ABSTRACT
To replicate efficiently, viruses must create favorable cell conditions and overcome cell antiviral responses. We previously reported that the reovirus protein μ2 from strain T1L, but not strain T3D, represses one antiviral response: alpha/beta interferon signaling. We report here that T1L, but not T3D, μ2 localizes to nuclear speckles, where it forms a complex with the mRNA splicing factor SRSF2 and alters its subnuclear localization. Reovirus replicates in cytoplasmic viral factories, and there is no evidence that reovirus genomic or messenger RNAs are spliced, suggesting that T1L μ2 might target splicing of cell RNAs. Indeed, RNA sequencing revealed that reovirus T1L, but not T3D, infection alters the splicing of transcripts for host genes involved in mRNA posttranscriptional modifications. Moreover, depletion of SRSF2 enhanced reovirus replication and cytopathic effect, suggesting that T1L μ2 modulation of splicing benefits the virus. This provides the first report of viral antagonism of the splicing factor SRSF2 and identifies the viral protein that determines strain-specific differences in cell RNA splicing.
IMPORTANCE Efficient viral replication requires that the virus create favorable cell conditions. Many viruses accomplish this by repressing specific antiviral responses. We demonstrate here that some mammalian reoviruses, RNA viruses that replicate strictly in the cytoplasm, express a protein variant that localizes to nuclear speckles, where it targets a cell mRNA splicing factor. Infection with a reovirus strain that targets this splicing factor alters splicing of cell mRNAs involved in the maturation of many other cell mRNAs. Depletion of this cell splicing factor enhances reovirus replication and cytopathic effect. Our results provide the first evidence of viral antagonism of this splicing factor and suggest that downstream consequences to the cell are global and benefit the virus.
KEYWORDS: RNA processing, SRSF, reovirus, splicing
INTRODUCTION
Viruses are obligatory intracellular pathogens that require a hospitable cell environment for their replication. However, viral infection induces cell innate responses that are antiviral. Accordingly, viruses have evolved numerous strategies to evade these responses. For example, viral infection of virtually any cell type induces a protective type I interferon (IFN-α/β) response, and many viruses use at least one mechanism to suppress this antiviral system (1, 2). Viral proteins that inhibit cell antiviral responses have been identified for most viruses, as have the cell proteins they target (1, 3). Not surprisingly, the most commonly identified targets are cell proteins involved in the IFN-α/β response (1, 2).
Mammalian reoviruses are double-stranded RNA nonenveloped viruses that replicate in membrane-associated cytoplasmic viral factories (VFs) (4, 5). Reovirus strain type 1 Lang (T1L) represses IFN-β signaling, while type 3 Dearing (T3D) does not (6), and this difference is determined by the reovirus μ2 protein (6, 7) encoded by the M1 gene. μ2 is a minor capsid protein but is expressed abundantly in infected cells and has RNA binding (8) and NTPase activities (9, 10). It also determines virus strain-specific differences in the morphology of VFs through its capacity to bind and stabilize microtubules (10, 11). Strain-specific differences in repression of IFN-β signaling and stabilization of microtubules are determined by a polymorphism in amino acid 208 of the μ2 protein (6, 7, 11). This may reflect the impact of amino acid 208 on μ2 stability (12) or on μ2 function. During infection, μ2 localizes predominantly to VFs (10–13) but can also be visualized diffusely in the cytoplasm and nucleus (10, 11, 14). However, the role of μ2 in the nucleus remains to be elucidated.
Here, we show that μ2 from T1L and reovirus recombinants that encode the T1L μ2 amino acid polymorphism form a complex with the pre-mRNA splicing factor SRSF2 (previously known as SC35 [15]) in nuclear speckles. Moreover, T1L but not T3D alters the splicing of transcripts for genes involved in RNA processing and maturation. Finally, depletion of SRSF2 enhances reovirus replication and cytopathic effect, suggesting that T1L μ2 modulation of splicing benefits the virus. We provide here the first report of viral antagonism of the splicing factor SRSF2 and suggest that the consequences to the cell are global.
RESULTS
μ2 undergoes constant nuclear shuttling, but the predominant intracellular localization is strain specific.
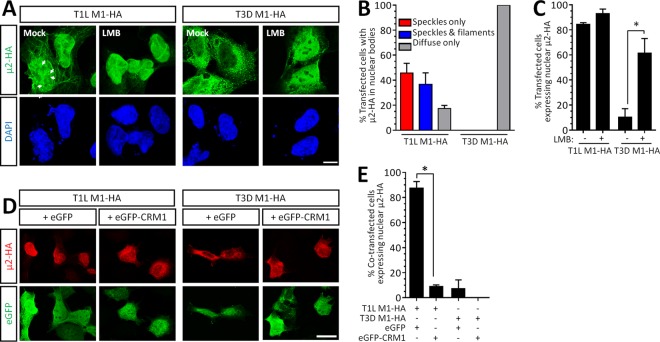
Reovirus replication is exclusively cytoplasmic (4), and yet the reovirus protein μ2 can translocate to the nucleus (10, 11, 14). Given that the capacity for reovirus μ2 to repress IFN-β signaling is virus strain specific (6, 7), we probed μ2 intracellular localization further. Notably, we found that the predominant intracellular localization of μ2 is also strain specific. T1L-μ2-HA localized primarily to the nucleus, whereas T3D-μ2-HA localized to the cytoplasm (Fig. 1A). Interestingly, nuclear T1L μ2 localized to intranuclear filamentous structures reminiscent of microtubules and also to discrete aggregates (Fig. 1A and B). T1L μ2 nuclear localization is dependent on the nuclear export signal receptor protein CRM1/exportin 1 (10). To determine whether T3D μ2 has the ability to transiently shuttle to the nucleus and associate with similar subnuclear structures, we inhibited CRM1 with the irreversible inhibitor leptomycin B (LMB) (16) for 5 h prior to immunostaining (Fig. 1A and C). Treatment with LMB revealed that T3D μ2 does indeed shuttle into the nucleus, but the predominant localization is cytoplasmic under normal nuclear protein export conditions. To further probe nuclear-cytoplasmic shuttling, eGFP-CRM1 or an eGFP empty vector control was coexpressed with μ2 (Fig. 1D and E). As expected, T1L μ2 was found mainly in the nucleus when coexpressed with eGFP but was found in the cytoplasm in cells overexpressing CRM1. Together, the results indicate that μ2 undergoes constitutive CRM1-dependent nuclear-cytoplasmic shuttling, but the predominant localization is reovirus strain specific. Interestingly, even when T3D μ2 accumulated at high levels in the nucleus upon CRM1 inhibition (Fig. 1A), it never associated with subnuclear structures, as seen for T1L μ2. T1L μ2 is known to bind and stabilize microtubules in the cytoplasm (11), but association with subnuclear aggregates has not been previously described. Although the role of intranuclear bodies in the life cycle of several viruses is of increasing interest (17), a role for μ2 in the nucleus has not been previously identified.
FIG 1.
μ2 undergoes constant CRM1-dependent nuclear shuttling, but the predominant intracellular localization is strain specific. (A) AD-293 cells were transfected with reovirus T1L- or T3D-M1-HA for 20 h and treated with medium alone (mock) or the CRM1 inhibitor LMB for 5 h. Arrows depict μ2 localization to intranuclear bodies. Scale bar, 20 μm. (B) The percentage of cells in panel A displaying the indicated nuclear bodies is presented for four independent experiments (means ± the SEM; n = 6 to 61 cells per condition). (C) The percentage of cells in panel A displaying nuclear μ2 is presented for two to four independent experiments (means ± the SEM; n = 46 to 197 cells per condition). (D) AD-293 cells were cotransfected with reovirus T1L- or T3D-M1-HA and either an eGFP or eGFP-CRM1 plasmid. Scale bar, 10 μm. (E) The percentage of cotransfected cells in panel D displaying nuclear μ2 is presented for two independent experiments (means ± the SEM; n = 18 to 71 cells per condition). *, Significantly different (P < 0.05).
Intranuclear μ2 localizes to nuclear speckles.
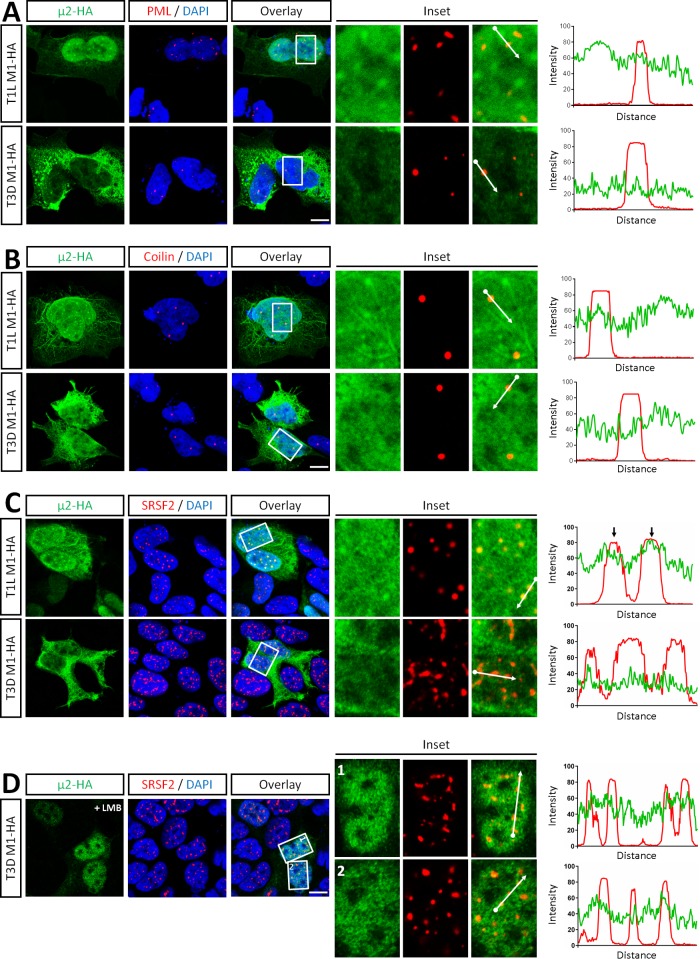
To determine the nature of the intranuclear T1L μ2 structures, AD-293 cells were transfected with μ2-HA and immunostained for markers of candidate nuclear bodies. Although viruses are known to modulate promyelocytic leukemia (PML) body function, particularly relating to the IFN system (reviewed in references 18 and 19), T1L μ2 did not colocalize with PML bodies (Fig. 2A). Similarly, μ2 did not colocalize with coilin (Fig. 2B), a marker of Cajal bodies (20). T1L μ2, however, colocalized with the serine/arginine-rich splicing factor 2 (SRSF2; previously known as SC35 [15]), a marker of nuclear speckles (Fig. 2C). In metazoans, nuclear speckles are enriched in mature snRNPs, non-snRNP pre-mRNA splicing factors, and polyadenylated RNA (21–23). Association of μ2 with nuclear speckles was specific to T1L μ2, since T3D μ2 failed to associate with SRSF2 even after treatment with LMB to induce its nuclear localization (Fig. 2D).
FIG 2.
Intranuclear T1L μ2 specifically localizes to nuclear speckles. AD-293 cells were transfected with reovirus T1L- or T3D-M1-HA for 20 h, fixed, and immunostained with antibodies against HA and markers of PML bodies (PML) (A), Cajal bodies (coilin) (B), or nuclear speckles (SRSF2) (C). (D) AD-293 cells were transfected with T3D M1-HA for 18 h, treated with LMB for 5 h, and immunostained as in panel C. Nuclei were counterstained with DAPI. Histograms display measured fluorescence intensity along the drawn line in the overlay inset panels. Scale bar, 10 μm.
Amino acid 208 in μ2 determines μ2 localization to nuclear speckles during infection.
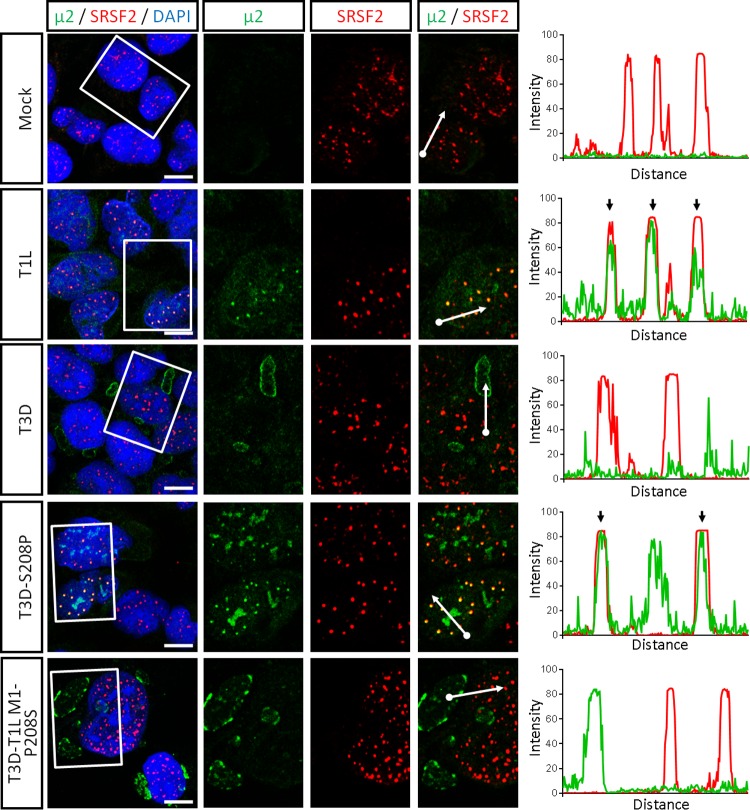
Amino acid 208 in the μ2 protein determines several strain-specific differences in μ2 function (7, 11, 12). To determine whether this amino acid is also a determinant of μ2 localization to nuclear speckles, AD-293 cells were infected with either the parental reovirus strains T1L or T3D or recombinant mutant viruses and then immunostained (Fig. 3). As expected (11), μ2 localized to cytoplasmic viral factories that displayed either a filamentous or globular morphology during T1L or T3D infection, respectively. In addition, T1L but not T3D μ2 colocalized with SRSF2 in nuclear speckles (in 61.5% ± 3.3% versus 0.0% of infected cells, respectively), a finding consistent with our results using plasmid-derived μ2 (Fig. 2). Remarkably, substitution of T3D μ2 amino acid 208 with that of T1L (T3D-S208P) induced μ2 localization to nuclear speckles (in 50.9% ± 2.7% of infected cells), whereas this localization was lost in a T3D virus expressing T1L μ2 with amino acid 208 reverted to that of T3D (T3D T1L-M1 P208S; 0.0% of infected cells). Together, results demonstrate that strain-specific differences in μ2 amino acid 208 are both required and sufficient for μ2 localization to nuclear speckles during reovirus infection.
FIG 3.
A single amino acid in μ2 determines μ2 localization to nuclear speckles during reovirus infection. AD-293 cells were infected with the indicated viruses for 20 h, fixed, and immunostained for μ2 and SRSF2. Nuclei were counterstained with DAPI. Histograms display measured fluorescence intensity along the drawn line in the overlay inset panels. The results are representative of at least two independent experiments. Scale bar, 10 μm.
μ2 alters the localization of SRSF2 in a microtubule-dependent manner.
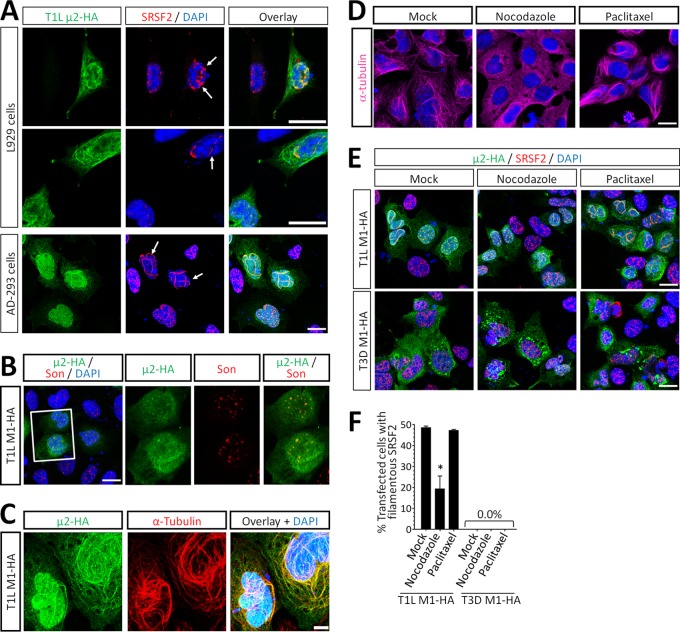
Interestingly, ectopic expression of T1L μ2 resulted in the mislocalization of SRSF2 to filamentous structures in approximately half of transfected L929 or AD293 cells (Fig. 4A, E, and F). This effect was specific for SRSF2, since T1L μ2 did not affect localization of the nuclear speckle matrix protein Son (Fig. 4B). T1L μ2 binds to and stabilizes cytoplasmic microtubules (11) and, not unexpectedly, T1L μ2 colocalized with nuclear alpha-tubulin in nuclear filaments (Fig. 4C). To determine whether T1L μ2-induced changes in SRSF2 localization were a consequence of μ2 association with nuclear microtubules, cells were treated with nocodazole and paclitaxel to inhibit polymerization of or to stabilize microtubules, respectively (24), and the cytoskeletal effects of these drugs were confirmed (Fig. 4D). Depolymerization of microtubules with nocodazole reduced the fraction of T1L μ2-expressing cells that displayed mislocalized SRSF2, demonstrating that this TL μ2 effect requires microtubules (Fig. 4E and F). However, stabilization of microtubules with paclitaxel did not alter SRSF2 localization in T3D μ2-expressing cells demonstrating that stabilized microtubules are not sufficient to support μ2 effects on SRSF2 localization (Fig. 4E and F). Together, results demonstrate that T1L μ2, but not T3D μ2, alters the localization of SRSF2 to filamentous nuclear structures, likely by interactions with both microtubules and SRSF2.
FIG 4.
T1L μ2 alters the localization of SRSF2 by a microtubule-dependent mechanism. (A) L929 and AD-293 cells were transfected with T1L M1-HA for 20 h, fixed, and immunostained using antibodies against HA and SRSF2. Nuclei were counterstained with DAPI. (B) AD-293 cells were transfected as in panel A and immunostained using antibodies against HA and Son. (C) Three-dimensional reconstruction of z-stacks were generated from T1L M1-HA-transfected AD-293 cells immunostained with antibodies against HA and alpha-tubulin. (D) AD-293 cells were treated with either media alone (Mock), 10 μM nocodazole, or 10 μM paclitaxel for 6 h. The morphology of microtubules was visualized by immunostaining against α-tubulin. (E) AD-293 cells were transfected with T1L M1-HA for 20 h and treated with either media alone (Mock), 10 μM nocodazole, or 10 μM paclitaxel for 6 h. The cells were immunostained as in panel A. (F) Quantitation of panel E. Results are means ± the SEM (n = 141 to 489 cells per condition) and are representative of at least two independent experiments. All scale bars, 10 μm.
μ2 forms a complex with the pre-mRNA splicing factor SRSF2.
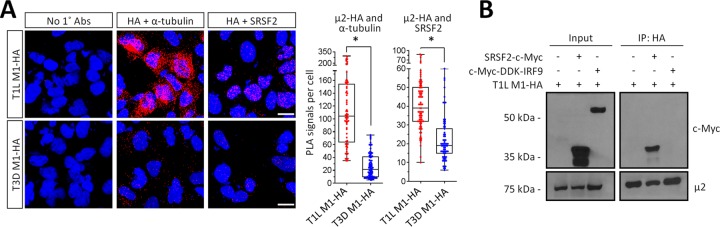
To determine whether T1L μ2 interacts with SRSF2, we first expressed T1L- and T3D-μ2-HA in AD-293 cells and performed an in situ proximity ligation assay (PLA [25]) (Fig. 5A). Antibody against tubulin was used as a control for the approach and, as expected, T1L μ2-HA generated much greater PLA signals than T3D μ2-HA did (Kruskal-Wallis nonparametric test; P < 0.001). Importantly, T1L μ2-HA also generated much greater PLA signals than T3D μ2-HA did with endogenous SRSF2 in the nucleus (Kruskal-Wallis nonparametric test; P < 0.001). Consistent with the lack of μ2-mediated changes in Son localization (Fig. 4B), PLA experiments demonstrated that T1L μ2-HA does not interact with Son (data not shown). Next, we confirmed formation of a complex between μ2 and SRSF2 biochemically by coimmunoprecipitation of cell lysates. AD-293 cells were cotransfected with T1L-M1-HA and myc-tagged SRSF2 (because endogenous SRSF2 is not expressed at sufficient levels) or myc-tagged IRF9 as a negative control (Fig. 5B). Myc-tagged SRSF2, but not myc-tagged IRF9, coimmunoprecipitated with T1L μ2-HA. Together, results demonstrate that T1L-μ2 but not T3D-μ2 forms a complex with both microtubules and SRSF2 in the nucleus.
FIG 5.
T1L μ2 forms a complex with SRSF2 in the nucleus. (A) AD-293 cells were transfected with T1L M1-HA for 20 h, fixed, and subjected to a proximity ligation assay (PLA) using the indicated primary antibodies. Cells were imaged by confocal microscopy to obtain z-stack images of PLA signals (red) and nuclei (blue) representative of the total volume of the cell in the z axis. Scale bar, 10 μm. Quantitation of PLA results expressed as number of PLA dots per cell (n = 66 to 82 cells per condition) for a representative of at least three independent experiments. *, Significantly different from T3D μ2-HA (P < 0.001). (B) AD-293 cells were transfected with the indicated plasmids, whole-cell lysates were immunoprecipitated using anti-HA-conjugated agarose beads, and lysates (input) and immunoprecipitated extracts (IP) were resolved by SDS-PAGE for immunoblotting. The results are representative of at least two independent experiments. It is unclear why overexpressed SRSF2 appears as two distinct bands, but the upper band corresponds to SRSF2 migration in specifications provided with the antibody.
μ2 nuclear localization requires SRSF2 expression.
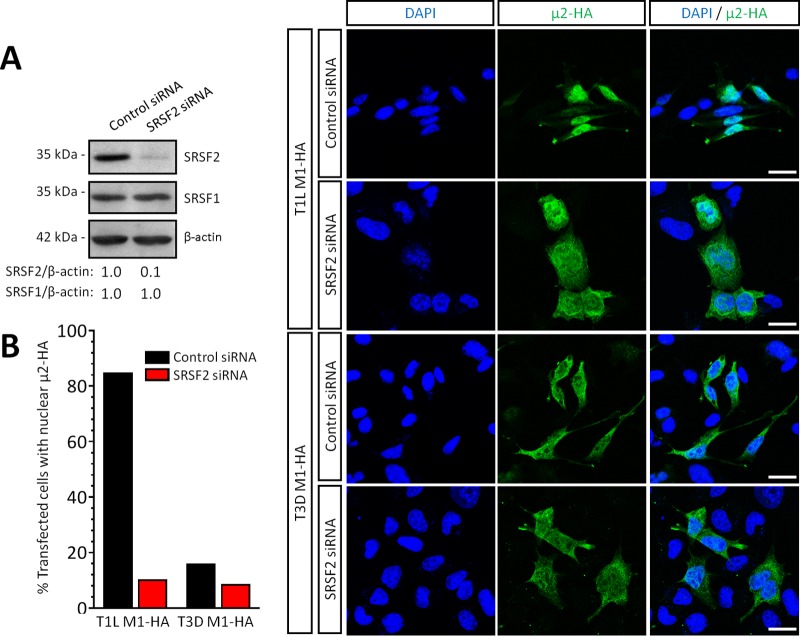
Given the impact of T1L μ2 on SRSF2 localization, we next sought to determine whether SRSF2 affects μ2 localization. SRSF2 was depleted by small interfering RNA (siRNA) in L929 cells (Fig. 6A), T1L- or T3D-M1-HA were transfected, and μ2 localization was assessed by immunofluorescence (Fig. 6B). As expected, in control siRNA-transfected cells, T1L μ2 and T3D μ2 localized primarily to the nucleus and the cytoplasm, respectively. However, T1L μ2 localization became primarily cytoplasmic in cells with reduced SRSF2 expression, mimicking T3D μ2. Thus, SRSF2 function is required for T1L μ2 nuclear localization where, given the exclusively nuclear location of SRSF2 (26), μ2 likely subsequently forms a complex with SRSF2.
FIG 6.
SRSF2 is required for μ2 nuclear localization. L929 cells were transfected with the indicated siRNA for 72 h and transfected with the indicated expression plasmids for 18 h. (A) Cells were lysed, and protein fractions were resolved by SDS-PAGE for transfer and immunoblotting with the indicated antibodies. SRSF1 (negative control) or SRSF2 band intensities are expressed relative to control siRNA-transfected cells. (B) The cells were fixed and immunostained using antibodies against HA and SRSF2. Nuclei were counterstained with DAPI. Quantitated results (n = 13 to 51 cells per condition) are representative of two independent experiments. Scale bar, 20 μm.
Depletion of SRSF2 increases reovirus replication and cytopathic effect and impact is strain specific.
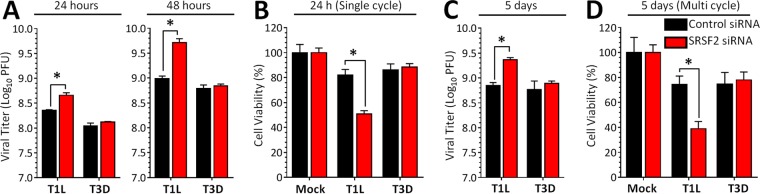
The interaction of T1L μ2 with SRSF2 suggested the possibility that reovirus modulates SRSF2 function to generate a more favorable cell environment. To determine whether SRSF2 affects viral replication and cytopathic effect, L929 cells were depleted of SRSF2 and then infected with T1L or T3D. SRSF2 depletion increased T1L but not T3D replication and cytopathic effect in both primary (Fig. 7A and B) and secondary infections (Fig. 7C and D). The absence of an effect of SRSF2 depletion on T3D infection suggests that SRSF2-independent events are the predominant determinants of T3D replication in L929 cells. The increased T1L replication and cytopathic effect suggest that T1L μ2 does not inhibit SRSF2 activity entirely and that further depletion of SRSF2 is beneficial to the virus.
FIG 7.
SRSF2 effects on reovirus replication and cytopathic effect are strain-specific. (A) L929 cells were transfected with the indicated siRNA for 72 h and infected with reovirus at an MOI of 5 PFU per cell. The viral titer was determined by plaque assay at 24 or 48 h postinfection (single cycle infection). The results are means of four replicate samples ± the standard deviations (SD). (B) L929 cells were transfected as in panel A and infected with the indicated virus at an MOI of 100 PFU per cell. Cell viability was measured using an MTT assay at 24 h postinfection. The results correspond to four replicate wells per infection and are expressed as means ± the SD relative to mock-infected cultures for the corresponding siRNA. (C) Same as for panel A, but the cells were infected at an MOI of 0.1 and harvested at 5 days postinfection. (D) Same as for panel B, but the cells were infected at an MOI of 0.1 and assessed at 5 days postinfection (multiple cycle infection). All results in all panels are representative of at least two independent experiments. *, Significantly different from the “control siRNA + T1L infected” group (P < 0.05).
Reovirus T1L alters cellular mRNA splicing.
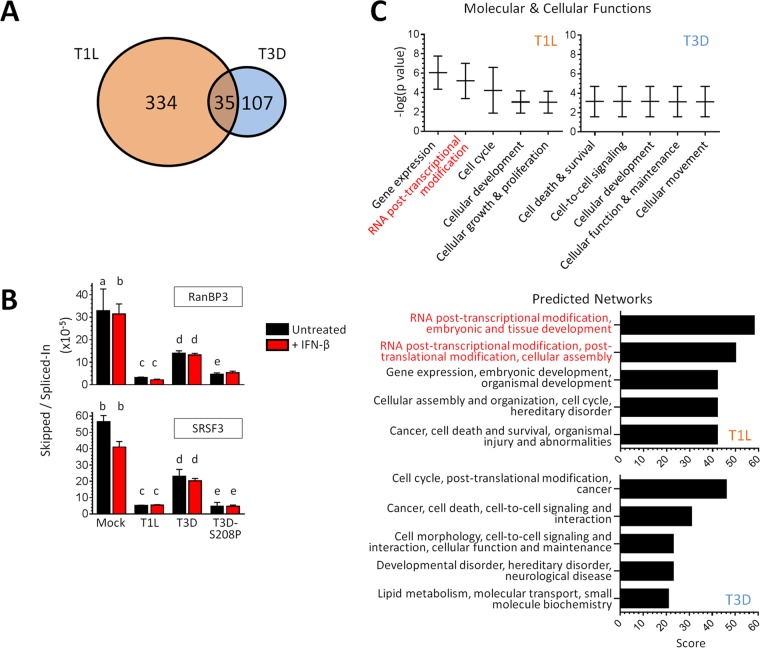
SRSF2 is a pre-mRNA splicing factor, but reovirus replication is exclusively cytoplasmic, and there is no evidence that any reovirus RNAs are spliced (4). We therefore used RNA-seq to determined whether reovirus T1L but not T3D alters cellular mRNA splicing. Because T3D induces significantly more IFN-β than T1L does (6, 7, 27) and because IFN-β alone alters mRNA splicing of some IFN-stimulated genes (28), we compared mock-infected, untreated cells to mock-infected or reovirus-infected cells stimulated with IFN-β. This identifies differences in T1L and T3D effects on splicing separate from differences in their induction of IFN-β and subsequent IFN-β effects on splicing. The results from RNA sequencing (RNA-seq) were analyzed by the mixture-of-isoforms (MISO) statistical model (29) to assess changes in mRNA alternative splicing (Fig. 8A; see also Tables S1 to S3 in the supplemental material). IFN-β stimulation generated 42 novel splicing variants in 41 different genes relative to untreated cells. Excluding those IFN-β-induced variants, T3D infection generated 142 novel splicing variants while, remarkably, T1L generated 369. Only 35 of these variants were found to overlap between the two reovirus strains, resulting in a total of 334 and 107 unique splicing events induced in 297 and 97 genes by T1L and T3D, respectively. Splicing events that were induced only in T1L were confirmed by quantitative reverse transcription-PCR (qRT-PCR) for two cases (RanBP3 and SRSF3) identified by MISO analysis (Fig. 8B). Whereas T3D also altered RanBP3 and SRSF3 splicing to some extent (2.4- and 2.5-fold, relative to the Mock group), T1L altered their splicing much more dramatically (10.4- and 10.8-fold, respectively). Moreover, these qRT-PCR results confirmed that IFN-β has minimal or no impact on viral effects on splicing and that the same μ2 amino acid that determines μ2 association with SRSF2 (Fig. 3) determines reovirus effects on splicing (S208P altered RanBP3 and SRSF3 splicing 7.0- and 11.5-fold, respectively, Fig. 8B). Thus, while both reovirus strains induce unique splicing variants, T1L induces more than three times more than T3D does (RNA-seq results: chi square, P < 0.001), and a single μ2 amino acid can determine reovirus effects on splicing.
FIG 8.
Reovirus infection alters cellular mRNA splicing. (A) Venn diagram indicating differences in splicing based on MISO analysis of RNA-seq results. (B) Confirmation of differential expression of novel splicing variants using qRT-PCR. L929 cells were infected and stimulated with IFN-β using conditions identical to those used for RNA-seq. Transcripts with a specific exon skipped or spliced-in (as identified by RNA-seq) were quantitated by qRT-PCR for two representative genes. The results are expressed as a ratio of the two splicing events to capture the events comprising MISO analysis. The results of statistical analyses (P < 0.05), indicated by lowercase letters, were as follows: a, mock different from T1L and S208P; b, mock or mock+IFN different from all viruses or viruses+IFN; c, T1L or T1L+IFN different from T3D or T3D+IFN; d, T3D or T3D+IFN different from S208P or S208P+IFN; and e, S208P or S208P+IFN not different from T1L or T1L+IFN. (C) IPA results for genes whose splicing was altered uniquely by either T1L (297 genes) or T3D (97 genes) and IFN-β stimulation, excluding genes whose splicing was altered by IFN-β stimulation alone.
To gain further insights into reovirus-induced global changes in host mRNA splicing, we used Ingenuity pathway analysis (IPA) and gene ontology (GO) analysis. Specifically, we performed IPA on the 297 and 97 genes whose splicing was altered uniquely by T1L or by T3D, respectively. The pathways most affected by T1L involved RNA posttranscriptional modifications (Fig. 8C). Indeed, T1L (but not T3D) altered the spicing of SRSF3, SRSF6, SRSF7, and SRSF11 (see Table S3 in the supplemental material), all of which have specialized roles in posttranscriptional regulation (30). Moreover, the top five predicted upstream regulators for altered splicing events induced by T1L (but not T3D) included SRSF1 (data not shown), another SR family member (31). Importantly, the minimal changes in splicing that occurred during T3D did not cluster in any of these categories, and there was no overlap with those obtained for T1L. Lastly, we used the network of genes affected by T1L with the highest statistical significance to create a web of known and predicted interactions. One-third of the genes in the network related to mRNA splicing or mRNA processing (data not shown). These IPA results were supported by GO analysis of the same data sets. Specifically, the top two processes for T1L were “mRNA processing” and “ribonucleoprotein complex assembly,” whereas those were 47th and 8th in priority for T3D, respectively (T1L different from T3D at P < 0.001 and P < 0.05, respectively).
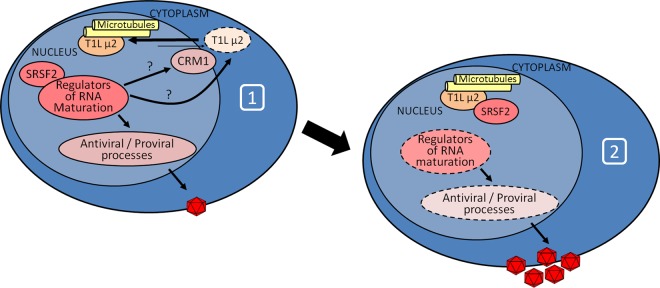
In sum, results suggest that SRSF2 is required for T1L μ2 nuclear localization whereupon T1L μ2 forms a complex with SRSF2 and antagonizes the organization of nuclear speckles, resulting in dysregulated splicing of other splicing factors and regulators of mRNA at the posttranscriptional level to benefit viral replication (Fig. 9).
FIG 9.
Model for T1L μ2 interactions with SRSF2 and impact on cell RNA splicing. Upon viral infection (diagram 1), cell splicing protein SRSF2 modulates T1L μ2 (possibly through effects on posttranslational modification enzymes) or nuclear protein transport effectors, resulting in translocation of T1L μ2 to the nucleus. Once in the nucleus (diagram 2), T1L μ2 forms a complex with SRSF2 and alters it function to modulate the cell response.
DISCUSSION
To replicate efficiently, viruses must create favorable cell conditions and overcome cell antiviral responses. Here, we report that the μ2 protein from strain T1L localizes to nuclear speckles, where it forms a complex with and alters the localization of the pre-mRNA splicing factor SRSF2. Moreover, infection with reovirus T1L alters mRNA splicing of genes involved in mRNA processing and maturation, likely as a consequence of T1L μ2 antagonistic effects on SRSF2. Finally, depletion of SRSF2 enhances reovirus replication and cytopathic effect, suggesting that T1L μ2 modulation of splicing benefits the virus. This provides the first report of viral antagonism of the splicing factor SRSF2 and suggests that by altering mRNA processing and maturation, the virus induces cell consequences that are global rather than in a single pathway.
Reovirus μ2 stability, repression of IFN-β signaling, and stabilization of microtubules are all determined by a single amino acid polymorphism in the μ2 protein (6, 7, 11). Here, we demonstrate that this amino acid similarly determines μ2 localization to nuclear speckles (Fig. 3). The common dependence on a single amino acid could reflect its impact on μ2 stability or its impact on function. The N terminus of μ2 includes a polybasic region that can function as a nuclear localization signal (10) and, interestingly, domains that are rich in basic amino acids have been identified as nuclear speckle-targeting signals (32). However, this μ2 polybasic region is conserved between T1L and T3D strains, indicating that the primary protein sequence alone does not predict its localization and that other factors contribute to the differential localization of μ2. This complexity in determinants of μ2 localization to intracellular compartments is highlighted by the observation that SRSF2 is required for μ2 nuclear localization (Fig. 6) and that T3D μ2 does not associate with nuclear speckles even when concentrated in the nucleus after inhibition of nuclear export (Fig. 2D). The requirement for SRSF2 could reflect trapping of μ2 in a complex, an impact on nuclear import or export processes, or an impact on cell proteins that affect μ2 itself (Fig. 9). Posttranslational modifications of μ2, including acetylation (33) and phosphorylation (34), have been reported, and yet the impact(s) of these modifications on μ2 function has not been fully elucidated. Future studies will address the roles of SRSF2 and posttranslational modifications on μ2 subcellular localization.
Multiple viruses regulate the cell splicing machinery to generate alternative viral RNA products. Several RNA viruses (35–38) and DNA viruses (39) use extensive alternative splicing of their mRNAs. The influenza A virus NS1 protein disorganizes nuclear speckles and Cajal bodies and binds members of the snRNP family of splicing factors and proteins required for 3′-terminal polyadenylation of host transcripts to facilitate splicing of viral RNAs (35). Retroviruses regulate cell splicing machinery to balance generation of viral mRNAs with maintenance of unspliced RNA for production of viral progeny (40, 41). There is no evidence for splicing of viral RNA for the cytoplasmically replicating reovirus (4). Reovirus RNAs also lack modifications at their 3′ termini (4); another RNA modification function associated with nuclear speckles (42–44). Finally, we did not detect any redistribution of splicing factors to cytoplasmic VFs during reovirus infection (data not shown). Thus, there is no evidence that reovirus modulation of SRSF2 affects viral RNAs. Instead, reovirus likely modulates the cell transcriptome to its benefit.
Alternative splicing of cell mRNAs can affect cell function and disease (45–47), and several viruses modulate cell splicing machinery to alter the cell transcriptome. The adenovirus E4-ORF4 antagonizes the function of SRSF1 and SRSF9 by promoting their dephosphorylation (48, 49). The herpes simplex virus ICP27 protein binds the SR protein kinase 1 (SRPK1), resulting in its relocalization from the cytoplasm to the nucleus to dephosphorylate SR proteins and regulate host mRNA splicing events (50–52). The vaccinia virus VH1 phosphatase appears to directly dephosphorylate SR proteins (53). Epstein-Barr virus (EBV) itself encodes a splicing factor, SM, that influences splicing and processing of host pre-mRNAs (54, 55). Splice variants, including those that encode dominant negative proteins, have been described for several genes involved in the IFN-α/β response (56–63). Notably, the EBV SM protein skews the mRNA splicing patterns of STAT1, enhancing production of the dominant-negative STAT1β isoform (55). Very recently, it has been shown that the NS5 protein of dengue virus binds to and interferes with components of the U5 snRNP particle and alters mRNA splicing of several antiviral factors (64). In contrast to our results for reovirus, dengue NS5 does not appear to affect the intranuclear localization of its splicing targets (64). Finally, in a recent report reovirus T3D was shown to alter cell mRNA splicing, but the impact on viral replication and the underlying mechanism were not investigated (65). It is interesting that a single reovirus protein, μ2, can modulate both IFN signaling (7) and SRSF2 function to benefit the virus. The results suggest that this protein has evolved to target both a specific antiviral pathway and mRNA maturation for more global effects.
Several SR proteins play roles in addition to those in splicing that affect mature mRNA expression and nuclear export (30). For example, SRSF1 participates in the organization of nuclear speckles and in the promotion of decay of aberrantly spliced transcripts (31, 66, 67), whereas SRSF2 influences transcriptional elongation of some mRNAs (68). With the exception of SRSF2, mammalian SR proteins constitutively shuttle between the nucleus and cytoplasm (26). Once in the cytoplasm, SRSF1 impacts translation by modulating mRNA entrance into polyribosomes (69, 70). Lastly and perhaps most intriguingly, SRSF1 has been associated with RIG-I and RNA polymerase III-dependent sensing of transfected non-self cytosolic DNA to facilitate IFN-β production (71); however, its role during viral infection remains unclear. Similarly, the spliceosomal protein SNRNP200 can also translocate to the cytoplasm, where it modulates the viral induction of IFN-β (72). Although T1L did not affect splicing of SRSF1 or SNRNP200 mRNA, the loss of SRSF2 can affect SRSF1 function (73), and the regulation of snRNPs is complex, leaving open the possibility that T1L alters the cytoplasmic function of spliceosomal proteins to benefit the virus.
T1L infection alters SRSF3 splicing (see Table S3 in the supplemental material), suggesting another possible mechanism by which T1L μ2 interactions with SRSF2 could impact cell RNA function. SRSF3 (previously known as SRp20 [15]) is involved in 5′ cap-independent internal ribosome entry site (IRES)-mediated translation of picornavirus transcripts (74, 75). The 2A proteinase of several picornaviruses induce a change in localization of SRSF3 from the nucleus to the cytoplasm and stress granules (76), and depletion of SRSF3 impacts the efficiency of IRES-mediated translation (74). Reovirus transcripts are 5′ capped and undergo 5′-cap-dependent translation in ribosomes within membrane-associated VFs (4, 5, 77). Thus, it is unlikely that reovirus modulation of SRSF3 splicing is related to IRES-dependent translation. However, T1L infection induces novel splice variants of SRSF3, SRSF6, SRSF7, and SRSF11 (see Table S3 in the supplemental material), all of which have specialized roles in posttranscriptional regulation (30). Thus, reovirus modulation of the cell response may be indirectly mediated through a dysregulation of mRNA nuclear export, processing, nonsense-mediated decay, and/or efficient translation through the targeting of SRSF2 and changes in the isoforms of other SR proteins.
Lastly, reovirus μ2 repression of the IFN-β response is a determinant of reovirus induction of myocarditis (7, 27), and the same μ2 amino acid polymorphism that determines this repression and severity of myocarditis (7) determines μ2 association with SRSF2 (Fig. 3). Together, these results suggest that SRSF2 could participate in protecting the heart against viral myocarditis. While this could reflect SRSF2 effects on the IFN response, it has been shown that mutations and/or decreased expression of splicing factors results in cardiac development defects (45), and cardiac tissue-specific ablation of SRSF2 results in dilated cardiomyopathy (78). The fact that viruses, such as adenoviruses, herpesviruses, and reoviruses, known to induce myocarditis (79) can modulate splicing factors raises the intriguing possibility that altered splicing represents yet another mechanism by which viruses induce cardiac damage.
MATERIALS AND METHODS
Cells.
Mouse L929 cells were maintained in minimal essential medium (MEM; SAFC Biosciences) as a suspension culture supplemented to contain 5% fetal calf serum and 2 mM l-glutamine. AD-293 cells were maintained in high glucose Dulbecco MEM (DMEM; Gibco) supplemented to contain 10% fetal calf serum and 1% of sodium pyruvate. Exponentially growing L cells or trypsinized AD-293 cells were plated and incubated for at least 3 h prior to transfection or infection.
Viruses.
All viruses were CsCl-purified, low-passage-number stocks originating from plaques of either cultured virus or virus generated by reverse genetics (80) as previously described (6, 7). The results were the same regardless of virus source.
Plasmids and plasmid transfections.
A plasmid expressing C-terminus hemagglutinin (HA)-tagged T1L μ2 in a pCAGGs backbone was described previously (34). The C-terminus HA-tagged T3D μ2 was generated from pCAGGs-M1-T3D (6) using a similar strategy and the primer 5′-CCCGGGTCAGCTAGCGTAATCTGGAACATCGTATGGGTACGCCAAGTCAGATCGGAAAGCTAGTC-3′. The eGFP-N1 and eGFP-HsCRM1 plasmids were obtained from Clontech Laboratories (catalog no. 6085-1) and GeneCopoeia (catalog no. EX-T0446-M29), respectively. The Myc-DDK-IRF9 plasmid was custom-cloned by OriGene Technologies, Inc., using the murine IRF9 cDNA (NM_001159417) in a pCMV6 expression vector. The HsSRSF2-c-Myc in a pcDNA3.1 backbone plasmid (81) was a gift from Kathleen Scotto (Addgene; plasmid 44721). Plasmids were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) for 20 to 24 h.
siRNAs and siRNA transfections.
L929 cells were transfected for a final 60 nM concentration of control nontargeting siRNA or siRNA against murine SRSF2 (GE Dharmacon; catalog no. D-001810-10 or L-044306) using the transfection reagent RNAiMAX (Thermo Fisher Scientific). At 24 h posttransfection, overlaying medium was replaced with fresh medium.
Antibodies and chemical treatments.
The following antibodies and dilutions were used for immunoblotting: anti-μ2 (generated against two μ2 peptides by Open Biosystems; 1:1,000), anti-β-actin (Santa Cruz Biotech, sc-1615-hrp; 1:3,000), anti-SRSF2 (Millipore, 04-1550; 1:1,000), anti-SRSF1 (Abcam, ab129108; 1:350), anti-c-Myc (catalog no. sc-40, 1:3,000; Santa Cruz Biotech), goat horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin (Ig; Millipore, 12-348; 1:2,000), and goat HRP-conjugated anti-mouse Ig (Millipore, 12-349; 1:2,000). For immunoprecipitation experiments, secondary antibodies were either TrueBlot anti-rabbit IgG-HRP or TrueBlot Ultra anti-mouse Ig-HRP (Rockland Immunochemicals, 18-8816-33 and 18-8817-33; 1:1,000). Primary antibodies used for immunofluorescence and PLA were anti-coilin (Cell Signaling Technology, 14168; 1:800), anti-SRSF2/SC35 (Abcam, ab11826; 1:1,000), anti-Son (Abcam, ab109472; 1:200), anti-HA epitope tag (GeneTex, 18181; 1:1200; or Sigma, H6908; 1:1,000 or 1:1,200 for PLA), rabbit anti-μ2 antisera (82) (1:1,000), and a mix of anti-reovirus T1L and anti-reovirus 8B mouse antisera (B. Sherry, unpublished; 1:5,000 each). The secondary antibodies were Alexa Fluor 488- or Alexa Fluor 594-conjugated goat anti-mouse or anti-rabbit IgG (Thermo Fisher Scientific; 1:1,000). The CRM1-inhibitor leptomycin B (LMB; Sigma, L2913) was used at a final concentration of 20 nM in supplemented DMEM. Nocodazole (Sigma, M1404) and paclitaxel (Sigma, T7191) in dimethyl sulfoxide were used at a final concentration of 10 μM in supplemented DMEM. Recombinant mouse interferon-β (PBL Assay Science, catalog no. 12410-1) was used at 1,000 U/ml diluted in supplemented MEM.
SDS-PAGE and immunoblotting.
Whole-cell protein extracts were obtained 2 days postplating using radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA) supplemented to contain 1% sodium dodecyl sulfate (SDS), a cocktail of protease and phosphatase inhibitors (Sigma, catalog nos. P8340 and P2850), and 1 mM phenylmethylsulfonyl fluoride (Sigma, P7626). Protein lysates were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare). Membranes were probed with the indicated antibodies, developed using Amersham enhanced chemiluminescence (ECL) or ECL Prime kits, exposed to film, and scanned using an HP Scanjet G4050. Band intensities were determined using the LI-COR Biosciences Image Studio Lite Software (v5.x).
Coimmunoprecipitation.
At 24 h posttransfection, AD-293 cells were harvested and lysed using a modified RIPA buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100) supplemented with 20 mM N-ethylmaleimide (Sigma, catalog no. E3786) and a cocktail of protease and phosphatase inhibitors as described above. Lysate supernatants were incubated with EZview Red anti-HA affinity gel (Sigma, E6779). Washed beads were boiled in Laemmli sample buffer. Supernatant was subjected to SDS-PAGE and immunoblotting as described above.
Indirect immunofluorescence.
Cells in poly-d-Lysine-coated chamber slides (BD Biosciences) were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline and permeabilized with 0.25% Triton X-100 (Sigma). Slides were blocked with normal goat serum (Sigma, catalog no. G9023), incubated in DAPI (4′,6-diamidino-2-phenylindole; Sigma, D8417), immunostained with the indicated primary and secondary antibodies, and preserved with ProLong Gold (Invitrogen). For immunostaining of μ2 and SRSF2 during reovirus infection, the cells were fixed in 100% methanol.
In situ proximity ligation assay.
Transfected cells in poly-d-lysine-coated chamber slides were fixed with 4% paraformaldehyde, permeabilized in 0.25% Triton X-100, blocked with Duolink blocking solution, incubated with primary antibodies, and then probed using a Duolink PLA kit (Sigma, catalog no. DUO92101). Dried slides were mounted on Duolink mounting medium with DAPI and imaged as z-stacks by confocal microscopy.
Quantitative (real-time) reverse transcription-PCR.
For confirmation of the MISO results, oligonucleotide primers were designed to amplify transcripts that either skipped or spliced-in a specific exon, as identified by RNA-seq. The primers were as follows: RanBP3 skipped exon (forward, 5′-CCGTCTGTCTTTGTGTTTCAA-3′; reverse, 5′-GCTGCTCTTCTCTTCTGAGTGA-3′), RanBP3 spliced-in exon (forward, 5′-CGACCGTCTGTCTTTGTGTTT-3′; reverse, 5′-AAACATGACCCCATCAAAA-3′), SRSF3 skipped exon (forward, 5′-TGATTACCGCAGGAGGAGTC-3′; reverse, 5′-GATCGAGACGGCTTGTGATT-3′), and SRSF3 spliced-in exon (forward, 5′-TGATTACCGCAGGAGGAGTC-3′; reverse, 5′-TGACGCTGAAAGGGCTAGTT-3′). The total RNA was harvested using an RNeasy kit (Qiagen, Inc.), treated with RNase-free DNase I (Qiagen, Inc.), converted to cDNA by reverse transcription, and used for real-time PCR on a LightCycler 480 fluorescence thermocyler (Roche Life Science). The reaction mixtures contained 1× Quantitech SYBR green master mix (Qiagen) and 0.3 μM (each) forward and reverse primers as previously described (83). The relative mRNA abundance of RanBP3 and SRSF3 were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) by the ΔΔCp method, and the fold induction was calculated relative to mock-treated cells for each primer pair.
Viral replication and cytopathic effect in siRNA-transfected cells.
L929 cells were transfected with siRNA as described above and infected 2 days later at the indicated multiplicity of infection (MOI). The siRNA-mediated depletion of SRSF2 was maintained throughout the experiment (data not shown). Viral replication and cytopathic effect were assessed by plaque assay (84) and MTT assay (27).
Confocal microscopy and image analysis.
A Zeiss LSM 710 confocal microscope equipped with a 40× C-Apochromat/1.1 NA water immersion objective from the Cellular and Molecular Facility at NC State University was used for all experiments. The pinhole diameter was maintained at 1 Airy unit, and all images were obtained using multitrack sequential scanning for each fluorophore to prevent bleedthrough. The excitation/emission wavelengths during micrograph acquisition were as follows: 488 nm/492 to 554 nm for Alexa Fluor 488, 561 nm/584 to 666 nm for Alexa Fluor 594 and PLA Duolink Red, and 405 nm/407 to 507 nm for DAPI. The images were processed for presentation using Photoshop CS4. Intensity plot profiles were generated using ImageJ software (82). Quantification of in situ PLA signals per cell was performed using the particle analysis tool of ImageJ.
RNA-seq.
L929 cells were infected at an MOI of 100 PFU per cell, and at 20 h postinfection overlays were replaced with supplemented media or mouse IFN-β diluted in supplemented medium to 1,000 U/ml. After 5 h of incubation, the overlays were removed, and the total RNA was harvested, the contaminating DNA was removed, and an aliquot of the RNA was converted to cDNA for qRT-PCR to confirm T1L repression of IFN-β signaling (data not shown). Total RNA samples were submitted to the North Carolina State University Genomic Sciences Laboratory (Raleigh, NC) for Illumina RNA library construction and sequencing. mRNA was purified using oligo(dT) beads [NEBNExt poly(A) mRNA magnetic isolation module (New England BioLabs, USA)], chemically fragmented, and primed with random oligonucleotides for first-strand cDNA synthesis (NEBNext Ultra Directional RNA library prep kit [NEB] and NEBNext Mulitplex Oligos for Illumina [NEB]). Second-strand cDNA synthesis was carried out with dUTPs to preserve strand orientation information. Double-stranded cDNA was “A-tailed” for adaptor ligation and selected for a final size of 250 to 400 bp, including adaptors (AMPure XP bead isolation; Beckman Coulter, USA). Library enrichment was performed, and indexes were added during PCR amplification. Libraries were pooled in equimolar amounts and sequenced on an Illumina NextSeq 500 DNA sequencer, and real-time analysis was used to generate raw base call files, which were then demultiplexed by sample into fastq files.
Annotation and clustering of changes in alternative splicing during reovirus infection.
Only those reads with a minimum of 50 bp were used, and all reads were trimmed to 50 bases (fastx_trimmer tool [http://hannonlab.cshl.edu/fastx_toolkit/]). Reads were mapped to the mm10 mouse reference genome using TopHat2 (85), and the results merged to create a single bam file for each sample. Picard Tools' CollectInsertMetrics (http://picard.sourceforge.net) was run on each of these bam files. MISO (mixture-of-isoforms) software (29) was run to calculate the isoform expression “percent spliced isoform” (ψ) value. The mm1v2 known skipped exon events provided on the MISO website were included in the run. The ψ values calculated for the exons for each sample were compared, and a Bayes factor was computed. The Bayes Factor results were filtered, requiring at least one read supporting each isoform and at least a total of 10 reads across all isoforms. In addition, the absolute difference in ψ values must be ≥0.20 and a Bayes factor of >10. Analyses were performed by IPA (Ingenuity) and a generic gene ontology (GO) term mapper (http://go.princeton.edu/cgi-bin/GOTermMapper).
Statistical analysis.
Unless stated otherwise, a Student two-sample t test (pooled variance) was applied (Systat software). Results were considered significant for P value of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shannon Chiera, Tiffany Benzine, and Nicole DeAngelis for insightful discussions and technical assistance. We thank David Andrew Baltzegar at the NC State University Genomic Sciences Laboratory for his guidance and expertise. We also thank John L. Parker (Cornell University) and Jennifer Luff (NC State University) and members of her laboratory for helpful suggestions.
This research was supported by National Institutes of Health (NIH) grant R01 AI083333 (B.S.), a Research Supplement to Promote Diversity in Health-Related Research through a parental NIH grant (B.S. and E.E.R-S.), and a U.S. Department of Education Graduate Assistance in Areas of National Need (GAANN) Fellowship (E.E.R-S.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02488-16.
REFERENCES
- 1.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signaling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann HH, Schneider WM, Rice CM. 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol 36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotwal GJ, Hatch S, Marshall WL. 2012. Viral infection: an evolving insight into the signal transduction pathways responsible for the innate immune response. Adv Virol 2012:131457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dermody T, Parker J, Sherry B. 2013. Orthoreoviruses, p 1304–1346. In Fields virology, 6th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Fernandez de Castro I, Zamora PF, Ooms L, Fernandez JJ, Lai CM, Mainou BA, Dermody TS, Risco C. 2014. Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes. mBio 5:e00931-13. doi: 10.1128/mBio.00931-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zurney J, Kobayashi T, Holm GH, Dermody TS, Sherry B. 2009. Reovirus μ2 protein inhibits interferon signaling through a novel mechanism involving nuclear accumulation of interferon regulatory factor 9. J Virol 83:2178–2187. doi: 10.1128/JVI.01787-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irvin SC, Zurney J, Ooms LS, Chappell JD, Dermody TS, Sherry B. 2012. A single-amino-acid polymorphism in reovirus protein μ2 determines repression of interferon signaling and modulates myocarditis. J Virol 86:2302–2311. doi: 10.1128/JVI.06236-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brentano L, Noah DL, Brown EG, Sherry B. 1998. The reovirus protein μ2, encoded by the M1 gene, is an RNA-binding protein. J Virol 72:8354–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Parker JS, Murray KE, Nibert ML. 2004. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. J Biol Chem 279:4394–4403. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Ooms LS, Chappell JD, Dermody TS. 2009. Identification of functional domains in reovirus replication proteins μNS and μ2. J Virol 83:2892–2906. doi: 10.1128/JVI.01495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker JS, Broering TJ, Kim J, Higgins DE, Nibert ML. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol 76:4483–4496. doi: 10.1128/JVI.76.9.4483-4496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller CL, Parker JS, Dinoso JB, Piggott CD, Perron MJ, Nibert ML. 2004. Increased ubiquitination and other covariant phenotypes attributed to a strain- and temperature-dependent defect of reovirus core protein μ2. J Virol 78:10291–10302. doi: 10.1128/JVI.78.19.10291-10302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broering TJ, Parker JS, Joyce PL, Kim J, Nibert ML. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J Virol 76:8285–8297. doi: 10.1128/JVI.76.16.8285-8297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mbisa JL, Becker MM, Zou S, Dermody TS, Brown EG. 2000. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology 272:16–26. doi: 10.1006/viro.2000.0362. [DOI] [PubMed] [Google Scholar]
- 15.Manley JL, Krainer AR. 2010. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. 1994. Leptomycin B targets a regulatory cascade of Crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem 269:6320–6324. [PubMed] [Google Scholar]
- 17.Zakaryan H, Stamminger T. 2011. Nuclear remodelling during viral infections. Cell Microbiol 13:806–813. doi: 10.1111/j.1462-5822.2011.01596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moller A, Schmitz ML. 2003. Viruses as hijackers of PML nuclear bodies. Arch Immunol Ther Exp 51:295–300. [PubMed] [Google Scholar]
- 19.Regad T, Chelbi-Alix MK. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 20.Morris GE. 2008. The Cajal body. Biochim Biophys Acta 1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Thiry M. 1995. The interchromatin granules. Histol Histopathol 10:1035–1045. [PubMed] [Google Scholar]
- 22.Hall LL, Smith KP, Byron M, Lawrence JB. 2006. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288:664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumontet C, Jordan MA. 2010. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov 9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koos B, Andersson L, Clausson CM, Grannas K, Klaesson A, Cane G, Soderberg O. 2014. Analysis of protein interactions in situ by proximity ligation assays. Curr Top Microbiol Immunol 377:111–126. doi: 10.1007/82_2013_334. [DOI] [PubMed] [Google Scholar]
- 26.Caceres JF, Screaton GR, Krainer AR. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev 12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherry B, Torres J, Blum MA. 1998. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J Virol 72:1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anko ML. 2014. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol 32:11–21. doi: 10.1016/j.semcdb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Tripathi V, Song DY, Zong X, Shevtsov SP, Hearn S, Fu XD, Dundr M, Prasanth KV. 2012. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol Biol Cell 23:3694–3706. doi: 10.1091/mbc.E12-03-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salichs E, Ledda A, Mularoni L, Albà MM, de la Luna S. 2009. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet 5:e1000397. doi: 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson MI, She YM, Ens W, Brown EG, Coombs KM. 2002. Mammalian reovirus core protein micro 2 initiates at the first start codon and is acetylated. Rapid Commun Mass Spectrom 16:2317–2324. doi: 10.1002/rcm.866. [DOI] [PubMed] [Google Scholar]
- 34.Stebbing RE, Irvin SC, Rivera-Serrano EE, Boehme KW, Ikizler M, Yoder JA, Dermody TS, Sherry B. 2014. An ITAM in a nonenveloped virus regulates activation of NF-κB, induction of beta interferon, and viral spread. J Virol 88:2572–2583. doi: 10.1128/JVI.02573-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubois J, Terrier O, Rosa-Calatrava M. 2014. Influenza viruses and mRNA splicing: doing more with less. mBio 5:e00070-14. doi: 10.1128/mBio.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell DF, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67:6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider PA, Schneemann A, Lipkin WI. 1994. RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus. J Virol 68:5007–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su TS, Lai CJ, Huang JL, Lin LH, Yauk YK, Chang CM, Lo SJ, Han SH. 1989. Hepatitis B virus transcript produced by RNA splicing. J Virol 63:4011–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez-Lopez HR, Graham SV. 2012. Alternative splicing in human tumour viruses: a therapeutic target? Biochem J 445:145–156. doi: 10.1042/BJ20120413. [DOI] [PubMed] [Google Scholar]
- 40.Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, Hagiwara M. 2006. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci U S A 103:11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavallari I, Rende F, D'Agostino DM, Ciminale V. 2011. Converging strategies in expression of human complex retroviruses. Viruses 3:1395–1414. doi: 10.3390/v3081395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter KC, Taneja KL, Lawrence JB. 1991. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol 115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E. 1993. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res 208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- 44.Cardinale S, Cisterna B, Bonetti P, Aringhieri C, Biggiogera M, Barabino SM. 2007. Subnuclear localization and dynamics of the pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit. Mol Biol Cell 18:1282–1292. doi: 10.1091/mbc.E06-09-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Hoogenhof MM, Pinto YM, Creemers EE. 2016. RNA splicing: regulation and dysregulation in the heart. Circ Res 118:454–468. doi: 10.1161/CIRCRESAHA.115.307872. [DOI] [PubMed] [Google Scholar]
- 46.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scotti MM, Swanson MS. 2016. RNA mis-splicing in disease. Nat Rev Genet 17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estmer Nilsson C, Petersen-Mahrt S, Durot C, Shtrichman R, Krainer AR, Kleinberger T, Akusjarvi G. 2001. The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO j 20:864–871. doi: 10.1093/emboj/20.4.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 50.Lindberg A, Kreivi JP. 2002. Splicing inhibition at the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology 294:189–198. doi: 10.1006/viro.2001.1301. [DOI] [PubMed] [Google Scholar]
- 51.Sciabica KS, Dai QJ, Sandri-Goldin RM. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J 22:1608–1619. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu B, Li X, Huo Y, Yu Y, Zhang Q, Chen G, Zhang Y, Fraser NW, Wu D, Zhou J. 2016. Cellular responses to HSV-1 infection are linked to specific types of alterations in the host transcriptome. Sci Rep 6:28075. doi: 10.1038/srep28075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang TS, Nilsson CE, Punga T, Akusjarvi G. 2002. Functional inactivation of the SR family of splicing factors during a vaccinia virus infection. EMBO Rep 3:1088–1093. doi: 10.1093/embo-reports/kvf217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruvolo V, Sun L, Howard K, Sung S, Delecluse H-J, Hammerschmidt W, Swaminathan S. 2004. Functional analysis of Epstein-Barr virus SM protein: identification of amino acids essential for structure, transactivation, splicing inhibition, and virion production. J Virol 78:340–352. doi: 10.1128/JVI.78.1.340-352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma D, Swaminathan S. 2008. Epstein-Barr virus SM protein functions as an alternative splicing factor. J Virol 82:7180–7188. doi: 10.1128/JVI.00344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lad SP, Yang G, Scott DA, Chao TH, da Correia JS, de la Torre JC, Li E. 2008. Identification of MAVS splicing variants that interfere with RIGI/MAVS pathway signaling. Mol Immunol 45:2277–2287. doi: 10.1016/j.molimm.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Hu X, Song Y, Lu Z, Ning T, Cai H, Ke Y. 2011. Identification of novel alternative splicing variants of interferon regulatory factor 3. Biochim Biophys Acta 1809:166–175. doi: 10.1016/j.bbagrm.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Koop A, Lepenies I, Braum O, Davarnia P, Scherer G, Fickenscher H, Kabelitz D, Adam-Klages S. 2011. Novel splice variants of human IKKεnegatively regulate IKKε-induced IRF3 and NF-κB activation. Eur J Immunol 41:224–234. doi: 10.1002/eji.201040814. [DOI] [PubMed] [Google Scholar]
- 59.Liu P, Lu M, Tian B, Li K, Garofalo RP, Prusak D, Wood TG, Brasier AR. 2009. Expression of an IKKγ splice variant determines IRF3 and canonical NF-κB pathway utilization in ssRNA virus infection. PLoS One 4:e8079. doi: 10.1371/journal.pone.0008079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y, Xu D, Jiang Y, Zhang L. 2010. Dual functions of interferon regulatory factors 7C in Epstein-Barr virus-mediated transformation of human B lymphocytes. PLoS One 5:e9459. doi: 10.1371/journal.pone.0009459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marie I, Smith E, Prakash A, Levy DE. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol Cell Biol 20:8803–8814. doi: 10.1128/MCB.20.23.8803-8814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samarajiwa SA, Mangan NE, Hardy MP, Najdovska M, Dubach D, Braniff SJ, Owczarek CM, Hertzog PJ. 2014. Soluble IFN receptor potentiates in vivo type I IFN signaling and exacerbates TLR4-mediated septic shock. J Immunol 192:4425–4435. doi: 10.4049/jimmunol.1302388. [DOI] [PubMed] [Google Scholar]
- 63.Deng W, Shi M, Han M, Zhong J, Li Z, Li W, Hu Y, Yan L, Wang J, He Y, Tang H, Deubel V, Luo X, Ning Q, Sun B. 2008. Negative regulation of virus-triggered IFN-β signaling pathway by alternative splicing of TBK1. J Biol Chem 283:35590–35597. doi: 10.1074/jbc.M805775200. [DOI] [PubMed] [Google Scholar]
- 64.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, Mammi P, Mancini E, Yanovsky MJ, Andino R, Krogan N, Srebrow A, Gamarnik AV. 2016. The dengue virus NS5 protein intrudes in the cellular spliceosome and modulates splicing. PLoS Pathog 12:e1005841. doi: 10.1371/journal.ppat.1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boudreault S, Martenon-Brodeur C, Caron M, Garant J-M, Tremblay M-P, Armero VES, Durand M, Lapointe E, Thibault P, Tremblay-Létourneau M, Perreault J-P, Scott MS, Lemay G, Bisaillon M. 2016. Global profiling of the cellular alternative RNA splicing landscape during virus-host interactions. PLoS One 11:e0161914. doi: 10.1371/journal.pone.0161914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Z, Krainer AR. 2004. Involvement of SR proteins in mRNA surveillance. Mol Cell 16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 67.Lejeune F, Maquat LE. 2005. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol 17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 68.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanford JR, Gray NK, Beckmann K, Caceres JF. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev 18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanford JR, Ellis JD, Cazalla D, Caceres JF. 2005. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci U S A 102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue F, Li X, Zhao X, Wang L, Liu M, Shi R, Zheng J. 2015. SRSF1 facilitates cytosolic DNA-induced production of type I interferons recognized by RIG-I. PLoS One 10:e0115354. doi: 10.1371/journal.pone.0115354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tremblay N, Baril M, Chatel-Chaix L, Es-Saad S, Park AY, Koenekoop RK, Lamarre D. 2016. Spliceosome SNRNP200 promotes viral RNA sensing and IRF3 activation of antiviral response. PLoS Pathog 12:e1005772. doi: 10.1371/journal.ppat.1005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M Jr, Fu XD. 2013. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell 50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bedard KM, Daijogo S, Semler BL. 2007. A nucleocytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO j 26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fitzgerald KD, Chase AJ, Cathcart AL, Tran GP, Semler BL. 2013. Viral proteinase requirements for the nucleocytoplasmic relocalization of cellular splicing factor SRp20 during picornavirus infections. J Virol 87:2390–2400. doi: 10.1128/JVI.02396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fitzgerald KD, Semler BL. 2011. Relocalization of cellular protein SRp20 during poliovirus infection: bridging a viral IRES to the host cell translation apparatus. PLoS Pathog 7:e1002127. doi: 10.1371/journal.ppat.1002127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Desmet EA, Anguish LJ, Parker JS. 2014. Virus-mediated compartmentalization of the host translational machinery. mBio 5:e01463-14. doi: 10.1128/mBio.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, Yeakley JM, Cheng H, Xiao RP, Ross J, Chen J, Fu XD. 2004. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J 23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feldman AM, McNamara D. 2000. Myocarditis. N Engl J Med 343:1388–1398. doi: 10.1056/NEJM200011093431908. [DOI] [PubMed] [Google Scholar]
- 80.Kobayashi T, Antar AA, Boehme KW, Danthi P, Eby EA, Guglielmi KM, Holm GH, Johnson EM, Maginnis MS, Naik S, Skelton WB, Wetzel JD, Wilson GJ, Chappell JD, Dermody TS. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi J, Hu Z, Pabon K, Scotto KW. 2008. Caffeine regulates alternative splicing in a subset of cancer-associated genes: a role for SC35. Mol Cell Biol 28:883–895. doi: 10.1128/MCB.01345-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stewart MJ, Smoak K, Blum MA, Sherry B. 2005. Basal and reovirus-induced beta interferon (IFN-β) and IFN-β-stimulated gene expression are cell type specific in the cardiac protective response. J Virol 79:2979–2987. doi: 10.1128/JVI.79.5.2979-2987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Virgin HW, Bassel-Duby R, Fields BN, Tyler KL. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J Virol 62:4594–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.