Abstract
Interspecific hybridisation between hexaploid and tetraploid wheat species leads to the development of F1 pentaploid hybrids with unique chromosomal constitutions. Pentaploid hybrids derived from bread wheat (Triticum aestivum L.) and durum wheat (Triticum turgidum spp. durum Desf.) crosses can improve the genetic background of either parent by transferring traits of interest. The genetic variability derived from bread and durum wheat and transferred into pentaploid hybrids has the potential to improve disease resistance, abiotic tolerance, and grain quality, and to enhance agronomic characters. Nonetheless, pentaploid wheat hybrids have not been fully exploited in breeding programs aimed at improving crops. There are several potential barriers for efficient pentaploid wheat production, such as low pollen compatibility, poor seed set, failed seedling establishment, and frequent sterility in F1 hybrids. However, most of the barriers can be overcome by careful selection of the parental genotypes and by employing the higher ploidy level genotype as the maternal parent. In this review, we summarize the current research on pentaploid wheat hybrids and analyze the advantages and pitfalls of current methods used to assess pentaploid-derived lines. Furthermore, we discuss current and potential applications in commercial breeding programs and future directions for research into pentaploid wheat.
Keywords: in situ hybridisation, interploidy crosses, pentaploid hybrids, Triticum aestivum, Triticum turgidium spp. durum
Introduction
The two major wheat species, hexaploid bread wheat Triticum aestivum L. (2n = 6x = 42) and tetraploid durum wheat T. turgidium spp. durum (2n = 4x = 28), are commercially important wheat species globally. Hexaploid wheat has three diploid sets of seven chromosomes belonging to the A-, B-, and D-genomes (AABBDD), whereas tetraploid wheat only has two diploid sets of seven chromosomes belonging to the A- and B-genomes (AABB). Hybridisation between these two species with different ploidy levels leads to a pentaploid hybrid (AABBD) that has the chromosomal constitution of 2n = 5x = 35 (Kihara, 1924). The genetic variability that is combined from hexaploid and tetraploid wheat into a pentaploid hybrid has great potential in crop improvement (Eberhard et al., 2010; Martin et al., 2013; Kalous et al., 2015). However, while several reviews have focussed on the successful establishment of interspecific wheat hybrids (Sharma and Gill, 1983; Jiang et al., 1993; Friebe et al., 1996), little emphasis has been placed on developing efficient methods to incorporate these pentaploid hybrids into commercial breeding practices.
Although pentaploid wheat hybrids can be efficiently used in crop improvement programs, pest and disease resistance have principally been transferred into hexaploid or tetraploid wheats through alien introgression. Sharma and Gill (1983) reviewed the status of wide hybridisation and listed successful crosses that had been established between wheat and its related genera. They also focussed on the genes that had been successfully transferred from related wild species into commercial wheat cultivars up to that time. Jiang et al. (1993) discussed further advances in successful alien gene transfer from related species into cultivated bread and durum wheat. Friebe et al. (1996) comprehensively reviewed a number of important wheat-alien translocations and their potential in plant breeding for developing pest and disease resistance. Complications occur when trying to introgress traits across different wheat species. These include incompatibility between different Triticum species and sterility of the F1 hybrids. Developing wheat hybrids through alien introgression is highly challenging when compared to hybridisation between domesticated inter-ploidy species such as bread and durum wheat. Recently, there has been a renewed interest in developing hexaploid/tetraploid wheat crosses to improve elite bread and durum wheat lines for a number of economically desirable characters (Martin et al., 2011, 2013; Han et al., 2014, 2016; Kalous et al., 2015).
This review first focusses on recent research into the production of hexaploid/tetraploid- and tetraploid/hexaploid-derived pentaploid hybrids, mainly between hexaploid T. aestivum and tetraploids T. durum, T. timopheevii, and T. dicoccoides, and then discusses current techniques for characterizing the chromosome composition of lines derived from them. In pentaploid wheat hybrids, the predominance of heterozygous loci present in their A and B genome, together with the retention of a haploid D genome, results in breeding material that has captured a high degree of genetic variation. Despite this rich source of genetic variation, there is still much to learn regarding the efficient screening, selection, and application of populations derived from pentaploid wheat hybrids in commercial breeding programs. This review will examine these challenges and consider the future potential of pentaploid wheat hybrids in crop improvement.
Genetic Variability in Pentaploid Wheat Hybrids
Chromosome Morphology
Combining two or more different genomes into one cell may cause changes in chromosome morphology, including differences in the size, thickening, or lengthening of chromosomes, a phenomenon referred to as genome shock (Navashin, 1934). Genome shock has been well-documented (Matsuoka, 2011) and, in addition to changes in morphology, includes chromosomal rearrangements, gain or loss of chromosomal segments, gene activation and suppression, variations in the epigenome especially with respect to the pattern of cytosine methylation, and activation of transposons (Matsuoka, 2011). Increases in genome dosage and genes in alloploid wheat lines causes chromosomal imbalance, genome instability, and incompatibility. For example, morphological changes were observed in the satellite D chromosome in an interploidy cross in Crepis species. These observations indicated that there were chromosomes that lacked satellites in the F1 progeny (Pikaard, 1999).
Chromosomal Constitution
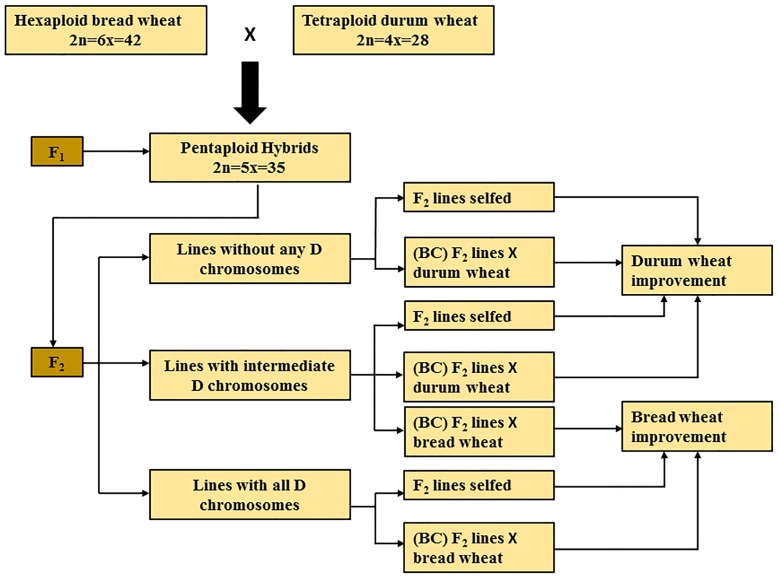
Chromosome elimination is an essential process that takes place in subsequent generations derived from F1 pentaploids, It may take a few generations to resolve the complex process of chromosome pairing and to give rise to a stable durum or bread wheat line. Subsequent generations derived from F1 pentaploid wheat hybrids can be broadly classified into three groups, based on the presence or absence of D-genome chromosomes (Figure 1). The progeny belonging to the first group have lost all seven D-genome chromosomes (2n = 4x = 28); the second group consists of progeny that have intermediate numbers of D-genome chromosomes (total chromosome numbers ranging from 2n = 29 to 41); while the third group have retained two copies of all seven D-genome chromosomes (2n = 6x = 42). Based on the objective of the breeding program that aims to develop bread or durum wheat lines, these three groups of pentaploid-derived wheat hybrids can by selfed or backcrossed with either parent. For example, lines belonging to the first group of hybrids can be selfed or backcrossed to a durum parent to develop elite durum lines (Figure 1).
FIGURE 1.
Summary of pentaploid wheat production for bread and durum wheat improvement. BC, back crossing.
To determine the general fate of D-genome chromosomes in the lines derived from F1 pentaploids, we analyzed previous studies of pentaploid-derived populations from different generations of hexaploid/tetraploid and tetraploid/hexaploid crosses. The number of D-genome chromosomes retained was determined using cytological characterisation (Kihara, 1982; Wang et al., 2005; Eberhard et al., 2010), or molecular markers (Lanning et al., 2008; Eberhard et al., 2010; Martin et al., 2011; Kalous et al., 2015). Molecular marker data indicates the presence or absence of unique chromosomes, but fails to determine the number of copies of unique chromosomes. The cytological studies used in the analysis identified the number of copies of unique chromosomes present, but, did not distinguish individual chromosomes. For this analysis the cytological and molecular data that represent D-genome retention were collected and divided into two main groups. Group-1 includes (i) lines without any D-genome chromosomes and (ii) lines with an intermediate number of D-chromosomes, including lines which had lost both copies of at least one unique D-genome chromosome. With the latter the assumption was made that these lines would loose their D chromosomes in later generations and revert back to a tetraploid constitution. Group-2 includes lines which retained at least one copy of all seven D-genome chromosomes.
The proportion of D-genome chromosomes retained within each group was calculated for each cross (Supplementary Table 1). The main finding from the D genome retention analysis is that there is a high probability that viable progeny derived from pentaploid wheat hybrids will lose their D-genome chromosomes in subsequent generations (P < 0.001), resulting in stable tetraploid lines (Supplementary Table 1). Differences were observed in the retention of D-genome chromosomes depending on the parents used in the cross. For example the hexaploid parent Choteau crossed with tetraploid parents Mountrail and Monroe, resulted in a larger number of D chromosomes being eliminated compared to the F8_Choteau/Avonlea cross (Figure 2). In the tetraploid/hexaploid cross involving tetraploid parent AS295 as female parent with hexaploid parents CN16 as male parent, a large proportion of D-genome chromosomes was eliminated (Figure 2).
FIGURE 2.
Summary of D-genome chromosome retention in hexaploid/tetraploid and tetraploid/hexaploid crosses. Durum wheat Group-1 (lines with no or intermediate number of D chromosomes) is indicated in black and bread wheat Group-2 (lines with at least one copy of all seven D chromosomes) in white. The cross which has a tetraploid maternal parent is indicated with a green box. The proportion of D-genome chromosomes retained in each cross is indicated on the X-axis.
Crosses involving Chinese Spring as hexaploid maternal parent and different tetraploid paternal parents retained high proportions of D-genome chromosomes. These lines are potentially useful for developing bread wheats. Conversely, Martin et al. (2011) observed that crosses involving the hexaploid variety Sunco and tetraploid breeder’s line 230604 and those involving the synthetic hexaploid CPI133814 lost their D-genome chromosomes in early generations. Thus, the parental combinations used in pentaploid crosses greatly influence the degeree to which D-genome chromosomes are retained.
Relationship between Retention of Tetraploid-Derived A and B Genome and Hexaploid-Derived D Genome
Genomic analysis of generations derived from pentaploid F1 lines indicated a significant relationship between the proportion of tetraploid-derived A and B genome retained and the retention of the D genome (Martin et al., 2011). These authors showed that the relative inheritance of chromosomes A and B alleles from bread and durum wheat differed among the hexaploid/tetraploid crosses. Lines with higher levels of durum-derived A and B chromosome segments tended to retain fewer D genome chromosomes. This implies that for characters inherited from the A and/or B genome of the hexaploid parent, pentaploid-derived lines with both low numbers of D chromosomes and the characters in question should be selected. Subsequent backcrossing will more rapidly yield elite tetraploid lines that have both lost all D genome material and inherited the targeted traits of the original hexaploid parent.
Applications of Pentaploid Wheat Hybrids
Pentaploid wheat hybrids are a potential source for developing resistance to pests and diseases and for improving the level of tolerance to abiotic stresses such as salinity and metal toxicity (Munns et al., 1999; Lopes et al., 2010; Han et al., 2016). Improving quality characteristics is one of the primary concerns when developing commercial varieties suitable for human and animal consumption. For example, grain protein content, protein quality, flour color, and grain texture are key quality criteria when developing wheat varieties for bread and pasta production (Sissons, 2008).
Disease Resistance
Crown rot, Fusarium head blight (FHB), leaf rust, stripe rust, stem rust, and powdery mildew are among the most devastating diseases of wheat that account for significant yield losses in terms of both quality and quantity (Rong et al., 2000; Bai and Shaner, 2004; Xu et al., 2013; Liu and Ogbonnaya, 2015). Crown rot disease caused by the fungus Fusarium pseudograminearum is considered to be one of the major constraints in durum production worldwide (Liu and Ogbonnaya, 2015). This is a chronic and severe disease in many semi-arid regions globally. In the Pacific Northwest of the USA, winter wheat crop yield reduction due to crown rot has been estimated at 35% (Smiley et al., 2005). To date, resistance to crown rot disease in durum and related tetraploid wheat species has not been identified. Partial resistance to crown rot has been identified in some bread wheat lines, such as Sunco, 2-49, and CPI133814 (Wildermuth et al., 2001; Bovill et al., 2006, 2010; Martin et al., 2015). This partial resistance in the hexaploid source is associated with multiple chromosomal regions, including 1A, 1B, 1D, 3B, and 4B in 2-49, and 2B in Sunco (Bovill et al., 2010; Martin et al., 2015). Subsequent hexaploid/tetraploid crosses were made using these partially resistant sources (2-49 and Sunco) with durum breeder’s line 950329. F6 lines were selected, based on the complete absence of D-genome chromosomes and crown rot score, and backcrossed with durum parent 950329. The BC2F2 progeny were assessed for crown rot severity and lines were identified with field-based resistance better than that of 2-49. The results of this study indicate that the hexaploid source of crown rot resistance was successfully introgressed into durum wheat (Martin et al., 2013).
Fusarium head blight is another important wheat disease caused by the fungus Fusarium graminearum, which results in a loss of yield and grain quality (Clear and Patrick, 1990). Losses due to reduced yield by FHB have been estimated to be US $2.7 billion (Matny, 2015). The production of mycotoxins by F. graminearum in cereal grain, particularly in wheat, is of great concern, rendering the grain unsuitable for human and animal consumption (Bottalico and Perrone, 2002). Chinese bread wheat variety Sumai3 and the two cultivars Ning8331 and 93FHB21 have been identified as resistant sources for FHB. They were crossed with the susceptible durum cultivars Stewart 63 and DT486, and the resulting F1 pentaploid hybrids showed improved resistance to FHB compared to the durum parent (Gilbert et al., 2000).
Stripe rust caused by the fungus Puccinia striiformis f. sp. tritici is an important disease that causes major damage in both durum and bread wheat and can cause severe yield losses of more than 90% in susceptible cultivars when the weather is favorable to disease development (Chen, 2013). The Ethiopian durum wheat line accession PI 480148 has a single dominant gene Yr53 that confers stripe rust resistance. Yr53 was transferred into the susceptible bread wheat genotype Avocet S through pentaploid crossing (Xu et al., 2013). The progeny derived from the crosses were cytologically selected based on the presence of all seven pairs of D chromosomes (2n = 6x = 42) and tested with stripe rust race PST100. The progeny of the F3 generation segregated in a 3:1 resistant: susceptible ratio, suggesting that a single dominant gene was responsible for the resistance.
Powdery mildew caused by the fungus Blumeria graminis f. sp. tritici is one of the major diseases that cause significant yield loss in wheat. Pentaploid hybrids derived from crossing susceptible hexaploid wheat cultivars Maris Nimrod and Norman with resistant tetraploid T. dicoccoides accession CLI060025 showed improved resistance to powdery mildew when the stable F3 progeny were back- and top-crossed to a second hexaploid wheat (Reader and Miller, 1991). In another study, Rong et al. (2000) transferred the powdery mildew resistance gene Pm26, present on chromosome 2B from an Israeli T. dicoccoides accession (TTD140), into the hexaploid cultivar Bethlehem.
Abiotic Stress Tolerance
Salt intolerance is a constraint that limits durum wheat production in Australia. Significant variability in saline tolerance exists in the tribe Triticeae, providing a considerable potential for transferring salt-tolerant traits into cultivated bread and durum wheat through pentaploid production (Colmer et al., 2006). Bread wheat is generally more tolerant to salinity than is durum wheat, due to the presence of salt-tolerant genes on the D-genome chromosomes (Munns et al., 1999). The major locus responsible for tolerance to salinity, Kna1, has been mapped to the distal end of 4DL (Lindsay et al., 2004). It has been shown that it is possible to improve the saline tolerance in durum wheat by introducing D chromosomes through wheat substitution lines. Langdon durum substitution lines, developed by Joppa and Williams (1988), have a pair of homoeologous chromosomes replaced by a pair of D chromosomes derived from the hexaploid wheat landrace Chinese Spring. These lines have opened up new avenues for developing durum wheat with improved tolerance to various abiotic stresses, including saline tolerance. A Langdon durum 4D (4B) substitution line was crossed with an Australian semi-dwarf durum wheat variety, Jandaroi, to incorporate aluminum tolerance. The chromosomal fragment of 4D was successfully introduced into the Jandaroi durum wheat, which substantially enhanced aluminum tolerance in the sister lines derived from three generations of backcrossing (Han et al., 2014). Two genes, TaALMT1 and TaMATE1B, were transferred using a Ph1 (pairing homoeologous) mutant of durum wheat through conventional breeding. The size of the 4D chromosomes introgressed from the bread wheat into durum wheat was estimated by markers, fluorescence in situ hybridisation (FISH), and real time quantitative PCR. The TaALMT1 and TaMATE1B genes increased Al3+ tolerance in durum wheat and, in contrast to bread wheat, the TaMATE1B gene was found to be more effective in increasing Al3+ tolerance in durum wheat grown on acid soil (Han et al., 2016).
Quality Improvement
Understanding the molecular, chemical, and functional aspects of the quality of bread and durum wheat has significantly improved in recent decades (Shepherd, 1988; Shewry et al., 1992; Liu et al., 1996; Troccoli et al., 2000; Zhang et al., 2008). Durum wheat is mainly used for the production of pasta, semolina, couscous, and some bread products (Palumbo et al., 2000; Sissons, 2012). Durum has the hardest texture of rich yellow starchy endosperm among all of the wheat species, which makes it the most suitable for pasta production. Protein content and gluten quality are important traits in defining pasta-cooking quality; thus, the quality of the durum grain is directly associated with pasta quality (Novaro et al., 1993).
Flatbread made of durum flour is popular in the Mediterranean region and has also become popular in other countries. The inextensible dough character of durum wheat flour results in a lower loaf volume than bread wheat flour. Introgression of certain traits associated with the dough quality from bread wheat into durum wheat might improve the loaf quality of durum wheat. Palumbo et al. (2000) transferred a high molecular weight gluten portion present on chromosome 1A (Glu-A1) that is absent in most of the durum wheat cultivars, through interspecific hybridisation with a bread wheat variety. The resultant interspecific lines showed improved bread loaf volume (cm3), ranging from 295 to 442.5, when compared to the tester bread wheat lines, which ranged from 390.0 to 437.5. Interspecific hybridisation involving hexaploid/tetraploid crosses has demonstrated that the resultant progeny have significantly improved grain weight, grain diameter, and grain yield (Kalous et al., 2015). Furthermore, it was suggested that introducing 1D chromosome segments and their associated protein products would improve the bread making quality of durum wheat (Sissons, 2008). Wang et al. (2005) indicated that recombinant allohexaploid lines, which had retained all seven copies of the D genome chromosomes, had enhanced protein content compared to their hexaploid parental lines. However, this study did not indicate which D genome regions are responsible for the improved bread making quality.
Allotetraploid lines derived from crossing of hexaploids/tetraploids that lack D-genome chromosomes showed improvement in storage protein content, indicating that several endosperm-protein genes on chromosomes A and B were activated in the absence of D genome chromosomes (Galili and Feldman, 1984). Furthermore they explained that the suppression of endosperm-protein genes might have occurred soon after the emergence of allohexaploid wheat, around 10,000 years ago, when the D-genome chromosomes were introduced into the tetraploid background. Potentially, these tetraploid specific endosperm-protein genes on chromosomes A and B can be re-activated in hexaploid lines that lose D chromosomes, while combining hexaploid/tetraploid and tetraploid/hexaploid crosses in pentaploid wheat production.
Techniques for Studying Genome Constitutions of Pentaploid Wheat Hybrids
Characterisation of pentaploid wheat hybrids requires techniques to identify chromosome number (e.g., of univalent D chromosomes), chromosome identity, and changes in chromosomal morphology (deletions or translocations). The following section will discuss the advantages and limitations of techniques currently available for characterizing pentaploid-derived wheat hybrids.
Cytological Characterisation of Pentaploid Wheat Hybrids
Fluorescence in situ hybridisation is a technique that can be used to identify chromosomes, and helps distinguish the 21 pairs of A-, B-, and D-genome chromosomes. The orginal technique involved hybridisation of radioactive-labeled DNA or RNA probes but this was subsequently replaced by fluroscence labeling of the probes (Gall and Pardue, 1969). In situ hybridisation enables the identification of deletions, translocations, introgressed chromatin fragments, and translocation breakpoints (Le et al., 1989; Schwarzacher et al., 1989; Friebe et al., 1993). The physical position of known DNA sequences on the chromosomes can be visualized with the help of labeled complementary DNA strands, i.e., probes (Rayburn and Gill, 1986; Zhang et al., 2007).
Multicolor fluorescence in situ hybridisation (MCFISH) has been widely used for simultaneous discrimination of different genomes in polyploids, incuding cereals. This technique uses probes with dispersed repetitive DNA sequences that preferentially hybridize to the A- (BAC676D4) and D- (BAC9M13) genome chromosomes, respectively. These FISH probes are labeled with fluorescent tags of different colors and together with counterstaining of the unlabeled B-genome will distinguish the three genomes under a fluorescent microscope (Zhang et al., 2004; Komuro et al., 2013). This method has been used in pentaploid wheat hybrid studies to distinguish the A, B, and D genome chromosomes and to determine the copy number of D-genome chromosomes present in F2 plants of hexaploid/tetraploid wheat hybrids (Eberhard et al., 2010). It is also possible to identify the individual chromosomes by using chromosome-specific repetitive probes. For instance, plasmid clone pAs1 can be used to discriminate between the individual D-genome chromosomes based on the signal patterns (Mukai et al., 1993; Koo et al., 2015). To differentiate the chromosomes of different genomes in polyploid individuals, the total genomic DNA of one parent is labeled and used as a probe, while a higher amount of unlabeled genomic DNA of the other parent is used to block the common repetitive sequences between the parents and to increase the specificity of DNA hybridisation. This technique is known as genomic in situ hybridisation (GISH) and can be used for studying intergenomic translocations and alien introgressions, and for discriminating genomes in polyploid cereals (Schwarzacher et al., 1989, 1992; Schubert et al., 2001; Silva and Souza, 2013).
Even though cytological techniques have improved over the past decades, there are still some disadvantages in utilizing these approaches for screening a large amount of samples. The genome size, homologous nature of diploid donor species and presence of large numbers of repetitive sequence is still challenging for wheat cytogenetics. Furthermore, cytological approaches are labor intensive, demand a high level of technical skill and require extended periods of time for assessment of multiple progeny. The time and skills required render these approaches unsuitable for use as a high-throughput screening method, as would be required for commercial breeding (Eberhard et al., 2010). However, these methods have an important role in fundamental research and in characterizing the chromosome constitution of elite pentaploid-derived lines.
Molecular Marker Technology
Unlike cytological techniques, molecular markers are not influenced by the environment or plant growth stages. Different types of molecular markers are available depending on their applications, such as hybridisation-based DNA markers, PCR-based markers, DNA chip and sequence-based markers. Applications of these markers in modern plant breeding efforts have been comprehensively discussed elsewhere (Gupta et al., 2008; Wang S. et al., 2014).
Simple sequence repeat (SSR) or microsatellite markers have been widely used and were considered as the marker of choice for many years by plant breeders analyzing interspecific hexaploid/tetraploid crosses (Lanning et al., 2008). SSR markers are readily reproducible and can be used to distinguish the presence or absence of unique chromosomes. Most SSRs are co-dominant markers and can be used to distinguish genotypes based on the size of alleles and are thus also useful for validating F1 hybrids. When co-dominant at a particular locus, SSR markers can also indicate whether one or two copies of a particular chromosome segment are present. Several high-density maps containing SSR markers have been constructed for bread and durum wheat (Roder et al., 1998; Somers et al., 2004; Marone et al., 2012).
In the past decade, a number of other marker platforms have been developed, including mircroarray or gene chips that contain 1000s of unique probes spanning the entire wheat genome (Gupta et al., 2008). For large-scale screening, SSR markers have been largely replaced with single nucleotide polymorphism (SNP) markers covering the entire wheat genome with high density (Wang H. et al., 2014). In addition, the DArT (Diversity Array Technology) markers are based on a microarray platform that hybridizes the sample genome to identify the presence or absence of 1000s of unique fragements covering the whole genome (Jaccoud et al., 2001; Akbari et al., 2006). These DArT markers have been used to study genome inheritance and chromosome structure in pentaploid-derived wheat hybrid crosses (Eberhard et al., 2010; Martin et al., 2011). Recently, the DArT genotyping system has been improved by combining next-generation sequencing (NGS) with existing DArT markers to develop DArTseqTM. DArTseqTM markers have the potential to significantly increase the number of markers on each chromosome (Von Mark et al., 2013). This technique has been successfully applied to high-throughput screening of genetically diverse plant materials (Ren et al., 2015).
The majority of existing genotyping systems are based on dominant markers, which fail to differentiate between the presence of a single or two copies of a particular locus. Thus, it is impossible to detect incomplete or partial chromosomes in the presence of a complete homologous chromosome. Hence, the information generated through molecular markers alone is not sufficient to validate the allotetraploid or allohexaploid lines that show chromosome deletions, additions, or translocations. In such instances, it is essential to apply cytological techniques, such as GISH or MCFISH, which provide a more systematic approach for analyzing complex chromosomal complements (Eberhard et al., 2010).
Challenges of Producing Pentaploid Wheat Hybrids
Pentaploid hybrid production can be complex and requires careful consideration before any crosses are commenced. Common difficulties encountered and important points to consider while developing pentaploid hybrids are summarized below.
Cross Direction (Hexaploid/Tetraploid or Tetraploid/Hexaploid)
To obtain the highest number of fertile F1 progeny from an interspecific cross, it has been proposed that the higher ploidy level species should be used as the maternal parent (Kihara, 1982). In most studies of pentaploid hybrids to date, a hexaploid wheat has been used as the maternal parent (Mesfin et al., 1999; Lanning et al., 2008; Eberhard et al., 2010; Martin et al., 2011; Kalous et al., 2015). Crosses using the lower ploidy level species as the female generally have been less successful and can lead to poor seed set and subsequent low levels of seed germination and seedling establishment (Sharma and Gill, 1983). However, successful pentaploid hybrid crosses combining tetraploid wheat as the maternal parent and hexaploid wheat as the paternal parent have been reported (Wang et al., 2005).
Seedling Abnormailities
The F1 seeds from inter-ploidy hybridisation between bread and durum wheat have relatively poor germination compared to seeds from crosses of parents of the same ploidy level (Kihara, 1982; Sharma and Gill, 1983) and can take several weeks to germinate. Complete failure of germination has been encountered in the reciprocal crosses between T. monococcum and T. aestivum (Kihara, 1982; Bhagyalakshmi et al., 2008). Unsurprisingly, a strong correlation between seed germination and seed morphology has been observed, with shriveled seeds showing poor germination compared to plump seeds (Kihara, 1982).
In interploidy crosses, normal seed development depends on the ploidy ratio of the maternal and paternal parent (Johnston et al., 1980; Carputo et al., 1999). Each species has been assigned a unique endosperm balance number (EBN). It is generally believed that the ploidy level of an embryo and its associated endosperm is critical for successful seed development (Ramsey and Schemske, 1998). Endosperm development is significant and has a major physiological and genetic relationship with the embryo of the newly formed allopolyploids. Johnston et al. (1980) explained the differences in EBN in an interspecific cross using tetraploid and diploid parent 4EBN(4x)/2EBN(2x) and its reciprocal cross. They indicated that progeny from the 4EBN/2EBN cross had a maternal:paternal ratio of endosperm of 4:1, while the reciprocal cross 2EBN/4EBN had a ratio of 1:1 which deviates from the normal 2:1 maternal:paternal ratio. EBN signifies the importance of endosperm development in inter ploidy crosses.
Even if F1 pentaploid seeds germinate normally, chlorophyll abnormalities can develop, such as striato-virescence, delayed virescence, and albino expression (Tsunewaki, 2004). Furthermore, Tsunewaki (2004) showed that chlorophyll abnormalities were observed in the tetraploid wheat species with two duplicated recessive genes controlling the abnormalities. Hexaploid wheat lines carry wild-type homoeoalleles Sv3, Dv3, and Abn3 for these abnormalities on chromosome 2D. Furthermore, abnormalities related to growth and development, such as stunted growth, grass clumping, and differences in flowering time, have been reported in interploidy wheat hybrids (Chen and Ni, 2006). While appropriate selection processes can rapidly remove these obvious abnormalities from subsequent pentaploid-derived generations, abnormalities with less obvious phenotypes may be harder to exclude.
Pollen Viability
Low pollen viability can also restrict seed set. The imbalanced chromosome number in the F1 individuals impacts pollen development and subsequent fertilization (Kihara, 1982). The affected pollen grains do not germinate, the pollen tubes fail to reach the ovary, or the male and female gametes fail to fuse (Sharma and Gill, 1983). Pentaploid hybrids derived from a cross between T. timopheevii (AAGG) and T. aestivum produced completely sterile white anthers with infertile pollen (Bhagyalakshmi et al., 2008). The tissue that forms the pollen grain has a lower threshold for respiratory deficiency than do other plant tissues, which can lead to a loss of pollen viability that has been associated with the expression of novel mitochondrial peptides (Leon et al., 1998). Nucleotide rearrangement in certain genes, including the atpa locus, is considered to be the major cause of cytoplasmic male sterility in many crops (Chase, 1994, 2007; Heazlewood et al., 2003).
Apart from pollen viability, several other mutations, such as infertility, leaf striping, and severe growth impairments, could possibly arise from certain combinations of nuclear cytoplasm due to loss of mitochondrial genes (Newton and Coe, 1986). This evidence suggests that the mitochondrial genome may play a vital role in mediating the viability of pollen grains in many species, including interspecific wheat hybrids.
Progressive Hybrid Necrosis
Progressive hybrid necrosis can affect F1 hybrids, resulting in prolonged chlorosis of plant leaf and sheath tissue. These symptoms lead to the premature death of leaves and tillers and eventually the whole plant in certain wheat hybrids (Caldwell and Compton, 1943; Hermsen, 1963a,b). Progressive necrosis is a lethal or semi-lethal condition that imposes a great barrier when trying to transfer desirable traits between species (Chu et al., 2006). This condition in F1 hybrids is predominantly controlled by two complementary genes, Ne1 and Ne2, located on the long arm of chromosome 5B and the short arm of chromosome 2B, respectively (Nishikawa et al., 1974). Both genes exist in bread and durum wheats (Tsunewaki, 1970, 1992, 2004). Recent genetic and mutational studies have found that the Ne2 gene is closely related to the leaf rust resistance gene Lr13 (Zhang et al., 2016). Knowledge of which bread and durum wheat genotypes carry alleles of the Ne1 and Ne2 genes will allow plant breeders to avoid the occurrence of progressive necrosis.
Nuclear-Cytoplasmic Interaction
Nuclear-cytoplasmic interaction (NCI) is the condition in which an interspecific hybrid possessing the nucleus of one parent interacts with cytoplasm inherited from the other parent (Simons et al., 2003). Pentaploid progeny with an unfavorable NCI exhibit a wide range of phenotypes, such as maternally inherited male sterility, female infertility, late maturity, reduced vigor, pigment deficiencies, and altered morphology of cotyledons, leaves, and flowers (Monika et al., 2013).
The NCI in an interspecific hybrid is expressed by species cytoplasmic specific (scs) genes (Monika et al., 2013). In favorable interactions, these scs genes maintain the NCI and provide sufficient vigor and viability to the alloplasmic lines (Maan, 1992). The scs genes are located on chromosomes 1DL of T. aestivum (scsae) and 1AL of T. timopheeviii (scsti). Triticum turgidum spp. durum has a segment in chromosome 1A carrying scsti transferred from T. timopheevii (Simons et al., 2003). The scs gene plays a major role only when the nucleus of T. aestivum or T. turgidum spp. durum is combined with the cytoplasm of a wild species; otherwise, the scs gene stays unexpressed in normal cells with a compatible nucleus and cytoplasm (Monika et al., 2013).
Cytoplasmic organelles such as chloroplast and mitochondria are uniparentally inherited, mainly through the maternal lineage. Inheritance of paternal and novel copies of mitochondrial genes has been witnessed in a number of inter-ploidy wheat crosses. The mitochondrial heteroplasmy in inter-ploidy hybrids between wheat (6x) and rye (2x) (Laser et al., 1997), wheat (6x) and Aegilops sp. (2x,4x,6x) (Hattori et al., 2002), wheat (6x) and T. timopheevi (4x) (Kitagawa et al., 2003) and between barley (2x) and wheat (6x) (Aksyonova et al., 2005) have all been the subject of study. However, no attention has been given to hexaploid/tetraploid wheat crosses.
Fourty different SNP’s have been identified between the hexaploid bread and tetraploid durum wheat mitochondrial genome. Five of these were present in known mitochondrial genes such as rps1, rps2, cox3, and ccmFN (Ogihara et al., 2005; Cui et al., 2009; Noyszewski et al., 2014). Developing SNP markers covering these identified genes would help to differentiate between bread and durum wheat mitochondrial genomes which might provide new insights regarding cytoplasmic inheritance in pentaploid wheat hybrids.
Conclusion and Future Directions
Although, the value of genetic variability generated from hexaploid/tetraploid or tetraploid/hexaploid crosses is vast this technique has generally not been taken up in plant breeding programs. It is evident from research conducted to date, that pentaploid derived wheat lines would be valuable in commercial plant breeding programs that aim to improve fungal disease resistance, abiotic stress tolerance, quality parameters and agronomic characters. There are a number of other candidate traits, that could be potentially incorporated through pentaploid-derived wheat lines. One of these is nematode tolerance/resistance as durum wheats in general are more resistant to root leison nematode (Pratylenchus thornei) disease than bread wheats (Owen et al., 2010; Sheedy et al., 2012, 2015; Thompson et al., 2012). The transfer of some stress tolerant characteristics have been successful, but incorporating drought, cold, and heat tolerance characteristics using pentaploid-dervied wheat lines has not been studied in detail. There is a great potential to improve these traits using interspecific hybridisation as there is a wide variation for stress adaptive traits in the Triticum gene pool (Reynolds et al., 2009).
Although there are many barriers that restrict the production of pentaploid wheat hybrids, choice of parental genotype and using the higher ploidy level species as the maternal parent can improve the success rate. Improving the molecular and cytological techniques used to screen recombinant progenies will increase the efficiency of the selection process and help breeders in accelerating pentaploid production. Hence, interploidy hybridisation may be a promising tool for developing wheat genotypes that can cope with changing climate conditions. Therefore, it is essential to initiate further research to incorporate such traits from bread into durum wheat or durum into bread wheat through pentaploid wheat hybrids to assist the sustainable and increased global wheat production which will be required in the future.
Author Contributions
SP conducted the literature survey and wrote the manuscript. PZ, RH, and MS improved the manuscript by comments and suggestions. AM initiated the project and contributed to manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was partly funded by the Grains Research and Devlopment Corportation, Australia. Sriram Padmanaban acknowledges the support of an Australian Postgraduate Award. The authors would like to thank Dr. Kathleen L. Farquharson (Plant Editors) for her critical comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00358/full#supplementary-material
References
- Akbari M., Wenzl P., Caig V., Carling J., Xia L., Yang S., et al. (2006). Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor. Appl. Genet. 113 1409–1420. 10.1007/s00122-006-0365-4 [DOI] [PubMed] [Google Scholar]
- Aksyonova E., Sinyavskaya M., Danilenko N., Pershina L., Nakamura C., Davydenko O. (2005). Heteroplasmy and paternally oriented shift of the organellar DNA composition in barley–wheat hybrids during backcrosses with wheat parents. Genome 48 761–769. 10.1139/g05-049 [DOI] [PubMed] [Google Scholar]
- Bai G., Shaner G. (2004). Management and resistance in wheat and barley to fusarium head blight. Annu. Rev. Phytopathol. 42 135–161. 10.1146/annurev.phyto.42.040803.140340 [DOI] [PubMed] [Google Scholar]
- Bhagyalakshmi K., Vinod K. K., Kumar M., Arumugachamy S., Prabhakaran A., Raveendran T. S. (2008). Interspecific hybrids from wild x cultivated Triticum crosses-a study on the cytological behaviour and molecular relations. J. Crop Sci. Biotechnol. 11 257–262. [Google Scholar]
- Bottalico A., Perrone G. (2002). Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur. J. Plant Pathol. 108 611–624. 10.1023/A:1020635214971 [DOI] [Google Scholar]
- Bovill W., Ma W., Ritter K., Collard B., Davis M., Wildermuth G., et al. (2006). Identification of novel QTL for resistance to crown rot in the doubled haploid wheat population ‘W21MMT70’ × ‘Mendos’. Plant Breed. 125 538–543. 10.1111/j.1439-0523.2006.01251.x [DOI] [Google Scholar]
- Bovill W. D., Horne M., Herde D., Davis M., Wildermuth G. B., Sutherland M. (2010). Pyramiding QTL increases seedling resistance to crown rot (Fusarium pseudograminearum) of wheat (Triticum aestivum). Theor. Appl. Genet. 121 127–136. 10.1007/s00122-010-1296-7 [DOI] [PubMed] [Google Scholar]
- Caldwell R. M., Compton L. (1943). Complementary lethal genes in wheat causing a progressive lethal necrosis of seedlings. J. Hered. 34 67–70. 10.1093/oxfordjournals.jhered.a105248 [DOI] [Google Scholar]
- Carputo D., Monti L., Werner J. E., Frusciante L. (1999). Uses and usefulness of endosperm balance number. Theor. Appl. Genet. 98 478–484. 10.1007/s001220051095 [DOI] [Google Scholar]
- Chase C. D. (1994). Expression of CMS-unique and flanking mitochondrial DNA sequences in Phaseolus vulgaris L. Curr. Genet. 25 245–251. 10.1007/BF00357169 [DOI] [PubMed] [Google Scholar]
- Chase C. D. (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends Genet. 23 81–90. 10.1016/j.tig.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Chen X. (2013). Review article: high-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 4 608–627. 10.4236/ajps.2013.43080 [DOI] [Google Scholar]
- Chen Z. J., Ni Z. (2006). Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays 28 240–252. 10.1002/bies.20374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.-G., Faris J., Friesen T., Xu S. (2006). Molecular mapping of hybrid necrosis genes Ne1 and Ne2 in hexaploid wheat using microsatellite markers. Theor. Appl. Genet. 112 1374–1381. 10.1007/s00122-006-0239-9 [DOI] [PubMed] [Google Scholar]
- Clear R., Patrick S. (1990). Fusarium species isolated from wheat samples containing tombstone (scab) kernels from Ontario, Manitoba, and Saskatchewan. Can. J. Plant Sci. 70 1057–1069. 10.4141/cjps90-128 [DOI] [Google Scholar]
- Colmer T. D., Flowers T. J., Munns R. (2006). Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 57 1059–1078. 10.1093/jxb/erj124 [DOI] [PubMed] [Google Scholar]
- Cui P., Liu H., Lin Q., Ding F., Zhuo G., Hu S., et al. (2009). A complete mitochondrial genome of wheat (Triticum aestivum cv. Chinese Yumai), and fast evolving mitochondrial genes in higher plants. J. Genet. 88 299–307. 10.1007/s12041-009-0043-9 [DOI] [PubMed] [Google Scholar]
- Eberhard F. S., Zhang P., Lehmensiek A., Hare R. A., Simpfendorfer S., Sutherland M. W. (2010). Chromosome composition of an F2 Triticum aestivum X T.turgidum spp. durum cross analysed by DArT markers and MCFISH. Crop Pasture Sci. 61 619–624. 10.1071/CP10131 [DOI] [Google Scholar]
- Friebe B., Jiang J., Raupp W., Gill B. (1993). “Molecular cytogenetic analysis of radiation-induced alien genetic transfers in wheat,” in Proceedings of the Eighth International Wheat Genetics Symposium Beijing: 20–25. [Google Scholar]
- Friebe B., Jiang J., Raupp W., Mcintosh R., Gill B. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91 59–87. 10.1007/BF00035277 [DOI] [Google Scholar]
- Galili G., Feldman M. (1984). Intergenomic suppression of endosperm protein genes in common wheat. Can. J. Genet. Cytol. 26 651–656. 10.1007/BF00281853 [DOI] [Google Scholar]
- Gall J. G., Pardue M. L. (1969). Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. U.S.A. 63 378–383. 10.1073/pnas.63.2.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J., Procunier J., Aung T. (2000). Influence of the D genome in conferring resistance to fusarium head blight in spring wheat. Euphytica 114 181–186. 10.1023/A:1004065620127 [DOI] [Google Scholar]
- Gupta P., Rustgi S., Mir R. (2008). Array-based high-throughput DNA markers for crop improvement. Heredity 101 5–18. 10.1038/hdy.2008.35 [DOI] [PubMed] [Google Scholar]
- Han C., Ryan P. R., Yan Z., Delhaize E. (2014). Introgression of a 4D chromosomal fragment into durum wheat confers aluminium tolerance. Ann. Bot. 114 135–144. 10.1093/aob/mcu070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Zhang P., Ryan P. R., Rathjen T. M., Yan Z., Delhaize E. (2016). Introgression of genes from bread wheat enhances the aluminium tolerance of durum wheat. Theor. Appl. Genet. 129 729–739. 10.1007/s00122-015-2661-3 [DOI] [PubMed] [Google Scholar]
- Hattori N., Kitagawa K., Takumi S., Nakamura C. (2002). Mitochondrial DNA heteroplasmy in wheat, Aegilops and their nucleus-cytoplasm hybrids. Genetics 160 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J., Whelan J., Millar A. (2003). The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 540 201–205. 10.1016/S0014-5793(03)00264-3 [DOI] [PubMed] [Google Scholar]
- Hermsen J. T. (1963a). The genetic basis of hybrid necrosis in wheat. Genetica 33 245–287. 10.1007/BF01725765 [DOI] [Google Scholar]
- Hermsen J. T. (1963b). Sources and distribution of the complementary genes for hybrid necrosis in wheat. Euphytica 12 147–160. 10.1007/BF00022351 [DOI] [Google Scholar]
- Jaccoud D., Peng K., Feinstein D., Kilian A. (2001). Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 29:e25 10.1093/nar/29.4.e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Friebe B., Gill B. S. (1993). Recent advances in alien gene transfer in wheat. Euphytica 73 199–212. 10.1038/hdy.2012.116 [DOI] [Google Scholar]
- Johnston S., Den Nijs T., Peloquin S., Hanneman R., Jr. (1980). The significance of genic balance to endosperm development in interspecific crosses. Theor. Appl. Genet. 57 5–9. 10.1007/BF00276002 [DOI] [PubMed] [Google Scholar]
- Joppa L., Williams N. (1988). Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30 222–228. 10.1139/g88-038 [DOI] [Google Scholar]
- Kalous J., Martin J., Sherman J., Heo H.-Y., Blake N., Lanning S., et al. (2015). Impact of the D genome and quantitative trait loci on quantitative traits in a spring durum by spring bread wheat cross. Theor. Appl. Genet. 128 1799–1811. 10.1007/s00122-015-2548-3 [DOI] [PubMed] [Google Scholar]
- Kihara H. (1924). Cytologische und genetische Studien bei wichtigen Getreidearten mit besondere Rüchsicht auf das Verhalten der Chromosomen und die Sterilität in den Bastarden. Mem. Coll. Sci. Kyoto Imp. Univ. B 1 1–200. [Google Scholar]
- Kihara H. (1982). Wheat Studies Retrospect and Prospects. Tokyo: Kodansha Ltd. [Google Scholar]
- Kitagawa K., Takumi S., Nakamura C. (2003). Selective transcription and post-transcriptional processing of the heteroplasmic mitochondrial orf156 copies in the nucleus-cytoplasm hybrids of wheat. Plant Mol. Biol. 53 609–619. 10.1023/B:PLAN.0000019075.31353.77 [DOI] [PubMed] [Google Scholar]
- Komuro S., Endo R., Shikata K., Kato A. (2013). Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56 131–137. 10.1139/gen-2013-0003 [DOI] [PubMed] [Google Scholar]
- Koo D.-H., Sehgal S. K., Friebe B., Gill B. S. (2015). Structure and stability of telocentric chromosomes in wheat. PLoS ONE 10:e0137747 10.1371/journal.pone.0137747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanning S. P., Blake N. K., Sherman J. D., Talbert L. E. (2008). Variable production of tetraploid and hexaploid progeny lines from spring wheat by durum wheat crosses. Crop Sci. 48 199–202. 10.2135/cropsci2007.06.0334 [DOI] [Google Scholar]
- Laser B., Mohr S., Odenbach W., Oettler G., Kück U. (1997). Parental and novel copies of the mitochondrial orf25 gene in the hybrid crop-plant triticale: predominant transcriptional expression of the maternal gene copy. Curr. Genet. 32 337–347. 10.1007/s002940050286 [DOI] [PubMed] [Google Scholar]
- Le H. T., Armstrong K., Miki B. (1989). Detection of rye DNA in wheat-rye hybrids and wheat translocation stocks using total genomic DNA as a probe. Plant Mol. Biol. Rep. 7 150–158. 10.1007/BF02669631 [DOI] [Google Scholar]
- Leon P., Arroyo A., Mackenzie S. (1998). Nuclear control of plastid and mitochondrial development in higher plants. Annu. Rev. Plant Biol. 49 453–480. 10.1146/annurev.arplant.49.1.453 [DOI] [PubMed] [Google Scholar]
- Lindsay M. P., Lagudah E. S., Hare R. A., Munns R. (2004). A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct. Plant Biol. 31 1105–1114. 10.1071/FP04111 [DOI] [PubMed] [Google Scholar]
- Liu C., Ogbonnaya F. C. (2015). Resistance to Fusarium crown rot in wheat and barley: a review. Plant Breed. 134 365–372. 10.1111/mpp.12011 [DOI] [Google Scholar]
- Liu C.-Y., Shepherd K. W., Rathjen J. R. (1996). Improvement of durum wheat pastamaking and breadmaking qualities. Cereal Chem. 73 155–166. [Google Scholar]
- Lopes M., Reynolds M., Draye X., Foulkes J., Hawkesford M., Murchie E. (2010). Dissecting drought adaptation into its phenotypic and genetic components in wheat. Asp. Appl. Biol. 105 7–11. [Google Scholar]
- Maan S. (1992). A gene for embryo-endosperm compatibility and seed viability in alloplasmic Triticum turgidum. Genome 35 772–779. 10.1139/g92-118 [DOI] [Google Scholar]
- Marone D., Laido G., Gadaleta A., Colasuonno P., Ficco D. B., Giancaspro A., et al. (2012). A high-density consensus map of A and B wheat genomes. Theor. Appl. Genet. 125 1619–1638. 10.1007/s00122-012-1939-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Bovill W., Percy C., Herde D., Fletcher S., Kelly A., et al. (2015). Markers for seedling and adult plant crown rot resistance in four partially resistant bread wheat sources. Theor. Appl. Genet. 128 377–385. 10.1007/s00122-014-2437-1 [DOI] [PubMed] [Google Scholar]
- Martin A., Simpfendorfer S., Hare R., Sutherland M. (2013). Introgression of hexaploid sources of crown rot resistance into durum wheat. Euphytica 192 463–470. 10.1007/s10681-013-0890-6 [DOI] [Google Scholar]
- Martin A., Simpfendorfer S., Hare R. A., Eberhard F. S., Sutherland M. W. (2011). Retention of D genome chromosomes in pentaploid wheat crosses. Heredity 107 315–319. 10.1038/hdy.2011.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matny O. N. (2015). Fusarium head blight and crown rot on wheat & barley: losses and health risks. Adv. Plants Agric. Res. 2:39 10.15406/apar.2015.02.00039 [DOI] [Google Scholar]
- Matsuoka Y. (2011). Evolution of polyploid triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52 750–764. 10.1093/pcp/pcr018 [DOI] [PubMed] [Google Scholar]
- Mesfin A., Frohberg R. C., Anderson J. A. (1999). RFLP markers associated with high grain protein from Triticum turgidum L. var. dicoccoides introgressed into hard red spring wheat. Crop Sci. 39 508–513. 10.2135/cropsci1999.0011183X003900020035x [DOI] [Google Scholar]
- Monika K., Jimenez M. D., Bassi F. M., Ghavami F., Simons K., Dizon R., et al. (2013). A radiation hybrid map of chromosome 1D reveals synteny conservation at a wheat speciation locus. Funct. Integr. Genomics 13 19–32. 10.1007/s10142-013-0318-3 [DOI] [PubMed] [Google Scholar]
- Mukai Y., Nakahara Y., Yamamoto M. (1993). Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36 489–494. 10.1139/g93-067 [DOI] [PubMed] [Google Scholar]
- Munns R., Hare R., James R., Rebetzke G. (1999). Genetic variation for improving the salt tolerance of durum wheat. Crop Pasture Sci. 51 69–74. 10.1071/AR99057 [DOI] [Google Scholar]
- Navashin M. (1934). Chromosome alterations caused by hybridization and their bearing upon certain general genetic problems. Cytologia 5 169–203. 10.1508/cytologia.5.169 [DOI] [Google Scholar]
- Newton K. J., Coe E. (1986). Mitochondrial DNA changes in abnormal growth (nonchromosomal stripe) mutants of maize. Proc. Natl. Acad. Sci. U.S.A. 83 7363–7366. 10.1073/pnas.83.19.7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K., Mori T., Takami N., Furuta Y. (1974). Mapping of progressive necrosis genes, Ne1 and Ne2 of common wheat by the telocentric method. Jpn. J. Breed. 24 277–281. 10.1270/jsbbs1951.24.277 [DOI] [Google Scholar]
- Novaro P., D’egidio M., Mariani B., Nardi S. (1993). Combined effect of protein content and high-temperature drying systems on pasta cooking quality. Cereal Chem. 70 716–716. [Google Scholar]
- Noyszewski A. K., Ghavami F., Alnemer L. M., Soltani A., Gu Y. Q., Huo N., et al. (2014). Accelerated evolution of the mitochondrial genome in an alloplasmic line of durum wheat. BMC Genomics 15:67 10.1186/1471-2164-15-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y., Yamazaki Y., Murai K., Kanno A., Terachi T., Shiina T., et al. (2005). Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 33 6235–6250. 10.1093/nar/gki925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen K., Clewett T., Thompson J. (2010). Pre-cropping with canola decreased Pratylenchus thornei populations, arbuscular mycorrhizal fungi, and yield of wheat. Crop Pasture Sci. 61 399–410. 10.1071/CP09345 [DOI] [Google Scholar]
- Palumbo M., Spina A., Boggini G. (2000). “Agronomic and bread-making characteristics of durum wheat genotypes deriving from interspecific hybridisation with bread wheat,” in Durum Wheat Improvement in the Mediterranean Region: New Challenges, eds Araus J. L., Di Fonzo N., Nachit M., Royo C. (Zaragoza: CIHEAM; ) 515–518. [Google Scholar]
- Pikaard C. S. (1999). Nucleolar dominance and silencing of transcription. Trends Plant Sci. 4 478–483. 10.1016/S1360-1385(99)01501-0 [DOI] [PubMed] [Google Scholar]
- Ramsey J., Schemske D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29 467–501. 10.1146/annurev.ecolsys.29.1.467 [DOI] [Google Scholar]
- Rayburn A. L., Gill B. S. (1986). Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Rep. 4 102–109. 10.1007/BF02732107 [DOI] [Google Scholar]
- Reader S., Miller T. (1991). The introduction into bread wheat of a major gene for resistance to powdery mildew from wild emmer wheat. Euphytica 53 57–60. 10.1007/BF00032033 [DOI] [Google Scholar]
- Ren R., Ray R., Li P., Xu J., Zhang M., Liu G., et al. (2015). Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Genet. Genomics 290 1457–1470. 10.1007/s00438-015-0997-7 [DOI] [PubMed] [Google Scholar]
- Reynolds M., Manes Y., Izanloo A., Langridge P. (2009). Phenotyping approaches for physiological breeding and gene discovery in wheat. Ann. Appl. Biol. 155 309–320. 10.1111/j.1744-7348.2009.00351.x [DOI] [Google Scholar]
- Roder M. S., Korzun V., Wendehake K., Plaschke J., Tixier M.-H. N., Leroy P., et al. (1998). A microsatellite map of wheat. Genetics 149 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J., Millet E., Manisterski J., Feldman M. (2000). A new powdery mildew resistance gene: introgression from wild emmer into common wheat and RFLP-based mapping. Euphytica 115 121–126. 10.1023/A:1003950431049 [DOI] [Google Scholar]
- Schubert I., Fransz P. F., Fuchs J., De Jong J. H. (2001). “Chromosome painting in plants,” in Chromosome Painting, eds Sharma A. K., Sharma A. (Dordrecht: Springer; ) 57–69. 10.1007/978-94-010-0330-8_7 [DOI] [PubMed] [Google Scholar]
- Schwarzacher T., Anamthawat-Jonsson K., Harrison G., Islam A., Jia J., King I., et al. (1992). Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theor. Appl. Genet. 84 778–786. 10.1007/BF00227384 [DOI] [PubMed] [Google Scholar]
- Schwarzacher T., Leitch A., Bennett M., Heslop-Harrison J. (1989). In situ localization of parental genomes in a wide hybrid. Ann. Bot. 64 315–324. 10.1093/oxfordjournals.aob.a087847 [DOI] [Google Scholar]
- Sharma H. C., Gill B. S. (1983). Current status of wide hybridization in wheat. Euphytica 32 17–31. 10.1007/BF00036860 [DOI] [Google Scholar]
- Sheedy J., Mckay A., Lewis J., Vanstone V., Fletcher S., Kelly A., et al. (2015). Cereal cultivars can be ranked consistently for resistance to root-lesion nematodes (Pratylenchus thornei & P. neglectus) using diverse procedures. Australas. Plant Pathol. 44 175–182. 10.1007/s13313-014-0333-4 [DOI] [Google Scholar]
- Sheedy J., Thompson J., Kelly A. (2012). Diploid and tetraploid progenitors of wheat are valuable sources of resistance to the root lesion nematode Pratylenchus thornei. Euphytica 186 377–391. 10.1007/s10681-011-0617-5 [DOI] [Google Scholar]
- Shepherd K. (1988). “Genetics of wheat endosperm proteins-in retrospect and prospect,” in Proceedings of the Seventh International Wheat Genetics Symposium eds Miller T. E., Koebner R. M. D. (Oxford: Oxford University Press; ) 919–931. [Google Scholar]
- Shewry P., Halford N., Tatham A. (1992). High molecular weight subunits of wheat glutenin. J. Cereal Sci. 15 105–120. 10.1016/S0733-5210(09)80062-3 [DOI] [Google Scholar]
- Silva G., Souza M. (2013). Genomic in situ hybridization in plants. Genet. Mol. Res. 12 2953–2965. 10.4238/2013.August.12.11 [DOI] [PubMed] [Google Scholar]
- Simons K. J., Gehlhar S. B., Maan S. S., Kianian S. F. (2003). Detailed mapping of the species cytoplasm-specific (scs) gene in durum wheat. Genetics 165 2129–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissons M. (2008). Role of durum wheat composition on the quality of pasta and bread. Food 2 75–90. 10.1371/journal.pone.0050057 [DOI] [Google Scholar]
- Sissons M. (2012). Durum Wheat Chemistry and Technology. London: Academic Press. [Google Scholar]
- Smiley R. W., Gourlie J. A., Easley S. A., Patterson L.-M., Whittaker R. G. (2005). Crop damage estimates for crown rot of wheat and barley in the Pacific Northwest. Plant Dis. 89 595–604. 10.1094/PD-89-0595 [DOI] [PubMed] [Google Scholar]
- Somers D. J., Isaac P., Edwards K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109 1105–1114. 10.1007/s00122-004-1740-7 [DOI] [PubMed] [Google Scholar]
- Thompson J. P., Zwart R. S., Butler D. (2012). Inheritance of resistance to root-lesion nematodes (Pratylenchus thornei and P. neglectus) in five doubled-haploid populations of wheat. Euphytica 188 209–219. 10.1007/s10681-012-0689-x [DOI] [Google Scholar]
- Troccoli A., Borrelli G., De Vita P., Fares C., Di Fonzo N. (2000). Mini review: durum wheat quality: a multidisciplinary concept. J. Cereal Sci. 32 99–113. 10.1006/jcrs.2000.0322 [DOI] [Google Scholar]
- Tsunewaki K. (1970). Necrosis and chlorosis genes in common wheat and its ancestral species. Seiken Ziho 22 67–75. [Google Scholar]
- Tsunewaki K. (1992). Aneuploid analyses of hybrid necrosis and hybrid chlorosis in tetraploid wheats using the D genome chromosome substitution lines of durum wheat. Genome 35 594–601. 10.1139/g92-089 [DOI] [Google Scholar]
- Tsunewaki K. (2004). Aneuploid analyses of three chlorophyll abnormalities in Emmer wheat. Genes Genet. Syst. 79 95–104. 10.1266/ggs.79.95 [DOI] [PubMed] [Google Scholar]
- Von Mark V. C., Kilian A., Dierig D. A. (2013). Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS ONE 8:e64062 10.1371/journal.pone.0064062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Jiang J., Chen S., Qi X., Fang W., Guan Z., et al. (2014). Rapid genetic and epigenetic alterations under intergeneric genomic shock in newly synthesized Chrysanthemum morifolium ×Leucanthemum paludosum hybrids (Asteraceae). Genome Biol. Evol. 6 247–259. 10.1093/gbe/evu008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu D., Yan Z., Wei Y., Zheng Y. (2005). Cytological characteristics of F2 hybrids between Triticum aestivum L. and T. durum Desf. with reference to wheat breeding. J. Appl. Genet. 46 365–369. [PubMed] [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., Huang B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12 787–796. 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth G., Mcnamara R., Quick J. (2001). Crown depth and susceptibility to crown rot in wheat. Euphytica 122 397–405. 10.1023/A:1012947516161 [DOI] [Google Scholar]
- Xu L., Wang M., Cheng P., Kang Z., Hulbert S., Chen X. (2013). Molecular mapping of Yr53, a new gene for stripe rust resistance in durum wheat accession PI 480148 and its transfer to common wheat. Theor. Appl. Genet. 126 523–533. 10.1007/s00122-012-1998-0 [DOI] [PubMed] [Google Scholar]
- Zhang P., Friebe B., Gill B., Park R. (2007). Cytogenetics in the age of molecular genetics. Crop Pasture Sci. 58 498–506. 10.1071/AR07054 [DOI] [Google Scholar]
- Zhang P., Hiebert C. W., Mcintosh R. A., Mccallum B. D., Thomas J. B., Hoxha S., et al. (2016). The relationship of leaf rust resistance gene Lr13 and hybrid necrosis gene Ne2m on wheat chromosome 2BS. Theor. Appl. Genet. 129 485–493. 10.1007/s00122-015-2642-6 [DOI] [PubMed] [Google Scholar]
- Zhang P., Li W., Friebe B., Gill B. S. (2004). Simultaneous painting of three genomes in hexaploid wheat by BAC-FISH. Genome 47 979–987. 10.1139/g04-042 [DOI] [PubMed] [Google Scholar]
- Zhang W., Chao S., Manthey F., Chicaiza O., Brevis J. C., Echenique V., et al. (2008). QTL analysis of pasta quality using a composite microsatellite and SNP map of durum wheat. Theor. Appl. Genet. 117 1361–1377. 10.1007/s00122-008-0869-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.