Summary
Although human induced pluripotent stem cells (hiPSCs) hold great potential for the study of human diseases affecting disparate cell types, they have been underutilized in seeking mechanistic insights into the pathogenesis of congenital craniofacial disorders. Craniofrontonasal syndrome (CFNS) is a rare X-linked disorder caused by mutations in EFNB1 and characterized by craniofacial, skeletal, and neurological anomalies. Heterozygous females are more severely affected than hemizygous males, a phenomenon termed cellular interference that involves mosaicism for EPHRIN-B1 function. Although the mechanistic basis for cellular interference in CFNS has been hypothesized to involve Eph/ephrin-mediated cell segregation, no direct evidence for this has been demonstrated. Here, by generating hiPSCs from CFNS patients, we demonstrate that mosaicism for EPHRIN-B1 expression induced by random X inactivation in heterozygous females results in robust cell segregation in human neuroepithelial cells, thus supplying experimental evidence that Eph/ephrin-mediated cell segregation is relevant to pathogenesis in human CFNS patients.
Keywords: human induced pluripotent stem cells (hiPSCs), craniofrontonasal syndrome (CFNS), EFNB1, Eph/ephrin signaling, cell segregation, cell sorting, X chromosome inactivation (XCI), neuroepithelial cells, neural progenitor cells, craniofacial
Highlights
-
•
A novel iPSC line can effectively model a craniofacial condition
-
•
Eph/ephrin-mediated cell segregation underlies CFNS
-
•
Cell segregation occurs in CFNS neuroepithelial progenitor cells
-
•
Neuroepithelial progenitors are a possible cell of origin for CFNS dysmorphogenesis
Bush and colleagues established a novel human induced pluripotent stem cell model of craniofrontonasal syndrome (CFNS), an unusual X-linked craniofacial condition. CFNS patient hiPSCs were able to undergo differentiation into human neuroepithelial (hNE) cells. Mosaicism for EPHRIN-B1 expression in hNE cells caused aberrant cell segregation, providing experimental evidence in human cells that EPHRIN-B1-mediated cell segregation underlies CFNS pathology.
Introduction
Congenital craniofacial disorders represent over one-third of all birth defects (Global strategies to reduce the health care burden of craniofacial anomalies, 2004). While the genetic causes of many syndromes are known, how these mutations lead to abnormal cellular mechanisms underlying these disorders is incompletely understood. A better understanding of the underlying etiology of these disorders is needed to develop new treatment strategies targeted to the cellular and molecular basis of disease. The emergence of human induced pluripotent stem cells (hiPSCs) as a tool for human disease modeling (Takahashi et al., 2007, Takahashi and Yamanaka, 2006, Tiscornia et al., 2011, Yu et al., 2007) holds great promise for improving our cellular understanding of craniofacial diseases, as hiPSCs can be differentiated into patient-specific, disease-relevant cell types. However, perhaps due to the challenge of modeling structural aspects of craniofacial disease in two dimensions in cell culture, hiPSC models for these disorders are not yet widely used.
Craniofrontonasal syndrome (CFNS; OMIM no. 304110) is an X-linked disorder caused by mutations in EFNB1 and characterized by craniofacial, skeletal, and neurological anomalies (Twigg et al., 2004, Wieland et al., 2004). The most common clinical findings include hypertelorism, frontonasal dysplasia, coronal synostosis, bifid nasal tip, longitudinal splitting of the nails, and wiry or frizzy hair; other less frequent symptoms include cleft lip and palate, diaphragmatic hernia, agenesis of the corpus callosum, syndactyly, and polydactyly (Twigg et al., 2004, Twigg et al., 2006, Twigg et al., 2013, Wieacker and Wieland, 2005, Wieland et al., 2004). CFNS is an unusual X-linked disorder in that heterozygous females are more severely affected than hemizygous male patients, who are usually unaffected or mildly affected and often present only with hypertelorism (Wieacker and Wieland, 2005). This counterintuitive inversion of severity has been termed cellular interference, a phenomenon whereby random X chromosome inactivation (XCI) in heterozygous female CFNS patients results in mosaicism for EFNB1 expression, leading to abnormal cellular interactions (Twigg et al., 2013, Wieacker and Wieland, 2005). Consistent with this notion, rare severely affected male CFNS patients have somatic mosaic mutations in EFNB1 (Twigg et al., 2013), reinforcing mosaicism as an important aspect of CFNS pathogenesis.
EFNB1 encodes EPHRIN-B1, a member of the Eph/ephrin family of membrane-linked signaling molecules, and abnormal signaling between cells expressing wild-type EPHRIN-B1 and cells that are functionally EPHRIN-B1-null may occur in the mosaic state (Compagni et al., 2003, Wieacker and Wieland, 2005). During development, Eph/ephrin signaling plays an important role in boundary formation, an essential process that requires signaling between adjacent cells and often involves segregation between different cell types (Batlle and Wilkinson, 2012, Cayuso et al., 2015, Fagotto, 2014, Fagotto et al., 2014). Differential expression of Eph receptors and ephrins in vivo can restrict cell intermingling in the vertebrate hindbrain (Xu et al., 1999), limb bud (Compagni et al., 2003, Davy et al., 2004), eye (Cavodeassi et al., 2013), somites (Barrios et al., 2003, Durbin et al., 1998), cranial sutures (Merrill et al., 2006, Ting et al., 2009), and intestinal crypts (Holmberg et al., 2006), as well as in the Drosophila wing disc (Umetsu et al., 2014). In culture, expressing an Eph receptor in one population of cells and an ephrin in another restricts intermingling of cells from the two populations (Jorgensen et al., 2009, Mellitzer et al., 1999, Poliakov et al., 2008). Further, cell segregation occurs in developing Efnb1+/− mouse limb (Compagni et al., 2003) and secondary palate (Bush and Soriano, 2010), supporting the idea that XCI-induced mosaicism leads to segregation of Ephrin-B1 expressing and non-expressing cells.
The role of Eph/ephrin signaling in boundary formation and supporting data from mouse models suggest that mosaicism for EPHRIN-B1 expression may lead to aberrant cell segregation in human CFNS patients (Compagni et al., 2003, Twigg et al., 2004, Twigg et al., 2006, Twigg et al., 2013, Wieacker and Wieland, 2005, Wieland et al., 2004). However, it has proven difficult to determine the mechanism of cellular interference, and EPHRIN-B1-mediated cell segregation has not been demonstrated in CFNS patients. Here, we report the generation of an hiPSC model to study defects in morphogenesis in a congenital craniofacial disorder. We demonstrate that cell segregation is a consequence of EPHRIN-B1 mosaicism in CFNS, providing evidence that this cell behavior is relevant to CFNS pathogenesis in humans. The CFNS hiPSC model provides proof of principle that hiPSC-derived cell types can be used both to model structural anomalies and to gain valuable insights into fundamental cellular mechanisms of morphogenesis in patient cells.
Results
Isolation of CFNS Human Dermal Fibroblasts and Reprogramming to hiPSCs
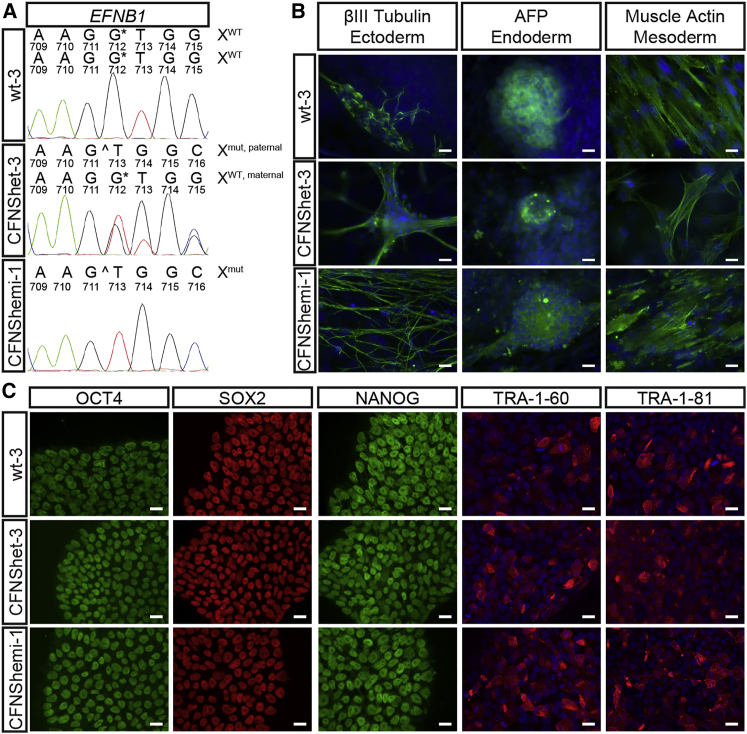
To investigate cellular mechanisms of CFNS, we established human dermal fibroblast (HDF) cultures from a female CFNS patient with a heterozygous mutation in exon 5 of EFNB1 (EFNB1+/c.712delG) (Byrne et al., 2009, Hogue et al., 2010). We also established HDF cultures from skin biopsies of the patient’s father, a hemizygous carrier of the mutation (EFNB1Y/c.712delG), and her unaffected mother (EFNB1+/+). We generated hiPSC lines from low-passage HDFs of each individual using non-integrating episomal vectors (Bershteyn et al., 2014, Okita et al., 2011) and selected three hiPSC lines from each individual for further analysis and experimentation (from the patient, lines CFNShet-1, -2, and -3; from the patient’s father, lines CFNShemi-1, -2, and -3; and from the patient’s mother, lines wt-1, -2, and -3). Sequencing of exon 5 of EFNB1 confirmed the expected genotypes (Figures 1A and S1A). All nine hiPSC lines were free of reprogramming plasmid integration by PCR (data not shown) and had normal G-banded karyotypes (Figure S1B).
Figure 1.
Reprogramming of Wild-Type, CFNShet, and CFNShemi HDFs to hiPSCs
(A) CFNShet-3 and CFNShemi-1 hiPSCs possess the EFNB1c.712delG mutation compared with wt-3 hiPSCs. See also Figure S1A.
(B) wt-3, CFNShet-3, and CFNShemi-1 hiPSCs possess differentiation potential to ectoderm (βIII-tubulin), endoderm (α-fetoprotein, AFP), and mesoderm (muscle actin). Samples were counterstained with DAPI (blue). Scale bars, 20 μm. See also Figure S1C.
(C) wt-3, CFNShet-3, and CFNShemi-1 hiPSCs express the endogenous pluripotency markers OCT4, SOX2, NANOG, TRA-1-60, and TRA-1-81. TRA-1-60- and TRA-1-81-labeled samples were counterstained with DAPI (blue). Scale bars, 20 μm. See also Figure S1D.
Characterization of CFNS hiPSC Pluripotency and Differentiation Potential
CFNShet and CFNShemi HDFs were reprogrammed to generate hiPSCs that possessed embryonic stem cell-like morphology similar to that of wild-type hiPSCs. All lines, regardless of genotype, possessed differentiation potential to ectoderm (βIII-tubulin), endoderm (α-fetoprotein), and mesoderm (muscle actin) in an embryoid body protocol (Figures 1B and S1C) and expressed the endogenous pluripotency markers OCT4, SOX2, NANOG, TRA-1-60, and TRA-1-81 (Figures 1C and S1D).
XCI-induced mosaicism in females plays a central role in CFNS (Twigg et al., 2004, Twigg et al., 2006, Twigg et al., 2013, Wieacker and Wieland, 2005, Wieland et al., 2004). However, XCI status of female hiPSCs can vary across different lines (Lessing et al., 2013), depending on conditions used for reprogramming (Tchieu et al., 2010, Tomoda et al., 2012, Wutz, 2012). To model CFNS, we used reprogramming conditions that favor maintenance of XCI rather than X reactivation (Wutz, 2012) and characterized XCI status of CFNShet HDFs and each CFNShet hiPSC line using the human androgen receptor assay (HUMARA) (Kiedrowski et al., 2011). CFNShet HDFs showed an expected XCI ratio close to 50%, as in earlier studies that indicated no skewed X inactivation in female CFNS patients (Table S1) (Twigg et al., 2004, Wieland et al., 2004, Wieland et al., 2007). All three CFNShet hiPSC lines showed complete inactivation of the maternal, wild-type X chromosome, consistent with the clonal XCI expected from female hiPSC lines derived from a single fibroblast under conditions that favor XCI maintenance (Tchieu et al., 2010). Therefore, only the paternal, mutant copy of EFNB1 is expressed in these lines (Table S1), and they are not expected to express functional EPHRIN-B1.
Differentiation and Characterization of CFNS Neuroepithelial Cells
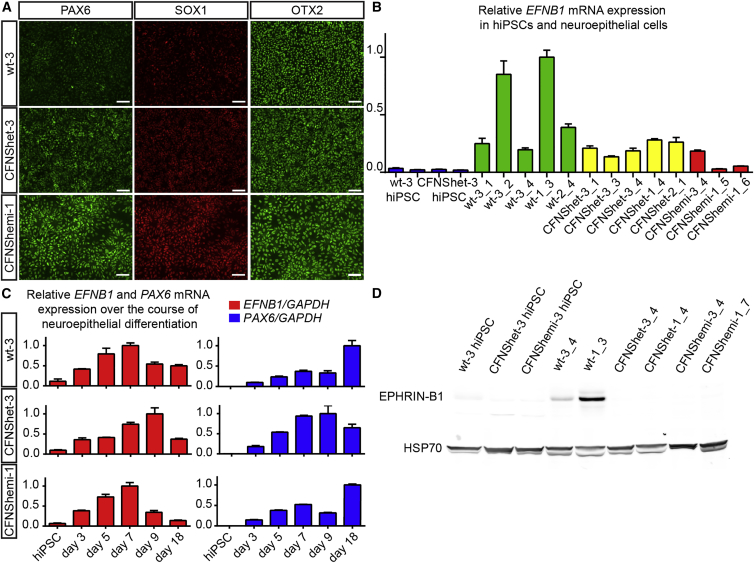
CFNS affects multiple structures derived from neural crest cells (NCCs), a multipotent population of stem cells that are induced at the neural plate border, delaminate, and migrate ventrolaterally to populate the craniofacial structures and contribute to skeletal, connective, neural, and vascular tissues. In mice, Ephrin-B1-mediated cell segregation occurs in the NE prior to NCC emigration (O’Neill et al., 2016). We therefore reasoned that NE cells are a good model for testing whether cell segregation occurs in CFNS, and we began by differentiating CFNS and control hiPSCs to human neuroepithelial (hNE) cells. We adapted a monolayer protocol that uses dual-SMAD inhibition to improve neural differentiation efficiency through inhibition of activin and nodal signaling (Chambers et al., 2009). All hNE cells, regardless of genotype, had neuroepithelial morphology and expressed the neural progenitor cell markers PAX6, SOX1, and OTX2 (Figure 2A). These data indicate that CFNS patient hiPSCs are able to differentiate into hNE cells independently of EPHRIN-B1 expression. hNE cells of all genotypes, but not hiPSCs, expressed EFNB1 mRNA (Figure 2B), and over the course of differentiation to hNE cells, expression of mRNA transcripts of both EFNB1 and PAX6 increased in a similar manner in cells of each genotype (Figure 2C). hNE cells of all three genotypes expressed EFNB2, a closely related B-type ephrin, as well as EPHB2 and EPHB3, signaling partners of EPHRIN-B1 that are involved in craniofacial development (Orioli et al., 1996) (Figures S2A–S2C). Wild-type hNE cells expressed EPHRIN-B1 protein, whereas CFNShet and CFNShemi EFNB1 mutant hNE cells did not (Figure 2D).
Figure 2.
Differentiation and Characterization of hNE Cells from hiPSCs
(A) Immunostaining reveals that wt-3, CFNShet-3, and CFNShemi-1 hNE cells express the hNE cell markers PAX6, SOX1, and OTX2. Scale bars, 50 μm.
(B) EFNB1 mRNA expression (normalized to GAPDH mRNA expression) in hiPSCs and wild-type, CFNShet, and CFNShemi hNE cells. Numbers following underscores represent separate hNE differentiations. Error bars represent the SD of n = 3 technical replicate qRT-PCR reactions per hiPSC or hNE line. See also Figure S2.
(C) Relative EFNB1 and PAX6 expression (normalized to GAPDH expression) increase over the course of hNE cell differentiation of wt-3, CFNShet-3, and CFNShemi-1 hiPSCs. Error bars represent the SD of n = 3 technical replicate qRT-PCR reactions per condition. Data shown are one of n = 2 biological replicates of each wt-3 and CFNShet-3 or one of n = 3 biological replicates of CFNShemi-1 (n = 2) and CFNShemi-3 (n = 1).
(D) Immunoblotting reveals expression of EPHRIN-B1 protein in wild-type but not CFNShet or CFNShemi hNE cells or in hiPSCs of any genotype.
EPHRIN-B1 Mosaicism Results in Cell Segregation in CFNS hNE Cells
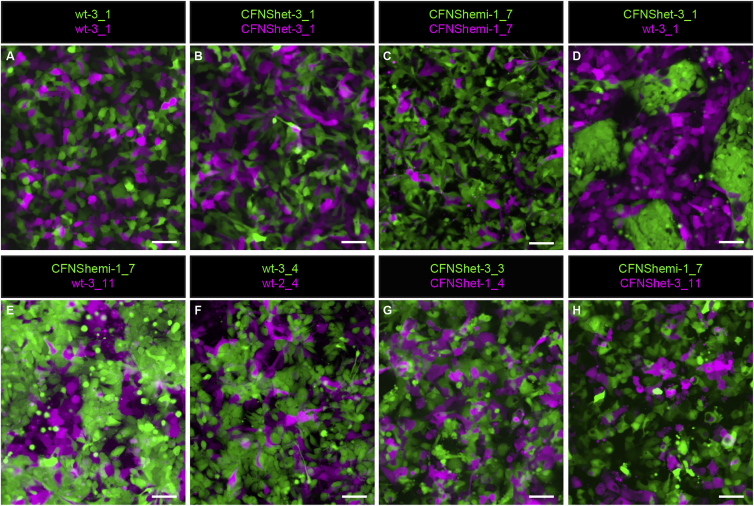
To model EFNB1 mosaicism and determine whether it results in segregation, we generated mixed cultures of fluorescently labeled hNE cells of different genotypes at a 1:1 ratio (Supplemental Experimental Procedures). Control mixed cultures in which all cells expressed EPHRIN-B1 (wt-3 + wt-3, N = 11/11 trials) or in which no cells expressed EPHRIN-B1 (CFNShet-3 + CFNShet-3, N = 7/7 trials; CFNShemi-1 + CFNShemi-1, N = 4/4 trials) resulted in hNE cells commingling freely, with no notable segregation after 48 hr (Figures 3A–3C). However, in hNE cell populations mosaic for EPHRIN-B1 expression, EPHRIN-B1-expressing cells segregated dramatically from EPHRIN-B1 non-expressing cells, forming distinct boundaries between the two different cell types by 48 hr (Figures 3D and 3E) (wt-3 + CFNShet-3, N = 10/10 trials; wt-3 + CFNShemi-1, N = 4/4 trials). To ensure that this segregation depended on EPHRIN-B1 expression and not on differences between independent hNE cell lines, we mixed hNE lines derived from different hiPSC lines of the same genotype and also observed intermixing of cells (wt-3 + wt-2, Figure 3F; CFNShet-3+CFNShet-1, Figure 3G) (N = 3/3 trials, each condition). Further, mixing hNE lines derived from different genotypes lacking EPHRIN-B1 expression (CFNShet-3 + CFNShemi-1, N = 3/3 trials) did not result in cell segregation (Figure 3H), demonstrating that hNE cells segregate based on the presence or absence of EPHRIN-B1 expression and not some other factor associated with individual variability.
Figure 3.
Robust Cell Segregation in Neuroepithelial Cells Mosaic for EPHRIN-B1 Expression
(A–C) Mixing two populations of wild-type EPHRIN-B1 expressing hNE cells (A), two populations of CFNShet hNE cells not expressing EPHRIN-B1 (B), or two populations of CFNShemi hNE cells not expressing EPHRIN-B1 (C) results in cell intermingling over 48 hr.
(D and E) Cell mixing to generate cultures mosaic for EPHRIN-B1 expression (wt-3 + CFNShet-3 (D); wt-3 + CFNShemi-1 (E)) results in robust segregation of EPHRIN-B1 expressing and non-expressing hNE cells over 48 hr. See also Figure S3.
(F–H) Mixing two different wild-type (EPHRIN-B1 expressing) hNE cell lines (wt-3 + wt-2) (F), two different CFNShet (EPHRIN-B1 non-expressing) hNE cell lines (CFNShet-3 + CFNShet-1) (G), or two EPHRIN-B1 non-expressing hNE cell lines of different genotypes (CFNShet-3 + CFNShemi-1) (H) results in cell intermingling without cell segregation. Adjustments to gamma were made to better visualize independent cell populations.
Scale bars, 50 μm.
To observe the process of cell segregation over time, we used live cell imaging to capture the first 25 hr after cell mixing in both mosaic and EPHRIN-B1 non-expressing cell mixtures. Both cell mixtures were intermingled at time of mixing (t = 0, Figures S3A and S3G). Cells in both mixtures continued to interact with each other over time, and EPHRIN-B1 non-expressing mixtures remained freely commingled at each time point (Figures S3B–S3E). However, the EPHRIN-B1 mosaic population of cells segregated progressively over 25 hr (Figures S3H–S3L), indicating that segregation is a continuous process that occurs over time. These data provide evidence that EPHRIN-B1-mediated cell segregation can occur in human CFNS.
Discussion
Here, we have generated an hiPSC model of a human craniofacial condition and have used it to address an outstanding question: does mosaicism for EFNB1 expression result in cell segregation in human CFNS? The c.712delG mutation found in this CFNS family occurred 5′ to the transmembrane domain-encoding region of EFNB1, and we found that EFNB1 mutant hNE cells did not express EPHRIN-B1, indicating that this mutation results in an unstable EPHRIN-B1 protein and most likely null loss of function. To enable us to model CFNS, it was essential that loss of EFNB1 function not prevent CFNS patient-derived HDFs from undergoing reprogramming to hiPSCs. We did not observe differences in reprogramming ability between EFNB1 mutant and control HDFs, leading us to conclude that EPHRIN-B1 expression is not necessary for reprogramming. Further, we found that both control and CFNS hiPSCs possessed differentiation potential to all three germ layers; loss of EPHRIN-B1 expression does not apparently prevent differentiation. This is consistent with our qRT-PCR data demonstrating that transcripts of EFNB1 and several other Eph/ephrin signaling family members are expressed at very low levels in hiPSCs relative to hNE cells, suggesting that these signaling molecules may not play critical roles in hiPSCs.
Previous human genetic studies have indicated that mosaicism for EFNB1 mutation is central to CFNS pathology, a phenomenon termed cellular interference suggested to result in cell segregation based on evidence from model organisms (Compagni et al., 2003, Twigg et al., 2004, Twigg et al., 2006, Twigg et al., 2013, Wieacker and Wieland, 2005, Wieland et al., 2004). Whether cell segregation occurs in CFNS, however, and in what cell types, was not known. Based on evidence of cell segregation in the neural plate NE in Efnb1+/− mice (O’Neill et al., 2016), we differentiated hiPSCs to hNE cells to address this question.
Both wild-type and CFNS patient-derived hNE cells expressed neural stem cell markers and several members of the Eph/ephrin gene family, including EFNB1. Expression of EFNB1 varied between hNE lines, as well as between independent differentiations of the same hiPSC line, indicating that there was inherent variability in the differentiations. Consistently, however, hNE cells expressed higher levels of EFNB1 than hiPSCs, indicating that increased EFNB1 expression is a characteristic of the hNE cell type. In addition, EFNB1 expression decreased as hNE cells were maintained over time, suggesting that higher levels of EFNB1 expression may mark a progenitor stage in the differentiation program.
As hiPSCs are clonally derived cell lines, CFNShet hNE lines are not mosaic for EFNB1 expression, necessitating a different approach to model cellular interference. Upon mixing wild-type and EPHRIN-B1 non-expressing hNE cells to generate EPHRIN-B1 mosaicism, the EPHRIN-B1 expressing and non-expressing cells segregated to form ectopic boundaries in culture. This robust segregation occurred in mosaic mixtures of wild-type + CFNShet cells and wild-type + CFNShemi cells, but not in mixtures of two different populations of EPHRIN-B1 expressing cells, or two different populations of EPHRIN-B1 non-expressing cells, even if these two populations were derived from different hiPSC lines. We therefore conclude that segregation is not an effect of mixing different hNE lines, but rather an effect of mosaicism for EPHRIN-B1 expression, and that cellular interference through EPHRIN-B1-mediated cell segregation occurs in CFNS cells. This finding informs our understanding of the etiology of CFNS and indicates that cell segregation contributes to cellular interference. How cell segregation leads to more severe disease phenotypes is not yet clear; the hiPSC model we have developed is a highly relevant system in which to answer remaining questions about CFNS pathology.
An hiPSC model is an important resource because patient cells can be differentiated into multiple disease-relevant cell types, overcoming the challenge of isolating primary cells from patients. It is notable that cell segregation occurs in the neuroepithelium in models of CFNS; recent studies have indicated that Treacher Collins syndrome, another neurocristopathy, also exhibits cellular defects originating in the neuroepithelium and ongoing in the NCC (Jones et al., 2008). Our hiPSC model of CFNS will facilitate further studies of cell segregation, such as investigating whether it occurs in hNCCs and their descendants. This will contribute to a better understanding of the developmental timing of CFNS etiology and will provide the ability to study aspects of the disease that are less well understood. For example, mutations in EFNB1 that cause CFNS are responsible for approximately 7% of cases of craniosynostosis in which a genetic cause is known (Johnson and Wilkie, 2011), and studies in mice have demonstrated that suture boundary formation is regulated by A-type Eph/ephrin signaling (Merrill et al., 2006, Ting et al., 2009). However, how EPHRIN-B1 affects boundary formation and maintenance at the suture is unknown, because EFNB1 mutant mice do not exhibit craniosynostosis. Further, in CFNS, other organ systems not derived from NCCs are also affected; patients exhibit limb anomalies and defects of the axial skeleton that may be attributable to cell segregation (Compagni et al., 2003, Davy et al., 2004, Davy et al., 2006). Differentiation of hiPSCs into these various cell types is a method for testing the importance of EPH/EPHRIN-mediated cell segregation in various tissues.
hiPSC models of congenital craniofacial disease will also facilitate targeted molecular therapies for these disorders. As we have shown that cell segregation resulting from EPHRIN-B1 mosaicism occurs in human CFNS cells, therapeutic benefits may be derived from preventing segregation. Additional research to determine the mechanism by which hNE cell segregation leads to craniofacial phenotypes in CFNS patients is likely to identify molecular candidates that could be targeted to achieve this goal. A human model system of EPHRIN-B1-mediated cell segregation could then serve as a high-throughput system for testing candidate therapeutic molecules. Finally, this CFNS hiPSC model system may encourage the use of hiPSC-based systems to model structural aspects of other congenital craniofacial anomalies in which self-organization of cells may play a role. Such studies have the potential to inform therapeutic approaches for congenital craniofacial anomalies, as well as to increase our understanding of cell self-organizing properties to facilitate tissue engineering and cell replacement therapies for patients with these disorders.
Experimental Procedures
hiPSC Generation, Culture, and Characterization
All human tissue collection, stem cell studies, procedures, and written consents were approved by the University of California, San Francisco (UCSF) Committee on Human Research and the UCSF Gamete, Embryo, and Stem Cell Research Committee. Prior to their inclusion in this study, written informed consent was obtained from all participants or from their parents. CFNS and control hiPSCs were established from primary human fibroblast cultures from each subject (Byrne et al., 2009) using episomal reprogramming (Bershteyn et al., 2014, Okita et al., 2011). hiPSC lines were genetically characterized using G-banded karyotype analysis (WiCell Research Institute) and sequencing of EFNB1 (SeqWright). hiPSCs were also assayed for episomal plasmid integration and for relative inactivation of each X chromosome with the HUMAR assay (Kiedrowski et al., 2011). See also the Supplemental Experimental Procedures.
hNE Cell Differentiation
Neural inductions were performed using a modified monolayer dual-SMAD inhibition protocol (Chambers et al., 2009) with STEMdiff Neural Induction Medium (STEMCELL Technologies) containing 10 μM SB-431542 (Santa Cruz Biotechnology) and 5 μM DMH1 (Sigma). See also the Supplemental Experimental Procedures.
Immunocytochemistry and Immunoblotting
For immunocytochemistry, cells were plated on Matrigel-coated glass coverslips. For immunoblotting, cells were lysed in NP-40 lysis buffer containing protease and phosphatase inhibitors. Both procedures were performed using standard protocols. See also the Supplemental Experimental Procedures.
qRT-PCR of hiPSCs and Neuroepithelial Cells
Total RNA was extracted from cells using TRIzol (Life Technologies). RNA was reverse transcribed using a SuperScript II First-Strand Synthesis System for RT-PCR (Life Technologies). qRT-PCR was performed using iTaq Universal SYBR Green and a CFX96 Real Time System (Bio-Rad), with primer pairs that span exon-intron boundaries (see Supplemental Experimental Procedures for primer sequences).
Statistics
For analysis of qRT-PCR data, GraphPad Prism 6 was used to plot mean expression ± SD of technical replicate reactions, indicated by error bars. Biological replicates (different hiPSC lines or different hNE differentiations), if applicable, are shown as separate bars on the graph.
Author Contributions
T.K.N., A.R.L., and J.O.B. designed experiments and analyzed data. O.D.K. and J.H.P. obtained patient material for generation of hiPSCs. A.R.L. and J.O.B. made and characterized hiPSC lines with technical expertise from E.C.H. T.K.N. differentiated, characterized, and performed experiments on hiPSC-derived neuroepithelial cells with technical expertise from M.B. and A.K.O. T.K.N. and J.O.B. wrote the manuscript.
Acknowledgments
This study was supported by a Graduate Research Fellowship (2013157314) to T.K.N. from the National Science Foundation, and by grant F31DE026059 to T.K.N., and grant R01DE02337 to J.O.B., both from the National Institute of Dental and Craniofacial Research. A.R.L. was a Howard Hughes Medical Institute Medical Research Fellow and completed this work as partial fulfillment of the requirements for an M.D. with Distinction in the UCSF Molecular Medicine Pathway to Discovery. Any opinions, findings, or conclusions in this work are those of the authors and do not necessarily reflect the views of the Howard Hughes Medical Institute, National Science Foundation, or NIH.
Published: February 23, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.01.017.
Supplemental Information
References
- Barrios A., Poole R.J., Durbin L., Brennan C., Holder N., Wilson S.W. Eph/Ephrin signaling regulates the mesenchymal-to-epithelial transition of the paraxial mesoderm during somite morphogenesis. Curr. Biol. 2003;13:1571–1582. doi: 10.1016/j.cub.2003.08.030. [DOI] [PubMed] [Google Scholar]
- Batlle E., Wilkinson D.G. Molecular mechanisms of cell segregation and boundary formation in development and tumorigenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a008227. doi: 10.1101/cshperspect.a008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershteyn M., Hayashi Y., Desachy G., Hsiao E.C., Sami S., Tsang K.M., Weiss L.A., Kriegstein A.R., Yamanaka S., Wynshaw-Boris A. Cell-autonomous correction of ring chromosomes in human induced pluripotent stem cells. Nature. 2014;507:99–103. doi: 10.1038/nature12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush J.O., Soriano P. Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes Dev. 2010;24:2068–2080. doi: 10.1101/gad.1963210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne J.A., Nguyen H.N., Reijo Pera R.A. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PLoS One. 2009;4:e7118. doi: 10.1371/journal.pone.0007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F., Ivanovitch K., Wilson S.W. Eph/Ephrin signalling maintains eye field segregation from adjacent neural plate territories during forebrain morphogenesis. Dev. Camb. Engl. 2013;140:4193–4202. doi: 10.1242/dev.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuso J., Xu Q., Wilkinson D.G. Mechanisms of boundary formation by Eph receptor and ephrin signaling. Dev. Biol. 2015;401:122–131. doi: 10.1016/j.ydbio.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagni A., Logan M., Klein R., Adams R.H. Control of skeletal patterning by ephrinB1-EphB interactions. Dev. Cell. 2003;5:217–230. doi: 10.1016/s1534-5807(03)00198-9. [DOI] [PubMed] [Google Scholar]
- Davy A., Aubin J., Soriano P. Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 2004;18:572–583. doi: 10.1101/gad.1171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A., Bush J.O., Soriano P. Inhibition of gap junction communication at ectopic Eph/ephrin boundaries underlies craniofrontonasal syndrome. PLoS Biol. 2006;4:e315. doi: 10.1371/journal.pbio.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin L., Brennan C., Shiomi K., Cooke J., Barrios A., Shanmugalingam S., Guthrie B., Lindberg R., Holder N. Eph signaling is required for segmentation and differentiation of the somites. Genes Dev. 1998;12:3096–3109. doi: 10.1101/gad.12.19.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F. The cellular basis of tissue separation. Development. 2014;141:3303–3318. doi: 10.1242/dev.090332. [DOI] [PubMed] [Google Scholar]
- Fagotto F., Winklbauer R., Rohani N. Ephrin-Eph signaling in embryonic tissue separation. Cell Adh. Migr. 2014;8:308–326. doi: 10.4161/19336918.2014.970028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global strategies to reduce the health care burden of craniofacial anomalies: report of WHO meetings on international collaborative research on craniofacial anomalies. Cleft Palate Craniofac. J. 2004;41:238–243. doi: 10.1597/03-214.1. [DOI] [PubMed] [Google Scholar]
- Hogue J., Shankar S., Perry H., Patel R., Vargervik K., Slavotinek A. A novel EFNB1 mutation (c.712delG) in a family with craniofrontonasal syndrome and diaphragmatic hernia. Am. J. Med. Genet. A. 2010;152A:2574–2577. doi: 10.1002/ajmg.a.33596. [DOI] [PubMed] [Google Scholar]
- Holmberg J., Genander M., Halford M.M., Annerén C., Sondell M., Chumley M.J., Silvany R.E., Henkemeyer M., Frisén J. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- Johnson D., Wilkie A.O.M. Craniosynostosis. Eur. J. Hum. Genet. 2011;19:369–376. doi: 10.1038/ejhg.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N.C., Lynn M.L., Gaudenz K., Sakai D., Aoto K., Rey J.-P., Glynn E.F., Ellington L., Du C., Dixon J. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen C., Sherman A., Chen G.I., Pasculescu A., Poliakov A., Hsiung M., Larsen B., Wilkinson D.G., Linding R., Pawson T. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science. 2009;326:1502–1509. doi: 10.1126/science.1176615. [DOI] [PubMed] [Google Scholar]
- Kiedrowski L.A., Raca G., Laffin J.J., Nisler B.S., Leonhard K., McIntire E., Mongomery K.D. DNA methylation assay for X-chromosome inactivation in female human iPS Cells. Stem Cell Rev. Rep. 2011;7:969–975. doi: 10.1007/s12015-011-9238-6. [DOI] [PubMed] [Google Scholar]
- Lessing D., Anguera M.C., Lee J.T. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu. Rev. Genomics Hum. Genet. 2013;14:85–110. doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- Mellitzer G., Xu Q., Wilkinson D.G. Eph receptors and ephrins restrict cell intermingling and communication. Nature. 1999;400:77–81. doi: 10.1038/21907. [DOI] [PubMed] [Google Scholar]
- Merrill A.E., Bochukova E.G., Brugger S.M., Ishii M., Pilz D.T., Wall S.A., Lyons K.M., Wilkie A.O.M., Maxson R.E. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum. Mol. Genet. 2006;15:1319–1328. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- O’Neill A.K., Kindberg A.A., Niethamer T.K., Larson A.R., Ho H.H., Greenberg M.E., Bush J.O. Unidirectional Eph/ephrin signaling creates a cortical actin differential to drive cell segregation. J. Cell Biol. 2016;215:217–229. doi: 10.1083/jcb.201604097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli D., Henkemeyer M., Lemke G., Klein R., Pawson T. Sek4 and Nuk receptors cooperate in guidance of commissural axons and in palate formation. EMBO J. 1996;15:6035–6049. [PMC free article] [PubMed] [Google Scholar]
- Poliakov A., Cotrina M.L., Pasini A., Wilkinson D.G. Regulation of EphB2 activation and cell repulsion by feedback control of the MAPK pathway. J. Cell Biol. 2008;183:933–947. doi: 10.1083/jcb.200807151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tchieu J., Kuoy E., Chin M.H., Trinh H., Patterson M., Sherman S.P., Aimiuwu O., Lindgren A., Hakimian S., Zack J.A. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting M.-C., Wu N.L., Roybal P.G., Sun J., Liu L., Yen Y., Maxson R.E. EphA4 as an effector of Twist1 in the guidance of osteogenic precursor cells during calvarial bone growth and in craniosynostosis. Development. 2009;136:855–864. doi: 10.1242/dev.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G., Vivas E.L., Izpisúa Belmonte J.C. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat. Med. 2011;17:1570–1576. doi: 10.1038/nm.2504. [DOI] [PubMed] [Google Scholar]
- Tomoda K., Takahashi K., Leung K., Okada A., Narita M., Yamada N.A., Eilertson K.E., Tsang P., Baba S., White M.P. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg S.R., Kan R., Babbs C., Bochukova E.G., Robertson S.P., Wall S.A., Morriss-Kay G.M., Wilkie A.O. Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc. Natl. Acad. Sci. USA. 2004;101:8652–8657. doi: 10.1073/pnas.0402819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg S.R.F., Matsumoto K., Kidd A.M.J., Goriely A., Taylor I.B., Fisher R.B., Hoogeboom A.J.M., Mathijssen I.M.J., Lourenço M.T., Morton J.E.V. The origin of EFNB1 mutations in craniofrontonasal syndrome: frequent somatic mosaicism and explanation of the paucity of carrier males. Am. J. Hum. Genet. 2006;78:999–1010. doi: 10.1086/504440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg S.R.F., Babbs C., van den Elzen M.E.P., Goriely A., Taylor S., McGowan S.J., Giannoulatou E., Lonie L., Ragoussis J., Akha E.S. Cellular interference in craniofrontonasal syndrome: males mosaic for mutations in the X-linked EFNB1 gene are more severely affected than true hemizygotes. Hum. Mol. Genet. 2013;22:1654–1662. doi: 10.1093/hmg/ddt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umetsu D., Dunst S., Dahmann C. An RNA interference screen for genes required to shape the anteroposterior compartment boundary in Drosophila identifies the Eph receptor. PLoS One. 2014;9:e114340. doi: 10.1371/journal.pone.0114340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieacker P., Wieland I. Clinical and genetic aspects of craniofrontonasal syndrome: towards resolving a genetic paradox. Mol. Genet. Metab. 2005;86:110–116. doi: 10.1016/j.ymgme.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Wieland I., Jakubiczka S., Muschke P., Cohen M., Thiele H., Gerlach K.L., Adams R.H., Wieacker P. Mutations of the Ephrin-B1 gene cause craniofrontonasal syndrome. Am. J. Hum. Genet. 2004;74:1209–1215. doi: 10.1086/421532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland I., Makarov R., Reardon W., Tinschert S., Goldenberg A., Thierry P., Wieacker P. Dissecting the molecular mechanisms in craniofrontonasal syndrome: differential mRNA expression of mutant EFNB1 and the cellular mosaic. Eur. J. Hum. Genet. 2007;16:184–191. doi: 10.1038/sj.ejhg.5201968. [DOI] [PubMed] [Google Scholar]
- Wutz A. Epigenetic alterations in human pluripotent stem cells: a tale of two cultures. Cell Stem Cell. 2012;11:9–15. doi: 10.1016/j.stem.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Xu Q., Mellitzer G., Robinson V., Wilkinson D.G. In vivo cell sorting in complementary segmental domains mediated by Eph receptors and ephrins. Nature. 1999;399:267–271. doi: 10.1038/20452. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.