ABSTRACT
Activation of signaling pathways ensuring cell growth is essential for the proliferative competence of human papillomavirus (HPV)-infected cells. Tyrosine kinases and phosphatases are key regulators of cellular growth control pathways. A recently identified potential cellular target of HPV E7 is the cytoplasmic protein tyrosine phosphatase PTPN14, which is a potential tumor suppressor and is linked to the control of the Hippo and Wnt/beta-catenin signaling pathways. In this study, we show that the E7 proteins of both high-risk and low-risk mucosal HPV types can interact with PTPN14. This interaction is independent of retinoblastoma protein (pRb) and involves residues in the carboxy-terminal region of E7. We also show that high-risk E7 induces proteasome-mediated degradation of PTPN14 in cells derived from cervical tumors. This degradation appears to be independent of cullin-1 or cullin-2 but most likely involves the UBR4/p600 ubiquitin ligase. The degree to which E7 downregulates PTPN14 would suggest that this interaction is important for the viral life cycle and potentially also for the development of malignancy. In support of this we find that overexpression of PTPN14 decreases the ability of HPV-16 E7 to cooperate with activated EJ-ras in primary cell transformation assays.
IMPORTANCE This study links HPV E7 to the deregulation of protein tyrosine phosphatase signaling pathways. PTPN14 is classified as a potential tumor suppressor protein, and here we show that it is very susceptible to HPV E7-induced proteasome-mediated degradation. Intriguingly, this appears to use a mechanism that is different from that employed by E7 to target pRb. Therefore, this study has important implications for our understanding of the molecular basis for E7 function and also sheds important light on the potential role of PTPN14 as a tumor suppressor.
KEYWORDS: human papillomavirus, E7, PTPN14, transformation, tyrosine phosphatases
INTRODUCTION
Human papillomaviruses (HPVs) are ubiquitous small DNA viruses that infect skin or mucosal epithelial cells and cause hyperproliferative lesions. Mucosal HPVs are considered to be one of the most common virus infections of the human reproductive tract, and the so-called high-risk mucosal types, such as HPV-16 and HPV-18, are the major etiological agents of cervical cancer, other anogenital cancers, and an increasing subset of head and neck cancers. Despite major advances in our understanding of the HPV life cycle and virus-induced malignancy (1–4), there are still major gaps in our understanding of the function of the viral oncoproteins. In addition, we still lack useful cellular markers for the accurate prognosis of precancerous lesions caused by oncogenic HPV infections, and pharmaceutical targets for the successful selective therapy of HPV-related cancers have yet to be found. Therefore, there remains an urgent need to understand the cellular processes driving malignant transformation of the HPV-infected cell.
It has been demonstrated that the malignant phenotype of high-risk HPV types depends mainly on the sustained expression of two early viral gene products, the E6 and E7 oncoproteins. These proteins interfere with the function of multiple cellular factors to keep the host epithelial cell in a replication-competent state, thereby supporting the amplification of the viral DNA genome (5–8), which can also ultimately promote tumorigenesis. A key element in this is the ability of the high-risk HPV E6 and E7 oncoproteins to target the tumor suppressor protein p53 and the retinoblastoma protein (pRb) family of pocket proteins, respectively. In both cases, E6 and E7 recruit essential elements of the ubiquitin proteasome pathway to ensure the efficient degradation of their cellular targets. In the case of E6 and p53, this requires the E6AP ubiquitin ligase (9), while in the case of E7 and the pocket proteins, this involves the recruitment of the cullin-2 ubiquitin ligase complex (10, 11). While these interactions are obviously important during cancer development, it is also clear that other functions of these viral oncoproteins contribute toward this multistep process (3, 9, 10, 12). In particular, a great deal of attention has been placed on understanding how E7 can modulate different cellular kinases, such as AKT and extracellular signal-regulated kinase (ERK) signaling (13–17). In contrast, there is little information available concerning the potential modulation of cellular phosphatases although E7 has been linked with the upregulation of the CDC25A tyrosine phosphatase and inhibition of protein phosphatase 2A (PP2A) (17–19). Interestingly, the cytoplasmic protein tyrosine phosphatase PTPN14 (PTPD2, Pez, or PTP36) was recently identified as a potential cellular target of E7 proteins derived from multiple HPV types (20, 21).
Protein tyrosine kinases and phosphatases are key regulators of cellular growth control pathways, and it is increasingly recognized that protein tyrosine phosphatases are as highly substrate specific as tyrosine kinases and have a major impact on the cellular microenvironment and disease pathophysiology (22, 23). PTPN14 (PTPD2, Pez, or PTP36) is particularly intriguing since this FERM (four-point-one, ezrin, radixin, moesin) domain-containing phosphatase can associate with the cytoskeleton and cell membrane and can affect actin cytoskeleton organization, cell adhesion, and cell growth (24, 25). PTPN14 has also been proposed to act as a tumor suppressor protein that influences multiple cellular processes. It has been shown to inhibit the oncogenic function of the Yes-associated protein (YAP) and other members of the Hippo signaling pathway (26–29). Furthermore, PTPN14 can decrease cell motility via dephosphorylation of β-catenin at adherens junctions (30). In a breast cancer model it has been recently demonstrated that PTPN14 also negatively regulates proteins involved in intracellular trafficking, such as RIN1 (Ras and Rab interactor 1) and protein kinase C-δ (PRKCD), thereby preventing metastasis by reducing the intracellular trafficking of both soluble and membrane-bound proteins (31).
In this study, we demonstrate that PTPN14 is a bona fide interacting partner of multiple HPV E7 oncoproteins. Furthermore, we show that cancer-causing high-risk HPV E7 oncoproteins can target PTPN14 for proteasome-mediated degradation, an activity which appears to require the UBR4/p600 ubiquitin ligase. Taken together, these studies indicate that E7 can profoundly affect the function of the PTPN14 tumor suppressor and thereby modulate processes relevant for the virus life cycle and for the development of malignancy.
RESULTS
HPV E7 interacts with PTPN14 both in vitro and in vivo.
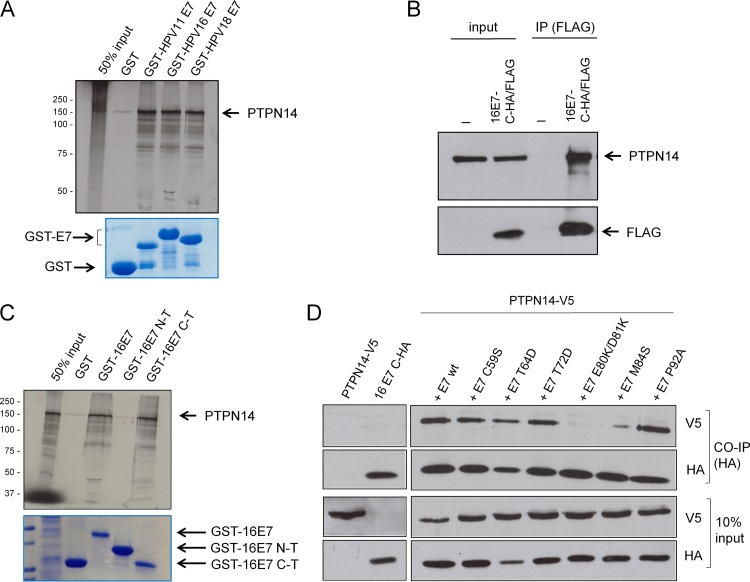
Recent proteomic analyses (20, 21) had highlighted the potential interaction between the E7 oncoproteins derived from diverse HPV types with the tyrosine phosphatase PTPN14. To determine whether PTPN14 is indeed a binding partner of both low-risk and high-risk E7 proteins, we performed an in vitro binding assay using in vitro-translated, radiolabeled PTPN14 and purified glutathione S-transferase (GST)-tagged E7 fusion proteins from both low-risk (HPV-11) and high-risk (HPV-16 and HPV-18) HPV types. After intensive washing, bound proteins were assessed by autoradiography. The results shown in Fig. 1A indicate a very strong physical association between PTPN14 and E7, which is conserved across different HPV types. To determine whether E7 could also interact with endogenously expressed PTPN14, C33a cells were transfected with FLAG-tagged HPV-16 E7 and then immunoprecipitated using anti-FLAG antibodies, and coimmunoprecipitating proteins were analyzed by Western blotting. The results shown in Fig. 1B demonstrate a specific interaction between HPV-16 E7 and PTPN14.
FIG 1.
HPV E7 interacts with PTPN14, and the interaction is conserved across different HPV types. (A) In vitro-translated, radiolabeled PTPN14 was incubated with GST (control), GST-HPV-11 E7, GST-HPV-16 E7, and GST-HPV-18 E7 fusion proteins bound to glutathione agarose beads. After a washing step, bound proteins were assessed by autoradiography (upper panel), and GST fusion proteins were visualized by Coomassie staining (lower panel). (B) C33a cells were transfected with empty plasmid or HA/FLAG-tagged HPV-16 E7. After 24 h the cells were incubated in the presence of the proteasome inhibitor CBZ for 3 h and were then harvested and immunoprecipitated (IP) using anti-FLAG antibody, and PTPN14 and HPV-16 E7 were detected by Western blotting. (C) Full-length HPV-16 E7 and the N-terminal (N-T) and C-terminal (C-T) truncations were expressed as GST fusion proteins, purified, and used in binding assays with in vitro-translated and radiolabeled PTPN14. The upper panel shows input and bound PTPN14, and the lower panel shows the Coomassie-stained gel. (D) The wild-type HPV-16 E7 and the indicated mutants (all HA tagged) were cotransfected in HEK293 cells with V5-tagged PTPN14, and after 24 h the cells were incubated in the presence of the proteasome inhibitor CBZ for 3 h and then harvested and immunoprecipitated with anti-HA antibody. Bound PTPN14 was then detected by Western blotting (V5), and E7 was detected using anti-HA antibody. The lower panels show the input proteins used in the immunoprecipitations. CO-IP, coimmunoprecipitation; wt, wild type.
To identify the region of E7 required for binding PTPN14, we performed in vitro interactions using in vitro-translated, radiolabeled PTPN14 and wild-type HPV-16 E7 and two deletion constructs of E7 comprising the N-terminal and C-terminal halves, expressed as GST fusion proteins. As can be seen from Fig. 1C, PTPN14 binds to the C-terminal domain of HPV-16 E7. Previous studies have highlighted the importance of multiple residues within the E7 carboxy-terminal region for potential protein-protein interactions and for their ability to contribute toward cell transformation based on their surface exposure (32), as predicted from the HPV-45 E7 structure (33). Therefore, in order to more accurately define the region of E7 involved in binding PTPN14, we performed a series of coimmunoprecipitation experiments using a panel of these previously described HPV-16 E7 mutants with a C-terminal hemagglutinin (HA) or FLAG tag. These were cotransfected with V5-tagged PTPN14 into HEK293 cells, and after 24 h the cells were harvested and subjected to immunoprecipitation using anti-HA-conjugated agarose beads. The immunoprecipitated E7 proteins were then detected by Western blotting using anti-FLAG antibodies, while coimmunoprecipitating PTPN14 was detected by Western blotting using anti-V5 antibodies. The results shown in Fig. 1D demonstrate that E7 amino acid residues 80, 81, and 84 are essential for the PTPN14 interaction.
High-risk HPV E7 degradation of PTPN14.
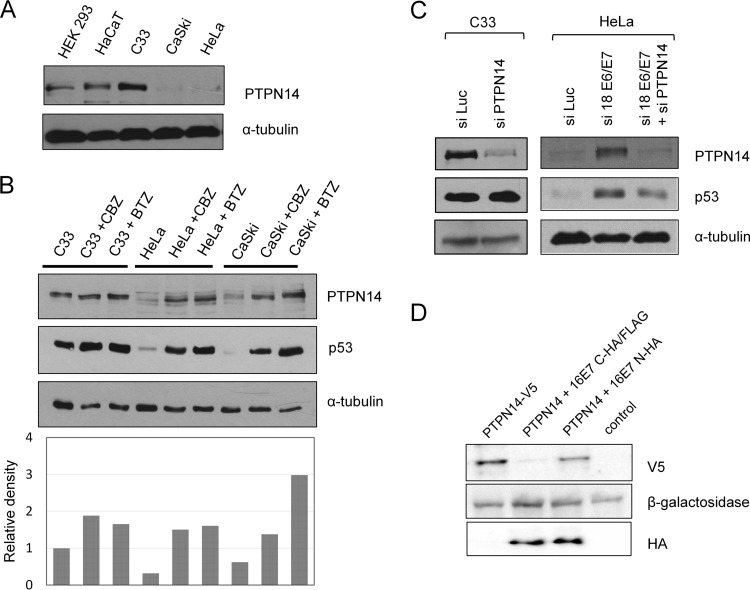
Having found that PTPN14 is a strong interacting partner of multiple HPV E7 proteins, we then wanted to investigate the potential relevance of this for HPV-induced malignancy. To do this, we first examined the levels of expression of PTPN14 in HPV-negative and HPV-positive cell lines by performing Western blot analysis of PTPN14 in HPV-16-positive CaSki and HPV-18-positive HeLa cells and in HPV-negative C33a, HaCaT, and HEK293 cells. The results (Fig. 2A) demonstrate much lower levels of PTPN14 protein in the two HPV-positive cell lines than in the non-HPV-containing cells, suggesting that PTPN14 protein levels are low in the presence of HPV. In order to then determine whether this low level of PTPN14 was due to proteasome-mediated degradation, HPV-negative C33a cells and HPV-positive HeLa and CaSki cells were grown in the presence of the proteasome inhibitor Z-Leu-Leu-Leu-al (CBZ) or bortezomib (BTZ) for 8 h. Cells were then harvested, and the levels of PTPN14 expression were analyzed by Western blotting. The results shown in Fig. 2B indicate that proteasome inhibition has no effect on the levels of PTPN14 in HPV-negative cells, while there is a dramatic increase in the levels of PTPN14 protein in HPV-positive cells following proteasome inhibition. These results indicate that PTPN14 is subject to proteasome-mediated degradation in HPV-positive cells derived from cervical tumors.
FIG 2.
PTPN14 is subject to proteasome-mediated degradation by HPV-16 E7. (A) The indicated cell lines were extracted, and levels of PTPN14 were analyzed by Western blotting. The lower panel shows the α-tubulin protein loading control. (B) The indicated cells were incubated in the presence of either DMSO or the two proteasome inhibitors, CBZ and BTZ, for 8 h. The cells were then harvested, and levels of PTPN14 were analyzed by Western blotting. A Western blot for p53 was included as a control for proteasome inhibition, and α-tubulin served as a protein loading control. The lower panel shows the quantitation of the Western blot. (C) C33a cells or HeLa cells were transfected with the indicated siRNAs (si- prefixes), and after 72 h the cells were harvested, and the levels of p53 and PTPN14 expression were analyzed by Western blotting, with α-tubulin used as the loading control. (D) HEK293 cells were transfected with V5-tagged PTPN14 and either C-terminally or N-terminally FLAG/HA-tagged HPV-16 E7. After 24 h the cells were harvested, and the levels of PTPN14 (V5) and E7 (HA) expression were analyzed by Western blotting. β-Galactosidase served as the control for transfection efficiency.
To investigate whether the low levels of PTPN14 were due to the presence of the HPV oncoproteins, we transfected HeLa cells with a small interfering RNA (siRNA) against HPV-18 E6/E7 or against luciferase as a control. In addition, to verify the correct identification of PTPN14, we also included an siRNA against PTPN14. After 72 h the cells were harvested, and the levels of protein expression were ascertained by Western blotting. As can be seen from Fig. 2C, ablation of E6/E7 expression results in a marked increase in the levels of PTPN14.
Since PTPN14 is a potential interacting partner of HPV E7, we then proceeded to investigate whether HPV-16 E7 could target PTPN14 for degradation in a transient-transfection assay. To do this, we used two different HPV-16 E7 expression constructs, one in which E7 was C-terminally tagged and one in which E7 was N-terminally tagged, as previous reports had suggested that the ability of E7 to direct proteins for proteasome-mediated degradation required an intact N terminus and could be blocked by addition of an epitope tag (34). HEK293 cells were transfected with a plasmid expressing N-terminally V5-tagged PTPN14 alone or together with plasmids expressing wild-type FLAG-tagged HPV-16 E7 (C-terminal tag) or HA-tagged HPV-16 E7 (N-terminal tag). A β-galactosidase expression plasmid was included in each transfection to control for transfection efficiency. After 24 h, cellular proteins were extracted and analyzed by Western blotting. As can be seen from Fig. 2D, overexpression of C-terminally tagged HPV-16 E7 caused a marked decrease in PTPN14 protein levels, while the N-terminally tagged HPV-16 E7 has little or no effect. Taken together, these results demonstrate that high-risk HPV E7 oncoproteins can target PTPN14 for proteasome-mediated degradation, an activity that requires a free N terminus on E7 and is blocked by the addition of an epitope tag.
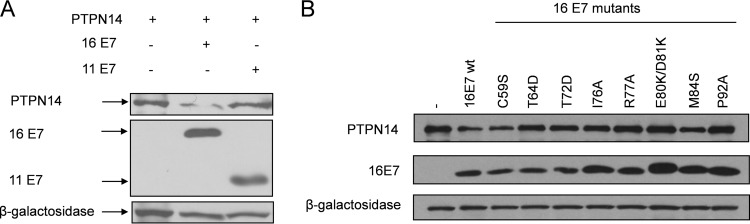
Having found that HPV-16 E7 can target PTPN14 for proteasome-mediated degradation, we were next interested in investigating whether the low-risk HPV-11 E7 could also affect the levels of PTPN14 expression. To do this, HEK293 cells were transfected with HA-tagged HPV-16 and HPV-11 E7, together with V5-tagged PTPN14, and the levels of expression were analyzed by Western blotting. The results shown in Fig. 3A demonstrate that whereas HPV-16 E7 can markedly reduce PTPN14 protein levels, HPV-11 E7 has a much weaker effect.
FIG 3.
Low-risk HPV-11 E7 does not degrade PTPN14. (A) HEK293 cells were transfected with PTPN14 together with HPV-16 E7 and HPV-11 E7 as indicated. After 24 h, the cells were harvested, and the levels of C-terminally HA-tagged E7 and V5-tagged PTPN14 were determined by Western blotting. β-Galactosidase served as a control for transfection efficiency. (B) The indicated HPV-16 E7 mutants were cotransfected with V5-tagged PTPN14 in HEK293 cells; after 24 h the cells were harvested, and proteins were analyzed by Western blotting, with β-galactosidase used a control for transfection efficiency. wt, wild type.
We were then interested in determining whether the ability of HPV-16 E7 to interact with PTPN14 correlates with its ability to target PTPN14 for proteasome-mediated degradation. To do this, we used a panel of carboxy-terminal point mutations in HPV-16 E7 and cotransfected these with V5-tagged PTPN14. The cells were harvested after 24 h, and the levels of E7 and PTPN14 expression were analyzed by Western blotting. The results obtained are shown in Fig. 3B and demonstrate a number of interesting features. In agreement with the coimmunoprecipitation analyses, the E80K/D81K and M84S point mutants, which show defects in interaction with PTPN14, also show corresponding defects in their abilities to target PTPN14 for degradation. However, it is also interesting that other mutants, such P92A, while retaining a wild-type ability to interact with PTPN14, are nonetheless defective in their ability to target PTPN14 for degradation. These results indicate that the efficient degradation of PTPN14 by E7 requires both a free E7 N terminus and an extended region within the E7 C terminus, not all residues of which appear to be involved in PTPN14 recognition, although they are presumably involved in the recruitment of additional cellular proteins required for the optimal targeting of PTPN14 for degradation.
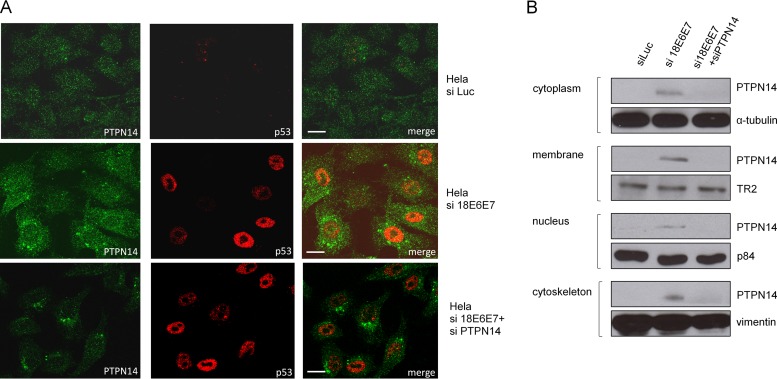
Having found that E7 could degrade PTPN14, we were next interested in investigating which cellular pools of PTPN14 were being targeted as previous studies have highlighted the importance of PTPN14 in the cytoplasm (26–31). To do this, we performed siRNA ablation of E6/E7 expression in HeLa cells and monitored the pattern of PTPN14 expression after 72 h using confocal microscopy. At the same time we also performed siRNA ablation of PTPN14 to confirm the specificity of the immunostaining of PTPN14. The results shown in Fig. 4A demonstrate a recovery in the PTPN14 levels following ablation of E6/E7. Interestingly, the recovery of PTPN14 appears to be found within both the cytoplasm and the nucleus. To confirm this, we also performed cell fractionations on HeLa cells following siRNA ablation of E6/E7 expression and then monitored the appearance of PTPN14 by Western blotting. The results shown in Fig. 4B demonstrate restoration of PTPN14 protein throughout the cell, which is consistent with the results in the immunofluorescence analyses.
FIG 4.
Different subcellular pools of PTPN14 are degraded by HPV-18 E7. (A) HeLa cells were transfected with the indicated siRNAs, and after 72 h the cells were fixed and stained for PTPN14 and for p53, which served as a control for the E6/E7 siRNA. Scale bar, 25 μm. (B) HeLa cells were transfected with the indicated siRNAs and after 72 h harvested and subjected to subcellular fractionation. The levels of PTPN14 were then analyzed by Western blotting, and each subcellular fraction was also probed for the indicated protein loading control.
UBR4/p600 ubiquitin ligase mediates the effect of E7 on PTPN14 levels.
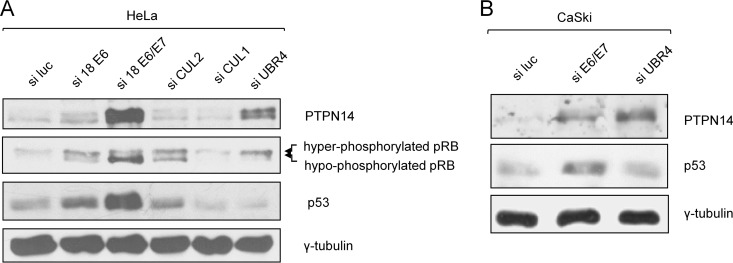
Previous studies have shown that PTPN14 can be targeted for ubiquitin-mediated degradation via the cullin-2 ubiquitin protein ligase (27). HPV E7 has also been shown to associate with the cullin-2 ubiquitin ligase complex, which mediates E7-induced Rb degradation (11). In addition, other ubiquitin ligases, namely, cullin-1 and UBR4/p600, have been found to interact with E7 (34, 35). In all cases an intact E7 N terminus would seem to be required for these interactions. Since E7 fails to degrade PTPN14 when N-terminally tagged, this led us to investigate which of the components of the ubiquitin proteasome machinery might be involved in E7-induced degradation of PTPN14. To do this, we transfected HeLa cells with siRNAs specific to cullin-1, cullin-2, and UBR4/p600. As controls we also included siRNAs against HPV-18 E6, HPV-18 E6/E7, and luciferase. After 72 h the cells were harvested and analyzed for p53, pRb, and PTPN14 by Western blotting. As can be seen from Fig. 5A, loss of E6, as expected, has no effect upon PTPN14 levels, while loss of E6/E7 results in a significant increase, confirming the role of E7 in the loss of PTPN14 protein. Furthermore, loss of cullin-1 or cullin-2 appears to have no effect on PTPN14 levels although loss of cullin-2 restores pRb levels, as expected (11). Most interestingly, only the loss of UBR4/p600 appears to rescue the PTPN14 protein, and, intriguingly, loss of UBR4/p600 in HeLa cells also results in a modest rescue of pRb levels. To investigate whether UBR4/p600 is also required for HPV-16 E7-induced degradation of PTPN14, we performed a similar analysis in CaSki cells, and the results obtained are shown in Fig. 5B, where it can be seen that loss of UBR4/p600 also results in a marked increase in PTPN14 protein levels. Taken together, these results demonstrate that HPV-16 and HPV-18 E7 can target PTPN14 for proteasome-mediated degradation in a manner that is in part dependent upon the UBR4/p600 ubiquitin ligase.
FIG 5.
UBR4/p600 is required for E7-induced degradation of PTPN14. (A) HeLa cells were transfected with the indicated siRNAs, and after 72 h the cells were harvested. The levels of PTPN14 were then analyzed by Western blotting; p53 served as a control for E6/E7 ablation, and pRb (different phospho-forms are indicated) served as a control for E7 and cullin-2 ablation. Overall protein loading was verified using γ-tubulin. (B) CaSki cells were transfected with the indicated siRNAs; after 72 h the cells were harvested, and the levels of p53 and PTPN14 were analyzed by Western blotting. γ-Tubulin served as the protein loading control.
PTPN14 inhibits HPV-16 E7-induced transformation.
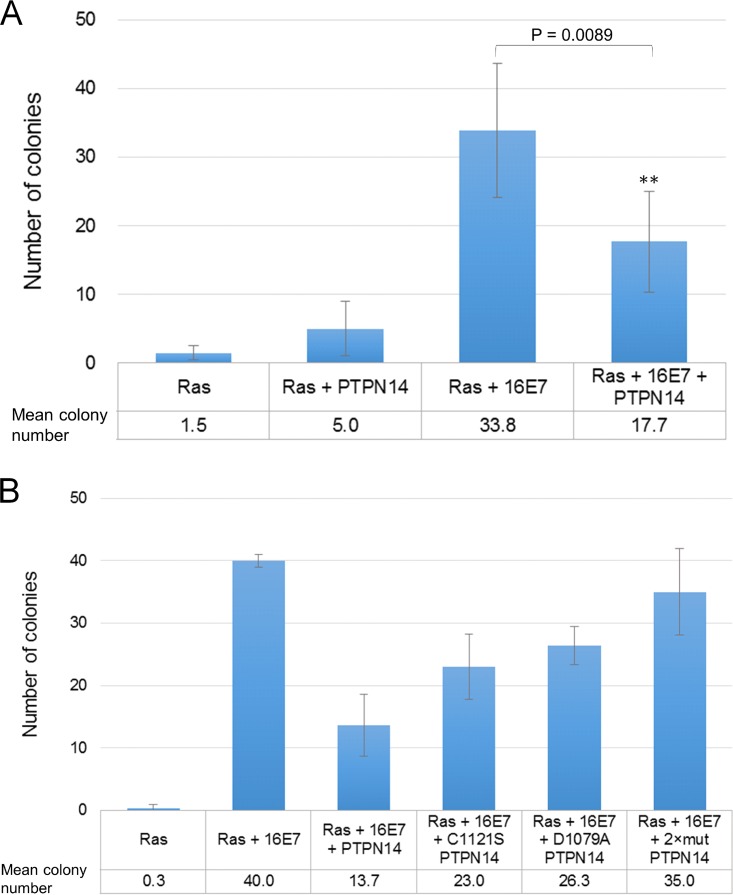
Previous studies have implicated a potential tumor-suppressive role for PTPN14 (26–30), and it has also been observed that PTPN14 overexpression reduces the motility and cell growth of HeLa cells (36). We were therefore interested in whether PTPN14 has any direct effects upon the transforming activity of E7. To examine this, we used primary BRK cells transfected with pJ4Ω:HPV-16 E7 and EJ-ras, with or without PTPN14. After 2 weeks of selection, the cells were fixed and stained, and the colonies were counted. The results are shown in Fig. 6A where it can be seen that PTPN14 is a potent inhibitor of the transforming activity of HPV-16 E7. To investigate which aspects of PTPN14 activity were required for this inhibitory function, we repeated these assays using two PTPN14 mutants, the catalytically inactive C1121S substitution and the substrate binding-defective D1079A substitution together with the corresponding double mutant. The results in Fig. 6B show that while the wild-type PTPN14 can inhibit E7 transforming activity, the two functionally defective mutants are compromised, while the double mutant has no effect on E7's transforming activity. Taken together, these results demonstrate that in the context of HPV-16 E7-induced cell transformation, PTPN14 is a potent tumor suppressor, an activity that requires its phosphatase and substrate recognition functions, while in the context of HPV-induced malignancy E7 can target PTPN14 for proteasome-mediated degradation in a manner requiring the p600 ubiquitin ligase.
FIG 6.
PTPN14 acts as a tumor suppressor in HPV-16 E7/EJ-ras cotransformation experiments. (A and B) Primary BRK cells were transfected with the indicated expression plasmids and subjected to selection with G418. After 2 to 3 weeks, the cells were fixed and stained, and the numbers of colonies were counted. The results show the mean number of colonies obtained from at least three separate assays, and standard deviations are also shown. A two-tailed Student's t test was carried out for the analysis of colony numbers to determine statistical significance (P < 0.05).
DISCUSSION
The data presented here show that E7 proteins of both low-risk and high-risk mucosal HPV types can associate with PTPN14, a FERM domain-containing nonreceptor protein tyrosine phosphatase. This phosphatase appears to have key regulatory roles in several signaling pathways that modulate cell growth, cell adhesion, and actin cytoskeleton organization (26–30). In this study, we have found that PTPN14 is particularly sensitive to HPV-16 and HPV-18 E7-induced degradation. This indicates a novel mechanism by which HPV E7 oncoproteins can subvert diverse cell signaling pathways. These results are in broad agreement with a recent study, which also found that PTPN14 was a degradation target of high-risk HPV E7 oncoproteins (37).
Previous studies using proteomic approaches had indicated that the E7 proteins from diverse HPV types could potentially interact with PTPN14 (20, 21). This included both high- and low-risk mucosal types as well as some cutaneous types. In agreement with these studies, we found a strong association between both high- and low-risk mucosal HPV E7 oncoproteins and PTPN14 in a variety of different assays both in vitro and in vivo. In support of a highly conserved interaction, mutational analysis of the HPV-16 E7 oncoprotein identified a domain within the carboxy-terminal half of E7 that plays an essential role in binding PTPN14. In particular, the double amino acid substitution E80K/D81K completely abolished the interaction with PTPN14, while the M84S mutation exhibited greatly reduced levels of interaction. Interestingly, the M84 residue is part of a very highly conserved region of the E7 carboxy terminus, and mutation of this residue significantly impairs the transforming activity of the HPV-16 E7 oncoprotein (32). However, interpretation of these HPV-16 E7 C-terminal mutations is complex. For example, the P92A substitution retains interaction with PTPN14 but is defective in its degradation, while at the same time it shows no defects in its ability to transform cells (32). Thus, while these studies suggest that E7 recognition of PTPN14 occurs through a conserved region of the E7 carboxy terminus, which may also contribute to the ability of E7 to induce cell transformation, it is also clear that other functions are involved. Indeed, a recent study found that residues affecting PTPN14 interaction within the E7 carboxy terminus could also affect p600 interactions (37), and this would be consistent with the results presented here. Nonetheless, considering the wide range of papillomavirus types that can interact with PTPN14, it is likely to be of considerable importance for the normal viral life cycles of multiple HPV types. Intriguingly, we also found that ectopic overexpression of PTPN14 significantly impaired the transforming activity of E7 in cooperation with EJ-ras in primary BRK cells, which was dependent upon PTPN14's substrate recognition function and its enzymatic activity.
To further investigate how E7 might modulate the activity of PTPN14, we first investigated the effects of E7 upon the levels of expression of PTPN14. We found that while PTPN14 protein was readily detectable in HPV-negative cells, the levels in HPV-positive cervical tumor-derived cell lines were very low. Furthermore, treatment with proteasome inhibitors indicated that PTPN14 was particularly susceptible to proteasome-mediated degradation in HPV-positive cells but not in HPV-negative cells. Using a series of siRNA knockdown assays, we confirmed that this low level of PTPN14 protein was a direct consequence of the continued expression of the HPV E7 oncoprotein. When combined with immunofluorescence analyses, we also found that PTPN14 exhibited a weak pattern of expression in HeLa cells, but upon siRNA ablation of E6/E7 expression, there was a dramatic increase in PTPN14 levels in both the cytoplasm and the nucleus. These results demonstrate that HPV E7 expression directly contributes toward the loss of PTPN14 protein throughout the cell and that this loss of PTPN14 involves the proteasome machinery. It is also interesting that any residual PTPN14 found within the HPV-positive cells appears to display a speckled nuclear location, consistent with a residual growth-promoting function of PTPN14 (30). In contrast, the major growth-inhibitory activities of PTPN14 have been associated with its cytoplasmic and cell-cell junction localization (31, 38), suggesting that these are the forms of the protein preferentially targeted by E7, although cell fractionation experiments indicate an increase in PTPN14 protein levels in multiple cellular compartments following loss of E7 expression.
A major activity of the high-risk HPV E7 oncoproteins is their ability to target a number of cellular targets for proteasome-mediated degradation, and this is particularly true for the members of the pRb family of pocket proteins. This activity of E7 appears to require the recruitment of the cullin-2 ubiquitin ligase complex, most likely via a direct association with elongin C (11). However, E7 has also been reported to interact with several other components of the proteasome machinery, including the S4 subunit of the proteasome, the cullin-1 ubiquitin ligase complex, and the p600 ubiquitin ligase (34, 39, 40). Therefore, we sought to determine how E7 might be directing the degradation of PTPN14. We first performed a series of transient cotransfection assays to confirm that E7 could directly drive the degradation of PTPN14. Interestingly, we found that only E7 protein that was C-terminally tagged could induce the degradation of PTPN14, with N-terminally tagged E7 being completely inactive. This demonstrates the critical requirement for an intact E7 N-terminal region, which is also required for the degradation of pRb and for recruitment of components of the cellular proteasome machinery (11, 35, 41).
We then proceeded to investigate which ligase might be required for E7-induced degradation of PTPN14. To do this, we used siRNA ablation of p600, cullin-1, and cullin-2 in HeLa cells and monitored the effects upon pRb and PTPN14 expression. To our surprise, we found that cullin-1 and cullin-2 do not seem to play a role in the ability of HPV-18 E7 to direct the degradation of PTPN14. Instead, the ability of E7 to degrade PTPN14 appears to require p600, since siRNA ablation resulted in a marked increase in the levels of PTPN14 protein. Similar results were also obtained in CaSki cells where again efficient rescue of the levels of PTPN14 expression were obtained following ablation of p600 expression. Again, these results are in broad agreement with those of a recent study also showing that p600 plays an important role in the ability of E7 to target PTPN14 (37).
These studies demonstrate that high-risk HPV E7 oncoproteins are potentially potent inhibitors of PTPN14. This has many important implications, both for the viral life cycle and for the ability of these viruses to contribute toward cell transformation and malignancy. There are now many reports of mutation of PTPN14 in several different human tumors (42–45) and clear evidence for a role for PTPN14 in the control of diverse signaling pathways. This includes regulation of the Hippo pathway, β-catenin signaling, and transforming growth factor β (TGF-β) and receptor trafficking (30, 31, 38, 46). Currently, we have no information on which of these pathways are likely to be affected as a result of E7-induced degradation of PTPN14, although our preliminary unpublished results would appear to indicate that the Hippo pathway is not directly affected by E7. Intriguingly, PTPN14 has been shown to induce TGF-β signaling, and several studies have shown that E7 is also a potent inhibitor of TGF-β (38, 47). Future studies will be aimed at investigating whether any of these pathways are directly affected as a result of E7-induced degradation of PTPN14. In addition, since the low-risk HPV E7 proteins can also interact with PTPN14, it will be important to ascertain whether this can also impact PTPN14 function, either as a result of altered enzymatic activity or subcellular distribution, and how this might thereby contribute toward a successful HPV life cycle.
Taken together, these studies identify PTPN14 as a novel interacting partner of the HPV E7 oncoprotein and demonstrate that it is highly susceptible to HPV E7-induced proteasome-mediated degradation, which most likely involves the recruitment of the p600 ubiquitin ligase.
MATERIALS AND METHODS
Plasmid constructs.
The wild-type and CR3 mutant HA-tagged pcDNA constructs have been described previously (32) as have the GST-tagged pGEX-2T HPV-16 E7 wild-type and deletion mutants (17). The HA/FLAG-tagged pCMV HPV-16 E7 (where CMV is cytomegalovirus) plasmid was a kind gift from Karl Münger (48), and the V5-PTPN14 pcDNA3 plasmid was kindly provided by Jianmin Zhang (26). The V5-PTPN14 pcDNA3 plasmid was used as a template for the generation of the catalytic site mutant C1121S, the substrate binding site mutant D1079A, and the C1121S/D1079A double mutant PTPN14 constructs using a GeneArt site-directed mutagenesis system (Invitrogen) and the following primers: C1121S forward primer 5′-CATCGTGGTCCACAGTAGTGCTGGGGTGGGAAG-3′ and C1121S reverse primer 5′-CTTCCCACCCCAGCACTACTGTGGACCACGATGA-3′; D1079A forward primer 5′-ACTGACTGGCCAGCTCACGGCTGTCCAGAAG-3′ and D1079A reverse primer 5′-CTTCTGGACAGCCGTGAGCTGGCCAGTCAGT-3′. The GST-tagged HPV-11 E7 and HPV-18 E7 pGEX-6P-1 plasmid constructs were generated by amplification and subcloning of the E7 coding sequences from previously made pRB322 plasmid constructs as templates (49, 50). The primers used were the following: HPV-11 E7 forward primer 5′-CCCGGATCCCTGGACAACATGCATGGAAGA-3′ and reverse primer 5′-CCCCTCGAGTCGTCCGCCATCCTTGTTA-3′; HPV-18 E7 forward primer 5′-GCACGGATCCCCAACGACGCAGAGAAACA-3′ and reverse primer 5′-GCACCTCGAGGTTGCTTACTGCTGGGATGC-3′. The amplified sequences were visualized by agarose gel electrophoresis, purified by using a QIAquick gel extraction kit (Qiagen), digested with BamHI and XhoI restriction enzymes, and ligated into pGEX-6P-1. The C-terminal FLAG-tagged HPV-11 E7 CMV plasmid construct was generated by amplification and subcloning of the E7 coding sequences from the HPV-11 E7 pGEX-6P-1 construct as templates to the pCMV Neo Bam (XhoI) empty vector (kindly provided by J. Mymryk). The primers used were the following: HPV-11 E7 forward primer 5′-CGACGGATCCGATTCGAGACCATGCATGGAAGA-3′ and reverse primer 5′-GCATCTCGAGCTACTTGTCATCGTCGTCCTTGTAGTCTGGTTTTGGTGC-3′. The amplified sequences were visualized by agarose gel electrophoresis, purified by using a QIAquick gel extraction kit (Qiagen), digested with BamHI and XhoI restriction enzymes, and ligated into pCMV Neo Bam (XhoI).
The HA/FLAG-tagged pCMV HPV-16 E7 plasmid was used as the template for the generation of 16 E7 mutant constructs by using a modification of a QuickChange site-directed mutagenesis system (Stratagene), according to the manufacturer's instructions, with the following primers: C59S forward primer 5′-TTGTAACCTTTTGTAGCAAGTGTGACTCTACGC-3′ and C59S reverse primer 5′-GCGTAGAGTCACACTTGCTACAAAAGGTTACAA-3′; T64D forward primer 5′-GCAAGTGTGACTCTGATCTTCGGTTGTGCGTAC-3′ and T64D reverse primer 5′-GTACGCACAACCGAAGATCAGAGTCACACTTGC-3′; T72D forward primer 5′-GGTTGTGCGTACAAAGCGACCACGTAGACATTCG-3′ and T72D reverse primer 5′-CGAATGTCTACGTGGTCGCTTTGTACGCACAACC-3′; I76A forward primer 5′-AGCACACACGTAGACGCTCGTACTTTGGAAGAC-3′ and I76A reverse primer 5′-GTCTTCCAAAGTACGAGCGTCTACGTGTGTGCT-3′; R77A forward primer 5′-CACACACGTAGACATTGCTTACTTTGGAAGACC-3′ and R77A reverse primer 5′-GGTCTTCCAAAGTAAGCAATGTCTACGTGTGTG-3′; E80K/D81K forward primer 5′-GACATTCGTACTTTGAAAAAGCTGTTAATGGGCAC-3′ and E80K/D81K reverse primer 5′-GTGCCCATTAACAGCTTTTTCAAAGTACGAATGTC-3′; M84S forward primer 5′-GGAAGACCTGTTAAGTGGCACACTAGGAATTGTG-3′ and M84S reverse primer 5′-CACAATTCCTAGTGTGCCACTTAACAGGTCTTCC-3′; P92A forward primer 5′-CTAGGAATTGTGTGCGCCATCTGTTCTCAGAAAC-3′ and P92A reverse primer 5′-GTTTCTGAGAACAGATGGCGCACACAATTCCTAG-3′. All constructs were verified by DNA sequencing.
Cell culture and transfections.
Cervical-tumor-derived HeLa (HPV-18-positive), CaSki (HPV-16-positive), and C33a (HPV-negative) cells as well as HEK293 and primary baby rat kidney (BRK) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, glutamine (300 μg/ml), and penicillin-streptomycin (100 U/ml).
Transient transfection of plasmid DNA in HEK293 cells was carried out using a standard calcium phosphate precipitation protocol as previously described (51). DNA transfections in C33a cells were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
For the delivery of all siRNAs (Dharmacon), the cells were seeded on 6-cm-diameter dishes and grown for 24 h until they were approximately 40% confluent and were transfected using Lipofectamine RNAiMAX (Invitrogen) with siRNA against either luciferase, HPV-18 E6/E7 (5′-CAUUUACCAGCCCGACGAG-3′), HPV-18 E6 (5′-CUCUGUGUAUGGAGACACAT-3′), PTPN14, or the different ubiquitin ligase proteins (relevant Dharmacon Smart Pools). For siRNA transfection followed by immunofluorescence analysis, HeLa cells were seeded at the same confluence on glass coverslips. After incubation for 72 h, the cells were fixed, and proteins were detected as described later.
Primary BRK cell transformation assays were performed as previously described (52).
Proteasome inhibitors.
The proteasome inhibitors Z-Leu-Leu-Leu-al ([CBZ] MG-132; Sigma) and bortezomib (BTZ; Sigma) were dissolved in dimethyl sulfoxide (DMSO) and used at 20 μM for 8 h unless stated otherwise.
Antibodies.
The following antibodies were used: mouse monoclonal anti-p53 (DO-1; Santa Cruz Biotechnology), mouse monoclonal anti-β-galactosidase (Promega), mouse monoclonal anti-V5 (Life Technologies), mouse monoclonal anti-Rb (G3-245; BD Pharmingen), mouse anti-transferrin receptor (Santa Cruz), mouse anti-p84 (Abcam), and mouse anti-vimentin (Santa Cruz). The following antibodies were purchased from Sigma: rabbit monoclonal anti-PTPN14 (HPA053864), rabbit polyclonal anti-PTPN14 (SAB2700311), mouse monoclonal anti-HA-peroxidase (clone HA-7), mouse monoclonal anti-FLAG-M2-peroxidase, mouse monoclonal anti-α-tubulin, and mouse monoclonal anti-γ-tubulin. Secondary anti-rabbit horseradish peroxidase (HRP) and anti-mouse HRP antibodies were purchased from Dako.
Western blotting and immunoprecipitation.
For Western blot sample preparation, whole-cell extracts were obtained by lysing the cells directly in 2× SDS-PAGE sample buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol [DTT], 4% SDS, 20% glycerol, 0.2% bromophenol blue), or alternatively cells were fractionated into cytoplasmic, membrane, nuclear, and cytoskeletal fractions using a ProteoExtract cell fractionation kit (Calbiochem) according to the manufacturer's instructions. Proteins were separated by SDS-PAGE and blotted on a 0.22-μm-pore-size nitrocellulose membrane (Schleicher & Schuell). Membranes were blocked for 1 h in 5% nonfat dry milk in Tris-buffered saline with Tween 20 ([TBST] 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2.7 mM KCl, 0.1% Tween 20) and probed with the appropriate primary antibodies. After washes with TBST, membranes were probed with appropriate HRP-conjugated secondary antibodies (Dako), if needed. Proteins were detected using ECL (GE Healthcare) according to the manufacturer's instructions.
For coimmunoprecipitation experiments, cells were grown in the presence of proteasome inhibitor for 8 to 10 h and then extracted on ice in lysis buffer (0.1% NP-40, 50 mM HEPES, pH 7.0, 250 mM NaCl) supplemented with protease inhibitor cocktail (Set 1; Calbiochem). Extracts from cells expressing FLAG-tagged HPV-16 E7 or HA-tagged HPV-16 E7 constructs were incubated with anti-FLAG beads or anti-HA beads (both from Sigma), respectively. Samples were incubated with the beads for 1 to 2 h on a rotating wheel at 4°C. The beads were then extensively washed, and the immunoprecipitated proteins were analyzed by Western blotting.
Fusion protein purification and in vitro binding assays.
GST-tagged fusion proteins were expressed as follows. An overnight culture of Escherichia coli strain DH5α transformed with the appropriate expression plasmid was passaged, and, once in log phase, the expression of recombinant protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) to a final concentration of 1 mM. After 3 h, the cells were harvested, resuspended in phosphate-buffered saline (PBS)–1% Triton X-100, and lysed by sonication for 30 s. The cleared supernatant was incubated with glutathione-conjugated agarose beads for 1 h at 4°C and washed extensively with PBS–1% Triton. SDS-PAGE and Coomassie blue staining were used to assess the purity of the fusion protein.
For in vitro binding assays, proteins were transcribed and translated in vitro in rabbit reticulocyte lysate using a Promega TNT coupled transcription-translation system according to the manufacturer's instructions. PTPN14 proteins were radiolabeled with [35S]methionine. Equal amounts of in vitro-translated, radiolabeled proteins were added to GST fusion proteins bound to glutathione resin and incubated for 1 h at room temperature. After extensive washing with PBS containing 0.2% NP-40, the bound proteins were analyzed by SDS-PAGE and autoradiography, and GST fusion proteins were visualized by Coomassie staining.
Immunofluorescence microscopy.
Cells were grown on glass coverslips, fixed with 4% paraformaldehyde in PBS for 15 min, and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Immunolabeling was carried out using an anti-PTPN14 rabbit monoclonal antibody (Sigma) and anti-p53 mouse monoclonal antibody (Santa Cruz Biotech) overnight at 4°C. Cells then were extensively washed in PBS and incubated for 30 min at 37°C with secondary anti-rabbit and anti-mouse antibodies conjugated to fluorescein or rhodamine (Molecular Probes). Samples were washed and mounted with Vectashield mounting medium (Vector Laboratories). Cells were viewed on a Zeiss LSM 510 confocal microscope under an oil immersion 60× objective lens. Images were analyzed using LSM imaging software.
ACKNOWLEDGMENTS
This work was supported partly by the Campus Hungary Fellowship for Higher Education Staff Mobility (B2/4R/19122 and B2/4H/13274), an ICGEB Arturo Falachi Postdoctoral Fellowship awarded to A.S., and a research grant from the Associazione Italiana per la Ricerca sul Cancro awarded to L.B.
We are grateful to Jianmin Zhang for kindly providing the PTPN14-V5 pcDNA plasmid construct, to Karl Münger for kindly providing the HPV-16 E7 pCMV plasmid construct, and to Miranda Thomas, Jayashree Thatte, and David Pim for help with experimental work and for comments on the manuscript.
REFERENCES
- 1.zur Hausen H. 2009. Papillomaviruses in the causation of human cancers—a brief historical account. Virology 384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 2.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V, WHO International Agency for Research on Cancer Monograph Working Group. 2009. A review of human carcinogens—part B: biological agents. Lancet Oncol 10:321–322. doi: 10.1016/S1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 3.Ghittoni R, Accardi R, Chiocca S, Tommasino M. 2015. Role of human papillomaviruses in carcinogenesis. Ecancermedicalscience 9:526. doi: 10.3332/ecancer.2015.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egawa N, Egawa K, Griffin H, Doorbar J. 2015. Human papillomaviruses, epithelial tropisms, and the development of neoplasia. Viruses 7:3863–3890. doi: 10.3390/v7072802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smotkin D, Wettstein FO. 1986. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc Natl Acad Sci U S A 83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamid NA, Brown C, Gaston K. 2009. The regulation of cell proliferation by the papillomavirus early proteins. Cell Mol Life Sci 66:1700–1717. doi: 10.1007/s00018-009-8631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng S, Schmidt-Grimminger DC, Murant T, Broker TR, Chow LT. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev 9:2335–2349. doi: 10.1101/gad.9.19.2335. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee NS, Genovese NJ, Noya F, Chien WM, Broker TR, Chow LT. 2006. Conditionally activated E7 proteins of high-risk and low-risk human papillomaviruses induce S phase in postmitotic, differentiated human keratinocytes. J Virol 80:6517–6524. doi: 10.1128/JVI.02499-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howie HL, Katzenellenbogen RA, Galloway DA. 2009. Papillomavirus E6 proteins. Virology 384:324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roman A, Munger K. 2013. The papillomavirus E7 proteins. Virology 445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, Harper JW, Munger K. 2007. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol 81:9737–9747. doi: 10.1128/JVI.00881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pim D, Banks L. 2010. Interaction of viral oncoproteins with cellular target molecules: infection with high-risk vs low-risk human papillomaviruses. APMIS 118:471–493. doi: 10.1111/j.1600-0463.2010.02618.x. [DOI] [PubMed] [Google Scholar]
- 13.Menges CW, Baglia LA, Lapoint R, McCance DJ. 2006. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res 66:5555–5559. doi: 10.1158/0008-5472.CAN-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charette ST, McCance DJ. 2007. The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene 26:7386–7390. doi: 10.1038/sj.onc.1210541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertelsen BI, Steine SJ, Sandvei R, Molven A, Laerum OD. 2006. Molecular analysis of the PI3K-AKT pathway in uterine cervical neoplasia: frequent PIK3CA amplification and AKT phosphorylation. Int J Cancer 118:1877–1883. doi: 10.1002/ijc.21461. [DOI] [PubMed] [Google Scholar]
- 16.Yuan H, Ito S, Senga T, Hyodo T, Kiyono T, Kikkawa F, Hamaguchi M. 2009. Human papillomavirus type 16 oncoprotein E7 suppresses cadherin-mediated cell adhesion via ERK and AP-1 signaling. Int J Oncol 35:309–314. [PubMed] [Google Scholar]
- 17.Pim D, Massimi P, Dilworth SM, Banks L. 2005. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 24:7830–7838. doi: 10.1038/sj.onc.1208935. [DOI] [PubMed] [Google Scholar]
- 18.Katich SC, Zerfass-Thome K, Hoffmann I. 2001. Regulation of the Cdc25A gene by the human papillomavirus type 16 E7 oncogene. Oncogene 20:543–550. doi: 10.1038/sj.onc.1204130. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen DX, Westbrook TF, McCance DJ. 2002. Human papillomavirus type 16 E7 maintains elevated levels of the cdc25A tyrosine phosphatase during deregulation of cell cycle arrest. J Virol 76:619–632. doi: 10.1128/JVI.76.2.619-632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabási AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Münger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White EA, Howley PM. 2013. Proteomic approaches to the study of papillomavirus-host interactions. Virology 435:57–69. doi: 10.1016/j.virol.2012.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Hertog J, Ostman A, Böhmer FD. 2008. Protein tyrosine phosphatases: regulatory mechanisms. FEBS J 275:831–847. doi: 10.1111/j.1742-4658.2008.06247.x. [DOI] [PubMed] [Google Scholar]
- 23.Tautz L, Critton DA, Grotegut S. 2013. Protein tyrosine phosphatases: structure, function, and implication in human disease. Methods Mol Biol 1053:179–221. doi: 10.1007/978-1-62703-562-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith AL, Mitchell PJ, Shipley J, Gusterson BA, Rogers MV, Crompton MR. 1995. Pez: a novel human cDNA encoding protein tyrosine phosphatase- and ezrin-like domains. Biochem Biophys Res Commun 209:959–965. doi: 10.1006/bbrc.1995.1591. [DOI] [PubMed] [Google Scholar]
- 25.Bosanquet DC, Ye L, Harding KG, Jiang WG. 2014. FERM family proteins and their importance in cellular movements and wound healing (review). Int J Mol Med 34:3–12. doi: 10.3892/ijmm.2014.1775. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Yang N, Figel SA, Wilson KE, Morrison CD, Gelman IH, Zhang J. 2013. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 32:1266–1273. doi: 10.1038/onc.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park JI, Chen J. 2012. PTPN14 is required for the density-dependent control of YAP1. Genes Dev 26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaloglou C, Lehmann W, Martin T, Delaunay C, Hueber A, Barys L, Niu H, Billy E, Wartmann M, Ito M, Wilson CJ, Digan ME, Bauer A, Voshol H, Christofori G, Sellers WR, Hofmann F, Schmelzle T. 2013. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS One 8:e61916. doi: 10.1371/journal.pone.0061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson KE, Li YW, Yang N, Shen H, Orillion AR, Zhang J. 2014. PTPN14 forms a complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J Biol Chem 289:23693–23700. doi: 10.1074/jbc.M113.534701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. 2003. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin. Mol Biol Cell 14:2520–2529. doi: 10.1091/mbc.E02-09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belle L, Ali N, Lonic A, Li X, Paltridge JL, Roslan S, Herrmann D, Conway JR, Gehling FK, Bert AG, Crocker LA, Tsykin A, Farshid G, Goodall GJ, Timpson P, Daly RJ, Khew-Goodall Y. 2015. The tyrosine phosphatase PTPN14 (Pez) inhibits metastasis by altering protein trafficking. Sci Signal 8:ra18. doi: 10.1126/scisignal.2005547. [DOI] [PubMed] [Google Scholar]
- 32.Todorovic B, Massimi P, Hung K, Shaw GS, Banks L, Mymryk JS. 2011. Systematic analysis of the amino acid residues of human papillomavirus type 16 E7 conserved region 3 involved in dimerization and transformation. J Virol 85:10048–10057. doi: 10.1128/JVI.00643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohlenschläger O, Seiboth T, Zengerling H, Briese L, Marchanka A, Ramachandran R, Baum M, Korbas M, Meyer-Klaucke W, Dürst M, Görlach M. 2006. Solution structure of the partially folded high-risk human papilloma virus 45 oncoprotein E7. Oncogene 25:5953–5959. doi: 10.1038/sj.onc.1209584. [DOI] [PubMed] [Google Scholar]
- 34.Huh KW, DeMasi J, Ogawa H, Nakatani Y, Howley PM, Münger K. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci U S A 102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. 2000. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- 36.Ogata M, Takada T, Mori Y, Oh-hora M, Uchida Y, Kosugi A, Miyake K, Hamaoka T. 1999. Effects of overexpression of PTP36, a putative protein tyrosine phosphatase, on cell adhesion, cell growth, and cytoskeletons in HeLa cells. J Biol Chem 274:12905–12909. doi: 10.1074/jbc.274.18.12905. [DOI] [PubMed] [Google Scholar]
- 37.White EA, Münger K, Howley PM. 2016. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. mBio 7:e01530-16. doi: 10.1128/mBio.01530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyatt L, Wadham C, Crocker LA, Lardelli M, Khew-Goodall Y. 2007. The protein tyrosine phosphatase Pez regulates TGFβ, epitheial-mesenchymal transition, and organ development. J Cell Biol 178:1223–1235. doi: 10.1083/jcb.200705035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berezutskaya E, Bagchi S. 1997. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. J Biol Chem 272:30135–30140. doi: 10.1074/jbc.272.48.30135. [DOI] [PubMed] [Google Scholar]
- 40.Oh KJ, Kalinina A, Wang J, Nakayama KI, Bagchi S. 2004. The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and cullin 1- and Skp2-containing E3 ligase. J Virol 78:5338–5346. doi: 10.1128/JVI.78.10.5338-5346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brokaw JL, Yee CL, Münger K. 1994. A mutational analysis of the amino terminal domain of the human papillomavirus type 16 E7 oncoprotein. Virology 205:603–607. doi: 10.1006/viro.1994.1688. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, Parmigiani G, Yan H, Wang TL, Riggins G, Powell SM, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. 2004. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas Research Network. 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 45.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareño C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science 333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson KE, Yang N, Mussell AL, Zhang J. 2016. The regulatory role of KIBRA and PTPN14 in Hippo signaling and beyond. Genes (Basel). 7:E23. doi: 10.3390/genes7060023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyatt L, Khew-Goodall Y. 2008. PTP-Pez: a novel regulator of TGFβ signaling. Cell Cycle 7:2290–2295. doi: 10.4161/cc.6443. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol 75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gáll T, Kis A, Tatár TZ, Kardos G, Gergely L, Szarka K. 2013. Genomic differences in the background of different severity in juvenile-onset respiratory papillomatoses associated with human papillomavirus type 11. Med Microbiol Immunol 202:353–263. doi: 10.1007/s00430-013-0297-y. [DOI] [PubMed] [Google Scholar]
- 50.Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. 1984. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J 3:1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C, Okayama H. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7:2745–2752. doi: 10.1128/MCB.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas M, Massimi P, Navarro C, Borg JP, Banks L. 2005. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24:6222–6230. doi: 10.1038/sj.onc.1208757. [DOI] [PubMed] [Google Scholar]