points (Figure S7). This suggests that deletion of mirC does not significantly alter the rate of uptake of TAFC+Fe or FSC+Fe.
Figure 3.
Extracellular siderophore production in A. fumigatus deletion strains. RP-HPLC analysis was used to measure extracellular siderophore levels in A. fumigatus WT and transporter knock-out strains grown in iron-deplete conditions for 72 h at 37°C (n = 3). Siderophore levels were normalized to biomass. Supernatant was ferrated at 1.5 mM FeSO4 before the analysis. Levels of (A) TAFC+Fe and (B) FSC+Fe were decreased in ΔimpC while no significant difference was observed in siderophore levels for (C,D) ΔmirC or other knock-out strains. *p < 0.05; **p < 0.01.
Intracellular ferricrocin level is significantly decreased in ΔmirC mycelia under iron limitation
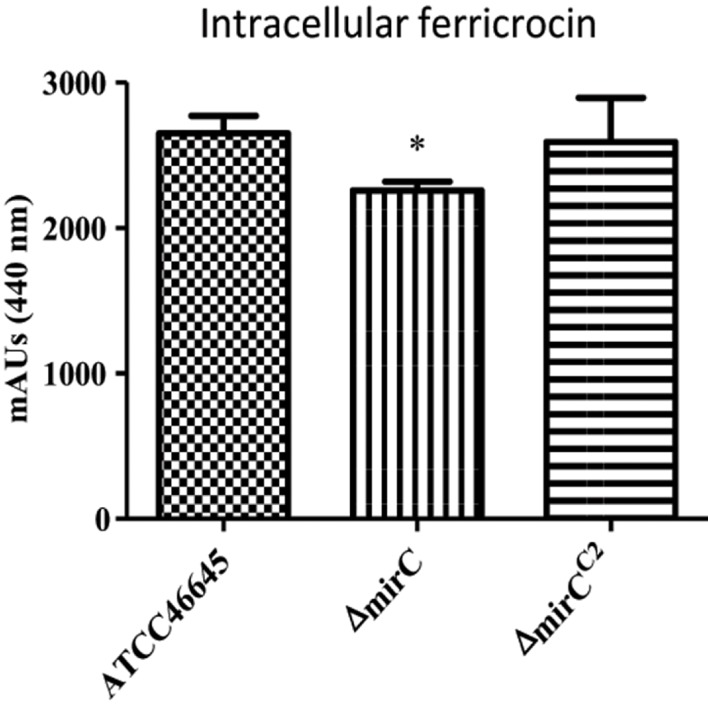
Mycelia from the WT, ΔmirC, and ΔmirCC2 were harvested after 72 h in iron-deplete conditions (n = 4). Lysed mycelial supernatants were ferrated to a final concentration of 1.5 mM FeSO4 before RP-HPLC analysis at 440 nm. The identity of the peak associated with FC identity was also confirmed by LC-MS/MS (Figure S5). Intracellular FC was found to be significantly reduced in the mycelia of ΔmirC vs. the WT and ΔmirCC2 under iron-deplete conditions (p < 0.05) (Figure 4). However, there was no significant difference in the amount of FC in Δsit2, Δncr1, and ΔimpA-E under iron-deplete conditions, compared to WT (Figures S3, S6).
Figure 4.
Intracellular siderophore content reduced in ΔmirC during iron-deplete growth. FC content of dried mycelia was analyzed by RP-HPLC. Ferricrocin was significantly reduced in ΔmirC vs. WT and ΔmirCC2 mycelia when grown in MM for 72 h at 37°C (p < 0.05). Statistical analyses were carried out using t-tests. (*p < 0.05).
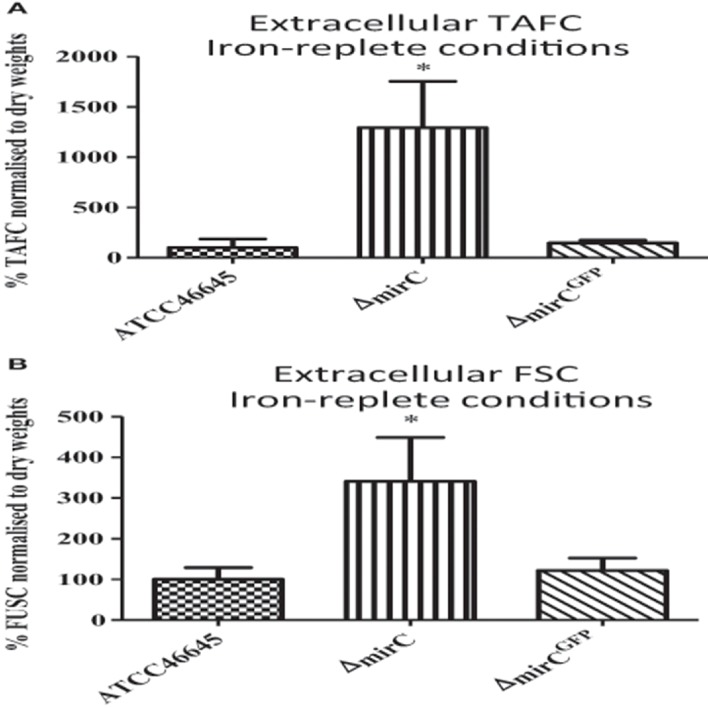
Increased extracellular siderophore levels in ΔmirC under iron sufficiency
Supernatants from WT, ΔmirC, and mirCGFP, grown under iron sufficiency for 72 h, were analyzed by RP-HPLC at 440 nm (n = 3). Prior to analysis supernatants were brought to 1.5 mM FeSO4. Siderophore levels were normalized to biomass. Surprisingly, there was a significant increase in the levels of TAFC+Fe (p < 0.05) and FSC+Fe (p < 0.05) produced by ΔmirC, compared to WT (Figure 5).
Figure 5.
Extracellular siderophore levels in ΔmirC are increased during iron-replete growth. RP-HPLC analysis was used to measure extracellular siderophore levels in A. fumigatus WT, ΔmirC, and ΔmirCGFP grown in iron-replete conditions for 72 h at 37°C (n = 4). Siderophore levels were normalized to biomass. Supernatant was ferrated at 1.5 mM FeSO4 before the analysis. There was a significant increase in the levels of (A) TAFC+Fe (p < 0.05) and (B) FSC+Fe (p < 0.05) in ΔmirC compared to WT. Statistical analyses were carried out using t-tests (*p < 0.05).
Label-free quantitative proteomic analysis reveals significant alterations to the proteome of ΔmirC compared to the WT during both iron-deplete and -replete growth
High sensitivity LFQ proteomic analysis revealed 511 proteins with differential abundance were identified in ΔmirC compared with WT during iron limitation (n = 4; 72 h; Figure 6); 251 showing increased abundance and 260 decreased abundance (p < 0.05, log2 fold change > 1) (Tables S4, S5). In addition, 297 proteins showed differential abundance in ΔmirC compared with WT during iron-replete growth (n = 3); 150 with increased abundance and 147 with decreased abundance (p < 0.05, log2 fold change > 1) (Tables S8, S9).
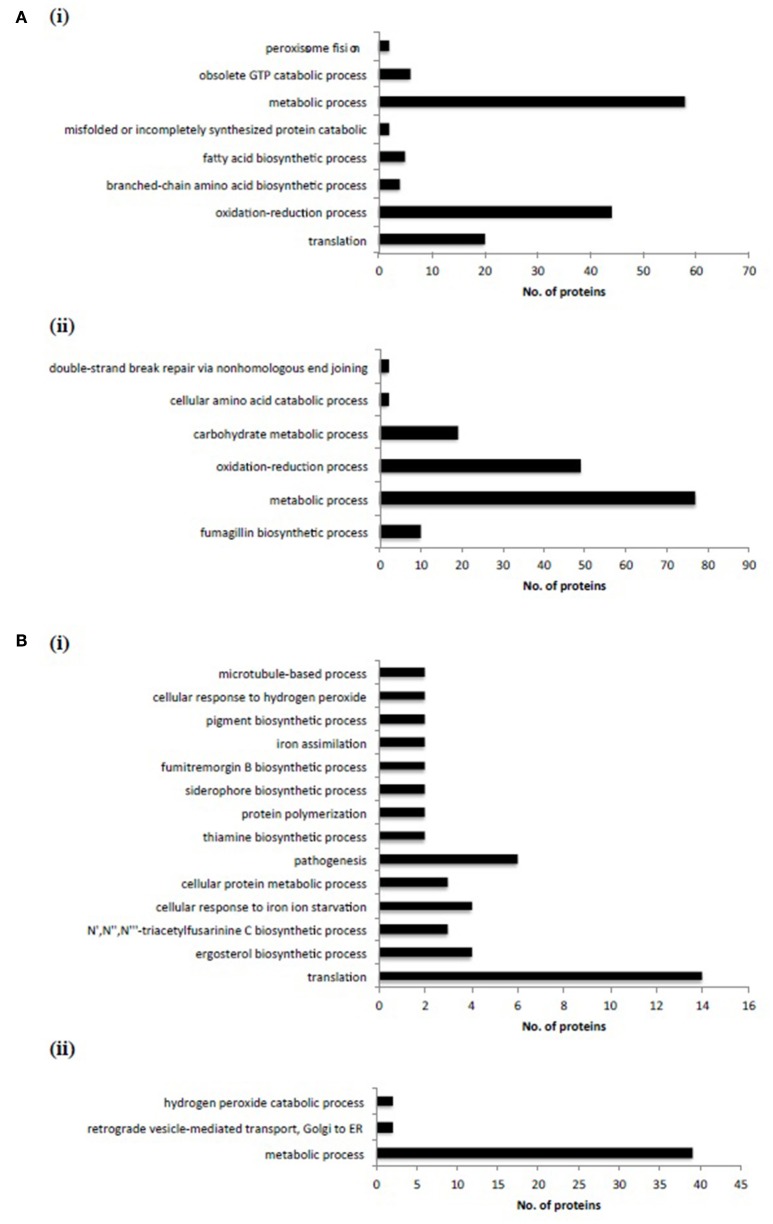
Figure 6.
Label-free proteomic analysis revealed significant proteomic remodeling in ΔmirC compared to WT during iron-replete and –deplete growth. (A) GO biological processes significantly enriched in proteins with (i) increased and (ii) decreased abundance during iron-replete growth in ΔmirC compared to WT. (B) GO biological processes significantly enriched in proteins with (i) increased and (ii) decreased abundance during iron-deplete growth in ΔmirC compared to WT. Notably, the GO biological process of “translation” was significantly represented in proteins with increased abundance during iron-replete and –deplete growth.
Differentially abundant proteins in ΔmirC compared with WT were analyzed for functional category enrichment (p < 0.05) using FungiFun2 with GO classification ontology (Priebe et al., 2015). GO biological process categories significantly represented by proteins with increased abundance in ΔmirC compared with WT during iron-deplete growth included; translation (n = 20), oxidation-reduction process (n = 44), branched-chain amino acid biosynthetic process (n = 4), fatty acid biosynthetic process (n = 5), misfolded, or incompletely synthesized protein catabolic process (n = 2), metabolic process (n = 58), obsolete GTP catabolic process (n = 6), and peroxisome fission (n = 2) (Figure 6). GO biological process categories significantly represented by proteins with decreased abundance in ΔmirC compared with WT during iron-deplete growth included; fumagillin biosynthetic process (n = 10), metabolic process (n = 77), oxidation-reduction process (n = 49), carbohydrate metabolic process (n = 19), cellular amino acid catabolic process (n = 2), and double-strand break repair via non-homologous end joining (n = 2) (Figure 6). Proteomic analysis also revealed increased abundance of proteins involved in siderophore biosynthesis in ΔmirC compared to WT during iron-deplete growth, including L-ornithine-N5-monooxygenase, SidA (AFUA_2G07680) and SidI (AFUA_1G17190) (Table 2). There was also a decreased abundance of TAFC transporter MirB (AFUA_3G03640) and putative transporter AFUA_3G03670 (ImpC) in ΔmirC, compared to WT (Table 3). GO biological process categories significantly represented by proteins with increased abundance in ΔmirC compared with WT during iron-replete growth included; translation (n = 14), ergosterol biosynthetic process (n = 4), N′,N″,N‴-triacetylfusarinine C biosynthetic process (n = 3), cellular response to iron starvation (n = 4), cellular protein metabolic process (n = 3), pathogenesis (n = 6), thiamine biosynthetic process (n = 2), protein polymerization (n = 2), siderophore biosynthetic process (n = 2), fumitremorgin B biosynthetic process (n = 2), iron assimilation (n = 2), pigment biosynthetic process (n = 2), cellular response to hydrogen peroxide (n = 2), and microtubule-based process (n = 2) (Figure 6). GO biological process categories significantly represented by proteins with decreased abundance in ΔmirC compared with WT during iron-replete growth included; metabolic process (n = 39), retrograde vesicle-mediated transport (n = 2), and hydrogen peroxide catabolic process (n = 2) (Figure 6).
Table 2.
Proteins associated with siderophore biosynthesis and transporter activity with increased abundance in A. fumigatus ΔmirC mycelia grown for 72 h in iron-deplete MM compared to A. fumigatus WT (See also Tables S3, S6, S7, S10 and S11).
| Protein description | Log2 (Fold Increase) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Ortholog(s) have iron ion transmembrane transporter activity | Unique | N/A | 4 | 33.7 | AFUA_6G12550 |
| Ortholog(s) have guanine nucleotide transmembrane transporter activity, role in cellular iron ion homeostasis | 1.54 | 5.53E-04 | 17 | 56.6 | AFUA_1G07450 |
| L-ornithine N5-oxygenase, first committed step in siderophore biosynthesis. SidA. | 1.47 | 4.72E-06 | 48 | 83.2 | AFUA_2G07680 |
| Cytochrome c; transcript derepressed during iron starvation in hapX mutant | 1.02 | 6.79E-03 | 5 | 14 | AFUA_2G13110 |
| Putative peroxisome biogenesis factor | 1.32 | 1.70E-03 | 5 | 26.8 | AFUA_6G07740 |
| Putative peroxin with a predicted role in peroxisome biogenesis | 1.14 | 4.34E-03 | 3 | 12 | AFUA_2G17100 |
| Putative long-chain-fatty-acid-CoA ligase. SidI. | 1.09 | 5.69E-04 | 38 | 75.8 | AFUA_1G17190 |
Table 3.
Proteins associated with siderophore biosynthesis and transporter activity with decreased abundance in A. fumigatus ΔmirC mycelia grown for 72 h in iron-deplete MM compared to A. fumigatus WT (See also Tables S3, S6, S7, S10 and S11).
| Protein description | Log2(Fold Decrease) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Putative siderophore iron transporter;SrbA-regulated during hypoxia. MirB. | −1.55 | 1.04E-03 | 9 | 26.1 | AFUA_3G03640 |
| ABC multidrug transporter with a predicted role in iron metabolism;part of an iron-regulated gene cluster. ImpC. | −1.84 | 5.55E-03 | 7 | 9.7 | AFUA_3G03670 |
Several GO-enriched categories representing proteins with increased abundance during iron-replete growth in ΔmirC compared with WT were also observed in WT with increased abundance in response to iron starvation. These categories included, translation, cellular protein metabolic process, siderophore biosynthetic process, ergosterol biosynthetic process, and N′,N″,N‴-triacetylfusarinine C biosynthetic process (Tables S8, S12). Interestingly, specific proteins associated with siderophore biosynthesis and transport functions were among these (Table 4).
Table 4.
Proteins associated with siderophore biosynthesis and transporter activity with increased abundance in A. fumigatus ΔmirC mycelia grown for 72 h in iron-replete MM compared to A. fumigatus WT (See also Tables S3, S6, S7, S10 and S11).
| Protein description | Log2 (Fold Increase) | p-value | Peptides | Sequence coverage [%] | Protein IDs |
|---|---|---|---|---|---|
| Putative long-chain-fatty-acid-CoA ligase. SidI. | Unique | N/A | 38 | 75.8 | AFUA_1G17190 |
| Putative siderophore biosynthesis lipase/esterase. EstB. | Unique | N/A | 9 | 58.9 | AFUA_3G03660 |
| L-ornithine N5-oxygenase, first committed step in siderophore biosynthesis. SidA. | 6.42 | 2.55E-04 | 48 | 83.2 | AFUA_2G07680 |
| Putative siderophore iron transporter;SrbA-regulated during hypoxia. MirB. | 3.48 | 8.17E-03 | 9 | 26.1 | AFUA_3G03640 |
| Putative siderophore transporter;expression upregulated under low iron conditions. MirD. | 2.35 | 1.71E-03 | 7 | 13.2 | AFUA_3G03440 |
| Hydroxyornithine transacylase;involved in extracellular siderophore biosynthesis;essential for triacetylfusarinine C biosynthesis. SidF. | 2.01 | 1.76E-04 | 38 | 90 | AFUA_3G03400 |
| Putative siderophore transporter;SrbA-regulated during hypoxia. Sit1. | 1.73 | 4.72E-02 | 5 | 11.4 | AFUA_7G06060 |
| Putative siderophore biosynthesis lipase/esterase. SidJ. | 1.15 | 4.65E-03 | 15 | 66.7 | AFUA_3G03390 |
hapX expression is up-regulated in ΔmirC under iron-replete but not iron-deplete conditions compared to WT
qRT-PCR analysis of A. fumigatus hapX expression in the ΔmirC and WT strains was carried out after growth in iron-deplete or iron-replete minimal media for 24–72 h (Figure 7). hapX expression was significantly up-regulated (p < 0.001) in ΔmirC (7.2-fold at 48 h and 13.8-fold at 72 h) under iron-replete conditions. hapX expression was not significantly different between the ΔmirC and WT strains under iron-deplete conditions (72 h; data not shown), or at 24 h under iron-replete conditions.
Figure 7.
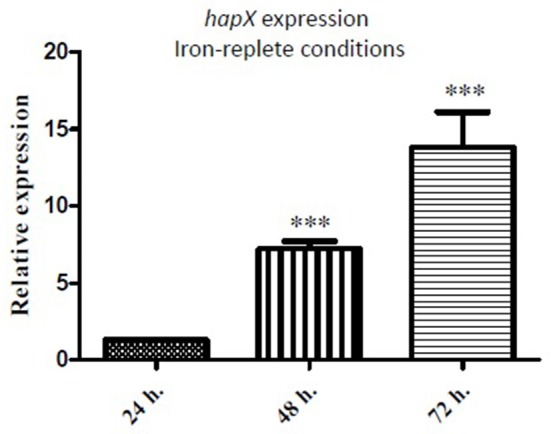
hapX is up-regulated in ΔmirC during iron-replete growth. qRT-PCR analysis of hapX expression in ΔmirC vs. WT after growth under iron-deplete or iron-replete minimal media for 72 h. hapX expression was significantly up-regulated (p < 0.001) in ΔmirC under iron-replete but not iron-deplete conditions. ***p < 0.001.
mirC appears to exhibit an intracellular localization
MirC-GFP fusion strain fluorescence was only evident under iron starvation, not iron sufficiency, in mirCGFP (Figure 8), and was localized intracellularly, possibly to vacuole-like structures. There was no evidence of MirC-GFP presence in the plasma membrane, although it is not possible to definitively assign the intracellular localization of the fusion protein, due to apparent degradation-even though intact MirC-GFP was also detected by Western blot analysis (Figure S8). Moreover, no fluorescence was observed in the WT control under iron-replete or iron-deplete conditions. mirCGFP was also transformed to create a MirC::GFP mRFP::Histone2A fusion strain to further elucidate the location of MirC-GFP expression. MirC-GFP fluorescence did not co-localize with mRFP::Histone2A fluorescence, which indicates that it is not localized to nuclei (Figure 8).
Figure 8.
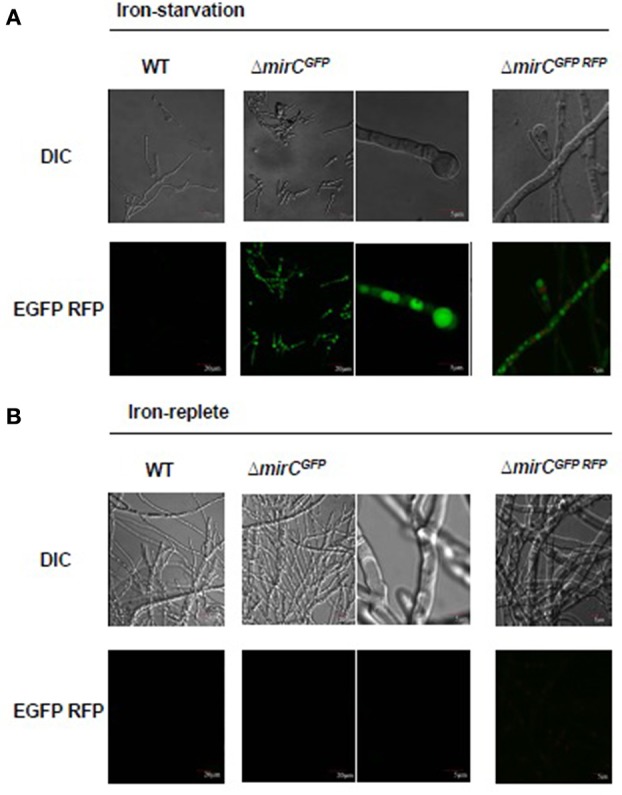
MirC exhibits intracellular localization during iron-deplete but not –replete growth. WT, ΔmirCGFP, and ΔmirCGFPRFP were grown in (A) iron-starved (400 μM BPS) and (B) iron–replete conditions for 48 h at 37°C. Hyphae were observed by differential interference microscopy (DIC). Fluorescence was localized intracellularly, possibly to vacuoles, in ΔmirCGFP, and ΔmirCGFPRFP during iron-starved but not –replete growth.
Disruption of mirC decreases virulence in G. mellonella
The virulence of ΔmirC in G. mellonella was significantly reduced compared to the WT strain (Figure 9). The Mantel-Cox test revealed that the survival curves of WT and ΔmirC were significantly different (p < 0.002).
Figure 9.
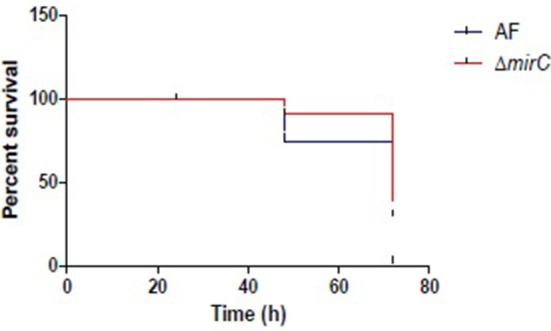
ΔmirC exhibits decreased virulence in G. mellonella. The survival curves of G. mellonella larvae when infected with 5 × 106 conidia per insect for ATCC46645 and ΔmirC. The Mantel-Cox test revealed that the virulence is significantly reduced in ΔmirC vs. the WT strain (p < 0.002).
Discussion
Despite significant efforts (Haas et al., 2003; Philpott and Protchenko, 2008; Moloney et al., 2016), little is known about the transporters involved in siderophore–mediated iron acquisition. Consequently, we functionally investigated putative siderophore transporters identified in previous proteomic analysis (Moloney et al., 2016). Deletion of mirC in A. fumigatus significantly decreased growth under iron-depletion and oxidative stress conditions, reduced intracellular ferricrocin levels, and attenuated virulence in the G. mellonella model. Additionally, LFQ proteomic analysis of ΔmirC revealed an increased abundance of proteins involved in siderophore biosynthesis in both iron-replete and -deplete conditions, indicating a dysregulated iron-sensing capacity. MirC appears to be located intracellularly in A. fumigatus. Interestingly, there was no decrease in growth under iron-deplete or oxidative conditions in the other deletion strains presented herein.
Significantly increased abundance of MirC was identified in the microsomal proteome of A. fumigatus under iron starvation (Moloney et al., 2016). The GATA-type transcription factor, SreA which is an important transcriptional regulator of iron homeostasis represses mirC expression in the presence of iron (Schrettl et al., 2008). Accordingly, it appears that mirC is involved in iron homeostasis since reduced growth was observed during iron starvation, following mirC deletion (p < 0.001) (Figures 1, 2). Compared to WT, no significant difference in extracellular siderophore production or uptake was observed for ΔmirC during iron starvation, thereby excluding a direct role for this transporter in siderophore uptake or efflux. This result is in accordance with previous work by Haas et al. (2003) in which Aspergillus nidulans mirC ortholog was expressed in an S. cerevisiae strain deficient in high affinity iron acquisition mechanisms (Haas et al., 2003). Analysis of iron uptake from siderophores TAFC, FC, ferrichrome, and enterobactin in this strain did not indicate any role for mirC in siderophore uptake.
In contrast, levels of intracellular FC decreased in ΔmirC compared to WT during iron-deplete growth (p < 0.05) (Figure 4). FC predominantly functions in the transport and storage of intracellular iron following ferri-siderophore uptake (Schrettl et al., 2007; Wallner et al., 2009; Blatzer et al., 2011). Depletion of intracellular siderophores, FC and HFC, has previously been implicated in impaired oxidative stress tolerance in hyphae and conidia (Schrettl et al., 2007; Blatzer et al., 2011). Iron and redox homeostasis mechanisms are tightly intertwined processes in A. fumigatus (Brandon et al., 2015) which is largely a result of the redox potential of free iron and its requirement as a co-factor for many antioxidant enzymes. Following the reduced intracellular FC content, it is logical that ΔmirC also displayed decreased tolerance to hydrogen peroxide compared to WT during iron-replete growth (p < 0.001) (Figure 2). Interestingly, ΔmirC exhibited significantly increased extracellular siderophore production during iron-replete growth suggesting a dysregulation of iron homeostasis (Figure 5). Thus, for a global and in-depth analysis of the impact of mirC deletion in A. fumigatus, large-scale LFQ proteomic analysis was employed to compare ΔmirC to WT during both iron-deplete and -replete growth.
Differential abundance of a total of 297 proteins (p < 0.05) was observed during iron-replete growth in ΔmirC compared to WT (Tables S8, S9). Moreover, with enrichment of GO biological process categories including translation (n = 14), ergosterol biosynthetic process (n = 4), N′,N″,N‴-triacetylfusarinine C biosynthetic process (n = 3), and siderophore biosynthetic process (n = 2), the iron-replete induced proteome of ΔmirC actually reflected that of WT during iron-deplete growth (Figure 6; Table 4; Table S8). In fact, 18 of the proteins observed in these categories also showed increased abundance in WT during iron-deplete growth compared to iron-replete. Proteins involved in siderophore biosynthesis with increased abundance in both protein datasets included L-ornithine N5-oxygenase SidA (AFUA_2G07680). Additionally, peroxisomally located siderophore biosynthetic enzymes required for FSC and TAFC biosynthesis were also shared; mevalonyl-CoA ligase SidI (AFUA_1G17190), transacylase SidF (AFUA_3G03400), and mevalonyl-CoA hydratase SidH (AFUA_3G03410). Strikingly, 27 proteins associated with the ribosome showed differential abundance in ΔmirC compared with WT during iron-replete growth (Tables S8, S9). Of the 19 ribosome-associated proteins with increased abundance, 14 were also increased in WT during iron-deplete growth compared to iron-replete. Translation and ribosome biogenesis are iron-consuming processes and HapX-mediated repression of ribosome biogenesis has been observed in A. fumigatus during iron starvation (Schrettl et al., 2010). So, the increased abundance of ribosomal proteins during iron starvation in WT may represent a ribosomal reprogramming necessary for maintaining viability during reduced iron availability. HapX-mediated regulation of enzymes related to cellular detoxification processes has also been previously observed (Schrettl et al., 2010). Enzymatic antioxidant mechanisms including peroxidases and catalases are dependent on iron as a heme constituent. Accordingly, ΔsidC conidia with reduced HFC and iron content also show reduced catalase (CatA; AFUA_6G03890) activity implicated in hydrogen peroxide sensitivity. In contrast, ΔsidC hyphae showed no reduced catalase activity in the form of catalase 1 (Cat1; AFUA_3G02270) and catalase 2 (Cat2; AFUA_8G01670) activity (Schrettl et al., 2007). Interestingly, Cat1 and Cat2 showed decreased abundance in ΔmirC during iron-replete growth compared to WT and thus may at least partially underlie the impaired oxidative stress tolerance observed (Figure 2). Though there was no difference in ΔsidC hyphae compared with WT, Schrettl et al. (2007) reported a reduction in Cat1 and Cat2 activity between iron-deplete and –replete growth (Schrettl et al., 2007). Given that ΔmirC displays an iron-starved phenotype during iron-replete growth, it is likely that the decreased abundance of these proteins is a result of an iron starvation response. This is supported by HapX-mediated repression of cat1 transcription during iron starvation (Schrettl et al., 2010). HapX is an iron-repressed bZip transcription factor that, in addition to SreA, governs iron homeostasis by inducing iron-acquiring pathways and repressing iron-requiring pathways. RT-qPCR revealed that hapX expression was up-regulated in ΔmirC between 7 and 13-fold (p < 0.001, at 48 and 72 h growth) during iron sufficiency (Figure 7). Thus, there is strong evidence that during iron-replete growth ΔmirC is sensing an iron-limiting environment.
During iron-deplete growth, differential abundance of a total of 511 proteins (p < 0.05) indicated significant phenotypic remodeling in ΔmirC compared with WT (Tables S4, S5; Figure 6; Tables 2, 3). Similar to the proteome during iron-replete growth, the GO biological process of translation was also significantly enriched (n = 20). A total of 35 proteins associated with the ribosome showed differential abundance in ΔmirC compared with WT during iron-replete growth. Of these, 29 proteins showed increased abundance, 13 of which were also increased in WT during iron-deplete growth compared to iron-replete. Interestingly, 3 translation elongation factor proteins were uniquely detected in ΔmirC EF-1 (AFUA_1G06390), EF-2 (AFUA_2G13530), and eEF-3 (AFUA_7G05660) compared with WT. Importantly several siderophore biosynthetic enzymes exhibited increased abundance in mirC compared with WT including SidA and SidI (Table 4). Together these data indicate a dysregulation of the response of A. fumigatus to iron limitation, following deletion of mirC. The consequence of this differential response to iron starvation is evident in the reduced virulence displayed by ΔmirC compared to WT in G. mellonella (p < 0.002) (Figure 9), where intact mechanisms for siderophore-mediated iron acquisition augment virulence (Slater et al., 2011). Significant insight into the iron-starved mycelial proteome of WT A. fumigatus is also apparent. Comparing iron-deplete to –replete growth in WT; 806 proteins showed increased abundance while 518 decreased (p < 0.05) (Tables S12, S13). Similarly, comparing iron-deplete to –replete growth in ΔmirC; 952 proteins showed increased abundance while 622 decreased (p < 0.05) (Tables S14, S15), indicating the magnitude of proteome remodeling induced under iron starvation in A. fumigatus.
Siderophore biosynthesis appears to be at least partially compartmentalized, with SidI-, SidH-, and SidF-catalyzed biosynthetic steps occurring in peroxisomes (Gründlinger et al., 2013b), while N5 transacylase, SidL is localized to the cytoplasm suggesting the following biosynthetic step occurs there (Blatzer et al., 2011). Cellular localization of the later biosynthetic steps awaits elucidation, including that of NRPSs, SidD, and SidC, although identification of both NRPSs from the microsomal fraction during iron starvation suggests that these proteins may either be localized at membranes or enclosed in membrane-bound compartments (Moloney et al., 2016). Such localization has been reported in Pseudomonas aeruginosa where the formation of “siderosomes” at membranes is postulated to increase biosynthetic efficiency (Imperi and Visca, 2013; Gasser et al., 2015). Recent work has also revealed the localization of siderophore procurement and processing mechanisms. Immunofluorescence studies with TAFC+Fe transporter, MirB indicated localization to vesicles at hyphal tips and the cytoplasm during iron starvation (Raymond-Bouchard et al., 2012; Moore, 2013). Finally, fluorescently derivatised FSC+Fe localized to vacuoles following re-uptake (Moloney et al., 2016). Herein, MirC-GFP showed intracellular fluorescence during iron-deplete but not –replete growth (Figure 8), moreover, mirCGFP/RFP revealed that MirC-GFP fluorescence did not co-localise to the nucleus (Figure 8). Though immunofluorescence indicated localization of MirB in vesicles at hyphal tips (Moore, 2013; Raymond-Bouchard et al., 2012), to the best of our knowledge MirC represents the first SIT to be localized to an intracellular location. While GFP localization studies did not exhibit fluorescence during iron-replete growth, it is clear from the observable phenotype that MirC also plays a role during iron sufficiency. Notably, MirC was not uniquely abundant in the iron-deplete microsomal proteome, but showed a 3.81-fold increase in abundance (p < 0.05) (Moloney et al., 2016). This indicates that MirC is present during iron-replete growth which suggests that it may function during iron sufficiency. Thus, labeled MirC levels may be too low such to facilitate detection in iron-replete ΔmirCGFP.
Interestingly, FC/HFC deficient strains generated by deletion of either sidL or sidC display a remarkably similar phenotype to ΔmirC. Deletion of sidL or sidC resulted in reduced growth consequent to iron starvation and oxidative stress, as well as up-regulation of hapX in iron-replete conidia (Schrettl et al., 2007; Blatzer et al., 2011). Furthermore, deficiency in intracellular siderophore content in ΔsidL and ΔsidC has been associated with reduced conidiation (Blatzer et al., 2011; Schrettl et al., 2007), which is also observed for ΔmirC during iron-deplete growth (Figure 2). Thus, many phenotypes observed in A. fumigatus following mirC deletion may be accounted for by a deficiency in FC/HFC content. Importantly, depletion of FC content accords with the iron-replete phenotype of ΔmirC. FC is utilized by A. fumigatus during iron-deplete conditions, and at lower levels during iron–replete growth. ΔsidL conidia harvested during the latter condition exhibited an iron-starved phenotype, whereby up-regulated hapX expression was evident (Blatzer et al., 2011). Thus, despite a low abundance, we propose that MirC functions in FC biosynthesis during iron-replete, as well as iron-deplete growth. The lack of complete abrogation of FC production in ΔmirC likely reflects redundancy in the siderophore-related proteome.
No specific phenotype was observed for A. fumigatus Δsit2, however it has recently been shown that Sit2 is involved in ferrioxamine B uptake (Park et al., 2016). ImpC (AFUA_3G03670) is a putative ABC transporter and mRNA expression of ImpC is induced in response to iron-deplete conditions (Kragl et al., 2007). Significantly increased abundance of ImpC was found in the microsomal fractions of iron-depleted mycelia in A. fumigatus (Moloney et al., 2016). Furthermore, ImpC has been found to be regulated by the iron-sensitive transcription factor SreA and was suggested to be involved in siderophore secretion (Schrettl et al., 2008). There was a significant reduction in extracellular TAFC and FSC at 72 h growth of ΔimpC under iron-deplete conditions, and suggests a role for this ABC transporter in the export of desferri-siderophores from the cell. However, growth of ΔimpC was unaffected under iron-deplete or oxidative stress conditions. This indicates that there may be a functional redundancy in the export of siderophores in A. fumigatus.
In conclusion, we have identified MirC as a putative vacuolar membrane protein, elucidated its role in ferricrocin biosynthesis and oxidative stress, and reveal dysregulation of siderophore biosynthetic systems in the absence of MirC under iron-replete conditions.
Ethics statement
Study exempt as animals used were insect larvae (Galleria mellonella).
Author contributions
EM, NM, SKD, and LR carried out experimental work. SD, EM, NM, and RO analyzed data. RO contributed methodology and proteomics expertise. EM, NM, and SD wrote the manuscript. All authors reviewed and approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by a Science Foundation Ireland Investigator Award to SD (12/IP/1695). Quantitative mass spectrometry facilities were funded by a competitive infrastructure award from Science Foundation Ireland [12/RI/2346 (3)]. qPCR facilities were funded by Science Foundation Ireland (SFI/07/RFP/GEN/F571/EC07). Southern analyses were facilitated by access to an Image Station funded by Science Foundation Ireland Career Development Award 13/CDA/2142 to Dr. Özgür Bayram. We acknowledge Aoife McHugh for technical input in the early stage of this work. The authors are also extremely grateful to Dr. Özgür Bayram and Professor Sven Krappmann for the gifts of the pME3857 and pSK275 plasmids respectively.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00418/full#supplementary-material
References
- Bayram Ö., Bayram Ö. S., Ahmed Y. L., Maruyama J.-I., Valerius O., Rizzoli S. O., et al. (2012). The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet. 8:e1002816. 10.1371/journal.pgen.1002816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatzer M., Schrettl M., Sarg B., Lindner H. H., Pfaller K., Haas H. (2011). SidL, an Aspergillus fumigatus transacetylase involved in biosynthesis of the siderophores ferricrocin and hydroxyferricrocin. Appl. Environ. Microbiol. 77, 4959–4966. 10.1128/AEM.00182-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon M., Howard B., Lawrence C., Laubenbacher R. (2015). Iron acquisition and oxidative stress response in Aspergillus fumigatus. BMC Syst. Biol. 9:19. 10.1186/s12918-015-0163-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C., Geraghty R., Neville C., Murphy A., Kavanagh K., Doyle S. (2005). Identification, cloning, and functional expression of three glutathione transferase genes from Aspergillus fumigatus. Fungal Genet. Biol. 42, 319–327. 10.1016/j.fgb.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Cassat J. E., Skaar E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C., Keane T. M., Turner D. J., O'Keeffe G., Fitzpatrick D. A., Doyle S. (2013). Genomic and proteomic dissection of the ubiquitous plant pathogen, Armillaria mellea: Toward a new infection model system. J. Proteome Res. 12, 2552–2570. 10.1021/pr301131t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais T. R. T., Keller N. P. (2009). Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22, 447–465. 10.1128/CMR.00055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan S. K., Bock T., Hering V., Owens R. A., Jones G. W., Blankenfeldt W., et al. (2017). Structural, mechanistic and functional insight into gliotoxin bis-thiomethylation in Aspergillus fumigatus. Open Biol. 7:160292. 10.1098/rsob.160292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser V., Guillon L., Cunrath O., Schalk I. J. (2015). Cellular organization of siderophore biosynthesis in Pseudomonas aeruginosa: Evidence for siderosomes. J. Inorg. Biochem. 148, 27–34. 10.1016/j.jinorgbio.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Gründlinger M., Gsaller F., Schrettl M., Lindner H., Haas H. (2013a). Aspergillus fumigatus SidJ mediates intracellular siderophore hydrolysis. Appl. Environ. Microbiol. 79, 7534–7536. 10.1128/AEM.01285-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründlinger M., Yasmin S., Lechner B. E., Geley S., Schrettl M., Hynes M., et al. (2013b). Fungal siderophore biosynthesis is partially localized in peroxisomes. Mol. Microbiol. 88, 862–875. 10.1111/mmi.12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H. (2014). Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 31, 1266–1276. 10.1039/C4NP00071D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., Eisendle M., Turgeon B. G. (2008). Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46, 149–187. 10.1146/annurev.phyto.45.062806.094338 [DOI] [PubMed] [Google Scholar]
- Haas H., Schoeser M., Lesuisse E., Ernst J. F., Parson W., Abt B., et al. (2003). Characterization of the Aspergillus nidulans transporters for the siderophores enterobactin and triacetylfusarinine C. Biochem. J. 371, 505–513. 10.1042/bj20021685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J. M. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219, 1–14. 10.1042/bj2190001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hissen A. H. T., Wan A. N. C., Warwas M. L., Pinto L. J., Moore M. M. (2005). The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N5-oxygenase, is required for virulence. Infect. Immun. 73, 5493–5503. 10.1128/IAI.73.9.5493-5503.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperi F., Visca P. (2013). Subcellular localization of the pyoverdine biogenesis machinery of Pseudomonas aeruginosa: a membrane-associated “siderosome.” FEBS Lett. 587, 3387–3391. 10.1016/j.febslet.2013.08.039 [DOI] [PubMed] [Google Scholar]
- Kragl C., Schrettl M., Abt B., Sarg B., Lindner H. H., Haas H. (2007). EstB-mediated hydrolysis of the siderophore triacetylfusarinine C optimizes iron uptake of Aspergillus fumigatus. Eukaryotic Cell 6, 1278–1285. 10.1128/EC.00066-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubodera T., Yamashita N., Nishimura A. (2002). Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 66, 404–406. 10.1271/bbb.66.404 [DOI] [PubMed] [Google Scholar]
- Lamb A. L. (2015). Breaking a pathogen's iron will: Inhibiting siderophore production as an antimicrobial strategy. Biochim. Biophys. Acta 1854, 1054–1070. 10.1016/j.bbapap.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder K., Philippe B., Jahn B., Latgé J. P., Brakhage A. A. (2001). Differential expression of the Aspergillus fumigatus pksP gene detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 69, 6411–6418. 10.1128/IAI.69.10.6411-6418.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M., Marahiel M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. 10.1128/MMBR.00012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney N. M., Owens R. A., Meleady P., Henry M., Dolan S. K., Mulvihill E., et al. (2016). The iron-responsive microsomal proteome of Aspergillus fumigatus. J. Proteomics 136, 99–111. 10.1016/j.jprot.2015.12.025 [DOI] [PubMed] [Google Scholar]
- Moore M. M. (2013). The crucial role of iron uptake in Aspergillus fumigatus virulence. Curr. Opin. Microbiol. 16, 692–699. 10.1016/j.mib.2013.07.012 [DOI] [PubMed] [Google Scholar]
- Nielsen M. L., Albertsen L., Lettier G., Nielsen J. B., Mortensen U. H. (2006). Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43, 54–64. 10.1016/j.fgb.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Oberegger H., Schoeser M., Zadra I., Abt B., Haas H. (2001). SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol. Microbiol. 41, 1077–1089. 10.1046/j.1365-2958.2001.02586.x [DOI] [PubMed] [Google Scholar]
- O'Hanlon K. A., Cairns T., Stack D., Schrettl M., Bignell E. M., Kavanagh K., et al. (2011). Targeted disruption of nonribosomal peptide synthetase pes3 augments the virulence of Aspergillus fumigatus. Infect. Immun. 79, 3978–3992. 10.1128/IAI.00192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keeffe G., Hammel S., Owens R. A., Keane T. M., Fitzpatrick D. A., Jones G. W., et al. (2014). RNA-seq reveals the pan-transcriptomic impact of attenuating the gliotoxin self-protection mechanism in Aspergillus fumigatus. BMC Genomics 15:894. 10.1186/1471-2164-15-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R. A., Hammel S., Sheridan K. J., Jones G. W., Doyle S. (2014). A proteomic approach to investigating gene cluster expression and secondary metabolite functionality in Aspergillus fumigatus. PLoS ONE 9:e106942. 10.1371/journal.pone.0106942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao S. S., Paulsen I. T., Saier M. H. J. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.-S., Kim J.-Y., Yun C.-W. (2016). Identification of ferrichrome- and ferrioxamine B-mediated iron uptake by Aspergillus fumigatus. Biochem. J. 473, 1203–1213. 10.1042/BCJ20160066 [DOI] [PubMed] [Google Scholar]
- Philpott C. C., Protchenko O. (2008). Response to iron deprivation in Saccharomyces cerevisiae. Eukaryotic Cell 7, 20–27. 10.1128/EC.00354-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Hemmons L. M., MacDonald K. D., Bufton A. W. J. (1953). The genetics of Aspergillus nidulans. Adv. Genet. 5, 141–238. 10.1016/s0065-2660(08)60408-3 [DOI] [PubMed] [Google Scholar]
- Priebe S., Kreisel C., Horn F., Guthke R., Linde J. (2015). FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31, 445–446. 10.1093/bioinformatics/btu627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond-Bouchard I., Carroll C. S., Nesbitt J. R., Henry K. A., Pinto L. J., Moinzadeh M., et al. (2012). Structural requirements for the activity of the MirB ferrisiderophore transporter of Aspergillus fumigatus. Eukaryotic Cell 11, 1333–1344. 10.1128/EC.00159-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Beatty J. T., Goffeau A., Harley K. T., Heijne W. H., Huang S. C., et al. (1999). The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1, 257–279. [PubMed] [Google Scholar]
- Schrettl M., Beckmann N., Varga J., Heinekamp T., Jacobsen I. D., Jöchl C., et al. (2010). HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 6:e1001124. 10.1371/journal.ppat.1001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Joechl C., Rogers T., Arst H. N., et al. (2004). Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200, 1213–1219. 10.1084/jem.20041242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Sabiha Y., Loss O., Eisendle M., et al. (2007). Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3, 1195–1207. 10.1371/journal.ppat.0030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Kim H. S., Eisendle M., Kragl C., Nierman W. C., Heinekamp T., et al. (2008). SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70, 27–43. 10.1111/j.1365-2958.2008.06376.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I., Lewis T. E., Cuff A., Das S., Ashford P., Dawson N. L., et al. (2015). CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 43, D376–D381. 10.1093/nar/gku947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater J. L., Gregson L., Denning D. W., Warn P. A. (2011). Pathogenicity of Aspergillus fumigatus mutants assessed in Galleria mellonella matches that in mice. Med. Mycol. 49(Suppl. 1), S107–S113. 10.3109/13693786.2010.523852 [DOI] [PubMed] [Google Scholar]
- Szigeti Z. M., Szaniszló S., Fazekas E., Gyémánt G., Szabon J., Antal K., et al. (2014). Optimization of triacetylfusarinine C and ferricrocin productions in Aspergillus fumigatus. Acta Microbiol. Immunol. Hung. 61, 107–119. 10.1556/AMicr.61.2014.2.2 [DOI] [PubMed] [Google Scholar]
- Wallner A., Blatzer M., Schrettl M., Sarg B., Lindner H., Haas H. (2009). Ferricrocin, a siderophore involved in intra- and transcellular iron distribution in Aspergillus fumigatus. Appl. Environ. Microbiol. 75, 4194–4196. 10.1128/AEM.00479-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelströter L. K., Dolan S. K., Fernanda Dos Reis T., Bom V. L. P., Alves de Castro P., Hagiwara D., et al. (2015). Systematic global analysis of genes encoding protein phosphatases in Aspergillus fumigatus. G3 (Bethesda) 5, 1525–1539. 10.1534/g3.115.016766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin S., Alcazar-Fuoli L., Gründlinger M., Puempel T., Cairns T., Blatzer M., et al. (2012). Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl. Acad. Sci. U.S.A. 109, E497–E504. 10.1073/pnas.1106399108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.