ABSTRACT
Rhinoviruses are the most common causes of the common cold. Their many distinct lineages fall into “major” and “minor” groups that use different cell surface receptors to enter host cells. Minor-group rhinoviruses are more immunogenic in laboratory studies, although their patterns of transmission and their cold symptoms are broadly similar to those of the major group. Here we present evolutionary evidence that minor-group viruses are also more immunogenic in humans. A key finding is that rates of amino acid substitutions at exposed sites in the capsid proteins VP2, VP3, and VP1 tend to be elevated in minor-group relative to major-group viruses, while rates at buried sites show no consistent differences. A reanalysis of historical virus watch data also indicates a higher immunogenicity of minor-group viruses, consistent with our findings about evolutionary rates at amino acid positions most directly exposed to immune surveillance. The increased immunogenicity and speed of evolution in minor-group lineages may contribute to the very large numbers of rhinovirus serotypes that coexist while differing in virulence.
IMPORTANCE Most colds are caused by rhinoviruses (RVs). Those caused by a subset known as the minor-group members of rhinovirus species A (RV-A) are correlated with the inception and aggravation of asthma in at-risk populations. Genetically, minor-group viruses are similar to major-group RV-A, from which they were derived, although they tend to elicit stronger immune responses. Differences in their rates and patterns of molecular evolution should be highly relevant to their epidemiology. All RV-A strains show high rates of amino acid substitutions in the capsid proteins at exposed sites not previously identified as being immunogenic, and this increase is significantly greater in minor-group viruses. These findings will inform future studies of the recently discovered RV-C, which also appears to exacerbate asthma in adults and children. In addition, these findings draw attention to the difficult problem of explaining the long-term coexistence of many serotypes of major- and minor-group RVs.
KEYWORDS: human rhinovirus, immunogenicity, major-group rhinoviruses, minor-group rhinoviruses, molecular evolution
INTRODUCTION
Cold infections are caused by more than 250 virus serotypes belonging to at least five different families. The most common cold-causing viruses are rhinoviruses (RVs) (10 to 50% of all colds), coronaviruses (10 to 15% of all colds), and influenza viruses (5 to 15% of all colds) (1, 2). Globally, RVs are the primary cause of acute upper respiratory tract infections. Although most such infections are mild, RVs can also replicate in the lower respiratory tract and exacerbate the severity of conditions such as chronic respiratory disease and asthma (3–7).
Rhinoviruses are nonenveloped, positive-sense, single-stranded RNA viruses belonging to the genus Enterovirus of the family Picornaviridae that infect epithelial cells that line the posterior nasopharynx (5). Like other positive-sense single-stranded RNA viruses, rhinoviruses encode an RNA-dependent RNA polymerase that lacks the ability to proofread and repair mismatches. The mutation rate has been estimated to range from 10−3 to 10−5 mutations per nucleotide per genome replication event (8). At ∼7,200 nucleotides long, the rhinovirus genome would be expected to accumulate roughly 1 mutation per replication cycle.
Unlike other respiratory tract viruses such as influenza viruses (Orthomyxoviridae), rhinovirus lineages are relatively stable in the sense that they are not serially replaced over time by newer, more fit serotypes (9). This group has accumulated a large diversity of genetically distinct lineages and now includes more than 165 recognized serotypes. Epidemiological studies have documented as many as 32 serotypes cocirculating in a given community (10–13).
These serotypes do not appear to turn over in predictable patterns during “the cold season,” and there are no data to suggest that they differ much in long-term average fitness; in particular, the major and minor serotype groups appear to be equally successful (14). In this respect, RVs contrast strikingly with other viruses such as influenza virus that are serially replaced each year (15). Rhinovirus infections can be asymptomatic and typically induce temporary, limited immunity (14), whereas influenza virus infections are often severe and induce long-term immunity (16). Attempts to predict which RV serotypes will become quantitatively more prevalent in future years have not been successful, and the extreme antigenic diversity of rhinoviruses has presented formidable barriers to the development of effective vaccines with broad epitope coverage (17).
Rhinovirus has been classified into three species: RV-A (80 serotypes), RV-B (32 serotypes), and RV-C (55 serotypes). Serotype and species classifications are determined primarily by nucleotide sequence identity (18, 19). The serotypes within RV-A and RV-B have been classified into major and minor groups based on their cell receptor specificity. Serotypes that use intercellular adhesion molecule 1 (ICAM-1) as a receptor (68 of the RV-A serotypes and all RV-B serotypes) belong to the major receptor group. The remaining 12 RV-A serotypes use members of the low-density lipoprotein (LDL) receptor family and belong to the minor receptor group (20–22). RV-C serotypes bind to CDHR3, which is a member of the cadherin family and is involved in susceptibility to childhood asthma (23, 24).
ICAM-1 is an important cell adhesion molecule that facilitates cellular communication (25). It is a transmembrane glycoprotein in the immunoglobulin superfamily and is expressed on epithelial cells such as those found in the adenoids and respiratory system and on a variety of immune cells (26). The expression of ICAM-1 on macrophages and dendritic cells is upregulated by inflammatory cytokines induced by rhinovirus infection (27). Although rhinoviruses cannot infect white blood cells, virus attachment makes these cells less likely to report the infection to cells required to make antibodies and immune memory (27). ICAM-1 binds to amino acids belonging to the RV capsid proteins VP1, VP2, and VP3, which line the floor of the cleft or “canyon,” which is a prominent feature on the surface of the capsid (20, 28).
Members of the LDL receptor family are involved in binding and transporting ligands with a variety of biological functions and are expressed on a diversity of cell types (29). The LDL receptor used by minor-group rhinoviruses binds around the top of the star-shaped mesa of the 5-fold axis and is thought to directly contact residues on the BC and HI loops of VP1 (22). LDL receptors are structurally and functionally unrelated to ICAM-1 (30) and are not known to interfere with immune responses (27, 31). Previous studies examining antibody responses in naturally infected human populations (11) and T-cell immunogenic responses in murine lymphoid dendritic cells (25) demonstrate that minor-group viruses are more effective than major-group viruses in stimulating an immune response.
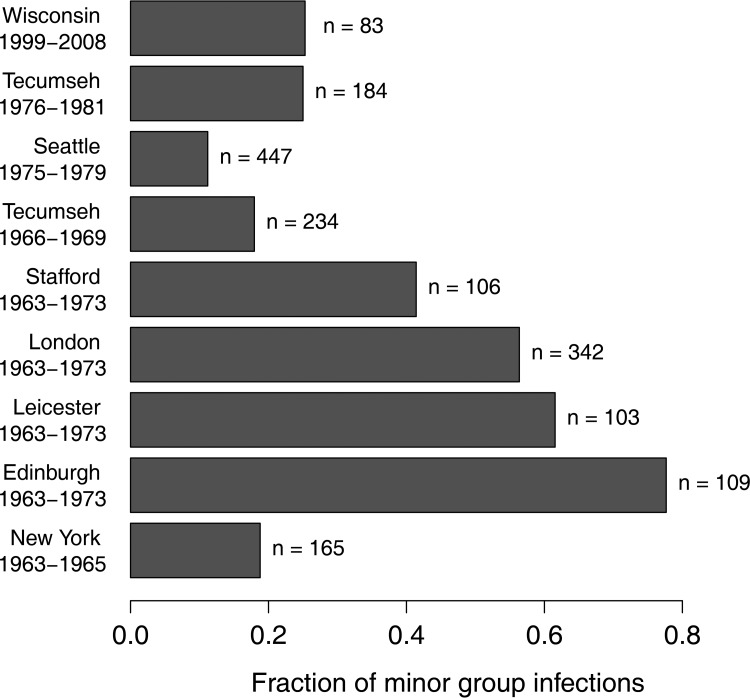
The greater immunogenicity of minor-group viruses might be expected to put them at an evolutionary disadvantage. Nonetheless, they have evolved independently three times (32), although recombination could have played a role generating these polyphyletic relationships (33). This suggests that the evolutionary change from major- to minor-group functionality might be relatively easy to accomplish and that the minor-group phenotype, although immunogenic, has a long-term average fitness equal to that of the major-group phenotype. Longitudinal studies of rhinovirus epidemiology in the 1960s, 1970s, and 2000s documented intermediate frequencies of minor-group infections over time and space (Fig. 1) (10–13, 34, 35; W. M. Lee, personal communication). The fraction of minor-group rhinoviruses in the four United Kingdom samples is significantly larger than in the five North American samples (P < 0.001, t test), but there is no evidence for seasonal trends (12). None of these data shed light on the mechanisms influencing minor-group prevalence or transmission (36).
FIG 1.
The fraction of minor-group viruses, among all rhinoviruses detected, in nine longitudinal infection studies from New York (34), Edinburgh (12), Leicester (12), London (12), Stafford (12), Tecumseh from 1966 to 1969 (35), Seattle (11), Tecumseh from 1976 to 1981 (13), and Wisconsin (W. M. Lee, personal communication).
Minor-group viruses have a strictly conserved lysine residue in the HI loop of the VP1 capsid protein that is essential but not sufficient for attachment to LDL receptors, as several “K-type” major-group viruses also have a lysine in the same position on VP1 but cannot use LDL receptors for cell entry (37). The amino acid residues within the LDL receptor footprint are primarily basic. In major-group viruses, residues located at similar positions tend to be more acidic (37, 38), implying that electrostatic interactions influence receptor-binding kinetics. Previous studies have reported different selection pressures acting on major- versus minor-group viruses (32, 39). Positive stabilizing selection (purifying selection that results in amino acids with similar biochemical properties) accounted for 93% of residues under selection in major-group viruses, while positive destabilizing selection (positive Darwinian selection that results in amino acids with different biochemical properties) dominated the evolution of minor-group viruses (78%) (32). In addition to the cell receptor footprint and antigenic sites, positive selection was inferred in some regions of the viral proteins for which the functional significance remains unknown.
The present study asks whether major- and minor-group viruses show distinct patterns of genetic change. We used previously reported data and the prototypic serotypes for major- and minor-group viruses within RV-A to test the following hypotheses: (i) minor-group serotypes elicit a stronger immune response than do major-group serotypes, and therefore, minor-group lineages should evolve more rapidly at antigenic sites; (ii) if the entire viral capsid surface interacts with the immune system rather than only at discrete antigenic sites, minor-group serotypes should evolve more rapidly at amino acid positions that are exposed on the surface of the intact capsid; and (iii) assuming that VP1 is the immunodominant protein for both major- and minor-group serotypes (40), a larger number of amino acid substitutions should occur in VP1 than in VP2 and VP3, and this difference should be more pronounced in minor-group viruses.
RESULTS
Phylogeny.
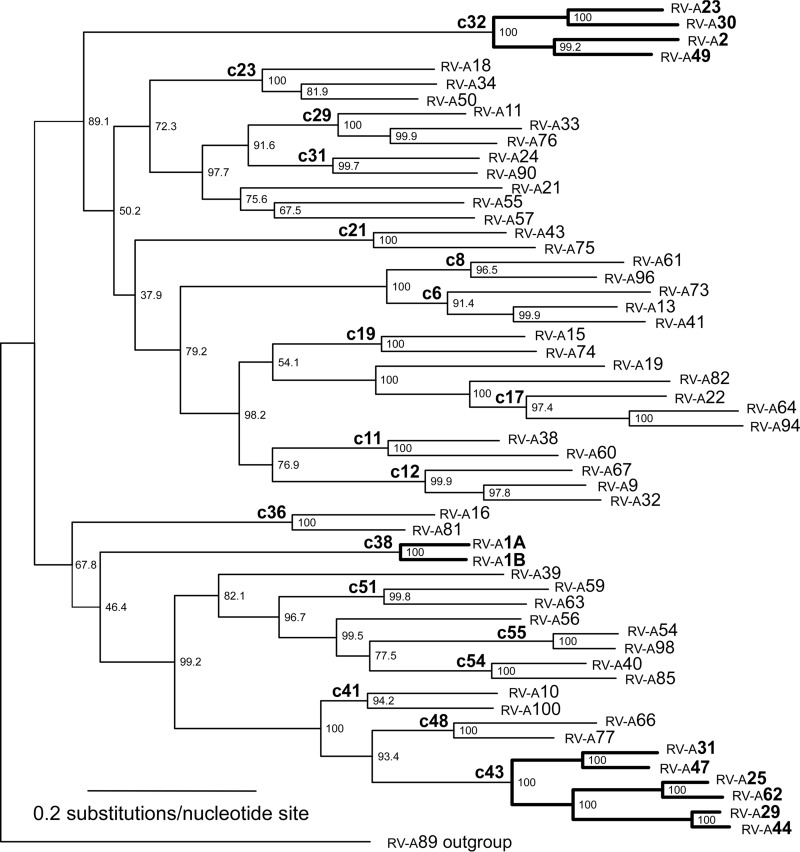
Phylogenetic relationships among sequences (Fig. 2) are similar to those described previously (32, 41–43). Small differences in topology probably result from the use of maximum likelihood (ML) methods in the present study and neighbor-joining methods in previous studies. The 12 minor-group sequences cluster into three monophyletic clades.
FIG 2.
Maximum likelihood estimate of phylogenetic relationships among minor-group (boldface type) and major-group serotypes using VP2, VP3, and VP1 capsid sequences concatenated in a single data set with the GTR+I+G model of evolution and being rooted with RV-A89. The 19 cladelets used for all statistical analyses are indicated by clade names in the form of cXX, where XX is the number derived from the numbering of internal nodes in the tree. Branch lengths are shown proportional to the amount of evolutionary change. Numbers to the right of the branch nodes indicate the bootstrap proportions. Labels at the tips of the branches are numbered according to each rhinovirus serotype.
Antibody response to infection caused by major- and minor-group viruses in Seattle Watch studies.
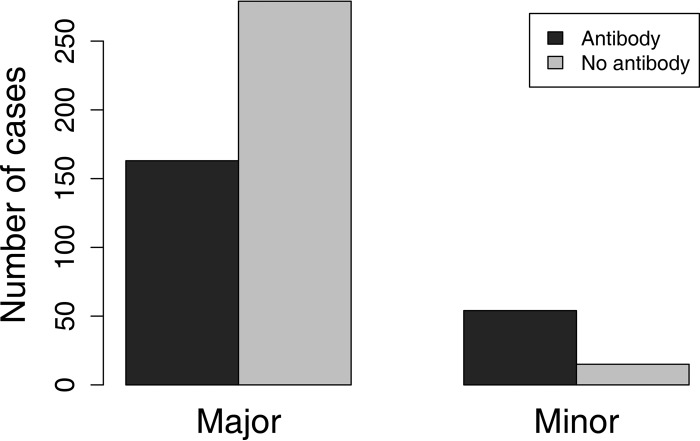
Infections caused by minor-group viruses stimulated an antibody response significantly more often (P < 0.0001) than those caused by major-group viruses (Fig. 3).
FIG 3.
Number of human rhinovirus infections caused by major-group and minor-group viruses that induced immunity (11). Minor-group infections were much more likely to raise a detectable antibody response (P < 0.001).
Accumulation of substitutions.
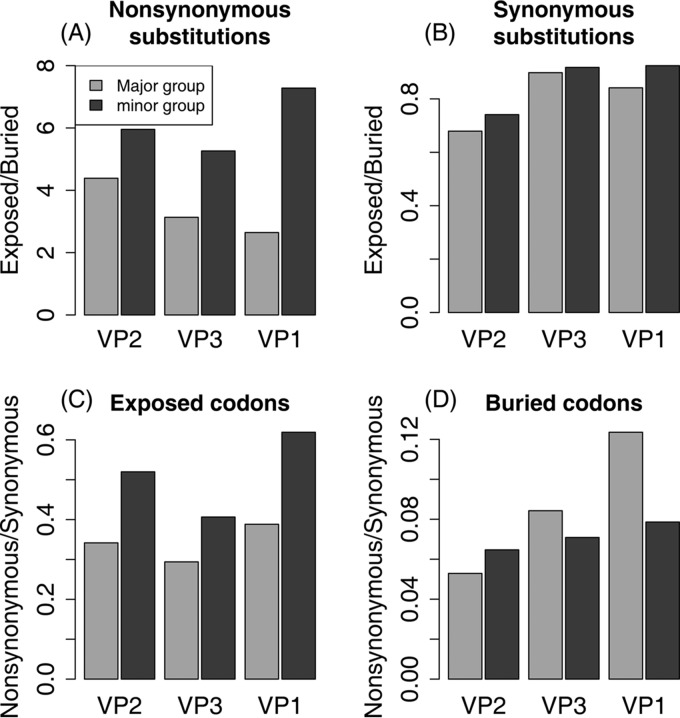
More than 9,200 nucleotide substitutions were detected on the included branches of the phylogeny; 1,655 of these substitutions are nonsynonymous, and 7,590 are synonymous. Most of the nonsynonymous substitutions (1,307; 79%) occur in exposed codons, but most of the synonymous substitutions (4,188; 55%) occur in buried codons, as expected, since 57% of the 781 codons are buried (Table 1). The much higher rate of nonsynonymous substitutions at exposed sites is extremely highly significant (χ2 = 634.1; P = 0). Major- and minor-group lineages show this pattern, but it is twice as strong in the minor group (ϕ2 = 0.111) as in the major group (ϕ2 = 0.055) (Table 1). This difference between the groups illustrates our main finding: an interaction between exposure status and cell receptor group, with a relative excess of nonsynonymous substitutions at exposed codons in the minor group (Fig. 4A).
TABLE 1.
Synonymous and nonsynonymous substitutions by cladelet and surface exposure, with interaction coefficients from fixed- and random-effects generalized linear modelsa
| Clade | Buried |
Exposed |
Interactions |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| xN | /cdn for xN | xS | /cdn for xS | xN/xS | xN | /cdn for xN | xS | /cdn for xS | xN/xS | Fixed | Random | |
| c32 (m) | 26 | 0.058 | 383 | 0.853 | 0.068 | 139 | 0.419 | 309 | 0.931 | 0.450 | 0.236 | 1.853 |
| c23 | 23 | 0.051 | 277 | 0.617 | 0.083 | 90 | 0.271 | 207 | 0.623 | 0.435 | −0.076 | 1.670 |
| c29 | 31 | 0.069 | 268 | 0.597 | 0.116 | 77 | 0.232 | 212 | 0.639 | 0.363 | −0.531 | 1.534 |
| c31 | 12 | 0.027 | 164 | 0.365 | 0.073 | 46 | 0.139 | 152 | 0.458 | 0.303 | −0.966 | 1.480 |
| c21 | 12 | 0.027 | 157 | 0.350 | 0.076 | 55 | 0.166 | 149 | 0.449 | 0.369 | 0.082 | 1.551 |
| c8 | 10 | 0.022 | 171 | 0.381 | 0.058 | 43 | 0.130 | 148 | 0.446 | 0.291 | 0.018 | 1.469 |
| c6 | 27 | 0.060 | 281 | 0.626 | 0.096 | 114 | 0.343 | 235 | 0.708 | 0.485 | 0 | 1.746 |
| c19 | 12 | 0.027 | 194 | 0.432 | 0.062 | 45 | 0.136 | 133 | 0.401 | 0.338 | −0.119 | 1.471 |
| c17 | 24 | 0.053 | 300 | 0.668 | 0.080 | 90 | 0.271 | 231 | 0.696 | 0.390 | −0.119 | 1.662 |
| c11 | 18 | 0.040 | 175 | 0.390 | 0.103 | 45 | 0.136 | 138 | 0.416 | 0.326 | −0.524 | 1.425 |
| c12 | 24 | 0.053 | 286 | 0.637 | 0.084 | 69 | 0.208 | 212 | 0.639 | 0.325 | −0.384 | 1.545 |
| c36 | 24 | 0.053 | 166 | 0.370 | 0.145 | 45 | 0.136 | 136 | 0.410 | 0.331 | −0.812 | 1.379 |
| c38 (m) | 3 | 0.007 | 115 | 0.256 | 0.026 | 41 | 0.123 | 96 | 0.289 | 0.427 | 1.175 | 1.502 |
| c51 | 16 | 0.036 | 196 | 0.437 | 0.082 | 42 | 0.127 | 143 | 0.431 | 0.294 | −0.475 | 1.415 |
| c55 | 8 | 0.018 | 117 | 0.261 | 0.068 | 20 | 0.060 | 105 | 0.316 | 0.190 | −0.524 | 1.226 |
| c54 | 11 | 0.024 | 173 | 0.385 | 0.064 | 34 | 0.102 | 130 | 0.392 | 0.262 | −0.312 | 1.376 |
| c41 | 12 | 0.027 | 170 | 0.379 | 0.071 | 33 | 0.099 | 147 | 0.443 | 0.224 | −0.429 | 1.359 |
| c48 | 18 | 0.040 | 167 | 0.372 | 0.108 | 46 | 0.139 | 132 | 0.398 | 0.348 | −0.502 | 1.434 |
| c43 (m) | 37 | 0.082 | 428 | 0.953 | 0.086 | 233 | 0.702 | 387 | 1.166 | 0.602 | 0.400 | 2.036 |
| Major | 282 | 3,262 | 0.086 | 894 | 2,610 | 0.343 | ϕ2 = 0.055 | |||||
| minor | 66 | 926 | 0.071 | 413 | 792 | 0.521 | ϕ2 = 0.111 | |||||
| Total | 348 | 4,188 | 0.083 | 1,307 | 3,402 | 0.384 | ||||||
Clades are in tree order as displayed in Fig. 2. xN, number of nonsynonymous (amino acid) substitutions; xS, number of synonymous nucleotide substitutions;/cdn, substitutions divided by codons in buried (449) or exposed (332) sites; fixed, interaction coefficients from the fixed-effects model (GLM); random, interaction coefficients from the random-effects model (GLMER); ϕ2, square of Pearson's phi coefficient (χ2/n). Minor-group clades (boldface) show elevated proportions of nonsynonymous substitutions.
FIG 4.
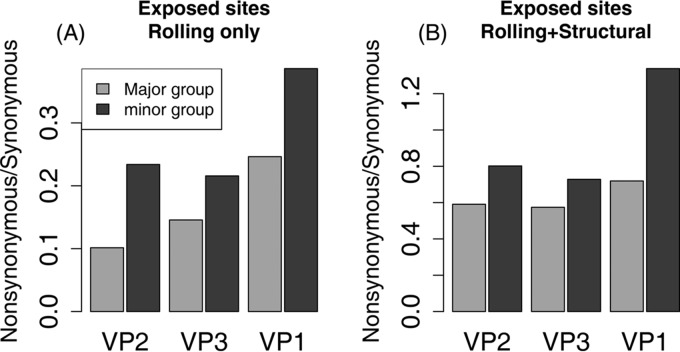
Ratios of substitutions using the combined characterization of exposed and buried codons. (A) Ratios of the number of nonsynonymous substitutions at exposed codons to the number at buried codons for each capsid protein for major and minor groups. Minor-group serotypes show consistently and significantly elevated rates at exposed positions. (B) Ratios of the number of synonymous substitutions at exposed codons to the number at buried codons for each capsid protein for major and minor groups. Minor-group serotypes show consistently and significantly elevated rates at exposed positions. (C) Ratios of nonsynonymous to synonymous substitutions at exposed codons. The minor-group substitution rate is significantly elevated for all three genes (Table 3). (D) Ratios of nonsynonymous to synonymous substitutions at buried sites. None of the differences are individually significant after Bonferroni correction (Table 3).
The basic generalized linear model (GLM) analysis shows that this pattern holds up, controlling for variation among the three genes (P = 0.006 for the exposure-group interaction) (Table 2), and that synonymous sites do not show any significant gene or interaction effects (Table 2 and Fig. 4B). In this analysis, branch length effects are captured so efficiently by group (M versus m) that the inclusion of synonymous substitutions (syn) as a covariate adds no significant explanatory power (P = 0.11) (not reported in Table 2). In the random-effects analysis (generalized linear mixed-effects regression [GLMER]), syn has a highly significant effect because group, which partitions branches into just two categories, cannot capture the branch length variation among 19 cladelets. With synonymous substitutions included as a covariate, the interaction between exposure status and group remains significant (Table 2). Analyses with antigenic rather than exposed codons show similar trends but no significant effects owing to the small numbers of antigenic sites (results not shown).
TABLE 2.
Results from GLM and GLMER analyses
| Effecta | GLM |
GLMER |
||||
|---|---|---|---|---|---|---|
| Coefficient | Standard error | P value | Coefficient | Standard error | P value | |
| Nonsynonymous changes using the combined characterization | ||||||
| Main effects | ||||||
| Intercept | −0.867 | 0.187 | <0.001 | −3.710 | 0.157 | <0.001 |
| Exposed | 1.842 | 0.208 | <0.001 | 1.833 | 0.127 | <0.001 |
| VP3 | 0.404 | 0.246 | 0.10 | 0.404 | 0.147 | 0.006 |
| VP1 | 0.694 | 0.224 | 0.002 | 0.693 | 0.134 | <0.001 |
| Minor | −1.452 | 0.228 | <0.001 | 0.096 | 0.310 | 0.76 |
| Syn | − | − | − | 0.253 | 0.033 | <0.001 |
| Two-way interactions | ||||||
| Exposed:VP3 | −0.557 | 0.275 | 0.043 | −0.557 | 0.165 | <0.001 |
| Exposed:VP1 | −0.602 | 0.250 | 0.016 | −0.602 | 0.150 | <0.001 |
| Exposed:minor | 0.690 | 0.249 | 0.006 | 0.676 | 0.154 | <0.001 |
| Main effects of synonymous changes using the combined characterization | ||||||
| Intercept | 1.976 | 0.0195 | <0.001 | −0.833 | 0.086 | <0.001 |
| Exposed | 0.094 | 0.0271 | <0.001 | 0.097 | 0.025 | <0.001 |
| Minor | −1.229 | 0.0322 | <0.001 | 0.361 | 0.223 | 0.11 |
| Nonsynonymous changes using the joint characterizationa | ||||||
| Main effects | ||||||
| Intercept | −0.867 | 0.176 | <0.001 | −3.826 | 0.150 | <0.001 |
| R | 0.749 | 0.261 | 0.004 | 0.682 | 0.173 | <0.001 |
| R+S | 2.442 | 0.202 | <0.001 | 2.427 | 0.131 | <0.001 |
| VP3 | 0.404 | 0.231 | 0.08 | 0.403 | 0.147 | 0.006 |
| VP1 | 0.693 | 0.210 | <0.001 | 0.695 | 0.134 | <0.001 |
| Minor | −1.452 | 0.214 | <0.001 | 0.045 | 0.284 | 0.88 |
| Syn | − | − | − | 0.238 | 0.034 | <0.001 |
| Two-way interactions | ||||||
| R:VP3 | −0.186 | 0.331 | 0.57 | −0.180 | 0.210 | 0.39 |
| R+S:VP3 | −0.480 | 0.269 | 0.75 | −0.472 | 0.171 | 0.006 |
| R:VP1 | 0.028 | 0.298 | 0.92 | 0.033 | 0.189 | 0.86 |
| R+S:VP1 | −0.528 | 0.248 | 0.034 | −0.502 | 0.158 | 0.002 |
| R:minor | 0.773 | 0.267 | 0.004 | 0.673 | 0.198 | <0.001 |
| R+S:minor | 0.662 | 0.245 | 0.007 | 0.644 | 0.160 | <0.001 |
| Main effects of synonymous changes using the joint characterization | ||||||
| Intercept | 1.976 | 0.0196 | <0.001 | −0.837 | 0.086 | <0.001 |
| R | 0.078 | 0.0312 | 0.012 | 0.078 | 0.028 | 0.006 |
| R+S | 0.115 | 0.0389 | 0.003 | 0.125 | 0.036 | <0.001 |
| Minor | −1.229 | 0.0324 | <0.001 | 0.385 | 0.220 | 0.08 |
The effects of exposed sites are relative to buried sites, those of VP3 and VP1 are relative to VP2, and those of the minor group are relative to the major group. Syn is the effect of synonymous substitutions at a codon position. “−” indicates that data are not reported because there are no significant effects. R, exposed by the rolling-only model; R+S, exposed by the rolling+structural model.
The relative excess of nonsynonymous changes in minor-group lineages could be caused by an increased substitution rate at exposed codons in the minor group (as we predicted) or by a reduced nonsynonymous substitution rate at buried codons in the major group. To distinguish between these possibilities, we plotted the ratio of nonsynonymous to synonymous changes at exposed and buried codons for the major and minor groups (Fig. 4C and D). At exposed codons, minor-group viruses have a significantly higher ratio for all three genes (chi-square tests with Bonferroni correction to reduce the probability of false-positive type I errors) (Table 3). However, major- and minor-group viruses show no significant differences at buried codons (Table 3), implying that the relative excess of nonsynonymous changes in minor-group viruses at exposed codons is not due to a reduced rate of nonsynonymous substitutions at buried codons in major-group viruses.
TABLE 3.
Differences in ratios of nonsynonymous to synonymous substitutions between major and minor groups in individual genesc
| Group | Gene | Characterizationa | NSyn | Syn | No. of codons | Ratio | Significanceb |
|---|---|---|---|---|---|---|---|
| Combined characterization of exposure status at codons | |||||||
| Major | VP2 | B | 62 | 1,172 | 162 | 0.053 | |
| Minor | VP2 | B | 22 | 340 | 162 | 0.065 | NS |
| Major | VP3 | B | 82 | 973 | 130 | 0.084 | |
| Minor | VP3 | B | 19 | 268 | 130 | 0.071 | NS |
| Major | VP1 | B | 138 | 1,117 | 157 | 0.123 | |
| Minor | VP1 | B | 25 | 318 | 157 | 0.071 | NSB χ2 = 3.65; P value = 0.056 |
| Major | VP2 | E | 254 | 777 | 97 | 0.363 | |
| Minor | VP2 | E | 123 | 248 | 97 | 0.496 | SB χ2 = 9.64; P value = 0.0019 |
| Major | VP3 | E | 257 | 847 | 107 | 0.294 | |
| Minor | VP3 | E | 100 | 246 | 107 | 0.406 | SB χ2 = 5.19; P value = 0.023 |
| Major | VP1 | E | 353 | 934 | 125 | 0.378 | |
| Minor | VP1 | E | 180 | 291 | 125 | 0.619 | SB χ2 = 18.5; P value = 0.000017 |
| Joint characterization of exposure status at codons | |||||||
| Major | VP2 | R | 42 | 414 | 53 | 0.101 | |
| Minor | VP2 | R | 29 | 124 | 53 | 0.234 | SB χ2 = 9.64; P value = 0.0019 |
| Major | VP3 | R | 82 | 563 | 69 | 0.146 | |
| Minor | VP3 | R | 33 | 153 | 69 | 0.216 | NS |
| Major | VP1 | R | 166 | 674 | 91 | 0.246 | |
| Minor | VP1 | R | 85 | 220 | 91 | 0.386 | SB χ2 = 8.12; P value = 0.0044 |
| Major | VP2 | R+S | 192 | 325 | 40 | 0.591 | |
| Minor | VP2 | R+S | 89 | 111 | 40 | 0.802 | NS |
| Major | VP3 | R+S | 174 | 303 | 37 | 0.574 | |
| Minor | VP3 | R+S | 67 | 92 | 37 | 0.728 | NS |
| Major | VP1 | R+S | 187 | 260 | 34 | 0.719 | |
| Minor | VP1 | R+S | 95 | 71 | 34 | 1.338 | SB χ2 = 10.9; P value = 0.00094 |
R, exposed by the rolling-only model; R+S, exposed by the rolling+structural model.
NS, not significant; SB, significant after Bonferroni correction; NSB, not significant after Bonferroni correction.
We show the number of nonsynonymous changes (NSyn), the number of synonymous changes (Syn), and the number of codons for each class of codons (exposed [E] or buried [B]) in major- and minor-group lineages for each gene, along with the ratio of nonsynonymous to synonymous changes. The significance is based on a chi-square test of the 2-by-2 contingency table of nonsynonymous and synonymous changes for these two rows. Note that we did not include the five sites identified only by the structural model because they could not have produced a significant interaction.
The identification of exposed amino acids by the rolling model was based on capsid protein structures for a minor-group virus, RV-A2. If some of these codons are not exposed on major-group viruses, this could bias our analyses toward the detection of more changes at exposed codons in minor-group viruses. To test this possibility, the joint characterization divides codons into four categories, for all three capsid genes combined, with the following numbers: 449 codons identified as being buried in both the rolling and structural models, 111 codons identified as being exposed in both the rolling and structural models (i.e., the rolling+structural or R+S model of exposed sites); 213 codons identified as being exposed in the rolling model and buried in the structural (i.e., the rolling-only or R model); and 5 codons identified as being buried in the rolling model and exposed in the structural model.
To test whether the 213 codons identified as being exposed only by the rolling-only (R) model show a disproportionate number of changes in minor-group viruses, we used GLM to model the numbers of substitutions as a function of group, gene, and three classes of sites: buried (449 codons), exposed by the rolling-only model (213 codons), and exposed by both models (111 codons). The five codons identified by only the structural model are not included in this analysis due to the small sample size. Both the rolling-only model and the rolling+structural model show an excess of amino acid changes at exposed sites but with a much larger coefficient for the codons identified by the rolling+structural model (Table 2). The magnitudes and significance of their interactions with the minor group are nearly identical.
Within the three protein subunits, VP3 and VP1 show excess changes at buried codons and codons identified as being exposed by the rolling-only (R) model but not by the rolling+structural (R+S) model. The ratio of nonsynonymous to synonymous changes is consistently higher in minor-group viruses for all three genes and for both types of exposed codons (Fig. 5), with individually significant differences in VP2 and VP1 for exposed codons identified by the R model and in VP1 for exposed codons identified by the R+S model (Table 3). The numbers of synonymous substitutions show small but significant excesses at codons identified as being exposed by both the R model and the R+S model (Table 2 and Fig. 4D).
FIG 5.
Ratios of nonsynonymous to synonymous substitutions using the joint characterization of exposed and buried codons. (A) At codons identified as being exposed rolling-only (R) model, minor-group viruses have consistently elevated ratios, significantly so for VP2 and VP1 (Table 3). (B) At codons identified as being exposed by the rolling+structural (R+S) model, minor-group viruses have consistently elevated ratios, significantly so in VP1 (Table 3).
To address the question of whether these effects occur throughout the major and minor groups or whether they arise from very strong effects in just a few lineages, we included the 19 cladelets (Fig. 2) as fixed effects and then as random effects in the generalized linear models. When analyzed as fixed effects, the minor-group clades (c32, c38, and c43) have the three highest coefficients for interaction with exposure status, indicating a consistently greater increase of the nonsynonymous substitution rate at exposed sites (Table 1). Treating clades as random effects yields similar results, although c38 (which includes only RV-A1A and RV-A1B) regresses toward the mean (Table 2). The significance and effect sizes of the interaction between group membership and exposure status for nonsynonymous substitutions remain nearly identical to those in the original GLM model, although the main effect of group is no longer significant in any model (Table 2). Exposure status explains a small portion of the variance in synonymous substitutions in the mixed-effects analysis, with the effect of group again no longer being significant (Table 2).
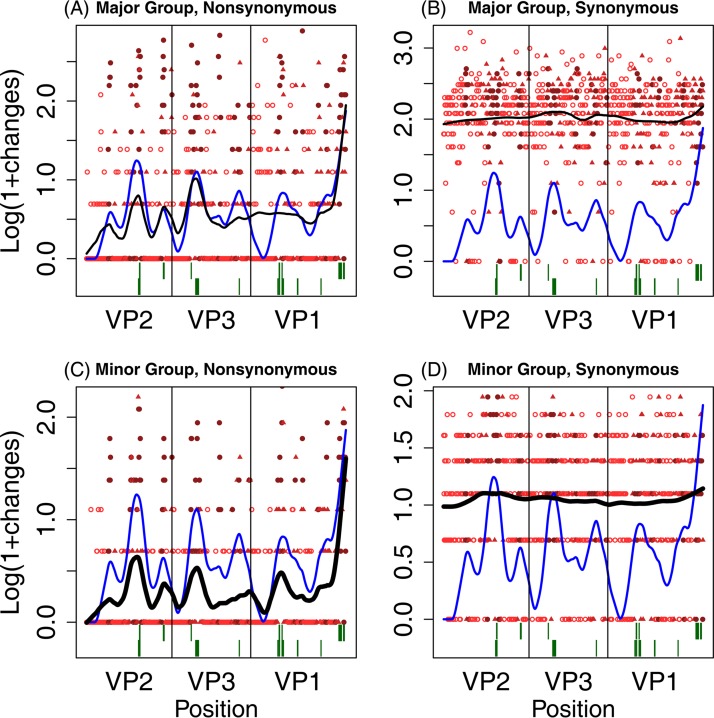
Figure 6 summarizes nonsynonymous and synonymous changes along the lengths of the VP2, VP3, and VP1 genes. The overall patterns of substitution are similar for both the major (Fig. 6A) and minor (Fig. 6B) cell receptor groups. The fitted curve shows that the major and minor groups tend to have elevated rates of substitution in the same genomic locations and that the peaks are generally well aligned with the known antigenic sites in the three proteins, although substitutions in VP1 fail to track exposed and buried regions in major-group viruses.
FIG 6.
Summary of locations of substitutions. For each codon in the three capsid protein genes, the log of 1 + the number of substitutions is plotted against position in the sequence. Solid red circles indicate buried sites; solid red triangles indicate sites identified as being exposed by either the rolling-only (R) model or the structural model, but not both; and open red circles indicate sites identified by both models, i.e., the rolling+structural (R+S) model. The solid black line is a smoothed fit of the log-transformed number of changes, and the solid blue line a smoothed fit through the number of models that identify sites as being exposed (with a value of 0, 1, or 2). The dark green lines at the bottom show the locations of antigenic sites, with major-group sites above minor-group sites. (A) Nonsynonymous changes in major-group viruses; (B) synonymous changes in major-group viruses; (C) nonsynonymous changes in minor-group viruses; (D) synonymous changes in minor-group viruses.
DISCUSSION
Major-group and minor-group rhinoviruses evolve at different tempos and with other thematic differences that may reflect the paths that they take to cell entry. Minor-group viruses enter cells after binding to an LDL receptor. This method of entering cells does not provide a way to downregulate the immune response, in contrast to the method used by major-group viruses, which bind to ICAM-1 (4). Minor-group viruses are more effective than major-group viruses in stimulating T-cell responses in vitro (25). We hypothesized that as a consequence, minor-group viruses would induce stronger antibody responses and thereby experience higher rates of amino acid substitutions at sites exposed to the immune system.
We considered three ways to characterize sites as being exposed to the immune system: (i) experimentally determined antigenic sites, (ii) sites determined to lie on the surface of the capsid protein of a minor-group rhinovirus based on proximity to the surface (rolling model), and (iii) sites determined to lie at the surface of a major-group rhinovirus based on a previously determined structure (structural model). At the 34 known antigenic sites, we found relatively large numbers of amino acid substitutions in both cell receptor groups, compared to nonantigenic sites, but the interaction between antigenicity and minor-group membership was not significant. The rolling model identified larger numbers of potentially antigenic sites. This approach identified 327 exposed sites, including all 34 sites previously identified as being antigenic. At these exposed sites, the amino acid substitution rate is increased, with a highly significant interaction between exposure status and cell receptor group and a greater increase in the minor group (Table 2 and Fig. 4A). The structural model (based on RV-A16 [44]) identifies fewer than half as many exposed sites, only five of which are not identified by the rolling model. These sites show an even higher nonsynonymous substitution rate than those identified only by the rolling model, and the interaction with the minor group is of a nearly equal magnitude.
These differences occur consistently throughout the phylogeny and do not result from large differences between just a few major-group and minor-group clades. We separately scored the synonymous and nonsynonymous substitutions in 16 major-group and 3 minor-group clades of roughly equal depths and treated clade identity as a random effect in the generalized linear mixed-model analyses. The interaction of the minor group with exposed sites remains significant and of a nearly identical magnitude (Table 2), and the two larger minor-group clades (c32 and c43) (Fig. 2) show the largest excess of substitutions at exposed sites. The small minor-group clade (c38) shows a large excess when treated as a fixed effect but regresses toward the mean of the full ensemble in the mixed-model analysis. Thus, the interaction between group and site occurs throughout the tree.
The relatively higher rate of nonsynonymous substitutions at exposed sites in minor-group viruses could be caused either by an absolutely higher nonsynonymous rate at exposed sites in minor-group viruses or by a lower nonsynonymous rate at buried sites in major-group viruses. The ratio of nonsynonymous to synonymous changes at these sites argues strongly for the first explanation, the one consistent with immune escape. If the nonsynonymous substitution rates at buried sites were lower in major- than in minor-group lineages, we would expect the ratio of nonsynonymous to synonymous substitutions (dN/dS ratio) at buried sites to also be lower in major-group lineages. However, where major- and minor-group lineages differ, the ratio of nonsynonymous to synonymous changes is higher in the major group.
Analysis of substitution rates at synonymous sites reveals a weaker but significant and consistent excess of changes at exposed codons in both major- and minor-group viruses and in all three capsid proteins. Evidence for weak positive and negative (purifying) selection on synonymous mutations has been found in a variety of studies, including one on vesicular stomatitis virus (45). A number of possible explanations have been proposed, including preferences for translationally “optimal” synonymous codons, effects on mRNA secondary structure and stability (46), and technical inaccuracy in estimating the numbers of synonymous and nonsynonymous substitutions in codons with multiple substitutions on a branch.
The current hypothesis for picornavirus evolution, and rhinovirus evolution in particular, is that most of the genome is subject to purifying selection, with positive selection predominantly being restricted to antigenic sites and to certain regions within the nonstructural proteins 2A protease, 3C protease, and 3D polymerase (32, 42, 47, 48). We found that both major-group and minor-group viruses fix substitutions more frequently at many sites that are exposed on the surface of the capsid and not just those previously identified as being antigenic; this finding supports the hypothesis that the entire capsid surface has the potential to be antigenic (49).
Rhinovirus antigenic sites were identified by using monoclonal antibodies harvested from experimentally infected BALB/c mice that had been exposed to HeLa cell lines infected with RV (50–52). The highly polymorphic major histocompatibility complex (MHC) determines which peptides will be recognized as being foreign by the immune system (53). Different MHC molecules within an individual, among individuals in a population, and among species will present different regions of a given protein, leading to different immune responses (53). Thus, the antigenic sites identified in a mouse model might plausibly differ from those that would be identified in another species, such as humans. The evolutionary evidence developed here suggests that the huge immunological diversity of the global human population collectively detects a large fraction of the amino acids displayed on the surface of the rhinovirus capsid.
The cell receptor footprints of major- and minor-group viruses differ, but this difference does not appear to explain why minor-group viruses are less well conserved at exposed amino acid positions. The cellular receptor footprint covers only a small region of the capsid surface in both groups. In major-group viruses, ICAM-1 binds to amino acid residues belonging to VP2, VP3, and VP1 that line the interior of the canyon (20). Smith and colleagues demonstrated previously that although antibodies are able to attach to residues within the canyon in the same region where ICAM-1 binds, residues in this region are conserved, and mutations were shown to be deleterious (54). In contrast, minor-group viruses use members of the LDL receptor family to bind to amino acids located on the BC and HI loops in VP1. Successful attachment has been shown to depend on the presence of a conserved amino acid (K224 in RV-A2) that is located in the HI loop and on an overall basic charge on the amino acids within the receptor footprint (22, 38). Members of the LDL receptor family have multiple repeated binding domains, and different viruses can use different repeats to attach to the receptor (55–57). This diversity in available LDL-binding domains suggests that minor-group viruses may be able to accumulate a greater number of substitutions at exposed residues within the cellular footprint to escape immune detection while maintaining access to their cellular receptor. Thus, reduced constraint in LDL attachment might play a role in elevating the substitution rates in this region, but only a few sites would be affected in this way.
Finally, we hypothesized that a higher rate of amino acid substitutions should occur in the immunodominant capsid protein VP1 (52) and that this increased substitution rate should be accentuated in minor-group viruses. We found significantly more nonsynonymous substitutions per codon in VP1 and VP3 only at buried sites and no interaction between the virus receptor type and capsid protein subunits, providing no support for a specific effect in minor-group viruses (Table 2). The detailed substitution pattern in minor-group VP1 shows a peak in the shared antigenic region from positions 83 to 100, which is missing in the major group (Fig. 6), indicating that differences in patterns of substitution might occur over regions smaller than capsid proteins.
Beginning with the known immunological difference between major- and minor-group rhinoviruses, we found evidence that minor-group viruses elicit a stronger host immune response and evolve more rapidly at exposed amino acid sites. Rapid evolution might help to maintain minor-group viruses in the population, although we suspect that these serotypes have other selective advantages, through either increased transmission or more effective intrahost competition. On the continuum from mild, highly diverse, and weakly immunogenic rhinoviruses to severe, less diverse, and highly immunogenic influenza viruses, minor-group rhinoviruses may be more “influenza-like” than major-group viruses (58). As additional studies characterize the recently discovered RV-C, the patterns described here may provide a way to determine whether rhinoviruses are currently continuing to stably maintain a diversity of serotypes and strategies or whether they are evolving generally increased or decreased virulence.
MATERIALS AND METHODS
Antibody response to infection caused by major- and minor-group viruses.
In the Seattle Watch studies conducted from 1965 to 1969 and from 1975 to 1979, Fox and colleagues (11) reported the proportion of people infected with different RV serotypes who developed an antibody response. We classified these serotypes into major and minor groups within RV-A and used a chi-square test to assess whether antibody production differed significantly between infections caused by major- and minor-group viruses using data reported previously (see Table 5 in reference 11).
Data collection and sequence alignment.
Eighty-four complete nucleotide sequences for the RV-A capsid genes VP1, VP2, and VP3 were downloaded from GenBank. To maintain the correct reading frame during alignment, each gene was converted to amino acids by using the AlignmentHelper program (59), which preserves the original nucleotide sequence, and then aligned separately at the amino acid level. Alignments were conducted by using default parameters in MUSCLE (60) and by using the G-INS-I refinement strategy and default parameters in MAFFT (61). The amino acid alignments were then converted back to nucleotides by using AlignmentHelper. MUSCLE and MAFFT generated similar alignments, and the gene tree topologies based on these alignments were identical. The aligned sequences were concatenated in the standard genomic order VP2-VP3-VP1.
Phylogenetic analysis.
The aligned, concatenated nucleotide sequences were used to estimate a consensus phylogenetic tree. Model selection was determined by using the Akaike information criterion (62) as implemented in jModeltest 0.1 (63). The best-fit model of evolution for all genes was the general time-reversible model with an estimated proportion of invariant sites and a gamma distribution of rates (GTR+I+G). Phylogenies were estimated by PhyML (64). Branch support values were estimated by using nonparametric bootstrapping with 1,000 pseudoreplications.
Trees generated from analyses of the full set of 84 RV-A sequences were used to identify 57 sequences that include the three minor-group clades and their closest major-group relatives. This subset of sequences was used in the analyses described below. The major-group serotype RV-A89 was used to root the tree and establish character state polarity because of its sister taxon relationship with the 56 members of the ingroup (Table 4 and Fig. 2).
TABLE 4.
Sequence accession numbers, serotypes, and references used in this study
| GenBank accession no. | Serotype | Reference |
|---|---|---|
| Minor-group serotypes | ||
| FJ445111 | 1A | 33 |
| D00239 | 1B | 71 |
| X02316 | 2 | 72 |
| DQ473497 | 23 | 42 |
| FJ445123 | 25 | 33 |
| FJ445125 | 29 | 33 |
| FJ445179 | 30 | 33 |
| FJ445126 | 31 | 33 |
| DQ473499 | 44 | 42 |
| FJ445133 | 47 | 33 |
| DQ473496 | 49 | 42 |
| FJ445145 | 62 | 33 |
| Major-group serotypes | ||
| FJ445177 | 9 | 33 |
| FJ445178 | 10 | 33 |
| EF173414 | 11 | 43 |
| FJ445116 | 13 | 33 |
| DQ473493 | 15 | 42 |
| L24917 | 16 | 73 |
| FJ445118 | 18 | 33 |
| FJ445119 | 19 | 33 |
| FJ445121 | 21 | 33 |
| FJ445122 | 22 | 33 |
| FJ445190 | 24 | 33 |
| FJ445127 | 32 | 33 |
| FJ445128 | 33 | 33 |
| FJ445189 | 34 | 33 |
| FJ445180 | 38 | 33 |
| AY751783 | 39 | 74 |
| FJ445129 | 40 | 33 |
| DQ473491 | 41 | 42 |
| FJ445131 | 43 | 33 |
| FJ445135 | 50 | 33 |
| FJ445138 | 54 | 33 |
| DQ473511 | 55 | 42 |
| FJ445140 | 56 | 33 |
| FJ445141 | 57 | 33 |
| DQ473500 | 59 | 42 |
| FJ445143 | 60 | 33 |
| FJ445144 | 61 | 33 |
| FJ445146 | 63 | 33 |
| FJ445181 | 64 | 33 |
| FJ445148 | 66 | 33 |
| FJ445149 | 67 | 33 |
| DQ473492 | 73 | 42 |
| DQ473494 | 74 | 42 |
| DQ473510 | 75 | 42 |
| FJ445182 | 76 | 33 |
| FJ445154 | 77 | 33 |
| FJ445157 | 81 | 33 |
| FJ445160 | 82 | 33 |
| FJ445163 | 85 | 33 |
| FJ445184 | 89a | 33 |
| FJ445167 | 90 | 33 |
| FJ445185 | 94 | 33 |
| FJ445171 | 96 | 33 |
| FJ445173 | 98 | 33 |
| FJ445175 | 100 | 33 |
Human rhinovirus A89 was used as the outgroup to root the phylogenetic tree.
Detection of amino acid and nucleotide substitutions.
Nucleotide substitutions on all branches of the 57-sequence genealogy were inferred by maximum likelihood reconstruction of its history. Statistical analyses were restricted to substitutions within the 3 minor-group clades (Fig. 2, thick branches) and 16 major-group clades with similar maximum and average branch depths. The roots of these 19 analyzed “cladelets” are indicated in Fig. 2 by clade names in the form of “cXX,” where XX is a number derived from the numbering of internal nodes in the tree. To be included, a cladelet's ancestral node was required to be no more distant than 0.22 expected substitutions per site from the tip nodes (modern sequences), in an independent estimate of the tree that was derived under molecular-clock assumptions by BEAST (65). This tree was topologically identical to the tree shown in Fig. 2 except for the relationships among a few of the deepest nodes. The limit of 0.22 substitutions/site includes one major-group clade (c6), which is slightly deeper than the two deepest minor-group clades (c32 and c43), and one major-group clade (c55), which is slightly shallower than the shallowest minor-group clade (c38).
By limiting the analysis to substitutions occurring on short, recent branches in both the major and minor groups, we reduced the risks that artifactual differences might arise between the groups and also that “overprinting” might hide significant numbers of nucleotide substitutions. (Synonymous sites begin to approach saturation in the deeper parts of the tree.) Ancestral nucleotide sequences were reconstructed at each internal node by using the dnaml program in PHYLIP 3.695 (66) and then translated by a custom Python script that separately scored synonymous and nonsynonymous (amino-acid-changing) substitutions for each codon along each included branch. Sites with ambiguous nucleotide states (e.g., “Y” for pyrimidine) were dropped from the analysis on branches affected by the ambiguity.
Identification of surface-exposed and antigenic amino acid residues.
The three-dimensional structure of RV-A2 (Protein Data Bank [PDB] accession number 3DPR) was used as a reference for the general capsid structure. The PyMOL molecular visualization program (version 1.5.0.1; Schrödinger LLC, New York) was used to reconstruct the complete icosahedral capsid using the rotation matrices in the PDB data file. Surface area calculations were limited to the exposed surface of the virus by the following procedure. A single protomer made up of VP2, VP3, and VP1 and the minimal number of additional protein subunits that surround the central protomer were identified and saved as a new molecule. To visualize the exterior and interior surfaces, the accessible surface area was drawn with a 5-Å probe radius, which is approximately the size of an amino acid residue. The “rotate” command was used to rotate the molecule and modify the model coordinates such that the exterior and interior surfaces of the central protomer were perpendicular to the y axis. The exterior and interior surfaces of the virus capsid could then be distinguished by their y coordinates. A cutoff value was determined by inspection of the structure, looking for residues that were about halfway between the inner and outer surfaces. A Python script written by D. P. Goldenberg was used to identify the y coordinate values of all residues in the central protomer that were above the designated cutoff value. The accessible surface areas for the VP2, VP3, and VP1 proteins within the central protomer were calculated by using the algorithm described previously by Lee and Richards (68), as implemented in the ACCESS program by T. J. Richmond, and the group radii described previously by Chothia (69). The accessible surface area was calculated by modeling a molecule with a probe radius of 1.4 Å “rolling” over the surface of the viral capsid. All amino acids that came into contact with the rolling molecule were identified as being exposed, and we refer to this as the rolling model.
We used a three-dimensional structure of major-group RV-A16 (44) to create an alternative characterization of exposed and buried sites, which we call the structural model. The union of sites exposed in either the rolling or structural model defines an inclusive “combined” characterization of exposed sites. The contrasting “joint” characterization disaggregates sites into four categories based on which model(s) identifies them as being exposed (see Results).
The location of amino acid residues within antigenic sites in major- and minor-group viruses was based on descriptions reported previously (50, 51). Major-group rhinoviruses have four primary neutralizing immunogenic sites (NIm-IA, NIm-IB, NIm-II, and NIm-III) that have been mapped to protruding regions on the external capsid proteins VP1, VP2, and VP3 (50). Minor-group rhinoviruses have three neutralizing immunogenic sites, known as site A, site B, and site C, that are located in the vicinity of the NIm sites (51). Due to the relatively small number of sites identified as being antigenic, we base our analyses on a combined characterization that looks at the union of all identified immunogenic sites.
Statistical analysis.
The raw data are counts of synonymous and nonsynonymous (amino-acid-changing) nucleotide substitutions per codon on each branch in the included parts of the tree. Branches are classified as occurring within major- or minor-group clades. Our basic statistical model analyzes the sums of the synonymous and nonsynonymous substitutions at each codon position across all major-group branches and across all minor-group branches. Codons are classified by gene and by surface exposure of the encoded amino acid. We used generalized linear models (glm in R, with the quasipoisson family of link functions) to explain the variation in the summed substitutions using various combinations of the explanatory factors of interest. For example, the most basic model reported here is of the form glm[aax ∼ GUE * (gene + group), family = quasipoisson, …]. Here aax is the summed number of nonsynonymous substitutions at each codon position; GUE is the combined characterization of surface exposure, with state E for exposed or state B for buried codons; the gene is VP1, VP2, or VP3; and the group is M for major or m for minor. Synonymous substitutions (syn) were analyzed using the same framework.
We began by testing two-way and three-way interactions and used backward model selection to remove nonsignificant terms, with a P value of <0.05 being the threshold for statistical significance. Our central question was whether the interactions between exposure to the immune system and major-group versus minor-group identity (GUE * group in the model described above) would be significant and large. Owing to their greater immunogenicity, we predicted that minor-group rhinoviruses would show a large increase of amino acid substitution rates at exposed nonsynonymous sites (relative to their rates at buried nonsynonymous sites).
The number of substitutions is expected to be proportional to the branch length, so a significant main effect of group is expected even under the null hypothesis, owing to the much greater total branch length of the major-group clades. One way to remove this “nuisance” effect of phylogeny is to normalize the nonsynonymous substitutions by the synonymous substitutions on the same branches and analyze the resulting dN/dS ratios. This approach requires averaging over many codon positions, and it depends on the assumption that synonymous mutations are selectively neutral. An alternative is to include the number of synonymous substitutions per codon as a covariate in the model, thereby removing the synonymous component of the branch length effect statistically without averaging over codons or taking ratios of the Poisson-distributed primary events (i.e., small numbers of nucleotide substitutions at individual codons). We did this in some analyses, although for the present purposes, it is not necessary because the main effects of group and/or cladelet capture the nonsynonymous component of the branch length effect. To summarize the overall patterns, we constructed 2-by-2 contingency tables of synonymous and nonsynonymous substitutions categorized by group membership, with separate tables for each combination of gene and exposure status, and carried out chi-square tests. In all of our analyses, the effects of central interest are the strengths of the interactions between group and exposure and not the magnitudes of the main effects as such.
We asked whether the increased substitution rate at exposed minor-group nonsynonymous sites could be seen in each of the three minor-group clades or whether it resulted from the influence of one very-fast-evolving lineage or clade. To address this question, branches were categorized by their cladelet memberships, which were added as a fixed effect in the GLM analysis and as a random effect in a mixed-model analysis (using glmer in R with the Poisson family).
ACKNOWLEDGMENTS
This work was supported by a 21st Century Science Initiative grant from the James S. McDonnell Foundation and the Modeling the Dynamics of Life Fund of the University of Utah to F.R.A. and by the NSF-DEB (grant 1051491) to J.S. The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank David P. Goldenberg for valuable advice and for generating the accessible-surface-area calculations and Chae Kyung Yoo for assistance with developing Python scripts. We also thank Duke S. Rogers for reviewing earlier versions of this paper and the sLaM and eKoSystems groups for useful discussion. Cole Porter invented the subtitle in 1944, anticipating recognition of the minor group by 4 decades (70).
N.L.-R. and F.R.A. conceived of the study, N.L.-R. and J.S. carried out the phylogenetic analyses and ancestral sequence reconstructions, F.R.A. and J.S. carried out statistical analyses, and all three authors wrote the manuscript.
REFERENCES
- 1.Eccles R. 2005. Understanding the symptoms of the common cold and influenza. Lancet Infect Dis 5:718–725. doi: 10.1016/S1473-3099(05)70270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byington CL, Ampofo K, Stockmann C, Adler FR, Herbener A, Miller T, Sheng X, Blaschke AJ, Crisp R, Pavia AT. 2015. Community surveillance of respiratory viruses among families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) study. Clin Infect Dis 61:1217–1224. doi: 10.1093/cid/civ486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitkäranta A, Hayden FG. 1998. Rhinoviruses: important respiratory pathogens. Ann Med 30:529–537. doi: 10.3109/07853899709002600. [DOI] [PubMed] [Google Scholar]
- 4.Kirchberger S, Majdic O, Stöckl J. 2007. Modulation of the immune system by human rhinovirus. Int Arch Allergy Immunol 142:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Heikkinen T, Jarvinen A. 2003. The common cold. Lancet 361:51–59. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monto AS. 2002. Epidemiology of viral respiratory infections. Am J Med 112:4–12. doi: 10.1016/S0002-9343(01)01058-0. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser L, Aubert J-D, Pache J-C, Deffernex C, Rochat T, Garbino J, Wunderli W, Meylan P, Yerly S, Perrin L, Letovanec I, Nicod L, Tapparel C, Soccal PM. 2006. Chronic rhinoviral infection in lung transplant recipient. Am J Respir Crit Care Med 174:1392–1399. doi: 10.1164/rccm.200604-489OC. [DOI] [PubMed] [Google Scholar]
- 8.Domingo E, Holland J. 1994. Mutation rates and rapid evolution of RNA viruses, p 161–184. In Morse SS. (ed), Evolutionary biology of viruses. Raven Press, New York, NY. [Google Scholar]
- 9.Palese P, Wang TT. 2011. Why do influenza virus subtypes die out? A hypothesis. mBio 2(5):e00150–11. doi: 10.1128/mBio.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox JP, Cooney MK, Hall CE. 1975. The Seattle virus watch. V. Epidemiological observations of rhinovirus infections, 1965-1969, in families with young children. Am J Epidemiol 101:122–143. [DOI] [PubMed] [Google Scholar]
- 11.Fox JP, Cooney MK, Hall CE, Foy HM. 1985. Rhinoviruses in Seattle families, 1975-1979. Am J Epidemiol 22:830–846. [DOI] [PubMed] [Google Scholar]
- 12.Roebuck MO. 1976. Rhinoviruses in Britain 1963-1973. J Hyg 76:137–146. doi: 10.1017/S0022172400055029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monto AS, Bryan ER, Ohmit S. 1987. Rhinovirus infections in Tecumseh, Michigan: frequency of illness and number of serotypes. J Infect Dis 156:43–49. doi: 10.1093/infdis/156.1.43. [DOI] [PubMed] [Google Scholar]
- 14.Gwaltney JM., Jr 1975. Rhinoviruses. Yale J Biol Med 48:17–45. [PMC free article] [PubMed] [Google Scholar]
- 15.Hope-Simpson RE, Golubev DB. 1987. A new concept of the epidemic process of influenza A virus. Epidemiol Infect 99:5–54. doi: 10.1017/S0950268800066851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staneková Z, Varečková E. 2010. Conserved epitopes of influenza A virus inducing protective immunity and their prospects for universal vaccine development. Virology J 7:351–364. doi: 10.1186/1743-422X-7-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minor PD. 1990. Antigenic structure of picornaviruses. Curr Top Microbiol Immunol 161:121–154. [DOI] [PubMed] [Google Scholar]
- 18.Ledford RM, Patel NR, Demenszuk TM, Watanyar A, Herbertz T, Collett MS, Pevear DC. 2004. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol 78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McErlean P, Shackelton LA, Lambert SB, Nissen MD, Sloots TP, Mackay IM. 2007. Characterization of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol 39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson NH, Kolatkar PR, Oliverira MA, Cheng RH, Greve JM, McClelland A, Baker TS, Rossmann MG. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci U S A 90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. 1994. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci U S A 91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hewat EA, Neumann E, Conway JF, Moser R, Ronacher B, Marlovits TC, Blass D. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J 19:6317–6325. doi: 10.1093/emboj/19.23.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkov YA, Palmenberg AC, Lee W-M, Rathe JA, Amineva SP, Sun X, Pasic TR, Jarjour NN, Liggart SB, Gern JE. 2011. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med 17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, Palmenberg AC, Gern JE. 2015. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A 112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastings GZ, Francis MJ, Rowlands DJ, Chain BM. 1993. Antigen processing and presentation of human rhinovirus to CD4 T cells is facilitated by binding to cellular receptors for virus. Eur J Immunol 23:1340–1345. doi: 10.1002/eji.1830230623. [DOI] [PubMed] [Google Scholar]
- 26.van de Stolpe A, van der Saag PT. 1996. Intercellular adhesion molecule-1. J Mol Med 74:13–33. doi: 10.1007/BF00202069. [DOI] [PubMed] [Google Scholar]
- 27.Kirchberger S, Majdic O, Steinberger P, Bluml S, Pfistershammer K, Zlabinger G, Kuechler E, Knapp W, Stockl J. 2005. Human rhinoviruses inhibit the accessory function of dendritic cells by inducing sialoadhesin and B7-H1 expression. J Immunol 175:1145–1152. doi: 10.4049/jimmunol.175.2.1145. [DOI] [PubMed] [Google Scholar]
- 28.Kolatkar PR, Bella J, Olson NH, Bator CM, Baker TS, Rossmann MG. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J 18:6249–6259. doi: 10.1093/emboj/18.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beffert U, Stolt PC, Herz J. 2004. Functions of lipoprotein receptors in neurons. J Lipid Res 45:403–409. [DOI] [PubMed] [Google Scholar]
- 30.Vlasak M, Blomqvist S, Hovi T, Hewat E, Blaas D. 2003. Sequence and structure of human rhinoviruses reveal the basis of receptor discrimination. J Virol 77:6923–6930. doi: 10.1128/JVI.77.12.6923-6930.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuthill TJ, Papadopoulos NG, Jourdan P, Challinor LJ, Sharp NA, Plumpton C, Shah K, Barnard S, Dash L, Burnet J, Killington RA, Rowlands DJ, Clarke NJ, Blair ED, Johnston SL. 2003. Mouse respiratory epithelial cells support efficient replication of human rhinovirus. J Gen Virol 84:2829–2836. doi: 10.1099/vir.0.19109-0. [DOI] [PubMed] [Google Scholar]
- 32.Lewis-Rogers N, Bendall ML, Crandall KA. 2009. Phylogenetic relationships and molecular adaptation dynamics of human rhinoviruses. Mol Biol Evol 26:969–981. doi: 10.1093/molbev/msp009. [DOI] [PubMed] [Google Scholar]
- 33.Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB. 2009. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ketler A, Hall CE, Fox JP, Elveback L, Cooney MK. 1969. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VII. Rhinovirus infections: observations of virus excretion, intrafamilial spread and clinical response. Am J Epidemiol 90:244–254. [DOI] [PubMed] [Google Scholar]
- 35.Monto AS, Cavallaro JJ. 1972. The Tecumseh study of respiratory illness. IV. Prevalence of rhinovirus serotypes 1966-1969. Am J Epidemiol 96:352–360. [DOI] [PubMed] [Google Scholar]
- 36.Tapparel C, Junier T, Gerlach D, Van Belle S, Turin L, Cordey S, Muhlemann K, Regamey N, Aubert J-D, Soccal PM, Eigenmann P, Zdobnov E, Kaiser L. 2009. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis 15:719–726. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Querol-Audi J, Konecsni T, Pous J, Carugo O, Fita I, Verdaguer N, Blaas D. 2009. Minor group human rhinovirus-receptor interactions: geometry of multimodular attachment and basis of recognition. FEBS Lett 583:235–240. doi: 10.1016/j.febslet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 38.Vlasak M, Roivainen M, Reithmayer M, Goesler I, Laine P, Snyers L, Hovi T, Blaas D. 2005. The minor receptor group of human rhinovirus (HRV) includes HRV23 and HRV25, but the presence of a lysine in the VP1 HI loop is not sufficient for receptor binding. J Virol 79:7389–7395. doi: 10.1128/JVI.79.12.7389-7395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waman VP, Kolekar PS, Kale MM, Kulkarni-Kale U. 2014. Population structure and evolution of rhinoviruses. PLoS One 9:e88981. doi: 10.1371/journal.pone.0088981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith TJ. 2001. Antibody interactions with rhinovirus: lessons for mechanisms of neutralization and the role of immunity in viral evolution. Curr Top Microbiol Immunol 260:1–28. [DOI] [PubMed] [Google Scholar]
- 41.Savolainen C, Laine P, Mulders MN, Hovi T. 2004. Sequence analysis of human rhinovirus in the RNA-dependent RNA polymerase coding region reveals large within-species variation. J Gen Virol 85:2271–2277. doi: 10.1099/vir.0.79897-0. [DOI] [PubMed] [Google Scholar]
- 42.Kistler AL, Webster DR, Rouskin S, Magrini V, Credle JJ, Schnurr DP, Boushey HA, Mardis ER, Li H, DeRisi JL. 2007. Genome-wide diversity and selective pressure in the human rhinovirus. Virol J 4:40. doi: 10.1186/1743-422X-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tapparel C, Junier T, Gerlach D, Cordey S, Van Belle S, Perrin L, Zdobnov EM, Kaiser L. 2007. New complete genome sequences of human rhinoviruses shed light on their phylogeny and genomic features. BMC Genomics 8:224. doi: 10.1186/1471-2164-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira MA, Zhao R, Lee W-M, Kremer MJ, Minor I, Rueckert RR, Diana GD, Pevear DC, Dutko FJ, McKinlay MA, Rossmann MG. 1993. The structure of human rhinovirus 16. Structure 1:51–68. doi: 10.1016/0969-2126(93)90008-5. [DOI] [PubMed] [Google Scholar]
- 45.Novella IS, Zarate S, Metzgar D, Ebendick-Corpus BE. 2004. Positive selection of synonymous mutations in vesicular stomatitis virus. J Mol Biol 342:1415–1421. doi: 10.1016/j.jmb.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Resch AM, Carmel L, Mariño-Ramírez L, Ogurtsoc AY, Shabalina SA, Rogozin IB, Koonin EV. 2007. Widespread positive selection in synonymous sites in mammalian genes. Mol Biol Evol 24:1821–1831. doi: 10.1093/molbev/msm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haydon DT, Bastos AD, Knowles NJ, Samuel AR. 2001. Evidence for positive selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics 157:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmonds P, Welch J. 2006. Frequency and dynamics of recombination within different species of human enteroviruses. J Virol 80:483–493. doi: 10.1128/JVI.80.1.483-493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mateu MG. 1995. Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res 38:1–24. doi: 10.1016/0168-1702(95)00048-U. [DOI] [PubMed] [Google Scholar]
- 50.Sherry B, Mosser AG, Colonno RJ, Rueckert RR. 1986. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J Virol 57:246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Appleyard G, Russell SM, Clarke BE, Speller SA, Trowbridge M, Vadolas J. 1990. Neutralization epitopes of human rhinovirus type 2. J Gen Virol 71:1275–1282. doi: 10.1099/0022-1317-71-6-1275. [DOI] [PubMed] [Google Scholar]
- 52.Sherry B, Rueckert R. 1985. Evidence for a least two dominant neutralization antigens on human rhinovirus 14. J Virol 53:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sompayrac LM. 2008. How the immune system works, 3rd ed Blackwell Publishing, Malden, MA. [Google Scholar]
- 54.Smith TJ, Chase ES, Schmidt TJ, Olson NH, Baker TS. 1996. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature 383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esser V, Limbird LE, Brown MS, Goldstein JL, Russell DW. 1989. Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J Biol Chem 263:13282–13290. [PubMed] [Google Scholar]
- 56.Russell DW, Brown MS, Goldstein JL. 1989. Different combinations of cyesteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J Biol Chem 264:21682–21688. [PubMed] [Google Scholar]
- 57.Wruss J, Rünzler D, Steiger C, Chiba P, Köhler G, Blaas D. 2007. Attachment of VLDL receptors to an icosahedral virus along the 5-fold symmetry axis: multiple binding modes evidenced by fluorescence correlation spectroscopy. Biochemistry 46:6331–6339. doi: 10.1021/bi700262w. [DOI] [PubMed] [Google Scholar]
- 58.Adler FR, Kim PS. 2013. Models of contrasting strategies of rhinovirus immune manipulation. J Theor Biol 327:1–10. doi: 10.1016/j.jtbi.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 59.McClellan DA, Woolley S. 2004. AlignmentHelper ver.1.0. Brigham Young University Department of Integrative Biology, Provo, UT. [Google Scholar]
- 60.Edgar RC. 2004. MUCSLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach, 2nd ed Springer-Verlag, Inc, New York, NY. [Google Scholar]
- 63.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 64.Guindon S, Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 65.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUTi and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Felsenstein J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- 67.Reference deleted.
- 68.Lee B, Richards FM. 1971. The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55:379–400. doi: 10.1016/0022-2836(71)90324-X. [DOI] [PubMed] [Google Scholar]
- 69.Chothia C. 1976. The nature of the accessible and buried surfaces in proteins. J Mol Biol 105:1–14. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 70.Porter C. 1944. Ev'rytime we say goodbye. Chappell & Company, New York, NY. [Google Scholar]
- 71.Hughes PJ, North C, Jellis CH, Minor PD, Stanway G. 1988. The nucleotide sequence of human rhinovirus 1B: molecular relationships within the rhinovirus genus. J Gen Virol 69:49–58. doi: 10.1099/0022-1317-69-1-49. [DOI] [PubMed] [Google Scholar]
- 72.Skern T, Sommergruber W, Blaas D, Gruendler P, Fraundorfer F, Pieler C, Fogy I, Kuechler E. 1985. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res 13:2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee WM, Wang W, Rueckert RR. 1995. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes 9:177–181. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- 74.Harris JR, Racaniello VR. 2005. Amino acid changes in proteins 2B and 3A mediate rhinovirus type 39 growth in mouse cells. J Virol 79:5363–5373. doi: 10.1128/JVI.79.9.5363-5373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]