ABSTRACT
Cyclic GMP-AMP synthase (cGAS) is a newly identified DNA sensor that recognizes foreign DNA, including the genome of herpes simplex virus 1 (HSV-1). Upon binding of viral DNA, cGAS produces cyclic GMP-AMP, which interacts with and activates stimulator of interferon genes (STING) to trigger the transcription of antiviral genes such as type I interferons (IFNs), and the production of inflammatory cytokines. HSV-1 UL24 is widely conserved among members of the herpesviruses family and is essential for efficient viral replication. In this study, we found that ectopically expressed UL24 could inhibit cGAS-STING-mediated promoter activation of IFN-β and interleukin-6 (IL-6), and UL24 also inhibited interferon-stimulatory DNA-mediated IFN-β and IL-6 production during HSV-1 infection. Furthermore, UL24 selectively blocked nuclear factor κB (NF-κB) but not IFN-regulatory factor 3 promoter activation. Coimmunoprecipitation analysis demonstrated that UL24 bound to the endogenous NF-κB subunits p65 and p50 in HSV-1-infected cells, and UL24 was also found to bind the Rel homology domains (RHDs) of these subunits. Furthermore, UL24 reduced the tumor necrosis factor alpha (TNF-α)-mediated nuclear translocation of p65 and p50. Finally, mutational analysis revealed that the region spanning amino acids (aa) 74 to 134 of UL24 [UL24(74–134)] is responsible for inhibiting cGAS-STING-mediated NF-κB promoter activity. For the first time, UL24 was shown to play an important role in immune evasion during HSV-1 infection.
IMPORTANCE NF-κB is a critical component of the innate immune response and is strongly induced downstream of most pattern recognition receptors (PRRs), leading to the production of IFN-β as well as a number of inflammatory chemokines and interleukins. To establish persistent infection, viruses have evolved various mechanisms to counteract the host NF-κB pathway. In the present study, for the first time, HSV-1 UL24 was demonstrated to inhibit the activation of NF-κB in the DNA sensing signal pathway via binding to the RHDs of the NF-κB subunits p65 and p50 and abolishing their nuclear translocation.
KEYWORDS: HSV-1, DNA sensor, UL24, NF-κB, p65
INTRODUCTION
The innate immune system is the first line of defense against viral infection. The first step in innate immunity is the detection of the invading pathogen. Pathogen recognition receptors (PRRs) recognize pathogen-associated molecular patterns to trigger the production of type I interferons (IFNs) and other antiviral immune responses (1–3). Viral nucleic acids exposed in the cytoplasm are signs of foreign invasion and can be recognized by a subset of host cell PRRs. The membrane-bound Toll-like receptors recognize endosomal nucleic acids, the family of RIG-I-like receptors recognizes viral RNAs, and a broad class of putative DNA sensors recognizes viral DNA (4–6). A number of DNA sensors have been discovered in recent years, including cyclic GMP-AMP (cGAMP) synthase (cGAS), gamma interferon-inducible protein 16, DNA-dependent activator of IFN-regulatory factors (IRFs), absent in melanoma 2, RNA polymerase III, Mre11, and Ku70/DNA-PK (7–16). Most of these DNA sensors activate downstream signaling by the same signal transducer, stimulator of interferon genes (STING). STING, also known as MPYS, MITA, ERIS, and TMEM173, is localized to the endoplasmic reticulum. As an adaptor molecule in the DNA sensing signal pathway, STING plays an important role in the regulation of immune responses (17–19). Recently, cGAS was reported to play a more important role in detecting herpes simplex virus 1 (HSV-1) DNA (11). After recognizing DNA, cGAS produces cGAMP to stimulate STING activation, leading to the recruitment of TANK-binding kinase 1 (TBK1) to phosphorylate IRF3 and nuclear factor κB (NF-κB). IRF3 and NF-κB then translocate into the nucleus to induce the production of IFN-β as well as a number of inflammatory chemokines and interleukins (20, 21).
During the activation of NF-κB (22, 23), the IκB kinase (IKK) complex, which comprises IKKα, IKKβ, and NF-κB essential modulators, is activated by upstream signals. IκBs are phosphorylated by activated IKK. K48-linked polyubiquitin chains then target the phosphorylated IκBs for proteasome-dependent degradation. The NF-κB p50/p65 heterodimer is released, translocates to the nucleus, and then regulates the innate immune response (24–27). Recent evidence suggests that TBK1 is necessary for the activation of the NF-κB signaling pathway mediated by double-stranded DNA (dsDNA) and utilizes the IKKα/β activation loop to activate the NF-κB subunit p65 in response to STING (21).
HSV-1, an archetypal member of the herpesvirus family, is widespread in the population. HSV-1 causes lifelong infections, and there is no effective vaccine to prevent infection or drug to prevent latency. HSV infection increases the risk of acquiring and transmitting human immunodeficiency virus (28). HSV-1 can replicate productively in infected cells, or it can establish latent infections under specific conditions (29). During HSV-1 replication, viral DNA that leaks into the cytosol can be sensed by cGAS, which activates cGAS-dependent innate immune responses (11, 30, 31). However, the herpesvirus family has acquired the ability to subvert the cGAS-cGAMP signaling pathway to evade the innate immune response (32–35). Our recent study showed that the HSV-1 serine protease VP24 could block dsDNA-triggered IFN-β production by inhibiting IRF3 activation, and the HSV-1 ubiquitin-specific protease UL36 abrogates NF-κB activation in the DNA sensing signal pathway by deubiquitinated IκBα and restricted IκBα degradation (36, 37). HSV-1 UL24 is conserved among members of the herpesviruses family. UL24 is required for efficient viral replication during HSV-1 infection and plays an important role in reactivation from latency (38–40). UL24 also contributes to HSV-1-induced nucleolar modification (40–42). However, whether UL24 affects the host DNA sensing pathways is unknown.
RESULTS
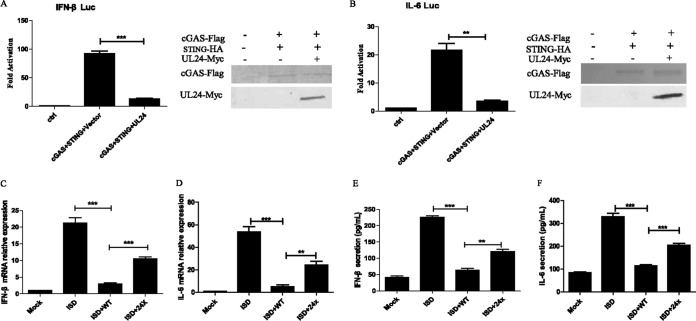
UL24 inhibits the activation of the IFN-β and IL-6 promoter induced by cGAS-STING and ISD-mediated production of IFN-β and IL-6.
Previous studies from our laboratory and other laboratories showed that the ectopic expression of a minimal amount of either cGAS or STING alone was unable to induce the activation of IFN-β in HEK293T cells, and the IFN-β promoter could be activated only when minimal amounts of cGAS and STING were cotransfected (11, 36, 37, 43). To investigate the role of UL24 in the cGAS-STING-mediated DNA sensing signal pathway, a Myc-tagged UL24 expression plasmid was transfected into HEK293T cells along with cGAS and STING expression plasmids and IFN-β promoter or interleukin-6 (IL-6) promoter constructs and then subjected to dual-luciferase reporter (DLR) assays to detect the promoter activity of IFN-β and IL-6. As shown in Fig. 1A and B, the ectopic expression of UL24 significantly inhibited the cGAS-STING-mediated activation of the IFN-β and IL-6 promoter.
FIG 1.
UL24 inhibits the activation of the IFN-β and IL-6 promoter induced by cGAS-STING and ISD-mediated production of IFN-β and IL-6. (A and B) HEK293T cells were cotransfected with an IFN-β–Luc (A) or IL-6–Luc (B) reporter plasmid, the pRL-TK control plasmid along with an empty vector, and plasmids encoding cGAS (15 ng) and STING (2.5 ng) with UL24. Luciferase activity was measured at 24 h posttransfection, and fold activation was determined compared to the empty vector. The expression of cGAS, STING, and UL24 was analyzed by WB using anti-Flag, anti-HA, and anti-Myc monoclonal antibodies. (C and D) HFF cells were infected with WT HSV-1 and the HSV-1 UL24X mutant at an MOI of 2 for 2 h and transfected with ISD (4 μg/ml) for another 10 h. Cells were harvested and subjected to RT-PCR to detect IFN-β (C) or IL-6 (D) mRNA. (E and F) HFF cells were infected with WT HSV-1 or the HSV-1 UL24X mutant and then transfected with ISD for another 16 h. Supernatants were subjected to an ELISA to detect IFN-β (E) or IL-6 (F). The data represent results from one of three independent experiments. Error bars represent standard deviations of data from three independent experiments. Statistical analysis was performed by using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns indicates that the comparison is not significant).
Human foreskin fibroblast (HFF) cells have the ability to respond to foreign DNA. When treated with exogenous DNA, such as an interferon-stimulatory DNA (ISD) fragment from the HSV-1 genome (HSV60), HFF cells mounted an IFN-β response. To further determine the role of UL24 in the regulation of IFN-β and IL-6 production during HSV-1 infection, HFF cells were infected with wild-type (WT) HSV-1 or UL24X at a multiplicity of infection (MOI) of 2, and quantitative PCR (qPCR) was performed to measure the accumulation of IFN-β and IL-6 mRNAs. As shown in Fig. 1C and D, transfection with ISD induced high levels of IFN-β and IL-6 mRNAs in HFF cells; however, infection with both WT HSV-1 and UL24X exhibited impaired IFN-β and IL-6 mRNA expression, whereas infection with UL24X partly restored IFN-β and IL-6 mRNA expression, indicating that UL24 inhibited the ISD-triggered activation of IFN-β and NF-κB. To further confirm this result, enzyme-linked immunosorbent assays (ELISAs) were performed to measure the secretion of IFN-β and IL-6 in the supernatant of HFF cells infected by WT HSV-1 or UL24X at an MOI of 2 after transfection with ISD. As a result, levels of both IFN-β and IL-6 were significant decreased in cells infected with WT HSV-1, while UL24X infection recovered them to a certain extent (Fig. 1E and F). These results indicated that the cGAS-STING-mediated DNA sensing signaling pathway was suppressed by UL24.
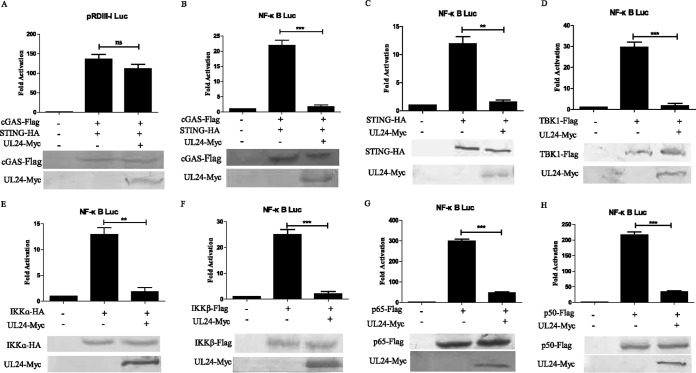
UL24 inhibits the cGAS-STING-mediated NF-κB signaling pathway via p65 and p50.
NF-κB, activator protein 1 (AP-1), and IRF family members are important transcription factors that regulate the expression of chemokines, proinflammatory cytokines, and type I IFNs. To investigate whether UL24 inhibited the cGAS-STING-mediated activation of IRF3 and NF-κB, reporter gene assays were performed in HEK293T cells in the presence of NF-κB–Luc and PRD(III-I)-Luc reporter genes. The results showed that the ectopic expression of UL24 inhibited NF-κB–Luc but not PRD(III-I)-Luc promoter activity induced by the transfection of trace amounts of cGAS and STING (Fig. 2A and B).
FIG 2.
UL24 inhibits the cGAS-STING-mediated NF-κB signaling pathway via p65 and p50. (A-B) HEK293T cells were transfected with 200 ng of the PRD(III-I)-Luc (A) or NF-κB–Luc (B) promoter reporter plasmid, renilla luciferase plasmid pRL-TK (50 ng), and expression plasmids for cGAS (15 ng) and STING (2.5 ng) along with pCMV-Flag or pCMV-UL24 (200 ng). (C to G) HEK293T cells were cotransfected with the NF-κB–Luc, pRL-TK, and STING (B), TBK1 (C), IKKα (D), IKKβ (E), or p65 (F) expression plasmids along with empty vector or a plasmid encoding UL24, and cells were harvested 24 h after transfection and subjected to a DLR assay. Cell lysates were analyzed by WB with tag-specific Abs (bottom) to detect the expression of cGAS-Flag, STING-HA, TBK1-Flag, IKKα-HA, IKKβ-Flag, p65-Flag, and p50-Flag, and UL24 expression was detected by using mouse anti-Myc MAb. The data represent results from one of three independent experiments. Error bars represent standard deviations of data from three independent experiments. Statistical analysis was performed by using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns indicates that the comparison is not significant).
To determine the step in the signaling pathway at which UL24 blocked the activation of NF-κB, UL24 and expression plasmids of NF-κB signaling pathway components, including STING, TBK1, IKKα, IKKβ, p65, and p50, were cotransfected into HEK293T cells. All expression constructs resulted in a 10- to 300-fold induction of NF-κB–Luc reporter activity (Fig. 2C to G). NF-κB promoter activation driven by all of the expression constructs was inhibited by UL24 (Fig. 2C to G). These results indicated that UL24 might inhibit the cGAS-STING-mediated NF-κB signaling pathway at or downstream of p65 and p50.
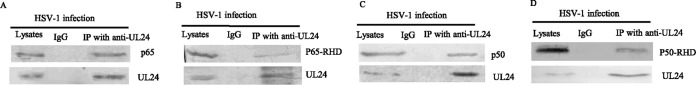
UL24 interacts with endogenous p65 and p50.
The above-mentioned results demonstrated that UL24 might inhibit the NF-κB signaling pathway at the level of p65 and p50. In order to clarify the underlying molecular mechanisms by which UL24 suppressed NF-κB activation, we analyzed the potential interaction between the NF-κB subunit p65 or p50 and UL24. HEK293T cells were infected with WT HSV-1, and coimmunoprecipitation (co-IP)/Western blot (WB) analysis was performed with anti-p65 and anti-UL24 antibodies (Abs). We found that the p65 or p50 protein was immunoprecipitated by UL24 (Fig. 3A and C). These results demonstrated that UL24 interacted with endogenous p65 and p50. Both p65 and p50 contain a DNA-binding Rel homology domain to determine if the UL24 protein bound to the Rel homology domain of p65 or p50. HEK293T cells were infected with WT HSV-1 24 h after transfection of the Flag-p65 RHD or Flag-p50 RHD expression plasmid. The results showed that Flag-p65 RHD or Flag-p50 RHD was efficiently coimmunoprecipitated by UL24 (Fig. 3B and D). Taken together, these results suggested that UL24 interacts with the RHD of p65 and p50 during viral infection.
FIG 3.
UL24 interacts with endogenous p65 and p50. (A and C) HEK293T cells were infected with WT HSV-1 at an MOI of 5 for 16 h. The cells were lysed, and the extracts were subjected to IP using anti-UL24 PAb or control IgG. Precipitates were analyzed by WB. (B and D) HEK 293T cells were transfected with the p65 RHD-Flag (B) or p50 RHD-Flag (D) expression plasmid. Thirty-six hours after transfection, cells were infected with WT HSV-1 at an MOI of 5 for 16 h. The cells were lysed, and the extracts were subjected to IP using anti-24 PAb or control IgG. Precipitates were analyzed by WB.
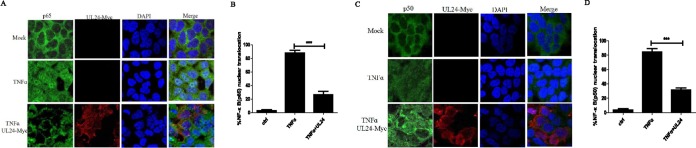
UL24 reduces the nuclear translocation of p65 and p50.
The nuclear translocation of p65 and p50 is crucial for the transcription of NF-κB-related genes. We hypothesized that the interaction between UL24 and the NF-κB subunit p65 or p50 might reduce its nuclear trafficking. To explore this hypothesis, an indirect immunofluorescence assay was performed. HeLa cells were transfected with or without the UL24-Myc expression plasmid and stimulated with TNF-α for 30 min, and the nuclear translocation of endogenous p65 and p50 was monitored. As shown in Fig. 4A and C, the ectopic expression of UL24 reduced the nuclear translocation of p65 and p50 induced by TNF-α. In addition, quantification of p65 or p50 nuclear translocation was also performed, and the results showed that the nuclear translocation of p65 and p50 was inhibited in cells expressing the UL24 protein (Fig. 4B and D). Taken together, these data suggested that the interaction between UL24 and p65 or p50 was sufficient to reduce their nuclear accumulation and thereby affect NF-κB transcription.
FIG 4.
UL24 prevents nuclear translocation of p65 and p50. (A and C) HeLa cells were transfected with an empty vector or a Myc-tagged UL24 expression plasmid. At 24 h posttransfection, cells were treated with or without recombinant human TNF-α (10 ng/ml) for 30 min, as indicated. Cells were stained with mouse anti-Myc MAb and rabbit anti-p65 PAb or anti-p50 PAb. FITC-conjugated goat anti-rabbit and TRITC-conjugated goat anti-mouse antibodies were used as secondary antibodies. Cell nuclei were stained with Hoechst 33258. DAPI, 4′,6-diamidino-2-phenylindole. (B and D) Quantification of p65 or p50 nuclear translocation. Percentages of p65 or p50 nuclear translocation were determined by counting 100 to 150 cells in 3 or 4 nonoverlapping fields. Results were evaluated by two-tailed paired Student's t test.
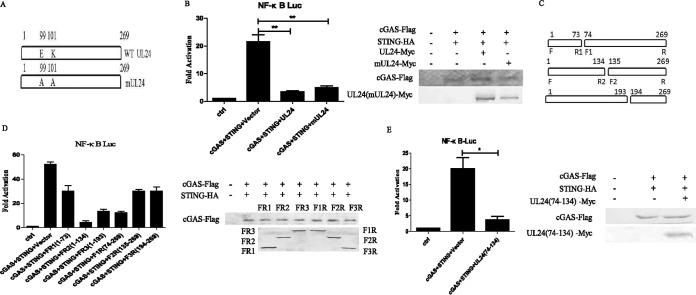
The endonuclease motif of UL24 is dispensable for its inhibition of NF-κB activation by cGAS-STING.
Previous reports have shown that UL24 can induce the distribution of nucleolin and affects the subcellular localization of the nucleolar protein B23, and the endonuclease activity of UL24 was important for the dispersal of nucleolin and B23. To examine whether the endonuclease motif of UL24 is required for its inhibition of the DNA sensor signaling pathway, the mUL24 mutant (E99A/K101A) containing substitutions of critical residues in the endonuclease motif was constructed (Fig. 5A). The mutated construct was cotransfected into HEK293T cells with reporter plasmids to examine its ability to inhibit cGAS-STING-induced NF-κB promoter activation. Interestingly, mUL24 still blocked the activation of the NF-κB reporter induced by the transfection of trace amounts of cGAS and STING (Fig. 5B). To determine which region of UL24 was responsible for the inhibition of cGAS-STING-mediated activation of the NF-κB promoter, a series of UL24 expression plasmids, including FR1(1–73)-Myc, FR2(1–134)-Myc, UL24-FR3(1–193)-Myc, F1R(74–269)-Myc, F2R(135–269)-Myc, and F3R(194–269)-Myc, were constructed (Fig. 5C). The expression of FR2(1–134)-Myc, FR3(1–193)-Myc, and F1R(74–269)-Myc, containing the domain spanning amino acids (aa) 74 to 134, blocked cGAS-STING-mediated NF-κB activation, while the expression of FR1(1–74)-Myc, F2R(135–269)-Myc, and F3R(193–269)-Myc only slightly inhibited cGAS-STING-mediated NF-κB activation (Fig. 5D). We next constructed UL24(74–134)-Myc, and the reduction of NF-κB–Luc activity confirmed that UL24(74–134) was sufficient for its inhibition (Fig. 5E). These results indicated that the region spanning aa 74 to 134 is responsible for UL24 blocking the production of IFN-β mediated by cGAS-STING, and the endonuclease motifs of UL24 are dispensable.
FIG 5.
The endonuclease motif of UL24 is dispensable for its inhibition of NF-κB activation by cGAS-STING. (A) Schematic representation of the wild type and the mUL24 (E99A/K101A) mutant. (B) Luciferase assays in HEK293T cells were performed as described in the legend of Fig. 1A to measure the activation of the NF-κB promoter following the expression of cGAS (15 ng) and STING (2.5 ng) in the presence of full-length UL24 and the mUL24 (E99A/K101A) mutant. (C) Schematic representation of deletion mutants of UL24, including FR1(1–73), F1R(74–269), FR2(1–134), F2R(135–269), FR3(1–193), and F3R(194–269). (D and E) Luciferase assays in HEK293T cells were performed as described in the legend of Fig. 1A to measure the activation of the NF-κB promoter following the expression of cGAS (15 ng) and STING (2.5 ng) in the presence of deletion mutants of UL24. The expression levels of cGAS, STING, UL24, mUL24, and deletion mutants of UL24 were analyzed by WB using anti-Flag, anti-HA, and anti-Myc monoclonal antibodies. The data represent results from one of three independent experiments. Error bars represent standard deviations of data from three independent experiments. Statistical analysis was performed by using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns indicates that the comparison is not significant).
DISCUSSION
During herpesviruses infection, herpesvirus DNA is released into the nucleus or the cytoplasm depending on the cell type (44). Herpesvirus infection has been shown to induce mitochondrial stress, and mitochondrial DNA (mtDNA) is then released into the cytoplasm (45). DNA in the cytoplasm triggers the activation of the cGAS-STING DNA sensing pathway, which results in the activation of IRF3 and NF-κB (11, 21). IFNs and inflammatory chemokines are then produced to suppress the replication of herpesviruses. It has been demonstrated that HSV-1 encodes a number of viral proteins to evade antiviral innate immunity. In this study, we have shown for the first time that UL24 plays an important role in immune evasion during HSV-1 infection.
The transcription factor NF-κB is critical for the regulation of innate immunity and inflammation in response to infections or tissue damage (46). Many cellular PRRs, including Toll-like receptors, RIG-I-like helicases, and NOD family proteins, can strongly activate the NF-κB signaling pathway (47). The mechanism of the NF-κB activation signaling pathway induced by PRRs has been extensively studied; however, current studies have not fully delineated the signal transduction mechanisms of NF-κB activation induced by DNA sensors (48). A recent study suggests that TBK1 utilizes the IKKα/β activation loop to activate NF-κB p65 in response to STING (21). In the present study, our results revealed for the first time that HSV-1 UL24 markedly impaired the activation of the NF-κB–Luc promoter induced by cGAS-STING. Furthermore, UL24 inhibited the TBK1, IKKα, and IKKβ p65- or p50-mediated activation of NF-κB–Luc promoter activity, indicating that UL24 might act at or downstream of p65 and p50.
Since NF-κB is a crucial component of host immunity involving the regulation of the antiviral response, HSV-1 has evolved various elaborate mechanisms to subvert it (47, 49). For example, the HSV-1 immediate early protein ICP0 interacts with p65 and p50 and degrades p50 through its E3 ubiquitin ligase activity (50). The HSV-1 protein kinase US3 was shown previously to hyperphosphorylate p65 at serine 75 and block its nuclear translocation, which inhibited NF-κB activity (51). HSV-1 ICP27 blocked the phosphorylation and ubiquitination of IκB to inhibit NF-κB activation (52). HSV-1 UL42, a DNA polymerase processivity factor, prevented NF-κB-dependent gene expression by retaining p65 and p50 in the cytoplasm (53). In this study, we show that UL24 interacts with p65 and p50, inhibits their translocation to the nucleus, and consequently impairs the production of IFN-β and inflammatory chemokines. The NF-κB signaling pathway is activated through multiple signaling pathways to reduce viral infection; meanwhile, HSV-1 can inhibit the major part of this innate response by using viral gene products expressed during infection (54). UL24 may be one of the regulators that maintains the balance between viral infection and the antiviral response.
UL24 is an important gene that has been reported to be a virulence determinant in several alphaherpesvirinae (38, 39, 55, 56). Based on data from previous reports, UL24 is involved in the dispersal of both nucleolin and B23, and the PD-(D/E)XK endonuclease motif of UL24 is important for this function, although nuclease activity has yet to be demonstrated (41, 42, 57). However, in this study, the endonuclease motif of UL24 was shown to be dispensable for the inhibition of NF-κB activation mediated by cGAS-STING. Collectively, these findings suggest that HSV-1 has evolved mechanisms to antagonize host cGAS-mediated DNA sensing signaling. Further studies in this area may reveal additional strategies of how HSV-1 subverts host DNA sensing antiviral immune responses.
In conclusion, we describe a new function of HSV-1 UL24 that inhibits the DNA sensor signaling pathway. Our results reveal that UL24 efficiently inhibited DNA sensor-mediated IFN-β and IL-6 production due to the inhibition of the activation of NF-κB. Furthermore, we provide convincing evidence that UL24 inhibited NF-κB subunit p65- and p50-mediated transactivation by interacting with p65 and p50 and preventing their nuclear translocation. These findings will help our understanding of the interaction between HSV-1 replication and the host DNA sensing signal pathway.
MATERIALS AND METHODS
Cells, viruses, antibodies, and reagents.
HEK293T cells, Vero cells, and HFF cells were grown in Dulbecco's modified Eagle medium (DMEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL) and 100 U/ml of penicillin and streptomycin. HeLa cells were maintained in Eagle's minimum essential medium (MEM) (Gibco-BRL) supplemented with 10% FBS (58, 59). The WT HSV-1 KOS strain and the UL24 mutant virus (UL24X) were originally from Don Coen's laboratory (39). These viruses were propagated in Vero cells, and their titers were determined as described previously (59). Mouse anti-Myc and anti-Flag monoclonal antibodies (MAbs) were purchased from ABmart (Shanghai, China). Mouse monoclonal IgG1 antibodies and rabbit polyclonal anti-p65 and anti-p50 were purchased from Proteintech (Wuhan, China). Rabbit anti-UL24 polyclonal antibody (PAb) was made by GL Biochem Ltd. (Shanghai, China). The protease inhibitor cocktail mixtures were purchased from CST (Boston, MA). Lipofectamine 2000 was purchased from Invitrogen (CA, USA).
Plasmid construction.
All enzymes used for cloning procedures were purchased from Thermo Fisher Scientific (MA, USA). The UL24 gene was amplified from the HSV-1 genome and cloned into the SalI and BamHI sites of pCMV-Myc vectors (Beyotime, Shanghai, China). Commercial reporter plasmids include the NF-κB–Luc (Stratagene, La Jolla, CA) and pRL-TK (Promega) plasmids. The construction of p65 RHD-Flag and p50 RHD-Flag was described in our previous studies (50). The UL24 E99A/K101A mutant and deletion mutants of the UL24 gene, including FR1(1–73), F1R(74–269), FR2(1–134), F2R(135–269), FR3(1–193), F3R(194–269), and UL24(73–134), were cloned into pCMV-Myc vectors to yield mUL24-Myc, FR1(1–73)-Myc, F1R(74–269)-Myc, FR2(1–134)-Myc, F2R(135–269)-Myc, FR3(1–193)-Myc, F3R(194–269)-Myc, and UL24(73–134)-Myc, respectively. Gifts of plasmids include pHA-IKKα (60), pFlag-IKKβ (61), pCMV-p65-Flag, pCMV-p50-Flag (62), IFN-β promoter reporter plasmid p125-Luc (63), a STING-hemagglutinin (HA) plasmid, pcDNA3.1-Flag-TBK1 (64), a cGAS-Flag plasmid (11), pGL3-pIL-6-Luc (65), and a PRD(III-I)-Luc plasmid (66).
RNA isolation and quantitative PCR.
Total RNA was extracted by using TRIzol (Invitrogen, CA) according to the manufacturer's instructions. Samples were digested with DNase I and subjected to reverse transcription (RT) as previously described (67). The cDNA was used as a template for quantitative PCR to investigate the expression patterns of human IFN-β and IL-6. Detailed protocols were described in our previous studies (68).
Transfection and dual-luciferase reporter assays.
HEK293T cells were cotransfected with IFN-β–Luc, IL-6–Luc, PRD(III-I)-Luc, and NF-κB–Luc reporter plasmids or an internal control plasmid, pRL-TK, with or without expression plasmids, by using standard calcium phosphate coprecipitation methods (69). At 24 h posttransfection, luciferase assays were performed with a dually specific luciferase assay kit (Promega, Madison, WI) as described in our previous studies (58).
ELISA.
ELISAs to quantify secreted IFN-β and IL-6 were carried out with culture supernatants collected from infected cells as previously described (70). Human IFN-β ELISA (PBL InterferonSource, Piscataway, NJ) and human IL-6 ELISA (RayBiotech, Norcross, GA, USA) kits were used to detect IFN-β and IL-6 according to the manufacturers' instructions.
Immunofluorescence assay.
Immunofluorescence assays were performed as described in our previous studies (71). In brief, HeLa cells were transfected with the indicated plasmids for 24 h and then fixed in 4% paraformaldehyde. Cells were incubated with rabbit anti-p65 or anti-p50 PAb (diluted 1:500) or anti-Myc MAb (diluted 1:2,000), followed by incubation with tetramethyl rhodamine isocyanate (TRITC)-conjugated goat anti-rabbit IgG (Pierce) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Sigma-Aldrich). After each incubation step, cells were washed extensively with phosphate-buffered saline (PBS). Samples were analyzed by fluorescence microscopy.
Co-IP assay and Western blot analysis.
Co-IP assays and WB analyses were performed as described in our previous study (59). HEK293T cells were cotransfected with 10 μg of each of the indicated expression plasmids carrying Flag tags. Transfected (36 h) or HSV-1-infected (16 h) cells were harvested, lysed on ice with 700 μl of lysis buffer, and then subjected to IP assays and WB analysis. Protein A/G Plus-agarose was purchased from Santa Cruz. All IP assays were repeated at least three times, and similar data were obtained.
Statistical analysis.
Data are presented as means ± standard deviations. Two-tailed unpaired Student's t test was used to determine differences. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Rongtuan Lin for the STING-HA plasmid, Zhijian Chen for the cGAS-FlagN1 plasmid, and Takashi Fujita for IFN-β–Luc. We also thank Slimane Dridi for help in optimizing the UL24 Western blots.
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (81371795 and 81571974), the Innovative Research Team in Soochow University (PCSIRT IRT1075), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (YX13400214).
REFERENCES
- 1.Ward CJ. 2010. Pathogen sensing in innate immunity. Expert Rev Vaccines 9:19–21. doi: 10.1586/erv.09.141. [DOI] [PubMed] [Google Scholar]
- 2.Cunha A. 2012. Innate immunity: pathogen and xenobiotic sensing—back to basics. Nat Rev Immunol 12:400. doi: 10.1038/nri3237. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Kawasaki T, Kawai T, Akira S. 2011. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev 243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishii KJ, Akira S. 2005. Innate immune recognition of nucleic acids: beyond Toll-like receptors. Int J Cancer 117:517–523. doi: 10.1002/ijc.21402. [DOI] [PubMed] [Google Scholar]
- 6.Goubau D, Deddouche S, Reis e Sousa C. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. 2008. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A 105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 11.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. 2013. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A 110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. 2012. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife 1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A 106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abe T, Barber GN. 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol 88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu YH, Zhao M, Chen ZJ. 2009. Ubiquitin in NF-kappaB signaling. Chem Rev 109:1549–1560. doi: 10.1021/cr800554j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonizzi G, Karin M. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol 8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 25.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. 1997. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 26.Iwai K. 2014. Diverse roles of the ubiquitin system in NF-kappaB activation. Biochim Biophys Acta 1843:129–136. doi: 10.1016/j.bbamcr.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 27.Hayden MS, Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell 132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 28.Corey L, Wald A, Celum CL, Quinn TC. 2004. The effects of herpes simplex virus-2 on HIV-1 acquisition and transmission: a review of two overlapping epidemics. J Acquir Immune Defic Syndr 35:435–445. doi: 10.1097/00126334-200404150-00001. [DOI] [PubMed] [Google Scholar]
- 29.Shimomura Y. 2008. Herpes simplex virus latency, reactivation, and a new antiviral therapy for herpetic keratitis. Nippon Ganka Gakkai Zasshi 112:247–264; discussion 265 (In Japanese.) [PubMed] [Google Scholar]
- 30.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF. 2016. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci U S A 113:E1034–E1043. doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F. 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, O'Hare P, Adler H, Sun R, Ploegh HL, Damania B, Upton JW, Fitzgerald KA, Paludan SR. 2015. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol 194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalamvoki M, Roizman B. 2014. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci U S A 111:E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D, Su C, Zheng C. 2016. Herpes simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J Virol 90:5824–5829. doi: 10.1128/JVI.00186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye R, Su C, Xu H, Zheng C. 2017. Herpes simplex virus 1 ubiquitin-specific protease UL36 abrogates NF-κB activation in DNA sensing signal pathway. J Virol 91:e02417-16. doi: 10.1128/JVI.02417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leiva-Torres GA, Rochette PA, Pearson A. 2010. Differential importance of highly conserved residues in UL24 for herpes simplex virus 1 replication in vivo and reactivation. J Gen Virol 91:1109–1116. doi: 10.1099/vir.0.017921-0. [DOI] [PubMed] [Google Scholar]
- 39.Jacobson JG, Chen SH, Cook WJ, Kramer MF, Coen DM. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161–169. doi: 10.1006/viro.1997.9012. [DOI] [PubMed] [Google Scholar]
- 40.Bertrand L, Leiva-Torres GA, Hyjazie H, Pearson A. 2010. Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin. J Virol 84:109–118. doi: 10.1128/JVI.01428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lymberopoulos MH, Pearson A. 2007. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363:397–409. doi: 10.1016/j.virol.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 42.Lymberopoulos MH, Bourget A, Ben Abdeljelil N, Pearson A. 2011. Involvement of the UL24 protein in herpes simplex virus 1-induced dispersal of B23 and in nuclear egress. Virology 412:341–348. doi: 10.1016/j.virol.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A 112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horan KA, Hansen K, Jakobsen MR, Holm CK, Soby S, Unterholzner L, Thompson M, West JA, Iversen MB, Rasmussen SB, Ellermann-Eriksen S, Kurt-Jones E, Landolfo S, Damania B, Melchjorsen J, Bowie AG, Fitzgerald KA, Paludan SR. 2013. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol 190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh S, May MJ, Kopp EB. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 47.Dev A, Iyer S, Razani B, Cheng G. 2011. NF-kappaB and innate immunity. Curr Top Microbiol Immunol 349:115–143. doi: 10.1007/82_2010_102. [DOI] [PubMed] [Google Scholar]
- 48.Kawai T, Akira S. 2007. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Oeckinghaus A, Ghosh S. 2009. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Wang K, Wang S, Zheng C. 2013. Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. J Virol 87:12935–12948. doi: 10.1128/JVI.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, Ni L, Wang S, Zheng C. 2014. Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-kappaB activation. J Virol 88:7941–7951. doi: 10.1128/JVI.03394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JC, Lee SY, Kim SY, Kim JK, Kim HJ, Lee HM, Choi MS, Min JS, Kim MJ, Choi HS, Ahn JK. 2008. HSV-1 ICP27 suppresses NF-kappaB activity by stabilizing IkappaBalpha. FEBS Lett 582:2371–2376. doi: 10.1016/j.febslet.2008.05.044. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Wang S, Wang K, Zheng C. 2013. Herpes simplex virus 1 DNA polymerase processivity factor UL42 inhibits TNF-alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. Med Microbiol Immunol 202:313–325. doi: 10.1007/s00430-013-0295-0. [DOI] [PubMed] [Google Scholar]
- 54.Amici C, Belardo G, Rossi A, Santoro MG. 2001. Activation of I kappa b kinase by herpes simplex virus type 1. A novel target for anti-herpetic therapy. J Biol Chem 276:28759–28766. doi: 10.1074/jbc.M103408200. [DOI] [PubMed] [Google Scholar]
- 55.Kasem S, Yu MH, Yamada S, Kodaira A, Matsumura T, Tsujimura K, Madbouly H, Yamaguchi T, Ohya K, Fukushi H. 2010. The ORF37 (UL24) is a neuropathogenicity determinant of equine herpesvirus 1 (EHV-1) in the mouse encephalitis model. Virology 400:259–270. doi: 10.1016/j.virol.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Blakeney S, Kowalski J, Tummolo D, DeStefano J, Cooper D, Guo M, Gangolli S, Long D, Zamb T, Natuk RJ, Visalli RJ. 2005. Herpes simplex virus type 2 UL24 gene is a virulence determinant in murine and guinea pig disease models. J Virol 79:10498–10506. doi: 10.1128/JVI.79.16.10498-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson A, Coen DM. 2002. Identification, localization, and regulation of expression of the UL24 protein of herpes simplex virus type 1. J Virol 76:10821–10828. doi: 10.1128/JVI.76.21.10821-10828.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J Virol 86:3528–3540. doi: 10.1128/JVI.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing J, Wang S, Lin F, Pan W, Hu CD, Zheng C. 2011. Comprehensive characterization of interaction complexes of herpes simplex virus type 1 ICP22, UL3, UL4, and UL205. J Virol 85:1881–1886. doi: 10.1128/JVI.01730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park KA, Byun HS, Won M, Yang KJ, Shin S, Piao L, Kim JM, Yoon WH, Junn E, Park J, Seok JH, Hur GM. 2007. Sustained activation of protein kinase C downregulates nuclear factor-kappaB signaling by dissociation of IKK-gamma and Hsp90 complex in human colonic epithelial cells. Carcinogenesis 28:71–80. doi: 10.1093/carcin/bgl094. [DOI] [PubMed] [Google Scholar]
- 61.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. 1997. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 62.Severa M, Coccia EM, Fitzgerald KA. 2006. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J Biol Chem 281:26188–26195. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
- 63.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol 80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, Servant MJ, Lin R, Meurs EF, Hiscott J. 2009. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol 29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao S, Song L, Li J, Zhang Z, Peng H, Jiang W, Wang Q, Kang T, Chen S, Huang W. 2012. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell Microbiol 14:1849–1866. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- 66.Ehrhardt C, Kardinal C, Wurzer WJ, Wolff T, von Eichel-Streiber C, Pleschka S, Planz O, Ludwig S. 2004. Rac1 and PAK1 are upstream of IKK-epsilon and TBK-1 in the viral activation of interferon regulatory factor-3. FEBS Lett 567:230–238. doi: 10.1016/j.febslet.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 67.Shen G, Wang K, Wang S, Cai M, Li ML, Zheng C. 2014. Herpes simplex virus 1 counteracts viperin via its virion host shutoff protein UL41. J Virol 88:12163–12166. doi: 10.1128/JVI.01380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol 87:9788–9801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL. 2011. Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J Virol 85:11079–11089. doi: 10.1128/JVI.05098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S, Wang K, Li J, Zheng C. 2013. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J Virol 87:11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xing J, Wu F, Pan W, Zheng C. 2010. Molecular anatomy of subcellular localization of HSV-1 tegument protein US11 in living cells. Virus Res 153:71–81. doi: 10.1016/j.virusres.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]