Abstract
Objective
We studied the role of the NC_000017.10:g.38051348A>G (rs8067378) single nucleotide polymorphism (SNP) located 9.5 kb downstream of gasdermin B (GSDMB), in the development and progression of cervical squamous cell carcinomas (SCC).
Methods
Using high-resolution melting curve analysis, we genotyped this SNP in patients with cervical SCC (n = 486) and controls (n = 511) from the Polish Caucasian population. Logistic regression analysis was used to adjust for the effect of confounders such as age, parity, oral contraceptive use, tobacco smoking, and menopausal status. The effect of this SNP on the expression of GSDMB was studied by reverse transcription and quantitative real-time polymerase chain reaction analysis of GSDMB transcript levels in SCC tissues.
Results
For all patients with SCC, the p trend value calculated for rs8067378 was statistically significant (p trend = 0.0019). The adjusted odds ratio for the G/G vs. A/A genotype was 1.304 (95% confidence interval 1.080–1.574, p = 0.0057) and the adjusted odds ratio for the G/A + G/G vs. A/A genotype was 1.444 (95% confidence interval 1.064–1.959, p = 0.0181). We also found a significant association of the rs8067378 SNP with tumor stages III, IV, and grade of differentiation G3, and with parity, oral contraceptive use, smoking, and women of postmenopausal age. We found increased GSDMB1 isoform transcripts in the cancerous and non-cancerous tissues from carriers of the G allele vs. carriers of the A/A genotype.
Conclusions
The rs8067378 SNP variants may increase the expression of GSDMB and the risk of the development and progression of cervical SCC.
Electronic supplementary material
The online version of this article (doi:10.1007/s40291-017-0256-1) contains supplementary material, which is available to authorized users.
Key Points
| Gasdermin B (GSDMB) NC_000017.10:g.38051348A>G (rs8067378) transition may upregulate transcription of GSDMB in tumoral and non-tumoral cervical tissues. |
| This transition may be associated with increased development and spread of cervical squamous cell carcinoma cells to the surrounding tissues. |
Introduction
Cervical cancer (CC) is one of the most common tumors in women and remains one of the main causes of death among women worldwide [1]. The precancerous state of cervical intraepithelial neoplasia, which may progress to CC, is usually the result of persistent infection with one of the oncogenic high-risk human papillomavirus (HPV) strains [2]. HPV E6 and E7 oncoproteins impair the function of p53 and pRb proteins and drive the cells to uncontrolled proliferation [3]. Additionally, E6 and E7 oncoproteins lead to chromosomal aberration [4]. It has been observed that the most high-risk HPV virions are eliminated by the hosts’ immune system and only a minority of infections lead to a cervical lesion [5, 6]. These studies indicate that long-term infection with high-risk HPV contributes to cervical carcinogenesis; however, this further depends on an individual’s genetic background [5, 6]. The risk factors for CC also include multiparity, cigarette use, sexual history, abnormalities in the immune system, and environmental pollutants [7–10]. Important risk factors for CC are heritable genetic components, including a family history of cancer, especially in first-degree relatives [11]. The recently carried out genome-wide association studies highlight the major histocompatibility complex region in particular as being associated with cervical carcinogenesis [12–14].
Additionally, genome-wide association studies in the Chinese population have demonstrated invasive CC to be associated with polymorphisms within two non-major histocompatibility complex loci: among them was the NC_000017.10:g.38051348A>G (rs8067378) single nucleotide polymorphism (SNP) [13]. The products of gasdermin B [GSDMB (OMIM *611221)] are the four protein isoforms GSDMB1–4 [15]. It has been demonstrated that the GSDMB protein may be linked with CC development and progression [15]. We replicated the prevalence of the rs8067378 SNP in patients with cervical squamous cell carcinomas (SCCs) in the Polish Caucasian population and we assessed the association of this SNP to different stages and grades of differentiation. We also evaluated the effect of rs8067378 genotypes on this gene’s transcript isoform GSDMB1–4 levels in primary SCC and non-cancerous tissues.
Patients and Methods
Study Population and Tissue Samples
The study population consisted of 486 patients with cervical SCC, with stage and grade of differentiation assessed by an experienced histopathologist based on the International Federation of Gynecology and Obstetrics classification system and the World Health Organization (Table 1). Patient data and primary cervical SCC tissue samples were obtained from patients enrolled between October 2007 and August 2015 at the Department of Radiotherapy of the Greater Poland Cancer Center in Poznań, Poland. The control group consisted of 511 unrelated healthy female volunteers selected during a medical examination at the University Hospital, Clinic of Gynecological Surgery at Poznań University of Medical Sciences (Table 1). Information regarding a parity of at least one, oral contraceptive use, active tobacco smoking at minimum within the last 12 months, and menopausal status was obtained as part of the control and patient history.
Table 1.
Clinical and demographic characteristics of patients with cervical squamous cell carcinoma and controls
| Characteristic | Patients (n = 486) | Controls (n = 511) | p valueb |
|---|---|---|---|
| Mean age (years)a ± SD | 51.5 ± 9.7 | 51.8 ± 9.5 | |
| Tumor stage, n (%) | |||
| IA | 67 (13.8) | ||
| IB | 65 (13.4) | ||
| IIA | 64 (13.2) | ||
| IIB | 56 (11.5) | ||
| IIIA | 155 (31.9) | ||
| IIIB | 54 (11.1) | ||
| IVA | 13 (2.7) | ||
| IVB | 12 (2.5) | ||
| Histological grade, n (%) | |||
| G1 | 99 (20.4) | ||
| G2 | 152 (31.3) | ||
| G3 | 104 (21.4) | ||
| Gx | 131 (27.0) | ||
| Parity, n (%) | |||
| Never | 58 (11.9) | 63 (12.3) | 0.849 |
| Ever | 428 (88.1) | 448 (87.7) | |
| Oral contraceptive pill use, n (%) | |||
| Never | 263 (54.1) | 289 (56.6) | 0.438 |
| Ever | 223 (45.9) | 222 (43.4) | |
| Tobacco smoking, n (%) | |||
| Never | 309 (63.6) | 337 (65.9) | 0.434 |
| Ever | 177 (36.4) | 174 (34.1) | |
| Menopausal status, n (%) | |||
| Premenopausal | 171 (35.2) | 197 (38.6) | 0.271 |
| Postmenopausal | 315 (64.8) | 314 (61.4) | |
| HPV genotypes, n (%) | |||
| 16 and 18 | 326 (67.1) | ||
| 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 | 368 (75.7) | ||
HPV human papillomavirus virus, SD standard deviation
aAge at first diagnosis
bChi-squared, p value
The primary SCC tissue samples were obtained from 47 patients with a mean age of 51.6 ± 10.2 years and classified as stage III at the time of surgery. The non-cancerous cervical tissue samples were obtained from 47 women with a mean age of 51.4 ± 9.8 years and with uterine fibroids undergoing uterine surgical resection in the Division of Gynecological Surgery, Poznań University of Medical Sciences, Poznań, Poland. A portion of the tissue sample was immediately snap frozen in liquid nitrogen and stored at −80 °C until RNA isolation. All the patients and controls participating in the study were Caucasians from the Wielkopolska area of Poland. Written consent was obtained from all participating individuals. The study procedures were approved by the Local Ethics Committee of the Poznań University of Medical Sciences (Reference no. of ethical approval: 1010/07).
Genotyping of the rs8067378 SNP
DNA was isolated from peripheral blood cells via a salting-out procedure. The primers were designated employing Oligo 7.6 software (DBA Oligo, Inc., Colorado Springs, CO, USA). The rs8067378 polymorphism DNA fragment (225 bp) was amplified using the primers forward 5′ GAAAGAAGAGCACAGATAAAC 3′ and reverse 5′ CGGATGACTGGTGAAATAAGC 3′. The rs8067378 SNP was then genotyped via high-resolution melting curve analysis using HOT FIREPol EvaGreen (Solis BioDyne, Tartu, Estonia) with a LightCycler 480 system (Roche Diagnostics, Mannheim, Germany). The presence of this SNP was reanalyzed by Sanger sequencing analyses of randomly selected samples comprising 10% of the samples from all of the participants. The concordance rate between high-resolution melting and sequencing was equal to 100%.
Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction Analysis of GSDMB Transcript Levels in SCC and Non-Cancerous Tissues
Frozen SCC and non-cancerous tissues were homogenized and total RNA was isolated according to the method of Chomczyński and Sacchi [16]. RNA quality was determined spectrophotometrically using a BioPhotometer® from Eppendorf AG (Hamburg, Germany) and agarose gel electrophoresis. RNA samples were treated with DNase I, quantified, and reverse transcribed into complementary DNA (cDNA) with the Moloney Murine Leukemia Virus from Invitrogen (Life Technologies, Carlsbad, CA, USA) [Online Resource 1].
Quantitative analysis of GSDMB1–4 cDNA isoforms (Online Resource 1) was performed by Light Cycler®480 II Real-Time PCR System (Roche Diagnostics GmbH, Mannheim, Germany), using SYBR Green I as the detection dye. GSDMB1–4 cDNA was quantified using the relative quantification method with a calibrator. The calibrator was prepared with a cDNA mix from all cDNA samples and consecutive dilutions were used to create a standard curve. For amplification, 1 µL of cDNA solution was added to 9 µL of LightCycler 480 SYBR Green I Master Mix (Roche Diagnostics GmbH, Mannheim, Germany) and primers (Online Resource 1).
The quantity of the GSDMB1–4 transcript in each sample was standardized by the geometric mean of reference transcript levels: hydroxymethylbilane synthase and β-2-microglobulin. The polymerase chain reaction amplification efficiency for target and reference cDNA was determined by different standard curves created by consecutive dilutions of the cDNA template mixture. The GSDMB1–4 cDNA, hydroxymethylbilane synthase, and β-2-microglobulin cDNA were amplified using the primer pairs presented in Online Resource 1. The GSDMB mRNA levels were expressed as multiples of these cDNA concentrations in the calibrator.
Data Analysis
The distinction in genotypic prevalence between the patients and controls and their genotype deviation from the Hardy–Weinberg equilibrium were evaluated using a χ 2 test. The polymorphism was tested for association with CC incidence using the Cochran–Armitage p trend test (p trend). The χ 2 and Fisher exact tests were used to determine the differences in genotypic distributions between the patients and controls. The odds ratio (OR) and 95% confidence intervals (CIs) were also calculated. A logistic regression analysis was used to adjust for the effect of confounders such as age, parity, oral contraceptive use, tobacco smoking, and menopausal status. A p value of <0.05 was considered statistically significant. Statistical analysis of comparing GSDMB1–4 isoform transcript levels between the G/G vs. A/A and A/G vs. A/A genotype carriers was evaluated by the Kruskal–Wallis test with either Dunn’s post-hoc test or Tukey’s post-hoc test. Statistical analyses were conducted using Statistica version 10, 2011 (Stat Soft, Inc., Tulsa, OK, USA).
Results
Prevalence of the rs8067378 SNP Among all Patients with SCC and Controls
The values for the χ 2 test of the Hardy–Weinberg equilibrium were 0.329 and 0.181 for the patients and controls, respectively. The statistical evaluation of the rs8067378 genotype prevalence in cases and controls are shown in Table 2. For all patients with SCC, the p trend value calculated for the rs8067378 transition was statistically significant (p trend = 0.0019). The logistic regression analysis, which adjusted for the effects of age, parity, oral contraceptive use, tobacco smoking, and menopausal status, also demonstrated a contribution of the rs8067378 SNP to cervical SCC development (Table 2). We observed that the G/G vs. A/A genotype is a risk factor of cervical SCC with an adjusted OR of 1.304 (95% CI 1.080–1.574, p = 0.0057). We observed the same risk effect of the G/A + G/G vs. A/A genotype, with an adjusted OR of 1.444 (95% CI 1.064–1.959, p = 0.0181). However, we did not find an association with cervical carcinogenesis for the G/A vs. A/A genotype, adjusted OR of 1.302 (95% CI 0.944–1.794, p = 0.1069).
Table 2.
Prevalence of the rs8067378 polymorphism among patients with squamous cell carcinoma and controls
| Genotype | Patients (frequency) | Controls (frequency) | Odds ratio (95% CI) | p valuea | Adjusted | p value | p trend |
|---|---|---|---|---|---|---|---|
| Odds ratio (95% CI)c | |||||||
| All | |||||||
| A/A | 91 (0.19) | 127 (0.25) | Referent | Referent | 0.0019 | ||
| G/G | 138 (0.28) | 108 (0.21) | 1.783 (1.233–2.578) | 0.002 | 1.304 (1.080–1.574) | 0.0057 | |
| G/A | 257 (0.53) | 276 (0.54) | 1.300 (0.9452–1.787) | 0.1063 | 1.302 (0.944–1.794) | 0.1069 | |
| G/A + GG | 395 (0.81) | 384 (0.75) | 1.436 (1.060–1.945) | 0.0193 | 1.444 (1.064–1.959) | 0.0181 | |
| MAFd | 0.55 | 0.48 | |||||
| Tumor stage | |||||||
| IA + IB | |||||||
| A/A | 34 (0.26) | 127 (0.25) | Referent | Referent | 0.5046 | ||
| G/G | 35 (0.26) | 108 (0.21) | 1.211 (0.7073–2.072) | 0.4855 | 0.998 (0.760–1.311) | 0.9906 | |
| G/A | 63 (0.48) | 276 (0.54) | 0.8526 (0.5344–1.360) | 0.5032 | 0.712 (0.417–1.217) | 0.2137 | |
| G/A + GG | 98 (0.74) | 384 (0.75) | 0.9533 (0.6147–1.478) | 0.8307 | 0.937 (0.597–1.469) | 0.7751 | |
| MAFd | 0.50 | 0.48 | |||||
| IIA + IIB | |||||||
| A/A | 32 (0.27) | 127 (0.25) | Referent | Referent | 0.8606 | ||
| G/G | 29 (0.24) | 108 (0.21) | 1.066 (0.6061–1.874) | 0.8251 | 0.985 (0.733–1.324) | 0.9202 | |
| G/A | 59 (0.49) | 276 (0.54) | 0.8484 (0.5255–1.370) | 0.5007 | 0.847 (0.524–1.371) | 0.4988 | |
| G/A + GG | 88 (0.73) | 384 (0.75) | 0.9095 (0.5789–1.429) | 0.6805 | 0.909 (0.577–1.432) | 0.6802 | |
| MAFd | 0.49 | 0.48 | |||||
| IIIA + IIIB | |||||||
| A/A | 23 (0.11) | 127 (0.25) | Referent | Referent | <0.0001 | ||
| G/G | 66 (0.32) | 108 (0.21) | 3.374 (1.967–5.788) | <0.0001 | 1.674 (1.260–2.225) | 0.0004 | |
| G/A | 120 (0.57) | 276 (0.54) | 2.401 (1.466–3.932) | 0.0004 | 2.180 (1.316–3.61) | 0.0024 | |
| G/A + GG | 186 (0.89) | 384 (0.75) | 2.675 (1.659–4.312) | <0.0001 | 2.696 (1.668–4.367) | 0.0001 | |
| MAFd | 0.60 | 0.48 | |||||
| IVA + IVB | |||||||
| A/A | 2 (0.08) | 127 (0.25) | Referent | Referent | 0.0452 | ||
| G/G | 8 (0.32) | 108 (0.21) | 4.704 (0.9776–22.631) | 0.0501b | 2.222 (0.990–4.990) | 0.0518 | |
| G/A | 15 (0.6) | 276 (0.54) | 3.451 (0.7773–15.323) | 0.1081b | NA | NA | |
| G/A + GG | 23 (0.92) | 384 (0.75) | 3.803 (0.8841–16.363) | 0.0568b | 3.654 (0.816–16.358) | 0.0894 | |
| MAFd | 0.62 | 0.48 | |||||
| Differentiation grade | |||||||
| G1 | |||||||
| A/A | 23 (0.23) | 127 (0.25) | Referent | Referent | 0.5261 | ||
| G/G | 24 (0.24) | 108 (0.21) | 1.227 (0.6555–2.297) | 0.5219 | 1.069 (0.775–1.474) | 0.6845 | |
| G/A | 52 (0.52) | 276 (0.54) | 1.040 (0.6099–1.775) | 0.8846 | 0.887 (0.486–1.617) | 0.6943 | |
| G/A + GG | 76 (0.77) | 384 (0.75) | 1.093 (0.6576–1.816) | 0.7318 | 1.036 (0.610–1.759) | 0.8947 | |
| MAFd | 0.49 | 0.48 | |||||
| G2 | |||||||
| A/A | 32 (0.21) | 127 (0.25) | Referent | Referent | 0.0829 | ||
| G/G | 43 (0.28) | 108 (0.21) | 1.580 (0.9350–2.670) | 0.0861 | 1.254 (0.960–1.638) | 0.0960 | |
| G/A | 77 (0.51) | 276 (0.54) | 1.107 (0.6970–1.759) | 0.6661 | 1.037 (0.641–1.677) | 0.8829 | |
| G/A + GG | 120 (0.79) | 384 (0.75) | 1.240 (0.7998–1.923) | 0.3353 | 1.218 (0.781–1.901) | 0.3841 | |
| MAFd | 0.54 | 0.48 | |||||
| G3 | |||||||
| A/A | 11 (0.10) | 127 (0.25) | Referent | Referent | 0.0004 | ||
| G/G | 34 (0.33) | 108 (0.21) | 3.635 (1.757–7.518) | 0.0003 | 2.004 (1.305–3.078) | 0.0014 | |
| G/A | 59 (0.57) | 276 (0.54) | 2.468 (1.254–4.858) | 0.0073 | 2.346 (1.180–4.666) | 0.0148 | |
| G/A + GG | 93 (0.89) | 384 (0.75) | 2.796 (1.450–5.391) | 0.0015 | 2.782 (1.437–5.389) | 0.0024 | |
| MAFd | 0.61 | 0.48 | |||||
| Gx | |||||||
| A/A | 25 (0.19) | 127 (0.25) | Referent | Referent | 0.0532 | ||
| G/G | 37 (0.28) | 108 (0.21) | 1.740 (0.9855–3.074) | 0.0545 | 1.232 (0.914–1.660) | 0.1692 | |
| G/A | 69 (0.53) | 276 (0.54) | 1.270 (0.7675–2.101) | 0.3514 | 0.672 (0.369–1.226) | 0.1940 | |
| G/A + GG | 106 (0.81) | 384 (0.75)) | 1.402 (0.8678–2.266) | 0.1658 | 1.167 (0.708–1.927) | 0.5436 | |
| MAFd | 0.54 | 0.48 | |||||
Significant results are highlighted in bold font
CI confidence interval, NA the number of genotypes is too small, therefore the logistic regression does not apply
a χ 2 test
bFisher exact test
cOdds ratios were adjusted by age, parity, oral contraceptive use, tobacco smoking, and menopausal status
dMinor allele frequency
Distribution of the rs8067378 SNP among SCC Patients with Different Tumor Stage and Grade of Differentiation
Stratified analyses revealed an association of the rs8067378 genotypes with tumor stages III, IV, and grade of differentiation G3 (Table 2). The p trend value calculated for the rs8067378 SNP in cervical SCC patients with stages III and IV was statistically significant (p trend < 0.0001 and p trend = 0.0452, respectively). Adjusting for the effect of age, parity, oral contraceptive use, tobacco smoking, and menopausal status in patients with stages III, the logistic regression analysis demonstrated that the G/G vs. A/A genotype is a risk factor of cervical carcinogenesis with an adjusted OR of 1.674 (95% CI 1.260–2.225, p = 0.0004). There was also a risk effect of SCC for the G/A vs. A/A genotype, with an adjusted OR of 2.180 (95% CI 1.316–3.61, p = 0.0024) and for the G/G + G/A vs. A/A genotype, with an adjusted OR of 2.696 (95% CI 1.668–4.367, p = 0.0001) in stage III patients.
In patients with grade of differentiation G3, the p trend value calculated for the rs8067378 SNP was statistically significant (p trend = 0.0004). We found a risk effect of the G/G vs. A/A genotype, with an adjusted OR of 2.004 (95% CI 1.305–3.078, p = 0.0014) for the G/A vs. A/A genotype, with an adjusted OR of 2.346 (95% CI 1.180–4.666, p = 0.0148), and for the G/G + G/A vs. A/A genotype, with an adjusted OR of 2.782 (95% CI 1.437–5.389, p = 0.0024). However, the logistic regression analysis did not show any association of the rs8067378 SNP with tumor stage I, II, and IV and grade of differentiation G1, G2, and GX. Moreover, there was no contribution of the rs8067378 SNP with HPV strains to either SCC or tumor stage I, II, and IV and grade of differentiation G1, G2, and GX (data not shown).
Distribution of the rs8067378 SNP among SCC Patients and Controls with a History of Parity, Oral Contraceptive Use, Tobacco Smoking, or Menopausal Status
The stratified analysis for the rs8067378 polymorphism revealed a risk role of this SNP among patients with a positive history of parity, oral contraceptive use, smoking, and among women of postmenopausal age (Table 3). The age-adjusted OR for women with a history of parity for G/G vs. A/A was 1.323 (95% CI 1.083–1.615, p = 0.0059) and for G/G + G/A vs. A/A was 1.484 (95% CI 1.071–2.055, p = 0.0174). The age-adjusted OR for women with a history of oral contraceptive use for G/G vs. AA was 1.376 (95% CI 1.043–1.816, p = 0.0232). The age-adjusted OR for women with a history of tobacco smoking for G/G vs. AA was 1.666 (95% CI 1.195–2.322, p = 0.0024) and for G/G + G/A vs. A/A was 1.958 (95% CI 1.133–3.384, p = 0.0157). The age-adjusted OR among women of postmenopausal age for G/G vs. A/A was 1.334 (95% CI 1.059–1.679, p = 0.0138).
Table 3.
The distribution of rs8067378 genotypes among squamous cell carcinoma risks: parity, oral contraceptive use, tobacco smoking, and menopausal status
| High risk exposure | Patients | Controls | Adjusted odds ratio (95% CI) | p value | Adjusted odds ratio (95% CI) | p value | ||
|---|---|---|---|---|---|---|---|---|
| Genotype | Ever | Never | Ever | Never | Ever | Never | ||
| Parity | ||||||||
| A/A | 79 | 12 | 112 | 15 | Referent | Referent | ||
| G/A | 228 | 29 | 241 | 35 | 1.347 (0.956–1.897) | 0.0881 | 1.129 (0.443–2.878) | 0.7954 |
| G/G | 121 | 17 | 95 | 13 | 1.323 (1.083–1.615) | 0.0059 | 1.289 (0.752–2.209) | 0.3447 |
| G/A + G/G | 349 | 46 | 336 | 48 | 1.484 (1.071–2.055) | 0.0174 | 1.274 (0.526–3.085) | 0.5876 |
| Oral contraceptive use | ||||||||
| A/A | 41 | 50 | 56 | 71 | Referent | Referent | ||
| G/A | 116 | 141 | 119 | 157 | 1.558 (0.944–2.573) | 0.0818 | 1.314 (0.853–2.024) | 0.2145 |
| G/G | 66 | 72 | 47 | 61 | 1.376 (1.043–1.816) | 0.0232 | 1.278 (0.991–1.648) | 0.0571 |
| G/A + G/G | 182 | 213 | 166 | 218 | 1.589 (1.001–2.523) | 0.0509 | 1.407 (0.934–2.121) | 0.1018 |
| Smoking | ||||||||
| A/A | 28 | 63 | 43 | 84 | Referent | Referent | ||
| G/A | 90 | 167 | 94 | 182 | 1.667 ().933–2.976) | 0.0829 | 1.220 (0.826–1.803) | 0.3169 |
| G/G | 59 | 79 | 37 | 71 | 1.666 (1.195–2.322) | 0.0024 | 1.196 (0.948–1.508) | 0.1288 |
| G/A + G/G | 149 | 246 | 131 | 253 | 1.958 (1.133–3.384) | 0.0157 | 1.306 (0.900–1.896) | 0.1586 |
| Menopausal status | ||||||||
| Premenopausal | Postmenopausal | Premenopausal | Postmenopausal | Premenopausal | Postmenopausal | |||
| A/A | 27 | 64 | 47 | 80 | Referent | Referent | ||
| G/A | 105 | 152 | 108 | 168 | 1.739 (1.000–3.023) | 0.0501 | 1.197 (0.800–1.791) | 0.3796 |
| G/G | 39 | 99 | 42 | 66 | 1.381 (0.981–1.939) | 0.0606 | 1.334 (1.059–1.679) | 0.0138 |
| G/A + G/G | 144 | 251 | 150 | 234 | 1.662 (0.977–2.826) | 0.0602 | 1.367 (0.938–1.992) | 0.1035 |
CI confidence interval
All p values were adjusted by age. Significant results are highlighted in bold font
Effect of the rs8067378 Polymorphism on GSDMB1–4 Transcript Isoform Levels in SCC and Non-Cancerous Tissues
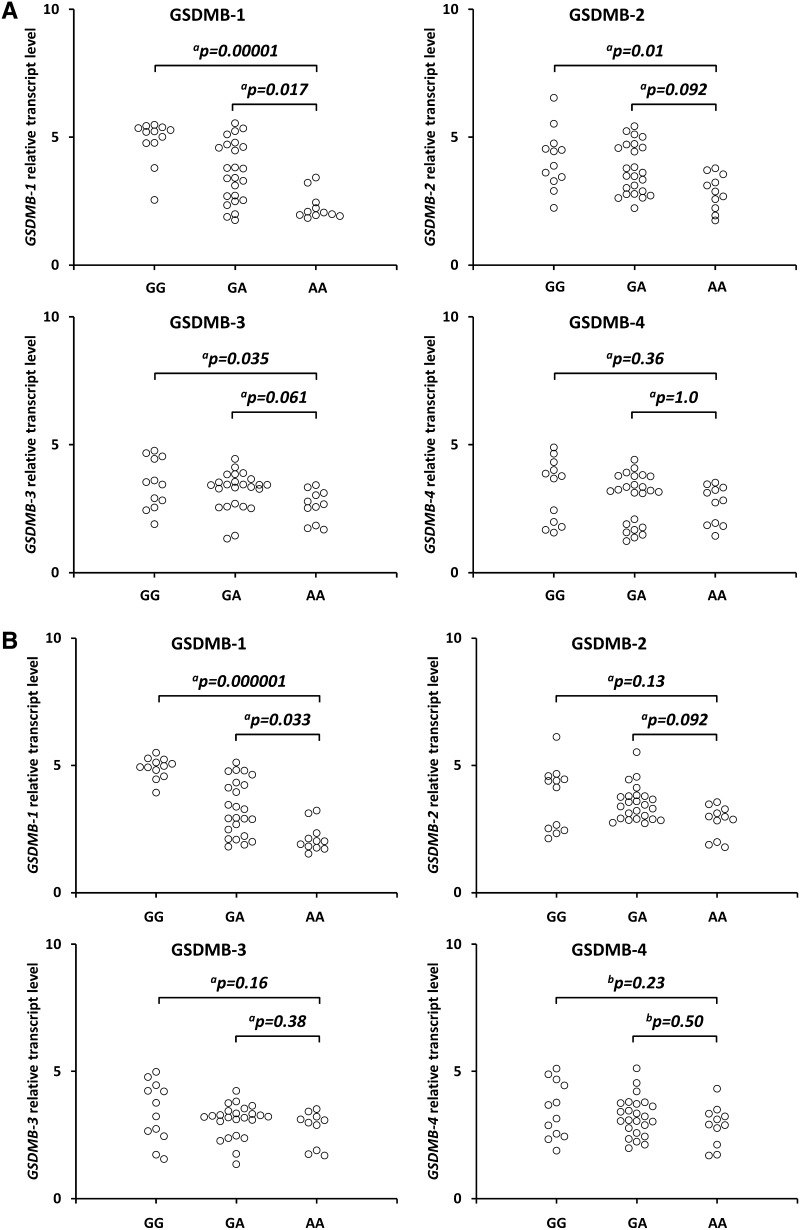
We observed statistically significant increased GSDMB1 isoform transcript levels in the SCC cervical tissues from carriers of the GG vs. A/A (p = 0.00001) and G/A vs. A/A (p = 0.017) (Fig. 1a). We also found a statistically significant increase in the GSDMB1 transcript levels in the non-cancerous cervical tissues from carriers of the GG vs. AA (p = 0.000001) and G/A vs. A/A (p = 0.033) (Fig. 1b). There were also statistically significant increased GSDMB2 and 3 isoform transcripts in the SCC cervical tissues from carriers of the G/G vs A/A genotype (p = 0.010, p = 0.035, respectively) (Fig. 1a).
Fig. 1.
Effect of the GSDMB rs8067378 polymorphism on GSDMB1–4 isoform transcript levels in cancerous cervical tissues (a) and non-cancerous tissues (b). Frozen tissue was homogenized, followed by total RNA isolation. Quantitative analyses of GSDMB transcript levels were performed by quantitative real-time polymerase chain reaction using the SYBR Green I system. The quantity of GSDMB transcript levels in each sample was standardized by the geometric mean of references using hydroxymethylbilane synthase and β-2-microglobulin cDNA levels. Kruskal–Wallis test with aDunn’s post-hoc or bTukey’s post-hoc test. GSDMB gasdermin B
However, we did not find significant differences in GSDMB2 and 3 isoform transcript levels in the SCC cervical tissues for the GA vs. AA genotype (Fig. 1a). There were also no significant differences in GSDMB2 and 3 isoform transcript levels in non-cancerous cervical tissues from carriers of the G allele as compared with carriers of the A/A genotype (Fig. 1b). We also did not find significant differences in GSDMB4 isoform transcript levels in SCC cervical tissues and non-cancerous cervical tissues from carriers of the G allele as compared with carriers of the A/A genotype (Fig. 1b).
Discussion
The GSDMB gene is located at 17q21.2, a region that contains the ERBB2 gene, and is frequently amplified in cancers [17–19]. GSDMB expression is driven by two promoters: the cellular promoter and the Long Terminal Repeats-derived promoter [20–22]. The studied NC_000017.10:g.38051348A>G (rs8067378) SNP is situated 9.5 kb downstream of the GSDMB, in a region suggested to be a functional polymorphism as part of the transcriptional regulatory element and/or modulator of chromatin structure in this domain [23, 24].
Human GSDMB is transcribed in proliferating normal epithelial cells and is overexpressed in various carcinomas, including those of the esophagus, breast, liver, stomach, colon, and uterine cervix [15, 25–29]. Sun et al. reported that GSDMB may also contribute to the development and progression of uterine CC [15]. They demonstrated that the GSDMB protein was present at increased levels in CC tissues as compared with the adjacent cancerous tissues and corresponding normal tissues [15].
We found the rs8067378 SNP to be a risk factor of cervical SCC among all patients from the studied Polish Caucasian population. To date, the minor allele G of rs8067378 has been found to be a significant risk factor for cervical carcinogenesis in Han Chinese and Japanese populations [13, 29].
We also observed that the rs8067378 polymorphism was associated with stages III and IV of cervical SCC. This suggests that there is a role of the G variant of rs8067378 in the increased extension and spread of malignant cells to neighboring tissues. In our studies, the rs8067378 variant was also observed to be associated with differentiation grade G3, which is prone to grow rapidly and spread faster than lower-grade cancerous cells. Our findings are similar to the recent replication study in a Japanese population, in which the authors demonstrated that Japanese women with the rs8067378 GG genotype belong to a high-risk group for invasive CC [30].
Moreover, there was an increased risk of cervical carcinogenesis for the rs8067378 SNP in patients with a positive history of parity, oral contraceptive use, smoking, and women of postmenopausal age. This is consistent with literature data indicating the causative influence of pregnancy, oral contraceptive use, tobacco smoking, and postmenopausal age in cervical carcinogenesis [7–10].
The rs8067378 SNP has also been associated with the development of asthma [24, 31, 32]. Functional studies conducted for the rs8067378 SNP demonstrated reduced expression of GSDMB for the G gene variant of rs8067378 in human bronchial epithelial and bronchial alveolar lavage cells from patients with asthma [24]. In contrast, we found significantly upregulated GSDMB1 isoform transcript levels in both the cervical SCC and non-cancerous tissues from carriers of the G allele as compared with carriers of the A/A genotype. Additionally, we observed significantly increased GSDMB2–3 isoform transcript levels in the cervical SCC tissue for carriers of the G/G vs. A/A genotype. These discrepancies may be owing to the use of different quantitate assays and analyses of different GSDMB isoform transcripts between the asthma study [24] and ours. These may also have resulted from distinct transcription and epigenetic factors regulating the expression of GSDMB in lung tissues and cancerous and non-cancerous cervical tissues.
The increased expression of GSDMB for carriers of the rs8067378 G gene variant confirm the observation of Sun et al. They demonstrated that ectopic GSDMB expression augmented the growth of CC cells in vitro, whereas silencing the endogenous expression of GSDMB reduced cancer cell proliferation [15].
Conclusions
Our genetic study is the first to report that the rs8067378 polymorphism can be a risk factor for cervical carcinogenesis in a Caucasian cohort. Moreover, we observed that the rs8067378 G variant upregulated the expression of GSDMB, which was associated with increased growth and spread of cancer cells to the surrounding tissues. However, our study contains some limitation, for instance, one Caucasian cohort with a relatively small sample size. This study is also limited to one tested SNP. To achieve full gene coverage, a number of other tag SNP needed to be covered in this study. Other SNPs not included in this study could account for alternation of GSDMB expression. Therefore, our genetic study should be replicated for rs8067378 and a number of other tag SNPs covering GSDMB in other independent ethnicities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully acknowledge the technical assistance of Ms. Agnieszka Mikuczewska.
Compliance with Ethical Standards
Conflict of interest
LA, RA, LM, SA, SE, and JPP have no conflicts of interest to report.
Funding
This study, presented by LA, RA, LM, SA, SE, and JPP, was funded by Grant No. 502-01-01124182-07474 from Poznań University of Medical Sciences. Open access was funded as part of the Open Access agreement between Springer and the ICM University of Warsaw, acting on behalf of the Polish Ministry of Science and Higher Education.
Ethics approval and consent to participate
The study procedures were approved by the local Ethics Committee of the Poznań University of Medical Sciences. Informed consent was obtained from all participating individuals.
References
- 1.Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/S0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen HP, Ramírez-Fort MK, Rady PL. The biology of human papillomaviruses. Curr Probl Dermatol. 2014;45:19–32. doi: 10.1159/000355959. [DOI] [PubMed] [Google Scholar]
- 4.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 5.Deligeoroglou E, Giannouli A, Athanasopoulos N, et al. HPV infection: immunological aspects and their utility in future therapy. Infect Dis Obstet Gynecol. 2013;2013:540850. doi: 10.1155/2013/540850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ault KA. Epidemiology and natural history of human papillomavirus infections in the female genital tract. Infect Dis Obstet Gynecol. 2006;2006(Suppl.):40470. [DOI] [PMC free article] [PubMed]
- 7.Kasap B, Yetimalar H, Keklik A, et al. Prevalence and risk factors for human papillomavirus DNA in cervical cytology. Eur J Obstet Gynecol Reprod Biol. 2011;159:168–171. doi: 10.1016/j.ejogrb.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 8.International Collaboration of Epidemiological Studies of Cervical Cancer. Appleby P, Beral V, Berrington de González A, et al. Cervical cancer and hormonal contraceptives: collaborative reanalysis of individual data for 16,573 women with cervical cancer and 35,509 women without cervical cancer from 24 epidemiological studies. Lancet. 2007;370:1609–1621. doi: 10.1016/S0140-6736(07)61684-5. [DOI] [PubMed] [Google Scholar]
- 9.Castellsagué X, Muñoz N. Chapter 3: cofactors in human papillomavirus carcinogenesis: role of parity, oral contraceptives, and tobacco smoking. J Natl Cancer Inst Monogr. 2003;2003:20–28. doi: 10.1093/oxfordjournals.jncimonographs.a003477. [DOI] [PubMed] [Google Scholar]
- 10.Almonte M, Albero G, Molano M, et al. Risk factors for human papillomavirus exposure and co factors for cervical cancer in Latin America and the Caribbean. Vaccine. 2008;26:L16–L36. doi: 10.1016/j.vaccine.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Hemminki K, Chen B. Familial risks for cervical tumors in full and half siblings: etiologic apportioning. Cancer Epidemiol Biomark Prev. 2006;15:1413–1414. doi: 10.1158/1055-9965.EPI-05-0933. [DOI] [PubMed] [Google Scholar]
- 12.Chen D, Juko-Pecirep I, Hammer J, et al. Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst. 2013;105:624–633. doi: 10.1093/jnci/djt051. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Li L, Hu Z, et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat Genet. 2013;45:918–922. doi: 10.1038/ng.2687. [DOI] [PubMed] [Google Scholar]
- 14.Miura K, Mishima H, Kinoshita A, et al. Genome-wide association study of HPV-associated cervical cancer in Japanese women. J Med Virol. 2014;86:1153–1158. doi: 10.1002/jmv.23943. [DOI] [PubMed] [Google Scholar]
- 15.Sun Q, Yang J, Xing G, et al. Expression of GSDML associates with tumor progression in uterine cervix cancer. Transl Oncol. 2008;1:73–83. doi: 10.1593/tlo.08112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 17.Saeki N, Sasaki H. Gasdermin superfamily: a novel gene family functioning in epithelial cells. In: Carrasco J, Mota M, editors. Endothelium and epithelium: composition, functions and pathology, vol 9. Nova Science Publishers, Inc., New York; 2012. p. 193–211.
- 18.Hergueta-Redondo M, Sarrio D, Molina-Crespo Á, et al. Gasdermin B expression predicts poor clinical outcome in HER2-positive breast cancer. Oncotarget. 2016;7(35):56295–56308. doi: 10.18632/oncotarget.10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh M, Katoh M. Evolutionary recombination hotspot around GSDML-GSDM locus is closely linked to the oncogenomic recombination hotspot around the PPP1R1B-ERBB2-GRB7 amplicon. Int J Oncol. 2004;24:757–763. [PubMed] [Google Scholar]
- 20.Komiyama H, Aoki A, Tanaka S, et al. Alu-derived cis-element regulates tumorigenesis-dependent gastric expression of GASDERMIN B (GSDMB) Genes Genet Syst. 2010;85:75–83. doi: 10.1266/ggs.85.75. [DOI] [PubMed] [Google Scholar]
- 21.Sin HS, Huh JW, Kim DS, et al. Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells. Arch Virol. 2006;151:1985–1994. doi: 10.1007/s00705-006-0764-5. [DOI] [PubMed] [Google Scholar]
- 22.Huh JW, Kim DS, Kang DW, et al. Transcriptional regulation of GSDML gene by antisense-oriented HERV-H LTR element. Arch Virol. 2008;153:1201–1205. doi: 10.1007/s00705-008-0105-y. [DOI] [PubMed] [Google Scholar]
- 23.Verlaan DJ, Berlivet S, Hunninghake GM, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Hastie AT, Hawkins GA, et al. eQTL of bronchial epithelial cells and bronchial alveolar lavage deciphers GWAS-identified asthma genes. Allergy. 2015;70:1309–1318. doi: 10.1111/all.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura M, Shiroishi T. GSDM family genes meet autophagy. Biochem J . 2015;469:e5–e7. doi: 10.1042/BJ20150558. [DOI] [PubMed] [Google Scholar]
- 26.Saeki N, Usui T, Aoyagi K, et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancers. 2009;48:261–271. doi: 10.1002/gcc.20636. [DOI] [PubMed] [Google Scholar]
- 27.Carl-McGrath S, Schneider-Stock R, Ebert M, et al. Differential expression and localisation of gasdermin-like (GSDML), a novel member of the cancer-associated GSDMDC protein family, in neoplastic and non-neoplastic gastric, hepatic, and colon tissues. Pathology. 2008;40:13–24. doi: 10.1080/00313020701716250. [DOI] [PubMed] [Google Scholar]
- 28.Hergueta-Redondo M, Sarrió D, Molina-Crespo Á, et al. Gasdermin-B promotes invasion and metastasis in breast cancer cells. PLoS One. 2014;9:e90099. doi: 10.1371/journal.pone.0090099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hergueta-Redondo M, Sarrio D, Molina-Crespo Á, et al. Gasdermin B expression predicts poor clinical outcome in HER2-positive breast cancer. Oncotarget. 2016;7:56295–56308. doi: 10.18632/oncotarget.10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura K, Mishima H, Yasunami M, et al. A significant association between rs8067378 at 17q12 and invasive cervical cancer originally identified by a genome-wide association study in Han Chinese is replicated in a Japanese population. J Hum Genet. 2016;61:793–796. doi: 10.1038/jhg.2016.50. [DOI] [PubMed] [Google Scholar]
- 31.Li FX, Tan JY, Yang XX, et al. Genetic variants on 17q21 are associated with asthma in a Han Chinese population. Genet Mol Res. 2012;11:340–347. doi: 10.4238/2012.February.10.5. [DOI] [PubMed] [Google Scholar]
- 32.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.