Abstract
Directed cell migration by contact guidance in aligned collagenous extracellular matrix (ECM) is a critical enabler of breast cancer dissemination. The mechanisms of this process are poorly understood, particularly in 3D, in part because of the lack of efficient methods to generate aligned collagen matrices. To address this technological gap, we propose a simple method to align collagen gels using guided cellular compaction. Our method yields highly aligned, acellular collagen constructs with predictable microstructural features, thus providing a controlled microenvironment for in vitro experiments. Quantifying cell behavior in these anisotropic constructs, we find that breast carcinoma cells are acutely sensitive to the direction and extent of collagen alignment. Further, live cell imaging and analysis of 3D cell migration reveals that alignment of collagen does not alter the total motility of breast cancer cells, but simply redirects their migration to produce largely one-dimensional movement. However, a profoundly enhanced motility in aligned collagen matrices is observed for the subpopulation of carcinoma cells with high tumor initiating and metastatic capacity, termed cancer stem cells (CSCs). Analysis of the biophysical determinants of cell migration show that nuclear deformation is not a critical factor associated with the observed increases in motility for CSCs. Rather, smaller cell size, a high degree of phenotypic plasticity, and increased protrusive activity emerge as vital facilitators of rapid, contact-guided migration of CSCs in aligned 3D collagen matrices.
Introduction
Cell migration is not only a critical process for embryonic development, wound healing, and the immune response (1, 2, 3) but is also essential for cancer cell invasion and metastasis that often leads to poor patient outcomes (4, 5, 6). Despite its vital role in physiologic and pathophysiologic processes, many aspects of cell migration, particularly directional cell migration, remain a complex enigma. Directional migration involves cells migrating along a defined path or gradient (7) and may arise as a consequence of either single or combinatorial factors such as chemical gradients (i.e., chemotaxis (8)), extracellular matrix (ECM) adhesion or substrate-bound chemoattractant gradients (i.e., haptotaxis (9)), stiffness gradients (i.e., durotaxis (10)), or anisotropic organization of the ECM creating directional migration cues (i.e., contact guidance (11)).
Utilizing multiphoton excitation microscopy and second harmonic generation (SHG) imaging of live mammary tumors, we identified unique tumor-associated collagen signatures (TACS) related to breast cancer progression (12). Included in these is TACS-3, comprising collagen fiber bundles perpendicularly aligned to the tumor boundary, and running throughout the tumor mass, which induce directional migration of tumor cells via contact guidance (12, 13). In human patient samples, the presence of these architectures indeed correlate with worse prognosis (14). Yet, although a large body of work exists elucidating the mechanisms of random cell motility on two-dimensional (2D) surfaces, mechanisms of directed migration, particularly in three-dimensions (3D), are poorly understood (7). This lack of mechanistic understanding both in terms of physical and molecular mechanisms is particularly striking for directed migration from contact guidance.
Our lack of understanding of this critical form of directed migration is in part because of a paucity of efficient techniques to mimic the anisotropic 3D ECM organization to robustly provide contact guidance cues that direct cell migration. To date, a handful of reports have been presented for fabrication of 3D microenvironments that successfully align collagen matrices in vitro including magnetic alignment (either by high-power magnets (15) or flow of magnetic beads embedded during formation of the matrix (13, 16), alignment in microfluidic channels (17, 18), alignment by mechanical stretching (18), or collagen fiber extrusion (19)). However, these methods may cause interference with imaging modalities (scattering from magnetic beads), introduce additional alignment cues by confinement (alignment in narrow channels), induce unwanted damage or prestress to the fibers (mechanical stretching), and overall may not be conducive to repeated benchtop production at a reasonably high throughput (e.g., alignment by ultra-high-power magnets). Also, it is important to recognize that in vivo, native collagen fibers are believed to be largely produced by fibroblasts and reorganized by cellular contractility to generate aligned collagen signatures (20). Thus, here we use a novel technique, to our knowledge, to harness this matrix reorganizing, contractile, property of fibroblasts and generate highly aligned collagen matrices for directed cell migration without external magnetic or mechanical stimulation.
In addition to extracellular cues that influence migration, heterogeneity in the carcinoma cell population is thought to influence invasion and the capacity to successfully form metastases. Indeed, accumulating evidence suggests that many solid tumors, including those of the breast, contain a subpopulation of carcinoma cells with stem/progenitor cell-like features, termed as cancer stem cells (CSCs) (21, 22, 23). This population is highly plastic with bidirectional interconversions between stem and nonstem (i.e., more differentiated) states (24, 25) and is known to possess robust tumor-sustaining properties, drug resistance, and increased metastatic potency (21, 22, 26). Further, the physical properties of the cell microenvironment, including stiffness and topology, have been demonstrated to influence (noncancerous) stem cell behavior (27). Thus we postulated that CSCs may respond more robustly to the biophysical features of the tumor ECM that promote disease progression such as aligned collagen. However, very little is known about the mechanoresponsiveness of CSCs to tumor microenvironmental features, particularly in 3D. We therefore established a robust system to evaluate cell dynamics in tissue-engineered 3D microenvironments and quantitatively evaluate the response of CSCs to cancer-relevant ECM architecture. Using long-term 3D migration analysis, analysis of cell and nuclear shape, and protrusion dynamics we provide previously uncovered insights, to our knowledge, into the mechanoresponse of cancer cells and CSCs to anisotropic ECM organization.
Materials and Methods
Cell culture
Validated MDA-MB-231 cells were freshly obtained from ATCC (Manassas, VA; 2013) and grown in high glucose DMEM (HyClone, Pittsburgh, PA) supplemented with 10% fetal bovine serum (FBS; HyClone). Stable GFP expression was induced in MDA-MB-231 cells using GFP-lentivirus particles (Gentarget, San Diego, CA) and subsequent positive selection using puromycin (Invivogen, San Diego, CA). Cancer stem cell populations of GFP-transfected MDA-MB-231 cells were obtained by plating 20,000 cells/well into ultra-low attachment six-well plates (Corning, Tewksbury, MA) in the presence of CSC-specific media where spheroids were allowed to form for 4 days. The CSC-specific media consisted of DMEM/F12 (1:1) (HyClone) supplemented with 2% B27 (Thermo Fisher, Waltham, MA), 20 ng/mL epidermal growth factor (BD Biosciences, San Jose, CA), 5 μg/mL insulin (Sigma-Aldrich, St. Louis, MO), and 10 ng/mL human basic fibroblast growth factor (Thermo Fisher). To obtain single cells of the CSC phenotype, these spheroids were collected and treated with 0.25% Trypsin-EDTA (Thermo Fisher) for 3–4 min with intermittent agitation.
Primary carcinoma-associated fibroblasts (mFb) were derived from invasive mammary carcinomas from MMTV-PyMT mice as described (28) using differential trypsinization and selective outgrowth, and maintained in high glucose DMEM with 10% FBS. Primary human fibroblasts (hFb) from normal adjacent human breast cancer tissues were obtained from Asterand Bioscience (Detroit, MI) and validated WI-38 lung fibroblast lines from ATCC. All human fibroblast lines were grown in high glucose DMEM supplemented with 10% FBS.
Fabrication of aligned and control collagen matrices
Aligned collagen matrices were engineered by constrained cellular compaction, using a method adapted from a recently reported technique to align microvessels in fibrin gels (29, 30). First, 2.5 × 1.0 cm rectangular regions were etched on six-well plates by partial melting of the polystyrene using the end of a heated spatula, and 1.0 × 0.5 cm autoclaved porous polyethylene spacers (Interstate Specialty Products) were glued to the far ends of the region using vacuum grease (Dow Corning). The spacers were cut out of hydrophobic polyethylene POR-4896 Porex sheets (http://www.interstatesp.com/materials/porex/porex-sheet-specs), which have a median pore size ranging from 55 to 120 μm according to the manufacturer’s specifications. High-density rat-tail collagen (Corning) was neutralized with an equal volume of 100 mM HEPES (Thermo Fisher) in 2× PBS, mixed with 300,000 mFb cells/ml and growth media to a final concentration of 3 mg/mL. For hFb and WI-38 cell lines, a 200,000 cells/mL seeding density was used. The mixture was allowed to sit at room temperature for 5 min, after which 350 μl was pipetted onto each demarcated rectangular region for aligned gels, making sure to allow the gel mixture to seep into the porous spacers. Equal volumes (350 μL) of the same gel mixture were pipetted into a standard 24-well plate for isotropic controls. The gels were allowed to polymerize slowly for 20 min at room temperature and then moved to 37°C, overlaid with growth media 3 h later and subsequently allowed to polymerize completely for another 16–24 h under physiologic growth conditions.
After polymerization, the rectangular collagen matrices, along with the polyethylene spacers connected at the two ends, were detached from the bottom and transferred to a fresh 3.5 cm plate, reanchoring the spacers on the new plate, while the disk-shaped control gels were detached from all sides to allow free contraction. Gels were incubated at 37°C for an additional 24–48 h in growth media for compaction. During this time, both the aligned and isotropic constructs compacted, with the aligned gel constructs typically assuming a dogbone shape.
Decellularization of the reorganized matrices was performed by following a well-established two-step process with minor modifications (31). Briefly, the constructs were rinsed in 1× PBS and incubated for 12–24 h at 4°C in a cell lysis solution comprising of 1.0% (volume/volume) Triton X-100 (Roche, Basel, Switzerland), 5 mM EDTA (Thermo Fisher), and 10 mM Tris-HCl (pH 7.6) in ddH2O, followed by washing and subsequent treatment at 37°C with a solution of 300 μg/ml DNase 1(Roche) + 300 μg/ml RNase A (Thermo Fisher) in 1× PBS for 12–16 h to digest residual nucleic acids. Finally, the cell-free matrices were rinsed and stored in 1× PBS at 4°C.
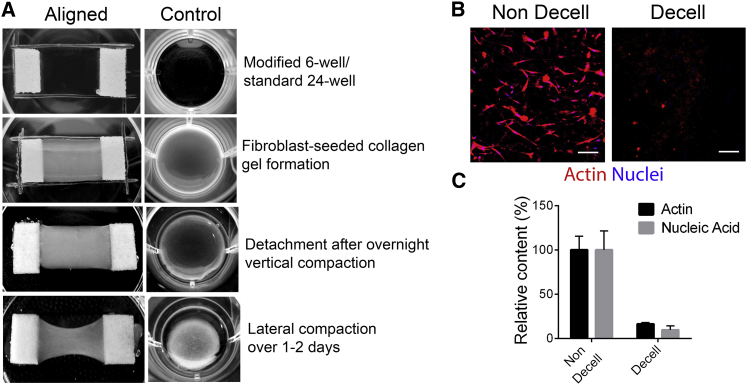
Decellularization efficiency was assessed by quantifying the amount of actin and nucleic acid retained within the matrices after decellularization. Aligned collagen constructs were fixed and stained for F-actin and nuclei (detailed method below) with or without undergoing decellularization. F-actin levels in decellularized and nondecellularized matrices were quantified from fluorescent micrographs by obtaining average intensities (32) after background correction. Nucleic acid was extracted from the collagen gels by proteinase K (Roche) digestion followed by phenol/chloroform extraction and ethanol precipitation. The amount of nucleic acid was quantified by measuring absorbance in a Nanodrop Spectrophotometer (Thermo Fisher). Data in both cases were normalized so that the average intensity for the nondecellularized group was at 100%.
Imaging and analysis of collagen fiber networks
Post-decellularization, collagen fibers were visualized using SHG imaging on a custom-built multiphoton laser scanning microscope (Prairie Technologies/Bruker, Middleton, WI) using a Mai Tai Ti:Sapphire laser (Spectra-Physics, Santa Clara, CA) at an excitation wavelength of 880 nm. Pore size measurements were performed using a methodology similar to that described by Friedl and coworkers (33). Here we improved on this approach by using higher-resolution micrographs and minimizing out-of-plane signal by employing optical sectioning inherent with two-photon imaging. We first obtained high-resolution Z-stacks (100–200 μm depth at 0.3 μm step size) at several locations in aligned and control matrices (≥2 locations/gel, ≥3 gels/group). XY pore sizes were obtained using the SHG images of the XY plane at different Z depths (∼3 per location) by measuring the dark areas (no signal) in between fibers within the field of view (32). XZ pore sizes were measured similarly from orthogonal projection of the Z-stacks (32).
Collagen fiber morphology was obtained using CT-FIRE (LOCI), which used curvelet transform and fiber extraction algorithms (34) to identify and analyze individual fibers. If σ denotes the SD of the distribution of fiber angles (ranging from −90 to 90) in a given field of view, then as a measure of fiber alignment, we defined the following:
| (1) |
where the numerator denotes the SD for a uniform distribution with a range from –90 to 90. Thus, this metric assumes a value of one for a perfectly uniform distribution of fiber angles and increases as the fibers become more and more aligned, i.e., as the value of σ reduces.
Live cell imaging, time-lapse microscopy, and analysis of cell migration
Multiphoton excitation (MPE) at 880 nm for simultaneous excitation of GFP and SHG imaging enabled visualization of GFP-expressing cells and collagen fibers, respectively, in aligned and control matrices. To study cell migration in 3D collagen matrices, GFP-expressing MDA-MB-231 or MDA-MB-231 CSCs were plated on immobilized control or aligned matrices at 100,000 cells/gel in growth media and incubated for 48 h to allow infiltration of cells into the matrix. Cell migration was captured by taking two-channel Z-stacks of 80–100 μm depth at 5 μm intervals at each stage position at 20 min intervals over 12–16 h for cells fully encased within the 3D volume. Temperature was maintained by a custom incubation chamber using a tunable plate heater (Bioscience Tools, San Diego, CA) and a miniature temperature probe (Bioscience Tools). During the time period of migration experiments, gels were overlaid with air-buffered L-15 medium supplemented with 10% FBS. Five-dimensional (5D) hyperstacks (x, y, z, t in two channels) were loaded into Fiji and drift-corrected using the 3D drift correction plugin (32). 3D tracking of cell migration was subsequently performed using TrackMate (32). The method of overlapping intervals (35) was used to fit the cell trajectories to a persistent random walk model (PRWM) (8, 36) using MATLAB (The MathWorks, Natick, MA) to interface with the cell tracking output. Briefly, the mean squared displacement (MSD) for a cell over time interval ti was obtained from the average of all squared displacements xik such that
| (2) |
| (3) |
where ni is the number of overlapping time intervals of duration ti, and N is the total number of time intervals for the experiment. Mathematically, the persistent random walk model can be written as follows:
| (4) |
where S is the migration speed and P is the persistence time. The motility coefficient is given as follows:
| (5) |
where nd is the dimensionality of the random walk. We fitted the model separately to the three orthogonal directions of motion, thus obtaining motility, speed, and persistence times for x, y, and z directions (therefore, nd = 1 in each case). Directionality of migration along any given axis was obtained by simply taking the ratio of the motility along that axis to the total motility of the cell, thus quantifying the fraction of total motility that is directed along that particular axis. For live cell tracking of cell nuclei, cells were incubated in 0.4 μg/mL Hoechst 33342 (Thermo Fisher) in growth media for 15 min and imaged by two-photon excitation at 750 nm.
To obtain the correlation between cell area and migration, cell movement on the XY plane was manually tracked (32) to find the total distance migrated with simultaneous measurement of cell shape at every other time point. Therefore, for this analysis, the cell shapes were measured at an interval of 40 min over 16 h (25 time points). Average circularity was calculated for each cell taking the mean of the cell shape circularities for all the time points in which it was measured. Similarly, the SD of circularity was calculated for each cell from the distribution of its cell circularities across the 25 time points. For cell volume measurement from Z-stacks, the 3D object counter (32) was used, including only cells encased entirely within the acquired image volume in the analysis.
To assess cellular response to alignment, Z-stacks of 20–50 μm depth were captured at different locations and fiber alignment indices were calculated for each from the maximum intensity projection images of collagen using the method described earlier. Identical to the fiber alignment index, if σcell denotes the SD of the distribution of cell angles (ranging from −90 to 90) at a given (x,y) location, then as a measure of cell alignment, we define as follows:
| (6) |
Cell projection areas and aspect ratios were calculated manually from maximum intensity projection images of cells at various (x,y) stage locations using Fiji.
Membrane protrusions were analyzed from time-lapse images manually by tracking well-defined extensions of the membrane that were distinctly different from the ellipsoid cell body. Most often these were thin, tapered protrusions extending along single fibers and occasionally broader and less-elongated ones. Protrusion frequency was defined as the number of new protrusions formed during the time of cell tracking divided by the total time.
F-actin and nuclear staining and morphometric analysis
For staining, collagen matrices with embedded cells were fixed in 4% paraformaldehyde (Sigma-Aldrich) for 20 min, quenched in 0.15 M glycine (Sigma-Aldrich) for 10 min, permeabilized with 0.1% Triton X-100 (Roche), and blocked with a solution of 2% fatty acid free-BSA (Fisher Scientific) and 2.5% goat serum (Sigma-Aldrich), followed by incubation with rhodamine phalloidin (1:500; Thermo Fisher) and 1:10,000 bisbenzamide (Sigma-Aldrich). Matrices were then washed and mounted with Prolong Gold (Thermo Fisher). Imaging was performed using either the multiphoton laser scanning microscopy setup described above in Imaging and Analysis of Collagen Fiber Networks (for simultaneous visualization of collagen by SHG along with F-actin and/or nuclei) or an inverted Olympus IX81ZDC spinning disk confocal microscope.
Nuclear shape analysis was performed in Fiji. Solidity, which measures the convexity of any shape, and is defined by the ratio of the shape area to the area of its convex hull, was used as a metric for nuclear deformation. Thus, whereas a perfectly ellipsoid convex (undeformed) nucleus has a solidity of one, with higher nuclear deformation the shape becomes more concave and its solidity decreases.
Results
Constrained cellular compaction generates aligned 3D collagen matrices
To generate aligned 3D collagen matrices, we adapted a protocol previously used for tissue-engineering aligned microvessels in fibrin gels (29, 30, 37). We modified this methodology employed by Tranquillo and coworkers (29, 30, 37) to account for the differences in mechanical and gelling properties between fibrin and collagen matrices as well as to meet our specific objective of generating acellular, aligned collagen matrices (see Materials and Methods for details). In an attempt to mimic the microenvironment of the breast tumor stroma, a primary carcinoma-associated fibroblast line obtained from an autochthonous murine mammary tumor (mFb) was used as the matrix reorganizing agent. Fibroblast-seeded collagen gels of equal volume, concentration, and cell density were allowed to form either in defined rectangular regions connected to porous polyethylene spacers at the two ends (aligned) or in a 24-well plate (control) (Fig. 1 A). Overnight incubation of the gels led to vertical compaction since the matrices were immobilized on all sides except the open top face (Fig. 1 A). Once the bottom of the aligned gels were detached from the plate, lateral compaction took place only along the short axis leading to a dog-bone shape that is desired for alignment of fibers along the primary axis connecting the two spacers. In contrast, detached control gels were able to contract from all sides, leading to a presumably isotropic, albeit reorganized collagen matrix (Fig. 1 A). Subsequent to the reorganization, we subjected the aligned and control tissues to a rigorous multistep decellularization protocol adapted from Hoshiba and others (31), comprising of cell lysis, nucleic acid digestion, and removal of debris. Successful decellularization was confirmed through fluorescent labeling of F-actin and the nuclei as well as nucleic acid extractions before and after decellularization. Intact cells or large cell fragments were not observed by fluorescent imaging of F-actin and nuclei. Further analysis confirmed a significant loss of DNA and actin where any retained material appeared as diffuse debris after decellularization (Fig. 1, B and C). Thus, with this protocol, we were able to generate acellular, 3D aligned collagen constructs and their corresponding control tissues in a repeatable manner, without external magnetic or mechanical perturbation using carcinoma-associated fibroblast-mediated collagen reorganization.
Figure 1.
Fabrication of acellular, aligned collagen constructs by cellular compaction and subsequent decellularization. (A) Step-by-step methodology is shown for generating aligned collagen matrices and corresponding control tissues: although the same starting volume of cells and matrix concoction are used for both, the aligned matrices (left panels) reorganize anisotropically whereas the control gels (right panels) are detached from all sides and allowed to compact freely. (B) Immunofluorescence images of nondecellularized and decellularized aligned gels showing efficient lysis and removal of cell debris from the matrix after alignment and (C) quantification of the amount of F-actin and DNA retained in the matrix post-decellularization are shown. Scale bar, 50 μm. Data in (C) are mean ± SE, n = 3 gels/group.
Structural characterization of aligned and control collagen matrices
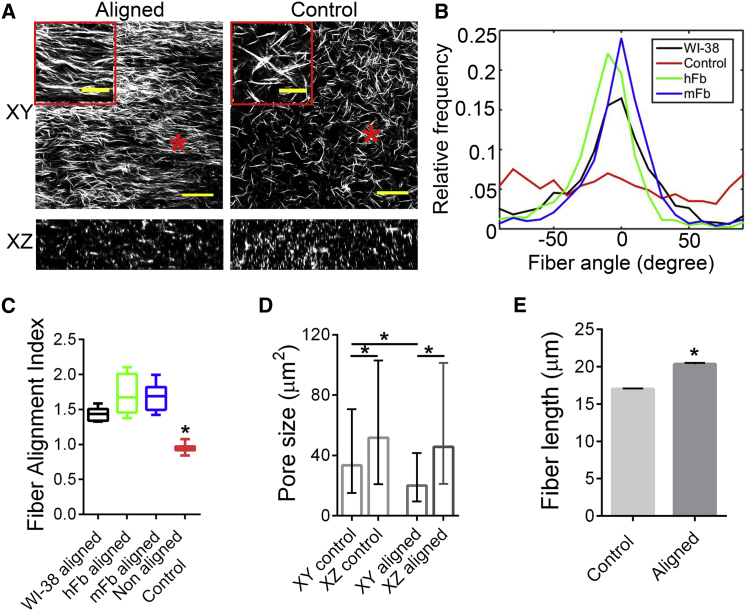
To characterize fiber structure in the collagen constructs, we employed high-resolution SHG imaging coupled to quantitative analysis of fiber structure and orientation, enabling direct visualization and analysis of the native collagen structure post-decellularization without extraneous probes or binding agents. SHG imaging revealed remarkable alignment of collagen fibers in the aligned constructs whereas the control matrices were randomly organized (Fig. 2 A). Indeed, quantitative analysis using a normalized SD metric, termed the Fiber Alignment Index, that is populated with fiber data extracted from curvelet transform analysis (see Materials and Methods) demonstrates robust and significant differences in the matrix architectures (Fig. 2, A–C). Further, we tested the robustness of our method by employing distinct primary and established fibroblast cell lines that include commercially available primary human breast fibroblasts (hFb) and the widely used WI-38 lung fibroblast cell line. We found that indeed, cell-induced constrained contraction of collagen matrices leads to robust alignment of fibers irrespective of the cell line, thus indicating that a contractile fibroblast cell type is sufficient to generate anisotropic matrices using our method (Fig. 2, B and C).
Figure 2.
Microstructural features of fibroblast-modified collagen matrices. (A) Second harmonic generation (SHG) imaging reveals collagen fiber morphologies in aligned and control matrices in the XY and XZ planes (scale bar, 50 μm); inset shows magnified images of regions marked with asterisks (scale bar, 20 μm). (B) Histograms of fiber orientations determined using curvelet transform and fiber extraction algorithm analysis are shown. (C) Corresponding quantification of fiber alignment for matrices aligned by distinct fibroblast lines are compared with an isotropic control (∗p < 0.0001, n ≥ 6 gels/group). (D) Quantification of pore sizes from SHG micrographs show larger XZ pore sizes than XY for both aligned and control matrices, whereas XY pore sizes in the aligned matrices were smaller than their control counterparts (∗p < 0.0001, n > 500 pores/group). (E) Fiber projection lengths on the XY plane for aligned and control tissues (∗p < 0.0001, n > 10 gels/group and >6000 individual fibers/group) are shown. Data are median with range (C), median with interquartile range (D), and mean ± SE (E).
We next characterized additional features of the 3D matrices that are known to influence cell migration. For instance, during 3D migration through fibrillar ECM, cells can navigate through the porous structure, thus making the cell:pore size ratio an important feature (33). Using 3D reconstruction of SHG data we quantified the areas of the pores as viewed from the XY plane (i.e., looking into the Z axis) and the XZ plane (i.e., looking into the Y axis) (Fig. 2, A and D). Interestingly, we found that even in the control gels, the XY pore sizes are significantly smaller than the XZ (Fig. 2, A and D). This is likely a consequence of the collagen gels being cast as thin disks (for the control) or rectangles (in case of the aligned) with a much smaller thickness (Z dimension) than length or width. Additionally, we found that the XY pore dimensions were significantly smaller in aligned constructs than in the control gels, demonstrating that the reorganization of fibers into aligned bundles causes redistribution of the pores within the fibrous matrix (Fig. 2 D).
In addition to pore size, fiber length can play a role in directed cell migration. Cells responding to contact guidance tend to track along individual or bundled parallel fibers, which greatly augment their directional motility. This effect is likely to be influenced by the planar reorganization of fibers in the aligned tissues. This can be examined by analyzing the fiber projection lengths in the XY plane, which were found to be significantly higher in the aligned than in the control tissues (Fig. 2 E). Overall, the characterization of the microstructure of these constructs provides key insights into the structural reorganization associated with matrix alignment and forebodes the disparities in migratory behavior of cells one might observe in such matrices.
Breast cancer cells are sensitive to the extent and direction of matrix alignment
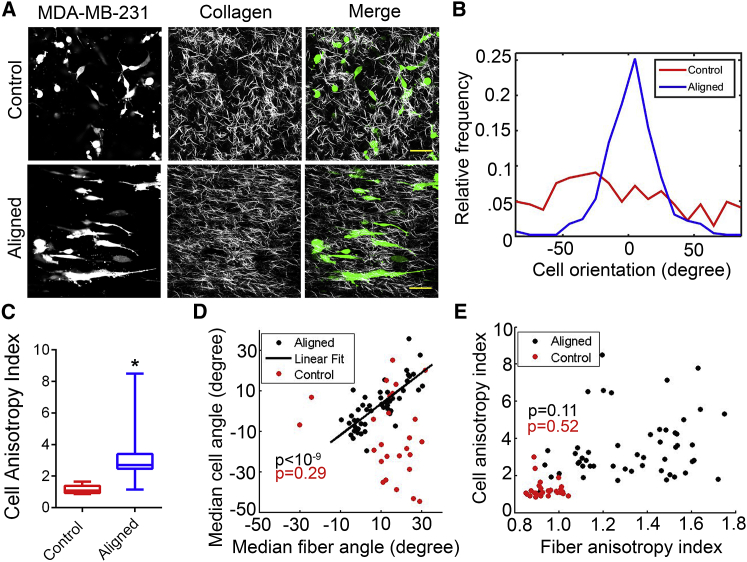
To ascertain the influence of 3D matrix alignment on carcinoma cells, we first characterized the shape and orientation of the highly metastatic MDA-MB-231 breast cancer cell line within our aligned and control matrices. We observed, perhaps not surprisingly, that these cells were elongated and oriented in the direction of fiber alignment in anisotropic matrices as opposed to random orientations in the isotropic controls (Fig. 3, A and B). In fact, quantitative analysis using a normalized SD metric termed the Cell Alignment Index (akin to Fiber Alignment Index; see Materials and Methods) clearly demonstrates isotropic orientation distributions in control matrices in stark contrast to the significantly distinct highly aligned cells in anisotropic matrices (Fig. 3, B and C). Further, aligned cells were more elongated (Fig. S1 A) and possessed greater spread area in the XY plane (Fig. S1 B), a consequence of the reorganization of fibers in the aligned matrices manifested by higher fiber projection lengths in XY and decreased XY pore sizes (Fig. 2, D and E). It is worth noting that in the isotropic control matrices infrequent patches of local alignment of fibers and cells were occasionally observed, but these were negligible compared with the large-scale global alignment in the anisotropic matrices, thus making the latter ideal for studying the responses of carcinoma cells to 3D fiber alignment. Generally, we found that fiber orientations in aligned constructs were quite homogeneous within a 300 × 300 μm field of view, even though there was some inherent variation in the direction and extent of fiber alignment at distant positions within the same matrix and between different matrices. We utilized this intrinsic variability to probe the sensitivity of breast carcinoma cells to the direction and extent of ECM fiber alignment.
Figure 3.
Breast carcinoma cells are acutely responsive to aligned collagen architecture in 3D. (A) Combined SHG and MPE imaging of GFP-expressing MDA-MB-231 cells in aligned collagen matrices show cellular morphology in response to anisotropic matrix organization (scale bar, 50 μm). (B) Histogram of cell orientations and (C) corresponding quantification of the degree of cell alignment (∗p < 0.0001, n ≥ 8/group) are shown. (D and E) Scatter plots display the correlation between the direction (D) and degree (E) of cell and fiber alignment. p-values for the null hypothesis that there is no correlation between the variables are noted in color-coded font (n > 25/group). Data are median ± range in (C).
Analysis of the fiber and cell alignment at numerous independent positions in multiple samples revealed a one-to-one correlation between the median cell and fiber orientations in aligned gels, whereas those in the control environments were not significantly correlated (Fig. 3 D). The extent of biased orientation, measured by the alignment indices, also showed similar, albeit less-pronounced trends (Fig. 3 E), thus demonstrating that breast cancer cells are exquisitely sensitive to the underlying matrix architecture. Certainly, we expect such “dose-response” relationships (Fig. 3, D and E) to contact guidance cues to be different across cell lines and hence it will be insightful to ascertain the sensitivity and tuning range of any particular cell type to this powerful mechanical cue.
3D matrix alignment reorients cell motility without altering its overall magnitude
To analyze 3D cell migration under various ECM architectures, cells were plated on acellular constructs and allowed to infiltrate into the gel for ∼48 h before tracking. Only cells completely embedded within the matrix were included in the analysis. Time-lapse SHG imaging of collagen fibers and multiphoton excitation of migrating GFP+ MDA-MB-231 cells in aligned and control matrices was performed over long periods (12–16 h) followed by 3D migration analysis. Fitting the PRWM (8, 35) to the trajectories demonstrated that the overall motility (sum of the motility coefficients in the three orthogonal directions: X, Y, and Z) was not significantly different between the aligned and control groups (Figs. 4, A and B, and S2 A; Movie S1). However, the presence of matrix alignment substantially biases motility in the direction of fiber anisotropy (Fig. 4, A and B; Movie S1). Indeed, quantification of migration directionality (i.e., the ratio of motility along a given axis and total motility) showed that ∼70% of the total motility of the cells was oriented along the direction of collagen alignment in the aligned constructs whereas the X or Y motility in control gels each only contributed ∼40% of the total motility (Fig. 4 D). Therefore, the motility in the direction of aligned ECM is almost twofold greater than the motility in any direction in the control matrices, exemplifying the directed migration of carcinoma cells in aligned collagen (Fig. 4 E). It is interesting to note that even in the control gels, the motility along the Z axis is only a fraction of that in X and Y and is further reduced in the aligned constructs (Fig. S2 B), in agreement with the XY pore size variations in these constructs (Fig. 2 D).
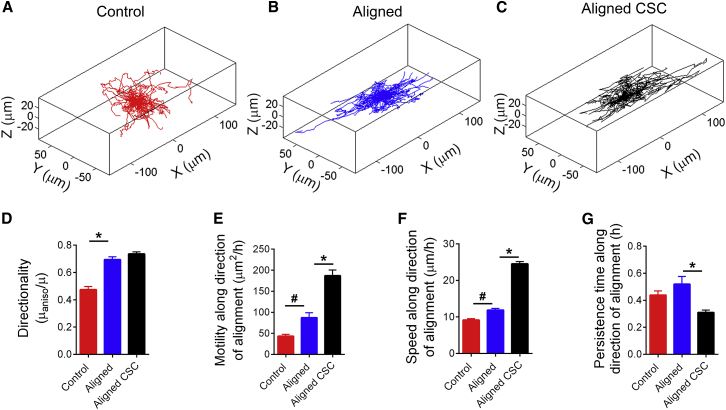
Figure 4.
CSCs exhibit enhanced directional motility in 3D aligned collagen. (A–C) 3D plots showing contrasting migration trajectories of breast carcinoma cells in control (A) and aligned ECM (B) and their CSC subpopulation in aligned ECM (C). (D) Directionality of migration increases in aligned matrices compared with the control and remains unchanged for the CSC subset versus the whole (more differentiated) population. (E) Total motility in the direction of alignment significantly increases between the control and aligned groups and is vastly enhanced in the CSCs, largely owing to concomitantly greater speeds of migration along the direction of alignment (F), whereas persistence times (G) remain the same between control and aligned or decrease between whole population and CSCs. Data are mean ± SE, ∗p < 0.0001, #p < 0.001, n = 130–140 cells/group for (D–G).
Consistent with our observations, Riching et al. (18) recently reported increased “persistence” of breast carcinoma cells in collagen matrices that were aligned by flow in a microchannel. However, Riching et al. (18) refers to “persistence” as the directional bias of cell movement that we have measured here as directionality rather than random walk persistence time. Here, by fitting the PRWM, we were able to investigate further which of the two components of random walk motility (that is, speed and/or persistence time) contributed to the increased motility along the fibers. Our analysis indicates that the increased motility was largely attributable to an increased speed of migration along the fiber tracks in the aligned matrices, whereas the persistence of migration was not significantly different (Fig. 4, F and G). Certainly, these findings provide interesting insights into the choreography of contact-guided cell migration, suggesting that whereas cells in aligned matrices still spend nearly as much time probing in other directions as they do in more random environments, after sampling their environment they subsequently move much faster on the guided path provided by the fiber alignment. Moreover, cells in aligned matrices are prone to moving back and forth along a fiber or fiber bundle. Such a trajectory would, in our analysis, have a high directionality (almost all of the motility is along one axis) but low persistence time (since the cell changes direction often). Indeed, it appears that in aligned matrices cells retain the same underlying potency to migrate as in the control; the anisotropic ECM simply redistributes the motility predominantly along a single axis. That the total motility of the cells remains unchanged by matrix alignment perhaps points toward a purely biophysical regulation of contact-guided migration.
Breast cancer stem cells exhibit enhanced directional motility in aligned collagen
Breast carcinoma cells grown on low attachment plates under specific media conditions readily formed large, multicellular spheroids, consistent with numerous previous reports (38, 39, 40). Inducing a population of cells to grow such mammospheres selectively enriches the stem or progenitor-like subpopulation, as demonstrated previously in mammary epithelial (41) as well as carcinoma cell lines (42). Indeed, several previous studies have used mammosphere formation from MDA-MB-231 cells as indicators of the stem-like phenotype (43, 44) since cancer cells capable of forming such mammospheres are well-known to represent a “cancer stem cell” (CSC) subpopulation (22, 42) that displays increased tumor-forming and metastatic potential (45, 46). As such, we tested the hypothesis that these CSCs exhibit enhanced contact-guided migration compared with the whole population (WP) largely comprised of carcinoma cells in a more differentiated state. Indeed, in aligned matrices, the CSCs displayed ∼2.5× higher total motility than the WP of cells (Figs. 4, B and C, and S2 A; Movie S1), suggesting that their inherent migratory mechanics may be distinct from more differentiated cells. In fact, whereas the directional bias in cell motility for the CSC population is very similar to that of the WP (Fig. 4 D), they exhibit greater than 2× enhanced motility in the direction of collagen alignment (Fig. 4 E). Further, time-lapse imaging clearly revealed that the CSCs are highly dynamic as they navigate through the fibrous matrix with frequent shape changes and very robust protrusive activity compared with more differentiated cells in either random or aligned ECMs (Movie S1). As analysis of migration by the PRWM confirmed, the higher motility of CSCs was the result of their much greater speed of migration even as the persistence time was lower than the WP (Fig. 4, F and G), suggesting an increased sampling of the microenvironment during directed migration but consistent commitment to directional movement with much greater cell speed between sampling intervals. Thus, our quantitative analysis of migration reveals that CSCs respond robustly to contact guidance with a response to architectural and mechanotransduction cues that is distinct from the whole population. As such, the enhanced directional motility (as well as total motility) is likely indicative of their increased metastatic potential and led us to critically examine the physical features that may be responsible for such behavior.
Smaller cell size may enable the enhanced migration of CSCs in aligned matrices
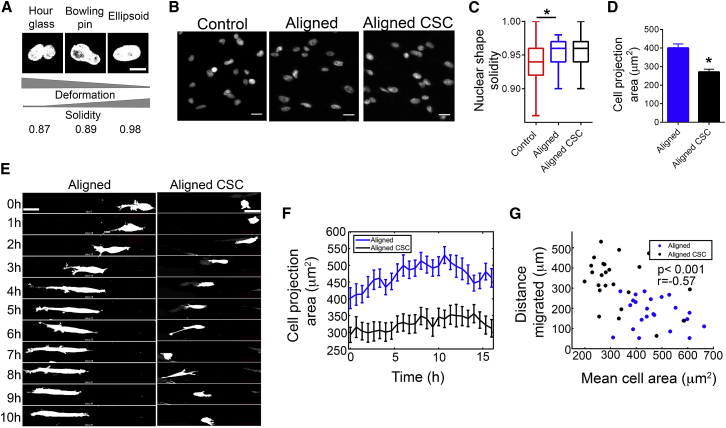
One of the established constraints for cell migration through fibrous ECM is the ability of cells to deform nuclei so as to be able to navigate the pores or tortuous networks in the matrix efficiently (33). In our matrices, time-lapse tracking of nuclei demonstrated the dynamic nature of nuclear shapes as MDA-MB-231 cells and their corresponding CSCs migrated through aligned collagen matrices (Movies S2 and S3). Nuclei switched between ellipsoid, “bowling pin” and “hourglass” shapes, which can be mathematically distinguished by measuring solidity of the shape (Fig. 5 A). Fluorescent staining of cells also captures the different shapes and extent of nuclear deformation in control and aligned gels for CSCs and the WP (Fig. 5 B). Our analysis of nuclear shapes revealed that there was no significant difference between the extent of nuclear deformation between the whole (i.e., more differentiated) population and CSCs migrating in aligned gels (Fig. 5 C), suggesting that ability to deform nuclei is not the differentiating factor driving the migratory tendencies of the two groups. Interestingly, the same cells in control matrices present with significantly more deformed nuclei (Fig. 5 C), likely because of the irregular structure and tortuosity of pores in such ECMs.
Figure 5.
Nuclear deformation and cell size dynamics of breast cancer cells and their CSC subpopulation in anisotropic collagen matrices. (A) Typical nuclear morphologies observed in 3D collagen matrices and their corresponding solidity values demonstrate the inverse relationship between deformation and solidity. (B) Nuclear morphology of the corresponding cells in fixed samples show the spectrum of nuclear shapes and (C) the corresponding quantification using nuclear shape solidity (data are median ± range, ∗p < 0.01, n ∼ 100 nuclei/group). (D) Cell areas of the whole population and the CSC subpopulation are measured from fixed samples (∗p < 0.0001, n > 70 cells/group). Data are mean ± SE. (E) Time-lapse montages show typical migratory behavior of breast carcinoma cells and their CSC subpopulation with the latter demonstrating smaller cell size and frequent formation of new protrusions leading to higher, yet still directional motility. (F) Cell areas are analyzed from migration data over time (n = 24 cells/group, data are mean ± SE). (G) Scatter plot shows the negative correlation between cell area and migration distance (p-value for the null hypothesis that no correlation exists between the variables and the value of the linear correlation coefficient r between the variables are noted, n = 24/group). Scale bars: 5 μm in (A), 20 μm in (B), and 25 μm in (E).
While investigating nuclei and cell shape during 3D migration, we found, quite intriguingly, that CSCs were smaller in size than the more differentiated population of cells in 3D. Existing evidence in literature does suggest that stem cells and CSC populations may be of smaller size than the more differentiated population (47, 48), potentially influencing their growth, differentiation, and ability to withstand harsh environments. In the context of 3D motility, the smaller cell size could provide a steric advantage to the CSCs when migrating through fibrous matrices. Inspection of time-lapse images and quantification of cell projection areas in the XY plane demonstrates the size discrepancy, with the average area of the non-CSC population being ∼50% higher than that of the CSCs (Fig. 5, D and E). Further, we analyzed cell volumes from 3D stacks in aligned gels to confirm that cell size, in the strictest sense of the term, was indeed significantly lower for the CSC population (Fig. S3 A).
Next, we used combined cell tracking with cell area measurement to investigate the relationship between cell size and motility of cells. Here, these measurements were carried out at every other time point in a 16 h migration data set. Because our findings have already shown that the majority of the motility in aligned gels (∼90%) is confined to the XY plane (Fig. S2 B), we used the total distance migrated in that plane as a simpler, surrogate measure of total cell motility. Along the same lines, we were able to use the projection area on the XY plane as a measure of cell volume, because both show similar variation between the two groups in question (Figs. 5 D and S3 A). In agreement with the analysis from single time point images, we find that over time CSCs, on an average, are much smaller in size than the WP (Fig. 5, E and F). Additionally, there exists a distinct negative correlation between cell size and motility in aligned gels for both groups (Fig. 5 G), pointing to the physical advantage conferred by the smaller size when migrating in 3D matrices. Indeed, it appears likely that the smaller size of CSCs, much like amoeboid cells, enables them to migrate faster along aligned ECM and contribute to their increased metastatic potential.
Phenotypic plasticity and increased protrusive activity drives increased directional migration in CSCs
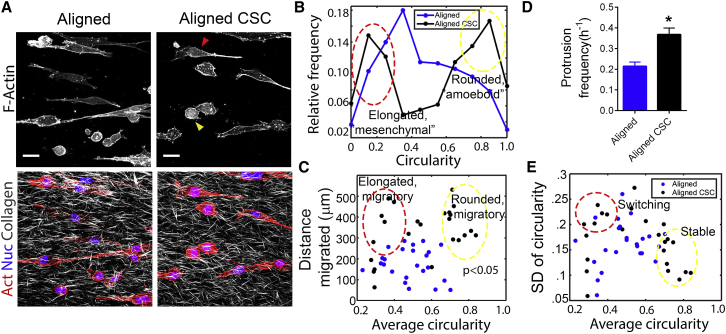
Inspection of time-lapse cell migration data shows that, both in the differentiated and CSC populations, cells display phenotypic plasticity and switch between an elongated, mesenchymal-like state and a rounded, amoeboid-like state (Movie S1), which is also confirmed by immunofluorescence staining (Fig. 6 A). To quantify such cell shape differences, we use circularity, which assumes a value of one for a perfectly circular shape and tends to zero as the shape becomes more and more elongated. We found a strong correlation between cell size and circularity (Fig. S3 B), which along with the data on migration and cell size led us to investigate the role of cell phenotypes (e.g., amoeboid/mesenchymal) in the altered migration of CSCs.
Figure 6.
Phenotypic plasticity and increased protrusion dynamics drive rapid directional CSC migration through aligned 3D matrices. (A) Immunofluorescence micrographs obtained from combined SHG and MPE imaging demonstrate collagen architecture along with F-actin morphology of breast carcinoma cells and their CSC-enriched subpopulation in aligned matrices. Yellow and red arrowheads identify typical cells of the amoeboid and mesenchymal phenotypes, respectively. (B) Histogram of cell circularity shows a bimodal distribution in the CSC group (aligned CSC), indicating the presence of distinct phenotypes, in contrast to the unimodal distribution with a peak at intermediate circularity for the whole (more differentiated) population (aligned). Histogram peaks corresponding to the two phenotypes are highlighted in red and yellow, corresponding to the cell morphologies shown in (A) with identically colored arrowheads (n = 600/group). (C) Scatter plot of average cell circularity versus distance migrated obtained from live cell imaging data demonstrates a general positive correlation (p-value for the null hypothesis that there is no correlation noted). Yellow and red highlighted regions denote the clusters of highly migratory amoeboid and mesenchymal cells respectively (n = 24/group). (D) Average protrusion frequency of the whole population of breast carcinoma cells and the CSC population is shown. Data are mean ± SE. (E) Scatter plot of average cell circularity versus SD of circularity from live cell imaging data shows phenotypic stability of migratory amoeboid-like cells (yellow highlighted region) and plasticity of a fraction of migratory elongated mesenchymal-like cells (red highlighted region) (n = 24/group). The red and yellow circles in parts (C) and (E) enclose approximately the same data points. Scale bar, 50 μm.
The CSC subpopulation exhibited a bimodal distribution with respect to circularity of cell shape, indicating that they exist predominantly in a highly elongated (circularity <0.4) mesenchymal-like or a very rounded (circularity >0.7) amoeboid-like phenotype (Fig. 6 B). In contrast, the majority of cells in the differentiated population of MDA-MB-231s exhibited an intermediate elongated phenotype (circularity 0.3–0.6) (Fig. 6 B). Overall, average circularity of the cell shape correlates loosely with the distance migrated (Fig. 6 C), indicating that the amoeboid-like cells are more motile on average than the elongated ones, consistent with the consensus in the field regarding amoeboid and mesenchymal migration in 3D. Thus, there exists a cluster of highly migratory cells with high average circularity, predominantly belonging to the CSC subpopulation representing these amoeboid-like cells (Fig. 6 C). Intriguingly, however, a substantial fraction of the elongated cells in the CSC subpopulation also exhibit enhanced motility, as compared with similarly elongated cells of the differentiated group (Fig. 6 C).
Our migration data showed that the CSCs that were mesenchymal-like (low circularity) on average, possess rapid protrusive activity, and frequently retracted to a more amoeboid-like state (with high circularity), as shown in the representative time-lapse montage (Fig. 5 E). In contrast, the more differentiated population of cells exhibited stable protrusions, often at the two ends of an elongated cell that can lock the cell into a nonmotile state for many hours (Fig. 5 E). This suggests that the stiffness of the local ligand area, the magnitude and/or frequency of the cell contractile forces, and the stability of the mechanical coupling regulate a bidirectional mechanical feedback control between the cell and matrix that influences phenotype and motility dynamics. Indeed, we find that the CSCs were more dynamic and their average protrusion rate, measured by the number of new protrusions formed per time, was significantly higher (Fig. 6 D). To capture the dynamic changes in cell phenotype from long-term time-lapse imaging data, the SD of circularity was calculated in addition to the average circularity of cell shape. Whereas the average informs predominantly which phenotype a cell assumed during the time course of an experiment, the SD provides an estimate of the rate and extent of its dynamic switching between phenotypes (Fig. 6 E). Interestingly, we found that the amoeboid-like highly migratory CSCs (high circularity) maintain this phenotype stably, as indicated by the low SD of circularity (<0.2) (Fig. 6 E). However, some of the highly motile, elongated, mesenchymal-like CSCs exhibit higher SD of circularity indicating that they tend to switch between phenotypes rapidly and transiently, whereas others were more stable (Fig. 6 E). The more differentiated population also showed an intermediate level of phenotypic switching (Fig. 6 E). This is certainly consistent with our migration data (Fig. 5 E; Movie S1) as well as the distribution of cell phenotypes in the two groups (Fig. 6 B).
Overall, our data suggest that smaller cell size, along with rapid phenotypic switching and increased protrusive activity enable CSCs to migrate swiftly through aligned 3D collagen matrices. Amoeboid-like CSCs were found to maintain this phenotype stably, enabling their highly migratory state, whereas more mesenchymal-like CSCs showed dynamic protrusive activity and in some cases, plasticity in switching between phenotypes that facilitated faster migration. In contrast, the more differentiated cells showed a more stable, intermediate phenotypic state with lower protrusive activity.
Discussion
Contact-guided cell migration of fibroblasts in 3D was reported by Dickinson et al. (11) more than two decades ago. Further, several applications, particularly in neural (15, 49, 50) and cardiovascular tissue engineering (51, 52, 53, 54) have made use of the phenomenon of contact guidance extensively in 3D in attempts to recapitulate key in vivo architectures. However, cell migration in response to such topographic cues has been largely studied in micropatterned systems using model fibroblast lines (55, 56). Over the past decade, the growing consensus on the role of contact guidance for cancer cell invasion (12, 13, 14, 57, 58) now necessitates detailed, systematic investigation of carcinoma cell responses to collagen alignment in 3D. Similar to matrix stiffness that has a profound impact on cell behavior (59, 60), accumulating evidence strongly suggests that matrix architecture is perhaps no less an important physical cue that may in fact direct cell migration by producing directional stiffness. Indeed, in both anisotropic collagenous matrices in vivo and in vitro, the modulus is highest in the direction of fiber alignment (18, 61, 62, 63, 64). This directional stiffness is likely to be perceived by cells and elicit anisotropic adhesion and traction force dynamics that regulate a mechanotransduction program that orients cells and maintains directional persistence. Thus, a robust exploration of this underlying physical mechanism using iterative experimental and modeling approaches is likely to provide insight into the mechanics of cell-matrix interactions and contact guidance. Yet, to accomplish this, platforms and metrics must exist to dissect fundamental cell behavior in engineered environments. Although different techniques exist to vary matrix stiffness of collagen matrices, generating aligned physiological ECMs that have stiffness anisotropy is equally, if not more, challenging. Herein, we introduce an inexpensive and accessible method to generate highly aligned collagen constructs, with a protocol that is easy to implement without the need for specialized equipment or microfabrication techniques while being amenable to high-resolution optical microscopy.
Similar to studies addressing sensitivity to directional cues (e.g., stiffness gradients, chemical gradients etc.), we asked how sensitive carcinoma cells are to alignment cues and find that the direction of cell alignment scales quite linearly with the direction of ECM alignment (e.g., Fig. 3 D), whereas the extent of cell alignment is associated weakly by the extent of ECM alignment (e.g., Fig. 3 E). Indeed, one of the telling features of control versus aligned 3D cell migration is the finding that total motility remains the same in both conditions. Yet, directional motility is profoundly altered with variability in the data well explained by heterogeneity in matrix architecture. Certainly, from the perspective of local-excitation-global-stimulation models of cell migration (65, 66) collagen fibers arranged in an isotropic fashion around a cell may create multiple, randomly distributed local excitation points to induce protrusion formation and engage associated migration machinery. In contrast, in the aligned matrices, these are still present in equal amounts but redistributed so that the local excitation signals are largely directed along a single axis. In fact, MDA-MB-231 cells, which display classic mesenchymal-like migration under many conditions, were found to occasionally elongate robustly and enter a nonmotile state, similar to that observed in fibroblasts when the latter switch between promigratory and procontractile phenotypes (67). Interestingly, CSCs rarely displayed such stalling, perhaps attributable to a more frequent sampling of their microenvironments and a robust program of versatile and frequent phenotypic switching during 3D cell migration. Moreover, our finding that CSCs have much higher motility in aligned matrices than the more differentiated population is consistent with the notion that CSCs have higher metastatic potential. It also suggests that CSCs may be the cells that lead the invasion front, where local TACS-3 involves streaming of individual or collections of cells along aligned collagen fibers (12, 13).
Our approach also allows us to examine the impact of navigation through finite pores, a defining feature of 3D migration, whose importance is well established in the field. It is interesting to note that although the relative differences in XZ and XY pore sizes were similar, the actual pore sizes we measured were larger than those reported by Wolf et al. (33). This disparity may be explained, in part, by the fact that SHG imaging using two-photon processes from a pulsed laser provides inherent optical sectioning, thereby eliminating out-of-plane signal, which could otherwise contribute to the underestimation of pore sizes. In addition, cell-reorganized ECMs such as the ones in here, similar to those in vivo, may possess different pore size distributions than corresponding cell-free matrices. Indeed, these matrices were found to possess a wide range of pore sizes. Although some of this is because of the inherent variation during the formation of the collagen gel, the reorganization of the matrix by cells also adds to the heterogeneity by widening some pores and constricting others. Following our nomenclature, fiber alignment was along the X axis, hence it was quite obvious that the distribution of fibers and hence pore structures would be very different when viewed on the YZ plane (i.e., looking into the X axis) than on other two planes in aligned matrices. But, perhaps not obvious is the indirect effect of alignment that causes redistribution of the fibers to extend mostly on the XY plane, thereby creating smaller XY pore sizes than in the control. As our analysis confirms, this leads to migration predominantly along one axis, recapitulating aspects of confined 2D migration (68), or of the one-dimensional (1D) migration phenotype (69), even in the absence of lateral confinement of cells.
The most striking, yet simple features that correlated with increased cell movement are cell size and the ability to rapidly switch phenotype. Like amoeboid cells, CSCs may utilize the steric advantage of smaller size to migrate through the pores within the matrix, or through endothelium in the case of intravasation, more efficiently. It is well known that migration in confined spaces can induce mesenchymal-to-amoeboid (MAT) transition (6, 70). Such shifts in phenotype for migrating cells are usually characterized by low adhesion and increased ROCK-dependent myosin II activation (6, 71). Interestingly, CSCs often switch back to the classical mesenchymal phenotype, which is reciprocally thought to be mediated by suppression of Rho and activation of Rac for some cell types (71). In fact, increased protrusive activity was one of the hallmarks of CSC migration compared with the more differentiated population in aligned matrices. Thus, CSCs appear likely to possess altered GTPase and focal adhesion signaling dynamics that make them amenable to rapid switching between phenotype/migration modes to move efficiently through fibrous matrices. Interestingly, their smaller cell size, which may alternatively be regulated by S6K in an mTOR-dependent manner (72), could also contribute to this effect by altering reaction-diffusion kinetics across the cytoplasm that influence adhesion and protrusion dynamics (73). Certainly, studies investigating the contextual and relative importance of the underlying physical mechanisms and mechanotransduction governing CSC migration through mechanically complex microenvironments are likely to greatly add to our understanding of cancer metastasis.
Overall the data presented in this study raise a number of important questions regarding the sequence of mechanical events and the mechanobiology of CSCs. For instance, are CSCs more mechanically active than their more differentiated counterparts with a robust ability to reorganize the matrix to facilitate invasion? Alternatively, do they act coordinately with other carcinoma cells and carcinoma-associated fibroblasts that instead organize matrices to provide mechanical cues that promote CSC invasion? Indeed, cell contractile forces that result in 3D matrix reorganization to generate contact guidance cues are known to promote invasion, yet these forces are more dispensable when the matrix is already aligned (13). Yet physically speaking, because cell migration in 3D is limited in part by the ability to navigate through the matrix, increased ability to rapidly generate matrix deformation by traction forces, or matrix degradation in some cases, allows cells to reorganize the fibers locally, and thus the pores, to enable more efficient navigation. However, a more dynamic state may allow cells such as CSCs to migrate more robustly in response to a physical cue already in place, i.e., the prealigned matrix. Interestingly, observations here demonstrate the capacity for robust phenotypic heterogeneity and plasticity, suggesting that either possibility is reasonable. In fact CSCs may have the ability to both act as a robust ECM-reorganizing mesenchymal-like cell as well as a lower traction amoeboid-like cell that rapidly responds to physical signals from the environment. Thus, it is clear that a deeper examination of the unique mechanobiology and cell and matrix mechanics, in both time and space, associated with CSC motility is needed concomitant with the growing consensus evidence of the importance of CSCs in disease progression.
Author Contributions
A.R. and P.P.P. designed the study. A.R., Z.M.S., R.K.M., and S.A.M. performed experiments to develop and analyze aligned 3D matrices. A.R. and R.K.M. performed cell migration experiments. A.R. performed quantitative analysis of data. A.R. and P.P.P. wrote the manuscript with input from all authors. P.P.P. oversaw the study.
Acknowledgments
We thank Jonathan H. Schrope for assistance with DNA extractions. We thank members of the Provenzano, Tranquillo, and Alford labs for technical assistance and insightful comments regarding this work.
P.P.P. was supported by a Research Scholar Grant, RSG-14-171-01-CSM from the American Cancer Society. This work was also supported by the NIH (R01CA181385, U54CA210190 University of Minnesota Physical Sciences in Oncology Center to P.P.P.), the UMN College of Science and Engineering, the Masonic Cancer Center, the UMN Institute for Engineering in Medicine, and the Randy Shaver Research and Community Fund (P.P.P.). A.R. is supported by a UMN Doctoral Dissertation Fellowship. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or other funding agencies.
Editor: Alissa Weaver
Footnotes
Three figures and three movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30046-2.
Supporting Material
Cells are shown in green and collagen in gray; the top panels represent XY projection images of the Z stack at each time point, whereas the bottom panels represents the XZ projection. Scale bar, 50 μm.
Scale bar, 50 μm.
Scale bar, 50 μm.
References
- 1.Daley W.P., Yamada K.M. ECM-modulated cellular dynamics as a driving force for tissue morphogenesis. Curr. Opin. Genet. Dev. 2013;23:408–414. doi: 10.1016/j.gde.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose D.M., Alon R., Ginsberg M.H. Integrin modulation and signaling in leukocyte adhesion and migration. Immunol. Rev. 2007;218:126–134. doi: 10.1111/j.1600-065X.2007.00536.x. [DOI] [PubMed] [Google Scholar]
- 3.Keller R. Cell migration during gastrulation. Curr. Opin. Cell Biol. 2005;17:533–541. doi: 10.1016/j.ceb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi H., Wyckoff J., Condeelis J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Provenzano P.P., Eliceiri K.W., Keely P.J. Shining new light on 3D cell motility and the metastatic process. Trends Cell Biol. 2009;19:638–648. doi: 10.1016/j.tcb.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedl P., Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Petrie R.J., Doyle A.D., Yamada K.M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harms B.D., Bassi G.M., Lauffenburger D.A. Directional persistence of EGF-induced cell migration is associated with stabilization of lamellipodial protrusions. Biophys. J. 2005;88:1479–1488. doi: 10.1529/biophysj.104.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy J.B., Palm S.L., Furcht L.T. Migration by haptotaxis of a Schwann cell tumor line to the basement membrane glycoprotein laminin. J. Cell Biol. 1983;97:772–777. doi: 10.1083/jcb.97.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo C.M., Wang H.B., Wang Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson R.B., Guido S., Tranquillo R.T. Biased cell migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann. Biomed. Eng. 1994;22:342–356. doi: 10.1007/BF02368241. [DOI] [PubMed] [Google Scholar]
- 12.Provenzano P.P., Eliceiri K.W., Keely P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provenzano P.P., Inman D.R., Keely P.J. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conklin M.W., Eickhoff J.C., Keely P.J. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceballos D., Navarro X., Tranquillo R.T. Magnetically aligned collagen gel filling a collagen nerve guide improves peripheral nerve regeneration. Exp. Neurol. 1999;158:290–300. doi: 10.1006/exnr.1999.7111. [DOI] [PubMed] [Google Scholar]
- 16.Guo C., Kaufman L.J. Flow and magnetic field induced collagen alignment. Biomaterials. 2007;28:1105–1114. doi: 10.1016/j.biomaterials.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Lee P., Lin R., Lee L.P. Microfluidic alignment of collagen fibers for in vitro cell culture. Biomed. Microdevices. 2006;8:35–41. doi: 10.1007/s10544-006-6380-z. [DOI] [PubMed] [Google Scholar]
- 18.Riching K.M., Cox B.L., Keely P.J. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys. J. 2014;107:2546–2558. doi: 10.1016/j.bpj.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornwell K.G., Downing B.R., Pins G.D. Characterizing fibroblast migration on discrete collagen threads for applications in tissue regeneration. J. Biomed. Mater. Res. A. 2004;71:55–62. doi: 10.1002/jbm.a.30132. [DOI] [PubMed] [Google Scholar]
- 20.Dumont N., Liu B., Tlsty T.D. Breast fibroblasts modulate early dissemination, tumorigenesis, and metastasis through alteration of extracellular matrix characteristics. Neoplasia. 2013;15:249–262. doi: 10.1593/neo.121950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charafe-Jauffret E., Ginestier C., Wicha M.S. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen L.V., Vanner R., Eaves C.J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 23.Al-Hajj M., Wicha M.S., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaffer C.L., Brueckmann I., Weinberg R.A. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. USA. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabrera M.C., Hollingsworth R.E., Hurt E.M. Cancer stem cell plasticity and tumor hierarchy. World J. Stem Cells. 2015;7:27–36. doi: 10.4252/wjsc.v7.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginestier C., Hur M.H., Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kshitiz, Park J., Kim D.-H. Control of stem cell fate and function by engineering physical microenvironments. Integr. Biol. 2012;4:1008–1018. doi: 10.1039/c2ib20080e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Provenzano P.P., Inman D.R., Keely P.J. Mammary epithelial-specific disruption of focal adhesion kinase retards tumor formation and metastasis in a transgenic mouse model of human breast cancer. Am. J. Pathol. 2008;173:1551–1565. doi: 10.2353/ajpath.2008.080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin K.T., Tranquillo R.T. Guided sprouting from endothelial spheroids in fibrin gels aligned by magnetic fields and cell-induced gel compaction. Biomaterials. 2011;32:6111–6118. doi: 10.1016/j.biomaterials.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Riemenschneider S.B., Mattia D.J., Tranquillo R.T. Inosculation and perfusion of pre-vascularized tissue patches containing aligned human microvessels after myocardial infarction. Biomaterials. 2016;97:51–61. doi: 10.1016/j.biomaterials.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshiba T., Yamada T., Chen G. Maintenance of cartilaginous gene expression on extracellular matrix derived from serially passaged chondrocytes during in vitro chondrocyte expansion. J. Biomed. Mater. Res. A. 2012;100:694–702. doi: 10.1002/jbm.a.34003. [DOI] [PubMed] [Google Scholar]
- 32.Schindelin J., Arganda-Carreras I., Frise E. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf K., Te Lindert M., Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bredfeldt J.S., Liu Y., Eliceiri K.W. Computational segmentation of collagen fibers from second-harmonic generation images of breast cancer. J. Biomed. Opt. 2014;19:16007. doi: 10.1117/1.JBO.19.1.016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickinson R.B., Tranquillo R.T. Optimal estimation of cell movement indices from the statistical analysis of cell tracking data. AIChE J. 1993;39:1995–2010. [Google Scholar]
- 36.Othmer H.G., Dunbar S.R., Alt W. Models of dispersal in biological systems. J. Math. Biol. 1988;26:263–298. doi: 10.1007/BF00277392. [DOI] [PubMed] [Google Scholar]
- 37.Morin K.T., Smith A.O., Tranquillo R.T. Aligned human microvessels formed in 3D fibrin gel by constraint of gel contraction. Microvasc. Res. 2013;90:12–22. doi: 10.1016/j.mvr.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S.Y., Kang J.W., Lee Y.J. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell. Signal. 2013;25:961–969. doi: 10.1016/j.cellsig.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Neill C.F., Urs S., Liaw L. Notch2 signaling induces apoptosis and inhibits human MDA-MB-231 xenograft growth. Am. J. Pathol. 2007;171:1023–1036. doi: 10.2353/ajpath.2007.061029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croker A.K., Goodale D., Allan A.L. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J. Cell. Mol. Med. 2009;13(8B):2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dontu G., Abdallah W.M., Wicha M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ponti D., Costa A., Daidone M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 43.Vazquez-Martin A., Oliveras-Ferraros C., Menendez J.A. Metformin regulates breast cancer stem cell ontogeny by transcriptional regulation of the epithelial-mesenchymal transition (EMT) status. Cell Cycle. 2010;9:3831–3838. [PubMed] [Google Scholar]
- 44.To K., Fotovati A., Dunn S.E. Y-box binding protein-1 induces the expression of CD44 and CD49f leading to enhanced self-renewal, mammosphere growth, and drug resistance. Cancer Res. 2010;70:2840–2851. doi: 10.1158/0008-5472.CAN-09-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korkaya H., Paulson A., Wicha M.S. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen D., Bhat-Nakshatri P., Nakshatri H. ANTXR1, a stem cell-enriched functional biomarker, connects collagen signaling to cancer stem-like cells and metastasis in breast cancer. Cancer Res. 2013;73:5821–5833. doi: 10.1158/0008-5472.CAN-13-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Paiva C.S., Pflugfelder S.C., Li D.Q. Cell size correlates with phenotype and proliferative capacity in human corneal epithelial cells. Stem Cells. 2006;24:368–375. doi: 10.1634/stemcells.2005-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu A., Oh S., Ohlfest J.R. Persistence of CD133+ cells in human and mouse glioma cell lines: detailed characterization of GL261 glioma cells with cancer stem cell-like properties. Stem Cells Dev. 2008;17:173–184. doi: 10.1089/scd.2007.0133. [DOI] [PubMed] [Google Scholar]
- 49.Dubey N., Letourneau P.C., Tranquillo R.T. Neuronal contact guidance in magnetically aligned fibrin gels: effect of variation in gel mechano-structural properties. Biomaterials. 2001;22:1065–1075. doi: 10.1016/s0142-9612(00)00341-0. [DOI] [PubMed] [Google Scholar]
- 50.Hoffman-Kim D., Mitchel J.A., Bellamkonda R.V. Topography, cell response, and nerve regeneration. Annu. Rev. Biomed. Eng. 2010;12:203–231. doi: 10.1146/annurev-bioeng-070909-105351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Black L.D., 3rd, Meyers J.D., Tranquillo R.T. Cell-induced alignment augments twitch force in fibrin gel-based engineered myocardium via gap junction modification. Tissue Eng. Part A. 2009;15:3099–3108. doi: 10.1089/ten.tea.2008.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson P.S., Johnson S.L., Tranquillo R.T. Functional tissue-engineered valves from cell-remodeled fibrin with commissural alignment of cell-produced collagen. Tissue Eng. Part A. 2008;14:83–95. doi: 10.1089/ten.a.2007.0148. [DOI] [PubMed] [Google Scholar]
- 53.Oliveira A.L., Sun L., Kaplan D.L. Aligned silk-based 3-D architectures for contact guidance in tissue engineering. Acta Biomater. 2012;8:1530–1542. doi: 10.1016/j.actbio.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kai D., Prabhakaran M.P., Ramakrishna S. Guided orientation of cardiomyocytes on electrospun aligned nanofibers for cardiac tissue engineering. J. Biomed. Mater. Res. B Appl. Biomater. 2011;98:379–386. doi: 10.1002/jbm.b.31862. [DOI] [PubMed] [Google Scholar]
- 55.Kim D.H., Seo C.H., Suh K.Y. Guided cell migration on microtextured substrates with variable local density and anisotropy. Adv. Funct. Mater. 2009;19:1579–1586. doi: 10.1002/adfm.200990041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D.H., Han K., Levchenko A. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials. 2009;30:5433–5444. doi: 10.1016/j.biomaterials.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedl P., Locker J., Segall J.E. Classifying collective cancer cell invasion. Nat. Cell Biol. 2012;14:777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 58.Sharma V.P., Beaty B.T., Eddy R.J. Reconstitution of in vivo macrophage-tumor cell pairing and streaming motility on one-dimensional micro-patterned substrates. Intravital. 2012;1:77–85. doi: 10.4161/intv.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Provenzano P.P., Inman D.R., Keely P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paszek M.J., Zahir N., Weaver V.M. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 61.Provenzano P.P., Martinez D.A., Vanderby R., Jr. Hindlimb unloading alters ligament healing. J. Appl. Physiol. 2003;94:314–324. doi: 10.1152/japplphysiol.00340.2002. [DOI] [PubMed] [Google Scholar]
- 62.Lynch H.A., Johannessen W., Elliott D.M. Effect of fiber orientation and strain rate on the nonlinear uniaxial tensile material properties of tendon. J. Biomech. Eng. 2003;125:726–731. doi: 10.1115/1.1614819. [DOI] [PubMed] [Google Scholar]
- 63.Stabile K.J., Pfaeffle J., Tomaino M.M. Bi-directional mechanical properties of the human forearm interosseous ligament. J. Orthop. Res. 2004;22:607–612. doi: 10.1016/j.orthres.2003.05.002. [DOI] [PubMed] [Google Scholar]
- 64.Hadi M.F., Barocas V.H. Microscale fiber network alignment affects macroscale failure behavior in simulated collagen tissue analogs. J. Biomech. Eng. 2013;135:021026. doi: 10.1115/1.4023411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Satulovsky J., Lui R., Wang Y.L. Exploring the control circuit of cell migration by mathematical modeling. Biophys. J. 2008;94:3671–3683. doi: 10.1529/biophysj.107.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma L., Janetopoulos C., Iglesias P.A. Two complementary, local excitation, global inhibition mechanisms acting in parallel can explain the chemoattractant-induced regulation of PI(3,4,5)P3 response in dictyostelium cells. Biophys. J. 2004;87:3764–3774. doi: 10.1529/biophysj.104.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grinnell F., Petroll W.M. Cell motility and mechanics in three-dimensional collagen matrices. Annu. Rev. Cell Dev. Biol. 2010;26:335–361. doi: 10.1146/annurev.cellbio.042308.113318. [DOI] [PubMed] [Google Scholar]
- 68.Nam K.H., Kim P., Kim D.H. Multiscale cues drive collective cell migration. Sci. Rep. 2016;6:29749. doi: 10.1038/srep29749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doyle A.D., Wang F.W., Yamada K.M. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y.J., Le Berre M., Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 71.Lämmermann T., Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr. Opin. Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 72.Fingar D.C., Salama S., Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyers J., Craig J., Odde D.J. Potential for control of signaling pathways via cell size and shape. Curr. Biol. 2006;16:1685–1693. doi: 10.1016/j.cub.2006.07.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells are shown in green and collagen in gray; the top panels represent XY projection images of the Z stack at each time point, whereas the bottom panels represents the XZ projection. Scale bar, 50 μm.
Scale bar, 50 μm.
Scale bar, 50 μm.