Summary
Zika virus (ZIKV) infection causes microcephaly in a subset of infants born to infected pregnant mothers. It is unknown whether human individual differences contribute to differential susceptibility of ZIKV-related neuropathology. Here, we use an Asian-lineage ZIKV strain, isolated from the 2015 Mexican outbreak (Mex1-7), to infect primary human neural stem cells (hNSCs) originally derived from three individual fetal brains. All three strains of hNSCs exhibited similar rates of Mex1-7 infection and reduced proliferation. However, Mex1-7 decreased neuronal differentiation in only two of the three stem cell strains. Correspondingly, ZIKA-mediated transcriptome alterations were similar in these two strains but significantly different from that of the third strain with no ZIKV-induced neuronal reduction. This study thus confirms that an Asian-lineage ZIKV strain infects primary hNSCs and demonstrates a cell-strain-dependent response of hNSCs to ZIKV infection.
Keywords: human neural stem cell, Zika virus, neuron, astrocyte, differentiation, proliferation, transcriptome, neurogenesis, innate immunity
Graphical Abstract
Highlights
-
•
Mexican ZIKV strain infects primary human fetal brain-derived neural stem cells
-
•
ZIKV inhibits neuronal differentiation in a cell-strain-dependent manner
-
•
Majority of differentiated ZIKV-infected cells are glial cells
-
•
ZIKV-mediated transcriptome alteration is cell-strain-dependent
In this article, Wu, Vasilakis, and colleagues demonstrate the infection of primary human fetal brain-derived neural stem cells by a 2015 Mexican strain of ZIKV. They show that ZIKV is cytotoxic and inhibits proliferation independent of different human origins, but inhibits neuronal differentiation and alters gene expression in a cell-strain-dependent manner.
Introduction
Zika virus (ZIKV) has recently emerged as a global public health threat with 70 countries and territories reporting active ZIKV transmission (Centers for Disease Control and Prevention, 2016a). ZIKV is a flavivirus transmitted by the Aedes aegypti and A. albopictus mosquitoes with recent outbreaks in the Americas, including in Brazil and Mexico (Kindhauser et al., 2016, Guerbois et al., 2016). Currently, there are two principal lineages of ZIKV, African and Asian (Haddow et al., 2012, Kindhauser et al., 2016, Weaver et al., 2016). Only the Asian lineage has been associated with fetal microcephaly and other neurological abnormalities such as Guillain-Barré syndrome, whereas few infections with the African lineage have been described (Broutet et al., 2016, Cao-Lormeau et al., 2016, Kindhauser et al., 2016, Paploski et al., 2016, Weaver et al., 2016). Recently, the Centers for Disease Control and Prevention officially confirmed the link between microcephaly and ZIKV infection, as well as the capability of ZIKV to be sexually transmitted (Hills et al., 2016). While ZIKV typically results in mild or asymptomatic infections in healthy adults or children, the risk of microcephaly and other serious neurological deficits in the developing fetus is an alarming consequence of ZIKV infection and thus is a serious health problem worldwide (Brasil et al., 2016).
Microcephaly is a neurodevelopmental disorder where the head is smaller than the typical size during fetal development and at birth, with the circumference less than 2 SDs below the mean (Centers for Disease Control and Prevention, 2016b). Babies with microcephaly can have a wide array of problems such as developmental delays, seizures, vision and hearing loss, and feeding difficulty. To date, little is known about the mechanism underlying ZIKV-associated microcephaly. Since a normal brain develops from neural stem cells (NSCs) and their differentiated neural cells, microcephaly is most likely associated with the abnormal function of these cells. Yet, many questions remain to be addressed, such as how human fetal brain NSCs or their progeny are susceptible to ZIKV infection, whether different strains of ZIKV infect NSCs with equal efficiency, if such infection affects functions of NSCs important in human brain development, and whether NSCs from different human origins respond to ZIKV equally. Recent evidence shows that ZIKV directly infects NSCs of the fetus and impairs growth in mice (Li et al., 2016, Wu et al., 2016). It has also been shown that ZIKV infects neural progenitors from human skin-cell-induced pluripotent stem cells (Garcez et al., 2016, Tang et al., 2016), but these studies used the murine neuro-adapted prototype strain (MR766) belonging to the African lineage. A more recent study showed that a 2015 Puerto Rico strain of ZIKV, PRVABC59, infects and kills primary human fetal neural progenitors (Hanners et al., 2016). However, none of these studies investigated the effect of ZIKV on neural stem cell functions, particularly their differentiation into neurons and glial cells, which is critical for brain development.
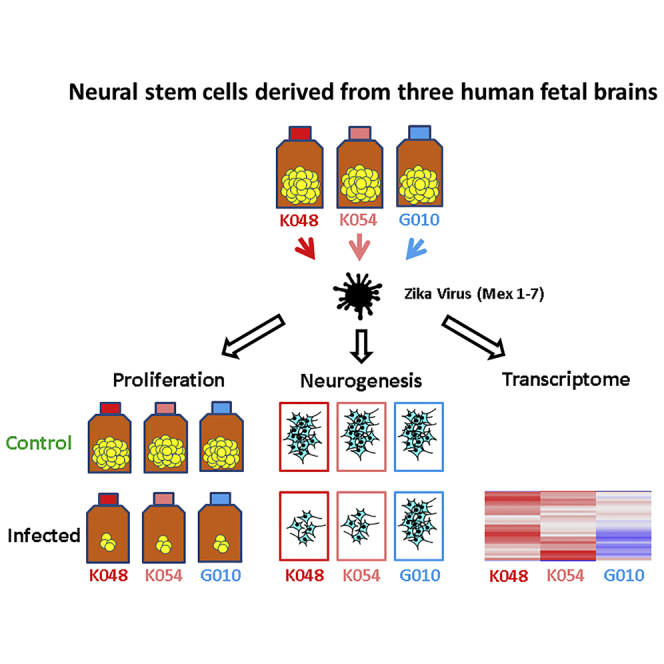
We used several strains of human fetal brain-derived NSCs (hNSCs) and evaluated the effects of recent ZIKV outbreak strains on their infection, proliferation, and differentiation. To distinguish from genetically modified cell lines, we refer to these non-genetically modified hNSCs as hNSC strains. We found that Mex1-7 was able to infect hNSCs and decrease proliferation in all strains but altered neuronal differentiation in only two of the hNSC strains. Transcriptome analyses also revealed cell-strain-dependent upregulation of genes associated with innate immunity and apoptotic cell death and downregulation of genes associated with cell-cycle progression and neurogenesis. The in vitro primary hNSC system will be critically important for further mechanistic studies of ZIKV strain and human neural damage and for providing insights into novel therapeutic developments.
Results
Infection of Primary hNSCs by Asian and African Lineages of ZIKV
Three primary strains of hNSCs were derived from three male donor brains at the gestational age of 9 weeks (K048) or 13 weeks (G010 and K054). ZIKV infectivity and all other analyses of these hNSCs were carried out at passage number 22. Fluorescent in situ hybridization (FISH) analysis revealed that the three cell strains maintained a normal cytogenetic profile with very few cells showing chromosomal trisomy (Figure S1). MOIs of 1 and 10 infectious units/cell were initially used to test the infectivity of the following three ZIKV strains: Mex1-7, FSS13025 (or FSS hereafter), and DakAr41525 (or DakAr). MOIs were calculated from Vero cell focus-forming units. Due to overall low infection rates, an MOI of 10 was chosen for all further experiments to ensure maximum infectivity. Mex1-7 and FSS (Asian-lineage strains) are a 2015 mosquito isolate from an outbreak in southern Mexico (Guerbois et al., 2016) and a 2010 human isolate from Cambodia (Haddow et al., 2012), respectively. DakAr, on the other hand, is a 1984 mosquito isolate from Senegal, belonging to the African lineage. These three ZIKV strains were propagated only in mosquito and African green monkey kidney Vero cells, and particularly Mex1-7 is a newly isolated Asian strain with very limited passage history in C6/36 mosquito cells (three passages).
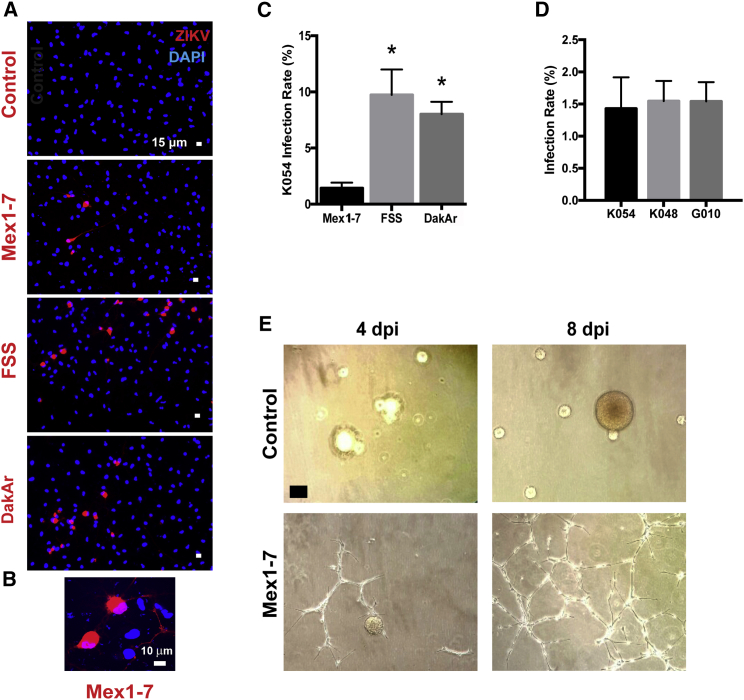
Figure 1A shows representative images from K054 hNSC infection with Mex1-7, FSS, and DakAr 4 days post infection (4 dpi), with adsorption for 1 hr. Both FSS and DakAr produced significantly higher titers 4 dpi compared with Mex1-7 in all three hNSC strains. One day of ZIKV infection resulted in cell-rounding cytopathic effects as shown in Figure 1B. Quantitative analyses of ZIKV infectivity revealed that 10% and 8% of K054 cells were infected by FSS and DakAr, respectively; whereas Mex1-7 resulted in only ∼1.5% cell infection in K054, K048, and G010 hNSCs (Figures 1C and 1D). Given the clinical relevance of Mex1-7 and the distinct difference of this strain from FSS and DakAr, Mex1-7 was chosen for further evaluation for its effects on hNSC survival, proliferation, and differentiation. As shown in Figure 1E, Mex1-7 infection resulted in a change in hNSC morphology. When cultured in flasks uncoated with adherent substances such as poly-D-lysine, hNSCs grew as neurospheres in suspension in the absence of ZIKV infection. However, upon the presence of ZIKV, cells developed an adherent network structure and gradually lost sphere formation (Figure 1E).
Figure 1.
Similar Infectious Rates of Mex1-7 ZIKV in Different Strains of hNSCs
(A) Representative fluorescent images stained for ZIKV (red) and cell nucleus (blue) showing co-localization of virus and cells 1 day after infection with various ZIKV strains.
(B) High magnification of Mex 1–7 infected cells.
(C) Quantification of infected cells from three ZIKV strains.
(D) Quantification of Mex1-7 infectivity in three hNSC strains.
(E) Representative phase contrast images taken 4 and 8 days after ZIKV infection in hNSCs.
Scale bars: 15 μm in (A), 10 μm in (B), and 60 μm in (E). ∗p < 0.05 compared with other strains of virus. Data are presented as means ± SEM, n = 3. See also Figure S1.
Proliferation and Survival of Primary hNSCs Altered by Mex1-7 Independent of Human Donor Origin
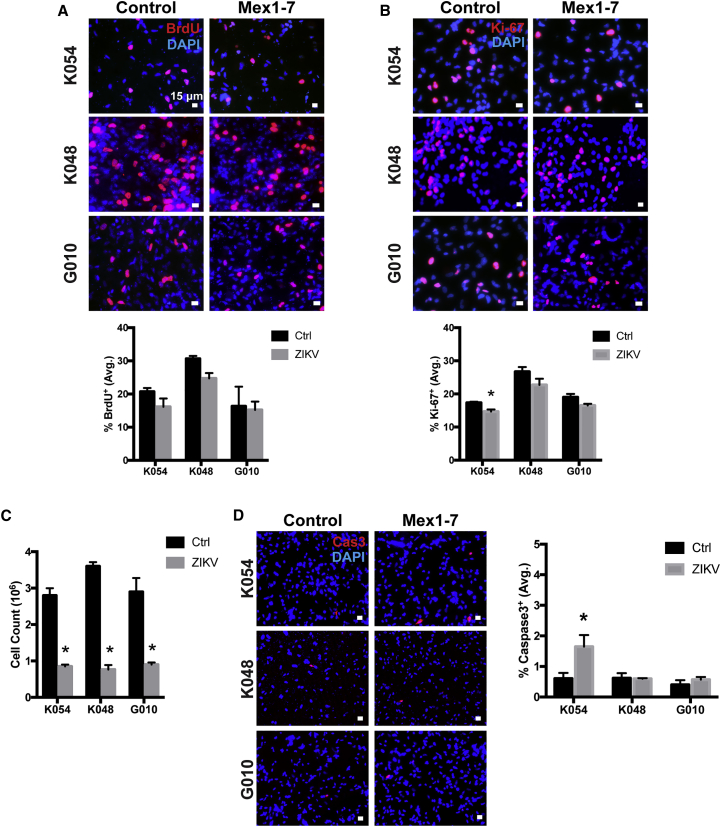
To measure the effect of ZIKV infection on hNSC proliferation, bromodeoxyuridine (BrdU) and Ki-67 staining was performed. BrdU incorporates into the DNA during the S phase, and thus labels dividing cells. In this study, cells were incubated with BrdU for 4 hr to ascertain the effect of Mex1-7 infection on proliferation 7 dpi. As shown in Figure 2A, no significant changes were observed in BrdU labeling in Mex1-7-infected cells compared with controls (Figure 2A). Ki-67, a marker for proliferating cells, showed a significant decrease of approximately 15% in K054 cells but not in K048 or G010 (Figure 2B). Considering that BrdU and Ki-67 provide information on cell proliferation at a certain point of time, proliferation following Mex1-7 infection was further evaluated by counting cells. Initially, 1 million cells with or without Mex1-7 infection were immediately plated into a T-25 flask and allowed to grow for up to 11 days, then passaged, and counted. Without ZIKV infection, the total cell number increased by about 2.8-fold in K054, 3.6-fold in K048, and 3-fold in G010 cells (Figure 2C). In contrast, Mex1-7 infection significantly reduced the total cell numbers compared with controls. Mex1-7-infected cells decreased from 1 million cells by 15% in K054 (0.85 million), 23% in K048 (0.77 million), and 10% in G010 (0.9 million) (Figure 2C). This reduction of cells may be due to loss of cells during the passaging procedure. Alternatively, it could be due to cytopathic effects of ZIKV infection. However, we found that there was no change in apoptosis in any of the cell strains following ZIKV infection in the proliferation stage, as determined by activated caspase-3 (Cas3) staining (Figure S2). On the other hand, following differentiation, ZIKV-infected K054 cells had a significant 3.5-fold increase in activated Cas3, whereas infected G010 and K048 showed no significant change in Cas3+ cells (Figure 2D).
Figure 2.
Reduction of hNSCs by Mex1-7
(A) Four hours of BrdU pulse labeling in three hNSC strains at 7 days post ZIKV infection.
(B) Immunocytochemical staining with Ki-67 proliferation marker at 7 days post infection.
(C) Total number of cells in three Mex1-7-infected hNSC strains after 11 days of proliferation, all starting at 1 million cells.
(D) Activated caspase 3 (Cas3, red) staining of hNSCs with or without Mex1-7 infection after differentiation.
DAPI, blue nuclear staining. Scale bars: 15 μm in (A), (B), and (D). Quantitative data are presented as means ± SEM. ∗p < 0.05 compared with control, n = 3. See also Figure S2.
Differentiation of Primary hNSCs Altered by Mex 1–7 in a Human Cell Strain-Dependent Manner
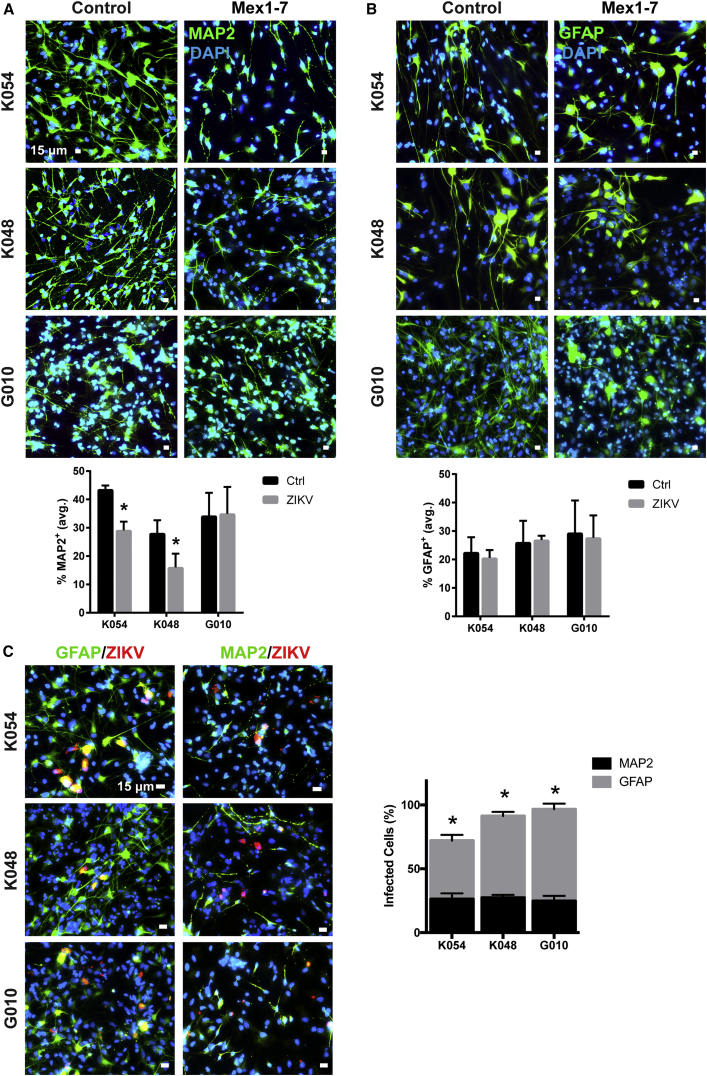
To evaluate the effect of ZIKV infection on hNSC differentiation, Mex1-7-innoculated G010, K048, and K054 hNSCs were allowed to differentiate for 9 days. At 15 dpi, cells were immunostained with markers for mature neurons (microtubule-associated protein 2 or MAP2), astrocytes (glial fibrillary acidic protein or GFAP), and ZIKV proteins. Mex1-7 infection significantly reduced MAP2+ neurons by approximately 33% in K054 cells and 43% in K048 cells (Figure 3A). Interestingly, G010 neuronal differentiation was not affected by ZIKV infection, although some morphological changes were visible in these as well as in K054 and K048 (Figure 3A). On the other hand, GFAP+ cell populations were not altered in any of the three cell strains, but there was a change in cell morphology (Figure 3B). These data suggest that hNSCs of different individuals vary in neuronal differentiation potential following ZIKV infection. When dual staining of ZIKV and either GFAP or MAP2 was conducted, the majority of Mex1-7-infected and surviving cells were GFAP+ astrocytes. Of the ZIKV+ population, approximately 46% in K054, 64% in K048, and 72% in G010 were double labeled with GFAP (Figure 3C). This was significantly higher than ZIKV+ cells dual-labeled with MAP2 for neurons which were about 26% in K054, 27% in K048, and 25% in G010 (Figure 3C). We do not believe apoptosis to be responsible for this decreased neurogenesis, considering less than 1.7% of cells showed activated Cas3 staining (Figure 2D).
Figure 3.
Cell-Strain-Dependent Inhibition of hNSC Neuronal Differentiation Following Mex1-7 Infection
(A) hNSCs differentiated into neuronal cells detected by MAP2 (green). ∗p < 0.05 compared with control.
(B) hNSCs differentiated into astroglia detected by GFAP (green).
(C) GFAP+ and MAP2+ cells (green) infected with ZIKV (red).
Scale bars: 15 μm in (A–C). Quantitative analyses are presented as means ± SEM, n = 3. ∗p < 0.05 comparing percent infection of MAP2+ with GFAP+.DAPI, blue nuclear staining.
ZIKV-Induced Changes of Gene Expression in a Human Cell Strain-Dependent Manner
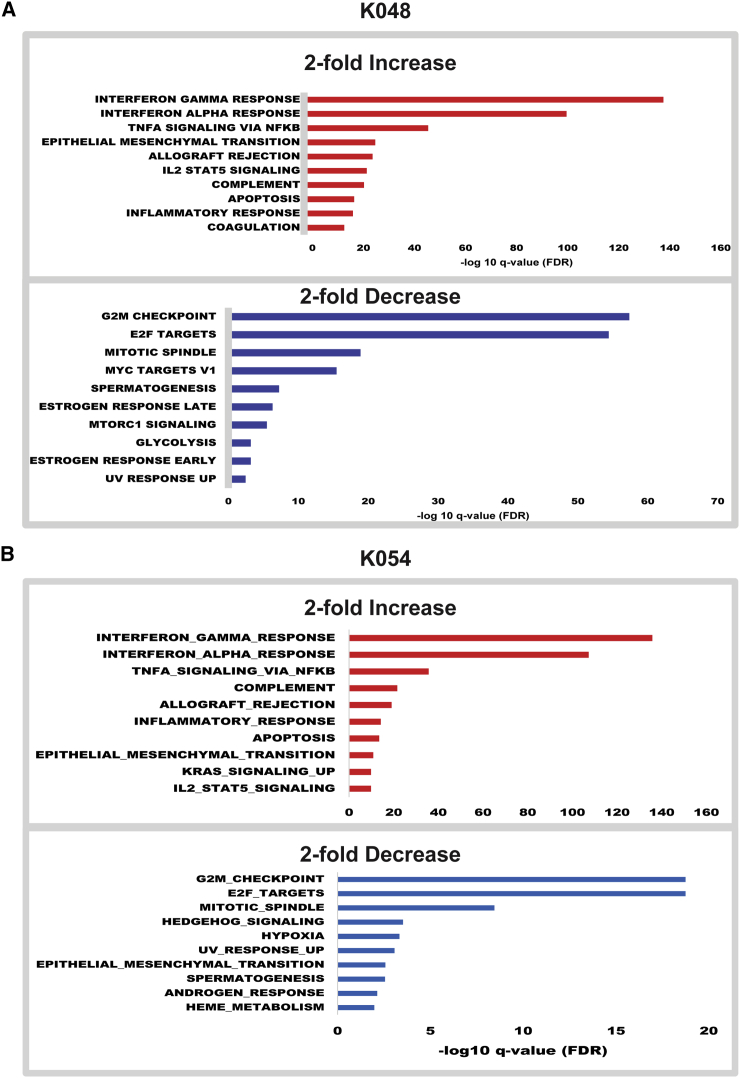
To investigate the underlying cause of ZIKV-infection-related changes in neuronal differentiation, deep sequencing of RNAs from all three strains was performed 7 dpi and after 1 day of differentiation. RNA analysis was conducted in triplicate for each cell strain. Figure 4 shows Venn diagrams and heatmaps of significantly changed genes based on a false discovery rate (FDR) cutoff of <0.05, with up- or downregulation by 2-fold or greater. Specifically, ZIKA upregulated 370 and 819 genes in K054 and K048 cells, respectively (Figure 4A); and downregulated 227 and 508 genes in K054 and K048, respectively (Figure 4B). In contrast, ZIKV-infected G010 had only 32 upregulated and 56 downregulated genes. Of the G010 upregulated genes, 11 were shared with K048, and only 2 shared with K054. In addition, 2 downregulated genes in G010 were shared with K048 and 1 gene with K054. A complete list of all ZIKV-altered genes can be found in Table S1. Gene set enrichment analysis (Subramanian et al., 2005) of Hallmark pathways on these changed genes confirmed that ZIKV-infected K048 and K054 had similarity in transcriptome alteration, which was remarkably different from that of G010 (Figure 4C). This trend was also held for the top hit pathways related to immune response, neurogenesis, cell cycle, and apoptosis (Figures 4D–4G, 5, and 6).
Figure 4.
Transcriptome Analysis of hNSCs Infected with Mex1-7 by RNA Sequencing
(A and B) Venn diagram showing genes that are upregulated or downregulated by 2-fold or greater in three hNSC strains.
(C–G) Heatmaps showing up- and downregulated genes of global expression (C), and specific genes associated with innate immune response (D), neurogenesis (E), cell cycling and proliferation (F), and apoptotic cell death (G). Results are the averages of n = 3 from each cell strain.
See also Table S1.
Figure 5.
Pathway Analyses of RNA Expression in hNSCs after Mex1-7 Infection
Hallmark pathway analysis bar graphs showing the top hits with up- or downregulation (2-fold or greater) of genes following ZIKV infection in K048 (A) and K054 (B) cells. n = 3 from each cell strains. See also Table S1.
Figure 6.
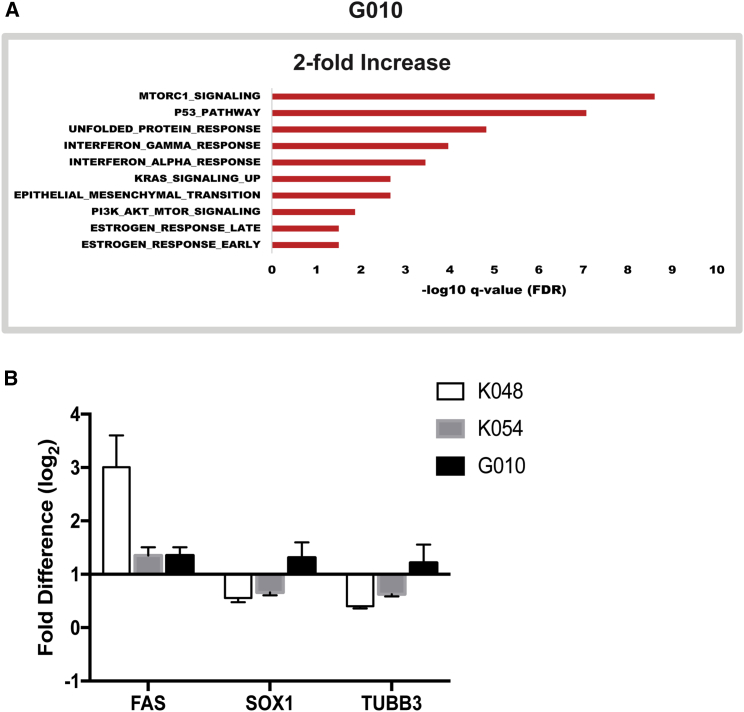
Pathway and qRT-PCR Analyses of RNA Expression in hNSCs after Mex1-7 Infection
(A) Hallmark pathway analysis bar graphs showing the top hits with up- or downregulation (2-fold or greater) of genes following ZIKV infection in G010 cells.
(B) Validation of selected genes by qRT-PCR, n = 3.
See also Table S1.
One of the most striking findings from the Hallmark analysis was that G010 had no significantly downregulated pathways (Figure 6). For example, doublecortin (DCX) is a protein that regulates microtubule stability and is commonly expressed in migratory neuroblasts or immature neurons. DCX was downregulated in both K048 and K054, but not G010, which correlates well with the immunocytochemical findings in Figure 3. Similar results were also detected in the expression of another neuronal marker, Tubulin Beta 3 class III (TUBB3). In addition, of the 200 genes implicated in the Hallmark pathway of the check point progression from G2 to mitotic phase in the cell cycle, K048 and K054 had 52 and 20 downregulated, respectively, while G010 showed no changes (Figures 5 and 6, Table S1). The histone cluster 1 family, particularly histone cluster 1 H4 family member A (HIST1H4A), is implicated in this cell-cycle progression process and was significantly downregulated in K048 and K054 (Figure 5, Table S1). Generally, this gene family is implicated in a wide variety of functions, including transcriptional regulation, DNA repair, and chromosomal architecture (Harshman et al., 2013, Zhou et al., 2013); and particularly HIST1H4A has been shown to be essential for DNA replication (Miotto and Struhl, 2010). It requires further studies to determine whether ZIKV-downregulated cell-cycling genes during the early differentiation stage affect neuronogenesis in the later stage. On the other hand, K048, K054, and G010 all had significantly upregulated genes associated with immune and inflammatory pathways such as interferon-gamma response; however K048 and K054 had much greater changes when compared with G010 (Figures 5 and 6A). Of the 200 genes associated with the interferon-gamma response pathway, K048 and K054 had 109 and 89 genes upregulated, respectively, compared with G010, which had only 4 upregulated genes (Figure 6A). One interesting gene upregulated in K048 and K054, but not G010, was tumor-necrosis-factor-alpha-induced protein 3 (TNFAIP3), which is involved in cytokine-mediated immune responses. It remains to be determined whether altered innate immune response genes contribute to ZIKV-mediated differential deficits in neuronogenesis.
qRT-PCR analysis further confirmed the similar cell strain-dependent trends of ZIKV-altered expression patterns in several genes such as apoptotic protein FAS, neural stem cell stemness marker SOX1, and neuron-specific TUBB3 (Figure 6B).
Discussion
Here, we established an in vitro system using primary human fetal brain-derived NSCs from three different donors to characterize the effect of ZIKV infection. We particularly focused on an Asian-lineage ZIKV strain, Mex1-7, that was involved in the first outbreak in North America in late 2015 in Chiapas State (Guerbois et al., 2016). Interestingly, we found that the Mex1-7 strain had a substantial lower infectivity (1.5%) in primary hNSCs when compared with other strains (10% by FSS and 8% by DakAr) tested in this study, as well as compared with the 1947 African lineage MR766 strain (20%–80% infection rate) and ArB41644 strain (40% infection rate) reported previously (Garcez et al., 2016, Tang et al., 2016, Simonin et al., 2016). The high infectivity of MR766 may be explained by the fact that it has been passaged for over 150 times in suckling mouse brains and developed a neuro-adapted phenotype, a concern addressed previously (Miner and Diamond, 2016). An alternative explanation is that African ZIKV strains may have a higher infectivity rate than Asian ZIKV strains. In this study, the FSS and DakAr strains used were passaged 3 and 7 times, respectively, only in mosquito and mammalian cell culture and not in neural cells. On the other hand, the source of NSCs may also contribute to the discrepant infectivity of ZIKV. For example, our observation of less than 10% cell infection rates are more in line with recent reports (Hanners et al., 2016). Both this, and the fact that our study used primary cultured human fetal brain NSCs may account for differences compared with Tang et al. (2016) and Garcez et al. (2016) who used neural progenitor cells derived from human skin-cell-induced pluripotent stem cells. However, all of these results indicate that the type of neural cells, the viral lineage/strain from different sources, and the viral propagation methods need to be considered when interpreting ZIKV infectivity and functional consequences in human cells.
Microcephaly may result from abnormal proliferation or differentiation of NSCs during early development (Homem et al., 2015). We thus established an in vitro system to characterize the effect of Mex1-7 on survival, proliferation, and differentiation of three primary human fetal NSC strains: G010, K048, and K054. These three cell strains were derived from three human fetal donors at gestational weeks 9 (K048) and 13 (K054 and G010), around the end of the first trimester of pregnancy and within the time frame with high risk of viral-mediated abnormal neural development, including ZIKV-induced microcephaly (Cauchemez et al., 2016). Further, Mex1-7 infections decreased the cell population in all three cell strains without significant changes in Cas3 or BrdU during the proliferation stage. The lack of change of Cas3 could indicate that, during the proliferative stage, cell-cycle progression is halted but cells are not undergoing apoptosis. Since only a small percentage of hNSCs are infected (1.5%), a bystander effect may be occurring to inhibit proliferation. Alternatively, a necrotic mechanism of cell death could account for the decreased population. The population decrease is in general accordance with the previous studies using either human embryonic stem cells or induced pluripotent stem cells (Dang et al., 2016, Garcez et al., 2016, Qian et al., 2016, Tang et al., 2016). In addition to these in vitro studies, two groups have also shown that ZIKV infection in pregnant mice leads to inhibition of neural progenitor proliferation in mouse embryos by cell-cycle arrest (Wu et al., 2016, Li et al., 2016). Further studies are warranted to dissect the mechanisms underlying ZIKV inhibition of NSC proliferation both in vitro and in animal models in vivo.
The most important contribution of our study to ZIKV research is the finding of ZIKV-mediated impairment of hNSC neuronal differentiation in a cell-strain-dependent manner. Specifically, ZIKV infection significantly reduced neuronal differentiation of K054 and K048 strains but not G010. Both RNA sequencing and qRT-PCR also confirm the similar trend of reduction in the expression of neuron-specific TUBB3 gene. This cell-strain-dependent impairment is unlikely related to the initial derivation of hNSC strains from different human gestational weeks: K048 at the week 9, and both K054 and G010 at 13. First, it is known that there are no significant developmental milestones between gestational weeks 9 and 13 (Stiles and Jernigan, 2010). Second, K054 and G010 were both derived from week 13 but respond differently to ZIKV in terms of neuronogenesis. On the other hand, both K054 and K048 showed significant reduction in the number of differentiated neurons despite being derived from different developmental stages. Third, the cell-strain-dependent impairment of neuronal differentiation is unlikely due to the long-term culture (passage 22) of hNSCs. Previous studies indicate that some hNSC strains, when cultured long-term in vitro, extensively accumulate chromosomal 7 and 19 trisomy that may affect neuronal differentiation (Sareen et al., 2009). However, our cytogenetic analyses identified only minimal if any chromosomal trisomy in all cell strains. Finally, the cell-strain-dependent neuronal deficit is unlikely attributed to ZIKV-induced apoptosis. This is because that K054 cells, although having a higher apoptotic death rate than K048, showed a smaller reduction of neurons than K048. Furthermore, the low rates (<1.7%) of apoptotic cell death in K048 and K054 do not match the magnitude of neuronal reduction (>33%) caused by ZIKV infection.
Interestingly, this cell-strain-dependent neuronal impairment seems well correlated with the ZIKV-induced alteration of global gene expression pattern; i.e., K054 and K048 share similar changes in transcriptome and neuronal phenotype that are distinct from G010. It remains to be determined what gene(s) is/are responsible for ZIKV-induced and cell-strain-dependent reduction of neuronal differentiation. One of the most obvious differences detected by transcriptome analyses is that K054 and K048 but not G010 share similar patterns in the innate immune response, including interferons, cytokines, and complements. A recent study also shows that cranial neural crest cells produce cytokines associated with decreased neurogenesis following infection with either MR-766 or H/PF/2013 ZIKV strains (Bayless et al., 2016). In addition, the findings related to downregulation of the Histone 1 gene family in K048 and K054 but not G010 also warrant further investigation due to the broad implications of histone modifications. Further studies with additional hNSC strains and ZIKV viral strains are required to investigate the correlation of these findings and the underlying mechanisms as to how different genetic backgrounds contribute to the cell-strain-dependent neuronal deficits following ZIKV infection.
Another important discovery of our study is that following 2 weeks of differentiation, Mex1-7 infection was mainly found in glial cells. This was true for all three cell strains. Possible explanations include that: (1) ZIKV does not interfere with astroglial differentiation from infected hNSCs; (2) hNSC-differentiated astroglia survive better than neurons; and (3) free virions released from the original target cells preferentially infect glial cells upon their differentiation. Studies to evaluate various infection time points during differentiation and to include other ZIKV strains are needed to investigate these hypotheses.
In summary, we demonstrate that the Asian-lineage ZIKV 2015 Mexican strain, Mex1-7, is neurotropic to human fetal brain-derived NSCs. It halts hNSC proliferation independent of human donor origin but elicits impairment of neuronal differentiation and alters the global gene expression pattern in a cell-strain-dependent manner. The primary hNSC culture system allows quantitative analyses of ZIKV affecting various perspectives of neural development, which lays a foundation for mechanistic investigation of ZIKV neuropathology and for therapeutic development.
Experimental Procedures
ZIKV Isolation and Culture
ZIKV Mex1-7 was isolated from an A. aegypti pool of mosquitoes collected in late 2015 in Mexico on a monolayer of Vero cells, an African green monkey kidney cell line. This virus was amplified 3× in Vero cell cultures prior to use and titrated on Vero cell monolayers as previously described (Rossi et al., 2016). Other ZIKV strains include FSS (2010 human isolate from Cambodia, Asian lineage) and DakAr (1984 mosquito isolate from Senegal, African lineage). FSS was passaged once in AP-1 cells, once in C6/36 mosquito cells, and five times in Vero cells. DakAR was passaged five times in Vero cells, once in C6/36 cells and another two times in Vero cells prior to use. All viruses were amplified to high titers with the final passage in Vero cells (Rossi et al., 2016).
Human Neural Stem Cell Culture and Proliferation
The G010, K048, and K054 strains of primary hNSCs, originally derived from the cortex of 9-week-old (K048) and 13-week-old (K054 and G010) human male fetuses, were propagated in vitro as described previously (Tarasenko et al., 2004, Wu et al., 2002). Cells were grown as neurospheres in DMEM/F12 basic medium supplemented with 20 ng/mL epidermal growth factor (EGF), 20 ng/mL fibroblast growth factor 2, 10 ng/mL leukemia inhibitor factor (LIF), and N2; and passaged every 10 days. Reagents were purchased from Invitrogen, Sigma-Aldrich, R&D Systems, or Fisher Scientific. All three cell strains were used at passage number 22. Cytogenetic analyses to determine chromosomal properties of these hNSC strains were performed by FISH. For details on FISH, see Supplemental Information.
ZIKV Infection
Three strains of ZIKV were used in this study, including Mex1-7 (2015 isolate in Mexico, Asian lineage), FSS (2010 human isolate from Cambodia, Asian lineage), and DakAr (1984 mosquito isolate from Senegal, African lineage). Primary hNSCs were infected for 1 hr in suspension with an MOI from 1 to 10 initially and then 10 for all experiments described here. Following ZIKV inoculation, cells were either kept in proliferation media or subjected to differentiation. Following infection, cells were pelleted by centrifugation and resuspended in the appropriate media, and then seeded into 24-well plates pre-coated with poly-D-lysine and laminin to facilitate adherence. To determine the effect of ZIKV infection on cell proliferation, uninfected or ZIKV-inoculated G010, K054, and K048 cells were seeded at the density of 1 million cells/flask into uncoated T-25 flasks to maintain cells in 3D free-floating spheres. Eleven days after infection, cells were dissociated and numbers counted.
Differentiation of Human Neural Stem Cells after ZIKV Infection
Dissociated hNSCs were plated into T25 flasks after Mex1-7 inoculation. After a 2-day incubation, 8 × 104 hNSCs (small spheres) were seeded into 24-well plates pre-coated with 0.01% poly-D-lysine and 1 μg/cm2 laminin (Invitrogen/Gibco) and incubated for 4 days with priming medium containing EGF (20 ng/mL), LIF (10 ng/mL), and laminin (1 μg/mL), followed by 9 days incubation with a differentiation medium consisting of N2 plus glutathione (1 μg/mL) (Sigma), biotin (0.1 μg/mL) (Sigma), superoxide dismutase (2.5 μg/mL) (Worthington), DL-α-tocopherol (1 μg/mL) (Sigma), DL-α-tocopherol acetate (1 μg/mL) (Sigma), and catalase (Sigma).
BrdU Labeling
BrdU was acquired from Sigma (catalog no. B9285-16) and prepared in a natural saline solution of 0.9% sodium chloride (Sigma). BrdU was added to the cells 7 dpi at a concentration of 10 μM for 4 hr. Following 4 hr of BrdU pulse labeling, cells were fixed with cold 4% paraformaldehyde and post fixed with cold methanol. Antigen unmasking was performed by adding 2 N hydrochloric acid to each well, then neutralizing with borate buffer. Cells were then blocked with 5% normal donkey serum and 0.3% BSA for 1 hr. Primary sheep antibody for BrdU (catalog no. ab1893) and secondary antibody donkey anti-sheep 568 (catalog no. A-21099) was used to stain and visualize BrdU.
Immunocytochemistry
Fixed cells were incubated with primary antibodies and then fluorophore-conjugated secondary antibodies according to the procedure described previously (Jordan et al., 2009). Particularly, ZIKV viral foci were detected by immunostaining using a polyclonal mouse hyperimmune ascites fluid (MHIAF) generated against ZIKV diluted 1:2,000 (Walker et al., 2015, Vasilakis et al., 2008). The MHIAF was obtained from the World Reference Collection for Emerging Viruses and Arboviruses at the University of Texas Medical Branch. Primary antibodies against MAP2 (Millipore, no. AB5622) and GFAP (Sigma, no. G9269) were used to stain for neurons and astroglia, respectively. Anti-active Cas3 antibody (Abcam, no. ab2303) was used to identify apoptotic cells. Anti-BrdU (Abcam, no. ab1893) and anti-Ki-67 antibodies (Abcam, no. ab15580) were used to determine proliferation. Nuclear marker, DAPI, was used to stain cell nuclei. Images were taken using a Nikon epifluorescent microscope.
RNA-Seq and qRT-PCR Analyses
An RNAqueous total RNA isolation kit (Thermo Fisher, no. AM1912) was used to extract total RNA from 1 day-differentiated hNSCs (or 5 days after ZIKV infection) in each of the three hNSC strains in independent triplicates, as recommended by the manufacturer. Uninfected cells served as controls. Total RNA (∼1 μg) was used for library construction for each of the three replicates of the control and infected cells. First- and second-strand synthesis, adapter ligation, and amplification of the library were performed using the Illumina TruSeq v2 RNA Sample Preparation kit under conditions prescribed by the manufacturer (Illumina). Samples were tracked using “index tags” incorporated into the adapters. Library quality was evaluated using an Agilent DNA-1000 chip on an Agilent 2100 Bioanalyzer. Quantification of library DNA templates was performed using qPCR and a reference standard of known size. Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster Kit v3 (Illumina) and the Illumina cBot workstation using conditions recommended by the manufacturer. Template input was adjusted to obtain a cluster density of 700–900 K/mm2. Paired-end 50 base sequencing by synthesis was performed using a TruSeq SBS Kit v3 (Illumina) on an Illumina HiSeq 1500 using the manufacturer’s protocols.
Reads were aligned to the human GRCh38 reference genome using STAR, an ultrafast splicing aware alignment program, version 2.5.1b (Dobin et al., 2013). Gene expression levels were quantified with the cuffquant function of the Cufflinks software (Trapnell et al., 2012), version 2.2.1, using the UCSC annotation file from the iGenomes website (Illumina). Differential expression analysis was performed with the Cufflinks function cuffdiff, using the default parameters, with the significance cutoff set to an FDR of <0.05. Hallmark pathways were identified using gene set enrichment analysis (Subramanian et al., 2005, Mootha et al., 2003), with genes that were upregulated more than 2-fold (781) or 4-fold (312), or downregulated more than 2-fold (448).
RNA-seq results were further validated by qRT-PCR on specific target genes. Duplicate CT values were analyzed in Microsoft Excel by using the comparative CT(ΔΔCT) method as described by the manufacturer (Applied Biosystems). Values of FAS, SOX1, and TUBB3 products (2−ΔΔCT) were normalized to the endogenous reference (18S rRNA) and then to a calibrator (one of the experimental samples). For details on qRT-PCR primer design and running conditions, see Supplemental Information.
Statistical Analyses
Experiments were performed in independent triplicate infections. For immunocytochemical analysis, 10–17 randomly chosen fields per sample were counted blindly from triplicate experiments by three independent investigators. Data were quantified using Student's t test or one-way ANOVA with Fisher’s least significant difference test. p < 0.05 was considered statistically significant.
Author Contributions
E.L.M., S.L.R., and J.G. contributed to this study equally. E.L.M. was involved in experimental design and execution, data analyses, and initial manuscript drafting and revising; S.L.R., in conceptual formulation, experimental design and execution, and manuscript drafting; J.G., in conceptual formulation, experimental design and execution, data analyses, and manuscript drafting. P.W., N.V., S.C.W., B.D.L., and Y.Y. designed the study, analyzed data, and drafted or edited the manuscript. S.R.A., D.J.P., and C.R. participated in experimental execution. I.F.-S. provided the original sample from which the Mexican isolate of ZIKV was made. T.J.D. and Y.X. provided technical support and discussion.
Acknowledgments
This work was supported in part by funds from the John S. Dunn Foundation (P.W.), the Brown Foundation of Houston (S.C.W. and N.V.), NIH grants R24AI120942 (N.V. and S.C.W.), R21AI129509-01 (P.W. and N.V.), and 4T32DA007287 (E.L.M.), and the Chief Research Office at the University of Texas Medical Branch (P.W.).
Published: February 16, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, two figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.01.008.
Contributor Information
Nikos Vasilakis, Email: nivasila@utmb.edu.
Ping Wu, Email: piwu@utmb.edu.
Accession Numbers
The GEO accession number for the RNA-seq data reported in this paper is GEO: GSE93385.
Supplemental Information
References
- Bayless N.L., Greenber R.S., Swigut T., Wysocka J., Blish C.A. Zika virus infection induces cranial neural crest cells to produce cytokines at levels detrimental for neurogenesis. Cell Host Microbe. 2016;20:1–6. doi: 10.1016/j.chom.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P., Pereira J.P., Raja Gabaglia C., Damasceno L., Wakimoto M., Ribeiro Nogueira R.M., Carvalho de Sequeira P., Machado Siqueira A., Abreu de Carvalho L.M., Cotrim da Cunha D. Zika virus infection in pregnant women in Rio de Janeiro- Preliminary report. N. Engl. J. Med. 2016 doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutet N., Krauer F., Riesen M., Khalakdina A., Almiron M., Aldighieri S., Espinal M., Low N., Dye C. Zika virus as a cause of neurologic disorders. N. Engl. J. Med. 2016;374:1506–1509. doi: 10.1056/NEJMp1602708. [DOI] [PubMed] [Google Scholar]
- Cao-Lormeau V.M., Blake A., Mons S., Lastere S., Roche C., Vanhomwegen J., Dub T., Baudouin L., Teissier A., Larre P. Guillain-Barre Syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchemez S., Besnard M., Bompard P., Dub T., Guillemette-Artur P., Eyrolle-Guignot D., Salje H., Van Kerkhove M.D., Abadie V., Garel C. Association between Zika virus and microcephaly in French Polynesia, 2013-15: a retrospective study. Lancet. 2016;387:2125–2132. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. (2016a). All Countries and Territories with Active Zika Virus Transmission. https://www.cdc.gov/zika/geo/active-countries.html.
- Centers for Disease Control and Prevention. (2016b). Facts About Microcephaly. https://www.cdc.gov/ncbddd/birthdefects/microcephaly.html.
- Dang J., Tiwari S.K., Lichinchi G., Qin Y., Patil V.S., Eroshkin A.M., Rana T.M. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;9:258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez P.P., Loiola E.C., Madeiro da Costa R., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., Rehen S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Guerbois M., Fernandez-Salas I., Azar S.R., Danis-Lozano R., Alpuche-Aranda C.M., Leal G., Garcia-Malo I.R., Diaz-Gonzalez E.E., Casas-Martinez M., Rossi S.L. Outbreak of Zika virus infection, Chiapas State, Mexico, 2015, and first confirmed transmission by Aedes aegypti mosquitoes in the Americas. J. Infect. Dis. 2016 doi: 10.1093/infdis/jiw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddow A.D., Schuh A.J., Yasuda C.Y., Kasper M.R., Heang V., Huy R., Guzman H., Tesh R.B., Weaver S.C. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl. Trop. Dis. 2012;6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanners N.W., Eitson J.L., Usui N., Richardson R.B., Wexler E.M., Konopka G., Schoggins J.W. Western Zika Virus in human fetal neural progenitors persists long term with partial cytopathic and limited immunogenic effects. Cell Rep. 2016;15:2315–2322. doi: 10.1016/j.celrep.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman S.W., Young N.L., Parthun M.R., Freitas M.A. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills S.L., Russell K., Hennessey M., Williams C., Oster A.M., Fischer M., Mead P. Transmission of Zika Virus through sexual contact with travelers to areas of ongoing transmission - Continental United States, 2016. MMWR Morb. Mortal. Wkly. Rep. 2016;65:215–216. doi: 10.15585/mmwr.mm6508e2. [DOI] [PubMed] [Google Scholar]
- Homem C.C., Repic M., Knoblich J.A. Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 2015;16:647–659. doi: 10.1038/nrn4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P.M., Ojeda L.D., Thonhoff J.R., Gao J., Boehning D., Yu Y., Wu P. Generation of spinal motor neurons from human fetal brain-derived neural stem cells: role of basic fibroblast growth factor. J. Neurosci. Res. 2009;87:318–332. doi: 10.1002/jnr.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindhauser M.K., Allen T., Frank V., Santhana R., Dye C. Zika: the origin and spread of a mosquito-borne virus. Bull. World Health Organ. 2016;9:675–686. doi: 10.2471/BLT.16.171082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Xu D., Ye Q., Hong S., Jiang Y., Liu X., Zhang N., Shi L., Qin C.F., Xu Z. Zika Virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;5:672. doi: 10.1016/j.stem.2016.10.017. [DOI] [PubMed] [Google Scholar]
- Miner J.J., Diamond M.S. Understanding how Zika Virus enters and infects neural target cells. Cell Stem Cell. 2016;18:559–560. doi: 10.1016/j.stem.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Miotto B., Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell. 2010;1:57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Paploski I.A., Prates A.P., Cardoso C.W., Kikuti M., Silva M.M., Waller L.A., Reis M.G., Kitron U., Ribeiro G.S. Time lags between exanthematous illness attributed to Zika Virus, Guillain-Barre Syndrome, and microcephaly, Salvador, Brazil. Emerg. Infect. Dis. 2016;22:1438–1444. doi: 10.3201/eid2208.160496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S.L., Tesh R.B., Azar S.R., Muruato A.E., Hanley K.A., Auguste A.J., Langsjoen R.M., Paessler S., Vasilakis N., Weaver S.C. Characterization of a novel murine model to study Zika Virus. Am. J. Trop. Med. Hyg. 2016;94:1362–1369. doi: 10.4269/ajtmh.16-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D., McMillan E., Ebert A.D., Shelley B.C., Johnson J.A., Meisner L.F., Svendsen C.N. Chromosome 7 and 19 trisomy in cultured human neural progenitor cells. PLoS One. 2009;4:e7630. doi: 10.1371/journal.pone.0007630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin Y., Loustalot F., Desmtz C., Foulongne V., Constant O., Fournier-Wirth C., Leon F., Moles J.P., Goubaud A., Lemaitre J.M. Zika Virus strains potentially display different infectious profiles in human neural stem cells. EBioMedicine. 2016;12:161–169. doi: 10.1016/j.ebiom.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles J., Jernigan T.L. The basics of brain development. Neuropsychol. Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M. Zika Virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasenko Y.I., Yu Y., Jordan P.M., Bottenstein J., Wu P. Effect of growth factors on proliferation and phenotypic differentiation of human fetal neural stem cells. J. Neurosci. Res. 2004;78:625–636. doi: 10.1002/jnr.20316. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Fokam E.B., Hanson C.T., Weinberg E., Sall A.A., Whitehead S.S., Hanley K.A., Weaver S.C. Genetic and phenotypic characterization of sylvatic dengue virus type 2 strains. Virology. 2008;377:296–307. doi: 10.1016/j.virol.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P.J., Widen S.G., Firth C., Blasdell K.R., Wood T.G., Travassos da Rosa A.P., Guzman H., Tesh R.B., Vasilakis N. Genomic characterization of Yogue, Kasokero, Issyk-Kul, Keterah, Gossas, and Thiafora viruses: nairoviruses naturally infecting bats, shrews, and ticks. Am. J. Trop. Med. Hyg. 2015;93:1041–1051. doi: 10.4269/ajtmh.15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S.C., Costa F., Garcia-Blanco M.A., Ko A.I., Ribeiro G.S., Saade G., Shi P.Y., Vasilakis N. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Tarasenko Y.I., Gu Y., Huang L.Y., Coggeshall R.E., Yu Y. Region-specific generation of cholinergic neurons from fetal human neural stem cells grafted in adult rat. Nat. Neurosci. 2002;5:1271–1278. doi: 10.1038/nn974. [DOI] [PubMed] [Google Scholar]
- Wu K.Y., Zuo G.L., Li X.F., Ye Q., Deng Y.Q., Huang X.Y., Cao W.C., Qin C.F., Luo Z.G. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res. 2016;6:645–654. doi: 10.1038/cr.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Feng H., Kato H., Dai L., Yang Y., Zhou Y., Bai Y. Structural insights into the histone H1-nucleosome complex. PNAS. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.