Abstract
Diverse microbial communities coordinate group behaviors through signal exchange, such as the exchange of acyl-homoserine lactones (AHLs) by Gram-negative bacteria. Cellular communication is prone to interference by neighboring microbes. One mechanism of interference is signal destruction through the production of an enzyme that cleaves the signaling molecule. Here we examine the ability of one such interference enzyme, AiiA, to modulate signal propagation in a spatially distributed system of bacteria. We have developed an experimental assay to measure signal transduction and implement a theoretical model of signaling dynamics to predict how the system responds to interference. We show that titration of an interfering strain into a signaling network tunes the spatial range of activation over the centimeter length scale, quantifying the robustness of the signaling network to signal destruction and demonstrating the ability to program systems-level responses of spatially heterogeneous cellular networks.
Introduction
Microbes use small-molecule signals to coordinate group behaviors. For example, many Gram-negative bacteria exchange acyl-homoserine lactones (AHLs) to regulate quorum sensing and related emergent behaviors such as biofilm formation, collective motility, bioluminescence, and virulence (1, 2, 3, 4). Activation of quorum sensing involves the production of a signal by a synthase, for example, LuxI, used here, which was isolated from Aliivibrio fischeri. The signals exit the cell either by passive diffusion through the membrane or by active transport (5), and they can be detected in neighboring cells by specific receptors, here the receptor LuxR. Binding of the signal to the receptor leads to global changes in gene expression. The signals enable bacteria to relay information about their physical, chemical, and biological environment (6, 7, 8, 9, 10). Communities that coordinate behavior through such mechanisms often have a fitness advantage (11, 12). Not surprisingly, other bacteria have evolved mechanisms to interfere with the signaling process.
Bacteria exhibit various signal interference mechanisms (13, 14); collectively, these mechanisms that interfere with bacterial cell communication have been termed quorum quenching (13, 15, 16). The two main mechanisms of quorum quenching found in natural communities are signal crosstalk and signal destruction. Signal crosstalk between related signals can result in competitive binding to the receptors (13, 17, 18), and some bacteria produce compounds whose only known function is receptor antagonism (13, 15, 16). Signal destruction often involves the production of an enzyme that chemically modifies or cleaves signaling molecules. The influence of signal destruction on signaling propagation through space has not been systematically explored.
Here, we develop an experimental assay to quantify multispecies pattern formation. The assay enables direct measurement of the robustness of signal exchange to the introduction of a variable amount of a signal-degrading strain. Since the initial work by Basu et al. (19), demonstrating the ability to program pattern formation through signal exchange, several studies have examined many aspects of bacterial pattern formation. Experimental works have examined scale-invariant patterns (20), the impact of signal removal on long-distance signaling (21), tuning the concentration dependence of the bacterial response to signal (22), multispecies pattern formation that was programmable and robust to environmental perturbations (23), the control of pattern formation coupled to motility (24), and the ability of signal degradation to impact wild-type behaviors such as biofilm formation and swarming (25). Theoretical advances have also deepened our understanding of the sensitivity of patterns to model parameters (26). Here, we build upon these previous studies to determine what level of interference can be tolerated by a microbial network exchanging AHLs, and we predict and experimentally validate changes in the signaling patterns in the presence of interference.
Interference by signal-destroying enzymes is a potential route by which to control bacterial community function in industrial and biomedical applications. For instance, signal destruction can be used to limit cell-cell communication in microbes, which has been directly linked to pathogenicity (13, 15, 16). The ability to precisely program and control emergent behaviors of microbial networks through molecular-level manipulation requires a thorough, quantitative understanding of signal exchange and interference in mixed communities. Although previous studies (13, 15, 16, 19) have identified many natural systems with signal interference, no quantitative measurements have been taken to understand the robustness of signal exchange in spatially heterogeneous systems in the presence of interference. Here, we developed an experimental assay to examine the influence of a signal-destroying enzyme, the lactonase AiiA, on the spatiotemporal dynamics of signal exchange in a multistrain synthetic signaling network. The assay measures the robustness of signaling to interference. We find that AHL signaling in the presence of an interference strain producing the enzyme AiiA results in a limited zone of activation. Using a theoretical model, we show that the size of this zone is tunable by adjusting the number of interfering cells and signal-producing cells. These results demonstrate that signal interference limits the spatial range of activation in the vicinity of a signal-producing colony and can be implemented to predictably program the response of multistrain bacterial communities.
Materials and Methods
Bacterial strains and plasmid
The sender strain is Escherichia coli (NEB 5-alpha) with the plasmid ptD103IuxI sfGFP(Kn) (Fig. S1) from (27). This strain produces the signal N-(3-oxohexanoyl)-HSL and the cognate receptor LuxR. The GFP reporter gene activates in response to quorum-sensing activation. The interfering strain consists of an E. coli host harboring the plasmid ptD103aiiA(Cm) (see Fig. S2 A and (27)).
The receiver strain is E. coli (NEB 5-alpha) with the plasmid pTD103LuxR RFP, shown in Fig. S3. To construct the receiver strain, the plasmid pTD103luxI sfGFP(Kn) was mutagenized by inserting mCherry in the position of sfGFP, using a Gibson assembly kit (New England Biolabs). The receiver strain plasmid expresses mCherry fluorescent protein in response to quorum-sensing activation.
Culturing conditions
Bacterial strains were taken from frozen stocks and grown in a 14 mL Falcon tube with 5 mL of lysogeny broth (LB) with appropriate antibiotics for plasmid maintenance. Cultures were grown in a shaker at 220 rpm and 37°C. After 16 h of growth, the culture in the late log-phase was taken out and 1 mL of the inoculum was transferred into a micro-centrifuge tube. The tube was centrifuged at 15,000 rpm for 1 min and the supernatant was discarded. The cells were then resuspended by vortexing in LB without antibiotics. Late log-phase cultures were used such that quorum sensing of the sender strain was activated before measurement in the plate assay, in which LB agar with 2.5% agar was used. During time-lapse experiments, the cells were imaged at 37°C.
Interference assay
Ten microliters of the receiver strain was mixed with the interference strain in a micro-centrifuge tube. The mixture was loaded onto an LB agar plate by pipetting, and cells were distributed evenly using 4-mm-diameter sterilized glass beads. After 10 min, 1 μL of the sender strain was pipetted to the middle of the plate. The plate was imaged using a fluorescent microscope for time-lapse experiments.
Microscopy measurements
The images were taken using a Nikon eclipse TI fluorescent microscope. Time-lapse experiments were carried out at 37°C using a temperature-controlled chamber. Samples were imaged with green (GFP) and red fluorescent protein (RFP) illumination at a magnification of 20×. RFP images were taken every 15 min for 16 h at 30 different distances from the sender colony, extending in two directions outward from the sender colony. Activation times were calculated at each position. Exposure time at each position was 1 s for RFP and 500 ms for GFP. No significant photobleaching was observed.
Each image taken was saved in .tiff format and analyzed using a custom MATLAB code. A low threshold was applied to the RFP images to identify the location of receiver cells within each image. An upper threshold was used to identify the receivers that had activated quorum sensing. For each time point and position, the fraction of cellular pixels above the quorum-sensing-activation threshold was calculated. If the fraction of activated pixels exceeded 10%, that position was included as part of the activated region.
Growth measurements
To obtain the growth curves, the cells were grown in LB media with appropriate antibiotics, as mentioned above. After 16 h growth, we diluted 5 μL of the cells in 4995 μL of LB. Dilutions of the culture were plated on selective media to measure cell density over time.
To test the growth influences on the receivers and senders when in coculture with the interferers, we diluted 5 μL of the senders (or receivers) with 5 μL of the interferers in 4990 μL of pure LB broth, and the previous procedure was repeated by selecting for the senders and receivers using proper antibiotic plates.
Plate-reader measurements
A Tecan Infinite m200 Pro plate reader was used to measure growth rates and fluorescence in well-mixed conditions. Cells were grown in LB media, as mentioned above. Cells were then diluted 1000-fold in LB media containing antibiotics and cultured for an additional 3 h. After 3 h of growth, 200 μL of these early log-phase cells were loaded into a flat bottom 96-well plate. The plate was inserted into the plate reader set to 37°C and the optical density and fluorescent intensity were measured every 15 min for 15 h. Optical density measurements were carried out at a wavelength of 600 nm. For GFP measurements, a wavelength of 485 nm was used for excitation and a wavelength of 515 nm was used for emission. For mCherry fluorescence measurements, a wavelength of 590 nm was used for excitation and a wavelength of 650 nm was used for emission. In the Supporting Material we introduce a method for analyzing the fluorescence of a single species from a mixed culture.
Results and Discussion
Quantifying signal exchange in spatially distributed microbial networks
An assay was developed to examine the dynamics of signal exchange in multistrain bacterial communities. The assay is based on previous reports that used solid agar plates to measure the response to the diffusive exchange of signaling molecules (19, 20, 21, 22). As shown in Fig. 1 a, our system consists of a 2.5% LB plate with a receiver strain evenly distributed on top of the plate. The receiver strain does not produce the signal (AHL); however, it produces the receptor LuxR and increases the expression of a fluorescent reporter gene, mCherry, in response to the AHL signal. A sender strain containing the synthase gene luxI produces the AHL signal and is pipetted onto the center of the plate. The sender strain makes the receptor LuxR, and as in the natural quorum-sensing system, the synthase is positively regulated by AHL. The sender strain produces GFP. Control measurements reported in Fig. S4 show that the fluorescence intensity of the receiver strain increases upon coculture with the sender strain. E. coli strains contained plasmids based on constructs reported in (27) (see Figs. S1–S3). Similar sender/receiver systems have been developed and investigated in previous work (19, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38).
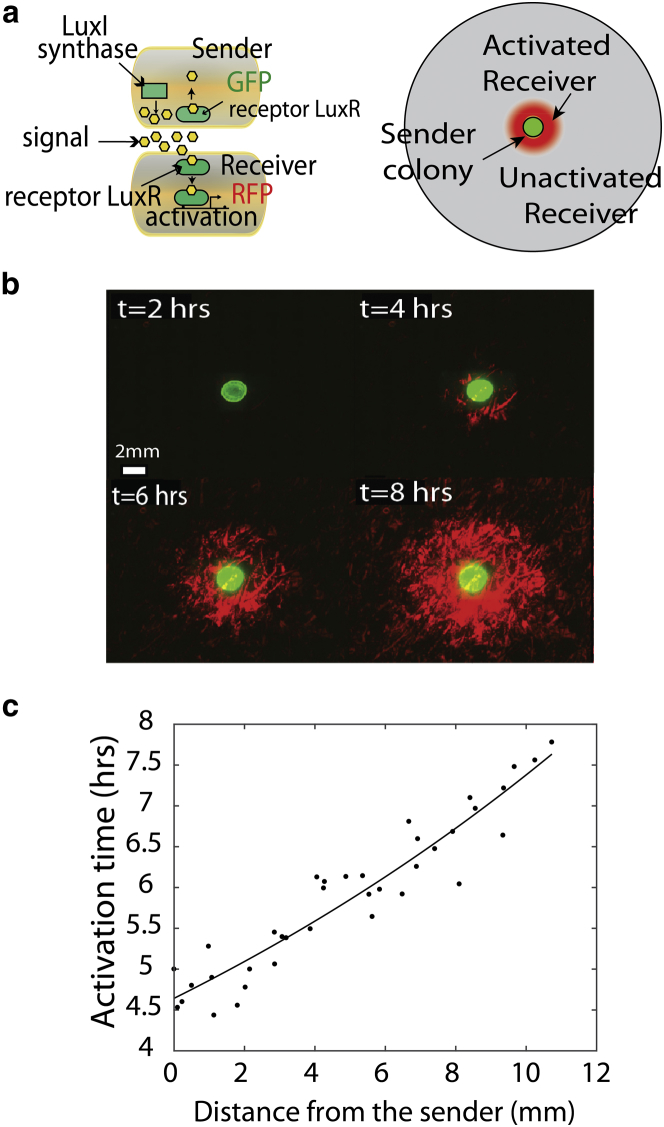
Figure 1.
(a) The two-strain signaling network consists of a sender strain (GFP) producing AHL signal and a receiver strain (mCherry) containing an AHL-regulated fluorescent reporter gene. In the plate assay, the receivers are evenly distributed on an LB-agar plate, and a colony of sender strains is added to the middle of the plate. (b) Time-lapse fluorescent imaging captures the activation of the receiver cells as AHL diffuses outward from the sender colony. The green fluorescence shows the location of the sender strain. (c) Activation time of the receivers as a function of distance from the sender colony. The data points have been fitted to an exponential curve. To see this figure in color, go online.
Upon adding the sender strain to the center of the plate, the sender strain excretes AHL signal, which diffuses into the lawn of receiver strains to activate expression of a fluorescent reporter gene (mCherry). As shown in Fig. 1 b, the receiver cells closest to the sender cells are activated first, and over time, this region of activation expands outward. Fluorescent images, shown in Fig. 1 b, represent the stitching together of images from several microscope positions taken at 20× magnification such that activation can be observed at the single-cell level over centimeter length scales. The dynamics of signal exchange in the system can be quantified by measuring the time at which receiver cells activate at a specific distance from the center of the sender cells. We set a threshold pixel intensity to indicate this transition from the basal to the activated rate of fluorescence expression (see Fig. S5). Quorum-sensing activation at a given position is defined as 10% of the pixels belonging to cells being above this threshold. In Fig. S6, we show that the activation dynamics are not strongly dependent on the value of the activation threshold used in the analysis. Fig. 1 c shows the dynamics of activation at distances of up to 12 mm from the sender colony, the size of the plate used in the assay.
AiiA inhibits quorum sensing in a synthetic microbial community
To extend the signaling assay to the examination of interference between strains, we first validated the interference capabilities of a strain producing the enzyme AiiA, a lactonase originally isolated from Bacillus thuringiensis (13, 15, 16). A plasmid containing aiiA, from (27), was transferred to E. coli to create the interference strain and added to the sender-and-receiver community, as depicted in Fig. 2 a.
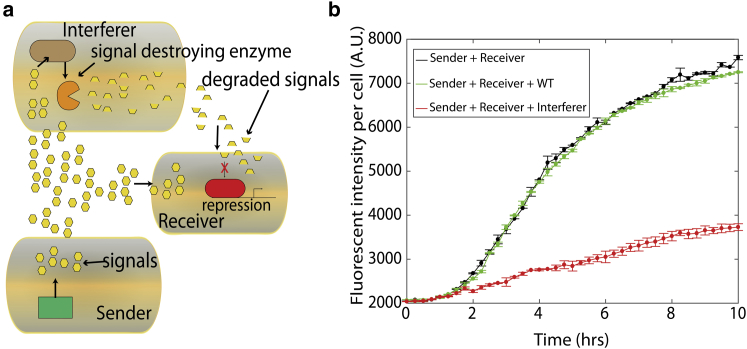
Figure 2.
(a) To quantify the impact of interference on signal exchange, a third strain is added to the system that produces the enzyme AiiA, a lactonase that cleaves AHL signal. (b) Addition of the interference strain to the sender-and-receiver community reduces the fluorescence intensity per cell of the receiver strain. Here, the negative control shows that the addition of the wild-type yields a curve similar to the case of no interference. Error bars show the standard deviation of three replicate measurements. To see this figure in color, go online.
Fig. 2 b shows the activation of quorum sensing for the sender strain alone and in coculture with the AiiA interference strain. Here, the fluorescence intensity of the sender strain was measured in a 96-well plate reader under well-mixed conditions. Coculture with the interference strain reduced the level of quorum-sensing activation, indicating that the AiiA producing strain interfered with AHL signal transduction. We did not observe significant growth influences that can be caused by the interfering strain on the senders or receivers (see Fig. S7). Next, we both experimentally and theoretically examine the influence of AiiA-mediated interference in spatially distributed networks of cells.
Signal destruction leads to finite and tunable zones of activation in spatial networks
The interference strain was added to the plate-based assay to quantify the influence of signal destruction on the signaling dynamics. The interference strain was added to the lawn of receiver cells, as depicted in Fig. 3 a. Different amounts of the interfering strains were titrated into the receiver cells to measure the robustness of signal propagation to a variable level of interference.
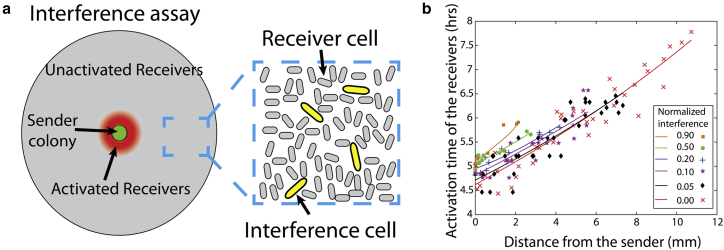
Figure 3.
(a) Introducing the AiiA cells in the plate-assay setup. (b) Activation time versus distance from the sender plot for variable levels of interference. The number of interference cells in the assay is normalized to the number of cells in 100 μL of cells in a culture with an absorbance of 1.0 at 600 nm. Curves show an exponential fit to demonstrate qualitative trends in the data. Experiments were run for 16 h, much longer than the time needed to observe the maximum radius of activation.
Fig. 3 b shows the impact of different levels of interference on signaling dynamics. The no-interference data from Fig. 1 c is shown for reference. As the level of interference increased, we observed a slight delay in quorum-sensing activation. Additionally, the system only activated up to a finite distance. For example, at a normalized level of interference of 0.9, receiver cells at a distance of 2.3 mm activated at ∼5.5 h. The experiment continued to run for a total time of 16 h, and activation beyond 2.3 mm was not observed (see Fig. S8). The radius of the activation zone was inversely proportional to the level of interference. As seen in Fig. S7, there are no significant growth influences between the strains. To confirm that this effect was due to production of the AiiA enzyme, in control experiments, a wild-type strain that does not produce AiiA was substituted for the interference strain (Fig. S9).
Modeling the impact of signal destruction on signal propagation
A model for signal exchange in the presence of interference was derived to further explore the ability of signal destruction to limit the zone of activation. Our model, based on previously reported models from (27) and (39) accounts for the growth of the sender strain, the receiver strain, and the interference strain. All three strains follow the logistic growth equation,
| (1) |
where μ is the growth rate constant, n is the cell density, is the total cell density (senders + receivers + interferers), and s is the maximal cell density in the system. All three strains use the same E. coli host and were shown to have similar growth characteristics (see Fig. S7). Fig. S7 shows that there are no growth influences on the sender and the receivers when cocultured with the interferer.
Several models have been used to describe quorum-sensing signaling (27, 28, 29, 30, 39, 40, 41), including signaling in the presence of signal destruction (27). The dynamics of the AHL signal, [AHL], by the sender strain can be described by the equation
| (2) |
where DAHL is the diffusion coefficient for the signal, is the cell density of the senders, is the maximal production rate of the AHL per cell, is the basal level of AHL production, represents the concentration of the signal at half maximum activity, mi represents the cooperative coefficients, is the basal rate of signal degradation in media (39, 42), is the rate of signal degradation due to the AiiA, and is the concentration of AiiA signal. The Hill’s function represents a threshold amount of signal needed to activate quorum sensing up to a maximum production rate.
The change in the amount of AiiA is described by
| (3) |
where, is the maximum production rate of the AiiA per cell, is the basal level of AiiA production, is the cell density of the AiiA cells, and is the basal rate of AiiA degradation. The values of all the parameters are given in Table 1. Given the timescale of our simulations, we assume that the degradation rate of the AiiA will be low and set it to the degradation rate of the AHLs. In the Supporting Material, we estimate the number of AiiA molecules per cell and conclude that the production rates used give a reasonable level of protein per cell.
Table 1.
Values of Parameters in Eqs. 1–3
| Parameter | Value |
|---|---|
| 1.50 h−1 (experimentally calculated) | |
| s | 109 cells per mL |
| DAHL | 1.764 mm2 h−1 (40) |
| 2.3 × 10−9 nmol h−1 per cell (39) | |
| 2.3 × 10−10 nmol h−1 per cell (39) | |
| 70 nM (39) | |
| 70 nM (32) | |
| m1 | 2.5 (32) |
| m2 | 2.5 (32) |
| 0.005545 h−1 (39) | |
| 0.01 h−1 (27) | |
| 2.3 × 10−9 nmol h−1 per cell | |
| 2.3 × 10−10 nmol h−1 per cell | |
| 0.005545 h−1 | |
| 70 nM (39) | |
| m3 | 2.5 (32) |
In the simulations, we solve the equations using the finite-difference method. The interference strain is evenly distributed in the simulation grid at various loading densities. Activation occurs at a distance X when the concentration of AHL signal at that point reaches the threshold concentration. Growth-rate constants were experimentally obtained, as shown in Fig. S7 A. Additional parameters were reported in previous studies.
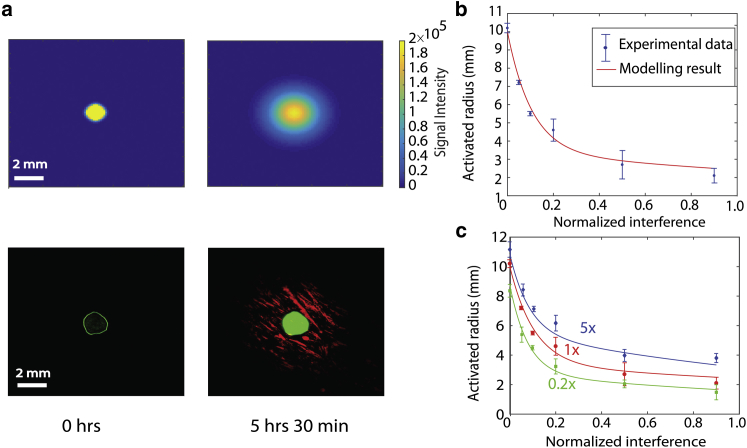
As shown in Fig. 4 a, simulations of the dynamics of activation in the plate-based assay qualitatively matched activation profiles from the experimental measurements at steady state. The transient behavior in Fig. 3 b was compared to the simulation results in Fig. S10. Predicted steady-state activation diameters were closed to experimental measurements, although simulations reached the steady-state diameter 20–30 min earlier than in experiments. The delayed activation in experiment could be due to processes such as fluorescent protein folding and maturation and time delays in signal processing, which were not included in the model. As shown in Fig. 4 b, model predictions for the plate-based assay are in close agreement with experimental results (see Fig. S11 for simulation predictions of the well-mixed system). As the percentage of interference strain increased, the activation zone shrank from 10 mm down to 2 mm, demonstrating that the number of interfering cells within the system tunes the radius of activation. As in experiments, simulations were run for 16 h, much longer than the time needed to reach steady-state activation profiles. In these experiments, expression of aiiA is regulated by the AHL quorum-sensing signal, although simulations and experiments shown in Fig. S12 demonstrate that strains without AHL-dependent aiiA expression exhibit similar activation dynamics.
Figure 4.
(a) A model using Eqs. 1–3 simulates receiver-cell activation in the plate-based assay (top), and the experimental comparison is shown below. Here, the normalized interference level is 0.9. (b) The maximal activated radius of the receivers versus the normalized number of interference cells. The experimental measurements (dots) agreed with model predictions (line). (c) Theoretical predictions and experimental data points for the activation radius when the number of sender cells is increased by 5× or decreased to 0.2×. Error bars show the standard deviation of three replicate measurements. To see this figure in color, go online.
In simulations and experiments we varied the number of cells in the sender colony and observed that when the number of cells in the sender strain was increased or decreased by a factor of 5, the activation range curve shifted (Fig. 4 c). Together, the ratio of sender to interference strains dictates the spatial extent of activation. To further explore the parameters that dictate the dynamics of the activation zone, in simulations, we varied multiple model parameters, including the diffusion coefficient and the degradation of AHL due to the media (Figs. S13 and S14). We observe that the major contributors to the activation zone are the diffusion of the AHLs and the degradation of the AHL by the interferer. The production rate of the AHLs by the senders weakly influences the activation radius, as shown in Fig. S14 A. In Fig. S15, through simulations, we demonstrate that the potential growth interactions between the strains and internalization of the signal by non-receiver strains can produce a similar limitation in the activation zone. Although theoretically hypothesized, the changes to the activation zone by these parameters would need to be experimentally validated.
Conclusions
We have demonstrated that signal interference with a bacterial communication pathway through signal destruction creates a finite region of activation around a colony of signal emitting cells. Similar spatial activity patterns have been programmed using external chemical gradients or synthetic signaling circuits (19, 22, 24, 43). Here a plate-based experimental assay was implemented to quantify the influence of interference on signal transduction in a spatially heterogeneous system. The assay measurements were supported by a reaction-diffusion simulation of signal dynamics, demonstrating the utility of such models in predicting signal exchange in systems with complex geometries that could not easily be analyzed experimentally.
Both the theoretical and experimental results revealed the capability of tuning the system-level parameters of a multistrain bacterial community. By adjusting the amount of interference strain, the zone of activation was tunable from 2 to 10 mm. Theoretical predictions and experimental results shown in Fig. 4 c demonstrate that parameters such as the number of sender cells also modulated the radius of activation. Parameters including the diffusion coefficient of the signal, degradation and production rates of the signal, and cell growth rates were modulated in simulations, predicting that the area of activation was most sensitive to signal transport and degradation rates. Experimental validation of these predictions is needed. Similar theoretical models of signaling have explored the network parameters that regulate bacterial pattern formation (20, 26). Such strategies for adjusting the dynamics of multispecies interactions might prove useful for programming the activity of synthetic communities.
Recent interest in engineering microbial communities to perform complex tasks (32, 37, 44), such as in nanoscale synthesis and biomedical applications (19, 45, 46, 47), call for new experimental techniques to control the network-level behavior of microbial communities (45, 48, 49, 50). Although characterization of diverse communities often focuses on well-mixed systems, nonlinearities in the interactions can result in unexpected outcomes, particularly in spatially heterogeneous systems (33, 51, 52). The plate-based assay developed here may prove useful to quantify the outputs of spatially distributed, multispecies networks (53). Such approaches identify control parameters, such as the level of signal destruction shown here, that strongly determine global behaviors. These experiments, in conjunction with theoretical models, will enable us to understand and predict the parameters that determine community behavior in multispecies bacterial communities.
Our results suggest that in natural systems even low levels of interference will modulate the dynamics of signal propagation, and hence the functional outputs of the community. Many natural systems are spatially heterogeneous, and the consequences of specific spatial distributions could be predicted in simulations. It remains unclear to what extent interference constrains signaling to local areas. Recent studies have examined quorum sensing that occurs locally within small populations (54, 55), but there are examples of long-distance coordination of cellular behaviors known to be coordinated by signal exchange (5, 56). Given the number of quorum-quenching and signal-interference mechanisms present in natural communities (13, 16, 17), future work should examine the contributions of different types of interference beyond signal destruction, such as receptor blocking through competitive binding, in shaping global signaling responses.
Author Contributions
K.P.S. performed the research, analyzed data, and wrote the article; P.C. performed the research and wrote the article; J.Q.B. designed the research, analyzed data, and wrote the article.
Acknowledgments
This work was supported by Office of Naval Research award number N00014-15-1-2573 and a Defense Advanced Research Projects Agency Young Faculty Award.
Editor: Reka Albert.
Footnotes
Supporting Materials and Methods and fifteen figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30105-4.
Supporting Material
References
- 1.de Kievit T.R., Iglewski B.H. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y.-H., Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel) 2012;12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Q., Defoirdt T. Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ. Microbiol. 2015;17:960–968. doi: 10.1111/1462-2920.12420. [DOI] [PubMed] [Google Scholar]
- 4.Boedicker J., Nealson K. INVITED: microbial communication via quorum sensing. IEEE Trans. Mol. Biol. Multi-Scale Commun. 2016;1:310–320. [Google Scholar]
- 5.Nealson K.H., Hastings J.W. Quorum sensing on a global scale: massive numbers of bioluminescent bacteria make milky seas. Appl. Environ. Microbiol. 2006;72:2295–2297. doi: 10.1128/AEM.72.4.2295-2297.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 7.Cornforth D.M., Popat R., Brown S.P. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc. Natl. Acad. Sci. USA. 2014;111:4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sifri C.D. Healthcare epidemiology: quorum sensing: bacteria talk sense. Clin. Infect. Dis. 2008;47:1070–1076. doi: 10.1086/592072. [DOI] [PubMed] [Google Scholar]
- 9.Heilmann S., Krishna S., Kerr B. Why do bacteria regulate public goods by quorum sensing?—How the shapes of cost and benefit functions determine the form of optimal regulation. Front. Microbiol. 2015;6:767. doi: 10.3389/fmicb.2015.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kavanaugh J.S., Thoendel M., Horswill A.R. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol. Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 11.Heurlier K., Dénervaud V., Haas D. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006;296:93–102. doi: 10.1016/j.ijmm.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Darch S.E., West S.A., Diggle S.P. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. USA. 2012;109:8259–8263. doi: 10.1073/pnas.1118131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rampioni G., Leoni L., Williams P. The art of antibacterial warfare: deception through interference with quorum sensing-mediated communication. Bioorg. Chem. 2014;55:60–68. doi: 10.1016/j.bioorg.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Williams P., Winzer K., Cámara M. Look who’s talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandclément C., Tannières M., Faure D. Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 2016;40:86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 16.Dong Y.H., Wang L.Y., Zhang L.-H. Quorum-quenching microbial infections: mechanisms and implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007;362:1201–1211. doi: 10.1098/rstb.2007.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez P.D., Weiss J.T., Hagen S.J. Noise and crosstalk in two quorum-sensing inputs of Vibrio fischeri. BMC Syst. Biol. 2011;5:153. doi: 10.1186/1752-0509-5-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClean K.H., Winson M.K., Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 19.Basu S., Gerchman Y., Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 20.Cao Y., Ryser M.D., You L. Collective space-sensing coordinates pattern scaling in engineered bacteria. Cell. 2016;165:620–630. doi: 10.1016/j.cell.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masiello C.A., Chen Y., Silberg J.J. Biochar and microbial signaling: production conditions determine effects on microbial communication. Environ. Sci. Technol. 2013;47:11496–11503. doi: 10.1021/es401458s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohka T., Heins R.A., Ostermeier M. An externally tunable bacterial band-pass filter. Proc. Natl. Acad. Sci. USA. 2009;106:10135–10140. doi: 10.1073/pnas.0901246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong W., Blanchard A.E., Lu T. Engineering robust and tunable spatial structures with synthetic gene circuits. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1045. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C., Fu X., Huang J.-D. Sequential establishment of stripe patterns in an expanding cell population. Science. 2011;334:238–241. doi: 10.1126/science.1209042. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Dai Y., Chen S. Effects of quorum sensing autoinducer degradation gene on virulence and biofilm formation of Pseudomonas aeruginosa. Sci. China C. Life Sci. 2007;50:385–391. doi: 10.1007/s11427-007-0044-y. [DOI] [PubMed] [Google Scholar]
- 26.Borek B., Hasty J., Tsimring L. Turing patterning using gene circuits with gas-induced degradation of quorum sensing molecules. PLoS One. 2016;11:e0153679. doi: 10.1371/journal.pone.0153679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danino T., Mondragón-Palomino O., Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langebrake J.B., Dilanji G.E., De Leenheer P. Traveling waves in response to a diffusing quorum sensing signal in spatially-extended bacterial colonies. J. Theor. Biol. 2014;363:53–61. doi: 10.1016/j.jtbi.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 29.Dilanji G.E., Langebrake J.B., Hagen S.J. Quorum activation at a distance: spatiotemporal patterns of gene regulation from diffusion of an autoinducer signal. J. Am. Chem. Soc. 2012;134:5618–5626. doi: 10.1021/ja211593q. [DOI] [PubMed] [Google Scholar]
- 30.Trovato A., Seno F., Squartini A. Quorum vs. diffusion sensing: a quantitative analysis of the relevance of absorbing or reflecting boundaries. FEMS Microbiol. Lett. 2014;352:198–203. doi: 10.1111/1574-6968.12394. [DOI] [PubMed] [Google Scholar]
- 31.Molina L., Constantinescu F., Défago G. Degradation of pathogen quorum-sensing molecules by soil bacteria: a preventive and curative biological control mechanism. FEMS Microbiol. Ecol. 2003;45:71–81. doi: 10.1016/S0168-6496(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 32.Kong W., Celik V., Lu T. Programming the group behaviors of bacterial communities with synthetic cellular communication. Bioresour. Bioprocess. 2014;1:24. [Google Scholar]
- 33.Servinsky M.D., Terrell J.L., Bentley W.E. Directed assembly of a bacterial quorum. ISME J. 2016;10:158–169. doi: 10.1038/ismej.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weitz M., Mückl A., Simmel F.C. Communication and computation by bacteria compartmentalized within microemulsion droplets. J. Am. Chem. Soc. 2014;136:72–75. doi: 10.1021/ja411132w. [DOI] [PubMed] [Google Scholar]
- 35.Choi W.S., Ha D., Kim T. Synthetic multicellular cell-to-cell communication in inkjet printed bacterial cell systems. Biomaterials. 2011;32:2500–2507. doi: 10.1016/j.biomaterials.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 36.Marchand N., Collins C.H. Peptide-based communication system enables Escherichia coli to Bacillus megaterium interspecies signaling. Biotechnol. Bioeng. 2013;110:3003–3012. doi: 10.1002/bit.24975. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Kim J.K., Bennett M.R. SYNTHETIC BIOLOGY. Emergent genetic oscillations in a synthetic microbial consortium. Science. 2015;349:986–989. doi: 10.1126/science.aaa3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang F., Kwan A., Süel G.M. A synthetic quorum sensing system reveals a potential private benefit for public good production in a biofilm. PLoS One. 2015;10:e0132948. doi: 10.1371/journal.pone.0132948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fekete A., Kuttler C., Hartmann A. Dynamic regulation of N-acyl-homoserine lactone production and degradation in Pseudomonas putida IsoF. FEMS Microbiol. Ecol. 2010;72:22–34. doi: 10.1111/j.1574-6941.2009.00828.x. [DOI] [PubMed] [Google Scholar]
- 40.Stewart P.S. Diffusion in biofilms. J. Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramalho T., Meyer A., Simmel F.C. Single cell analysis of a bacterial sender-receiver system. PLoS One. 2016;11:e0145829. doi: 10.1371/journal.pone.0145829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Englmann M., Fekete A., Schmitt-Kopplin P. The hydrolysis of unsubstituted N-acylhomoserine lactones to their homoserine metabolites. Analytical approaches using ultra performance liquid chromatography. J. Chromatogr. A. 2007;1160:184–193. doi: 10.1016/j.chroma.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 43.Payne S., Li B., You L. Temporal control of self-organized pattern formation without morphogen gradients in bacteria. Mol. Syst. Biol. 2013;9:697. doi: 10.1038/msb.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamsir A., Tabor J.J., Voigt C.A. Robust multicellular computing using genetically encoded NOR gates and chemical “wires”. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner K., You L., Arnold F.H. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Coker V.S., Telling N.D., Lloyd J.R. Harnessing the extracellular bacterial production of nanoscale cobalt ferrite with exploitable magnetic properties. ACS Nano. 2009;3:1922–1928. doi: 10.1021/nn900293d. [DOI] [PubMed] [Google Scholar]
- 47.Chen A.Y., Deng Z., Lu T.K. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shong J., Jimenez Diaz M.R., Collins C.H. Towards synthetic microbial consortia for bioprocessing. Curr. Opin. Biotechnol. 2012;23:798–802. doi: 10.1016/j.copbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Zengler K., Palsson B.O. A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol. 2012;10:366–372. doi: 10.1038/nrmicro2763. [DOI] [PubMed] [Google Scholar]
- 50.Johns N.I., Blazejewski T., Wang H.H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 2016;31:146–153. doi: 10.1016/j.mib.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratzke C., Gore J. Self-organized patchiness facilitates survival in a cooperatively growing Bacillus subtilis population. Nat. Microbiol. 2016;1:16022. doi: 10.1038/nmicrobiol.2016.22. [DOI] [PubMed] [Google Scholar]
- 52.Kastrup C.J., Boedicker J.Q., Ismagilov R.F. Spatial localization of bacteria controls coagulation of human blood by ‘quorum acting’. Nat. Chem. Biol. 2008;4:742–750. doi: 10.1038/nchembio.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Straight P.D., Willey J.M., Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: role of surfactants in raising aerial structures. J. Bacteriol. 2006;188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Connell J.L., Kim J., Whiteley M. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA. 2014;111:18255–18260. doi: 10.1073/pnas.1421211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim H.J., Boedicker J.Q., Ismagilov R.F. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl. Acad. Sci. USA. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prindle A., Samayoa P., Hasty J. A sensing array of radically coupled genetic “biopixels”. Nature. 2011;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.