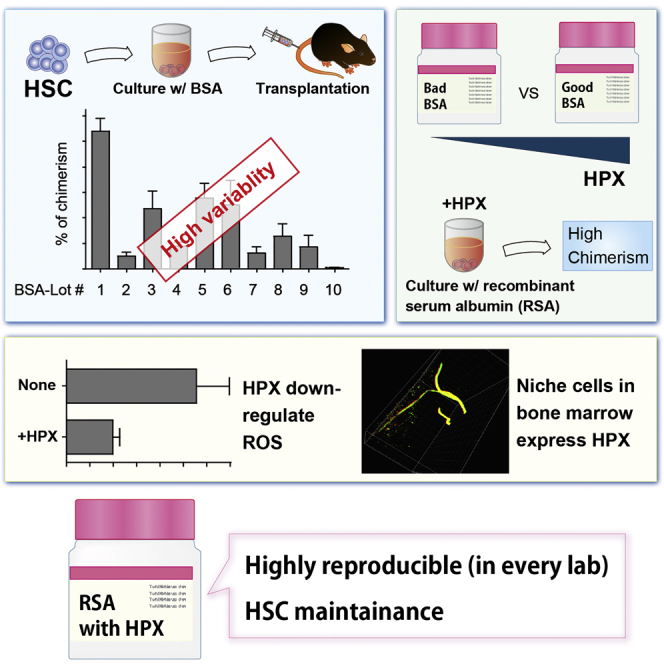
Summary
Hematopoietic stem cells (HSCs) are considered one of the most promising therapeutic targets for the treatment of various blood disorders. However, due to difficulties in establishing stable maintenance and expansion of HSCs in vitro, their insufficient supply is a major constraint to transplantation studies. To solve these problems we have developed a fully defined, all-recombinant protein-based culture system. Through this system, we have identified hemopexin (HPX) and interleukin-1α as responsible for HSC maintenance in vitro. Subsequent molecular analysis revealed that HPX reduces intracellular reactive oxygen species levels within cultured HSCs. Furthermore, bone marrow immunostaining and 3D immunohistochemistry revealed that HPX is expressed in non-myelinating Schwann cells, known HSC niche constituents. These results highlight the utility of this fully defined all-recombinant protein-based culture system for reproducible in vitro HSC culture and its potential to contribute to the identification of factors responsible for in vitro maintenance, expansion, and differentiation of stem cell populations.
Keywords: hematopoietic stem cell, BSA, FCS, all-recombinant protein-based culture system, hemopexin
Graphical Abstract
Highlights
-
•
Different BSA lots alter how HSCs respond to cytokines
-
•
RSA can replace BSA to provide HSC maintenance culture with minimal variability
-
•
By comparing the protein profiles of “good” and “bad” BSAs, HPX was identified
-
•
HPX reduces HSC intracellular reactive ROS and is expressed by BM Schwann cells
In this article, Yamazaki, Nakauchi, and colleagues demonstrate that BSA batches have unique protein profiles and vary widely in their ability to maintain mouse HSCs ex vivo. By replacing BSA with recombinant serum albumin, they developed a standardized HSC culture platform that can be used to identify novel maintenance and expansion factors.
Introduction
Hematopoietic stem cells (HSCs) maintain the ability to self-renew and differentiate within their in vivo microenvironment, the bone marrow (BM). From a clinical perspective, HSCs are important because they can generate the full blood cell repertoire upon transplantation (Eaves, 2015) and are therefore critical determinants of clinical BM transplant success. Additionally, in combination with gene therapy approaches HSCs also offer the significant potential to treat a range of inherited hematological disorders. However, our ability to maintain and expand HSCs outside of their in vivo microenvironment is currently limited.
The current protocols for ex vivo expansion of HSCs can be broadly divided into two groups, based on their use of cell-intrinsic or cell-extrinsic factors (Walasek et al., 2012). Cell-intrinsic factors include exogenous expression transcription factors such as HoxB4 (Sauvageau et al., 1995), and chromatin remodeling factors such as Bmi1 (Iwama et al., 2004). Such approaches have to date required genetic modification that limits their direct translational application. By contrast, cell-extrinsic factors such as cytokines are simply added to the culture media and act on unmodified HSCs.
Cytokines and other extrinsic factors are present in the specialized BM microenvironments, the so-called BM niche, and are thought to be involved in migration, quiescence, and differentiation of HSCs (Kiel and Morrison, 2008). Many different cell types have been proposed as the candidate for the BM niche, including osteoblasts (Calvi et al., 2003, Zhang et al., 2003), endothelial cells (Kiel et al., 2005), chemokine ligand 12 (CXCL12)-abundant reticular cells (Sugiyama et al., 2006), mesenchymal stem cells (Mendez-Ferrer et al., 2010), and non-myelinating Schwann glial cells (Yamazaki et al., 2006, Yamazaki et al., 2011).
BM niche cells are thought to secrete numerous factors such as stem cell factor (SCF) (Barker, 1994) and thrombopoietin (TPO) (Ku et al., 1996), which are generally necessary for HSC maintenance. These cytokines have long been added to culture media to investigate HSC proliferation and reconstitution ability. However, there are concerns about data reproducibility between laboratories, with such discrepancies often being ascribed to differences in experimental culture conditions.
HSCs have been widely analyzed using liquid or methylcellulose culture in the presence of fetal bovine serum (FBS). FBS contains myriad of growth factors, adhesion molecules, and other components, and also protects cells from rapid changes in pH. However, because of the high degree of unknown factors, FBS is now often replaced with serum-free medium containing BSA fraction V (BSA-FV; the fifth ethanol fraction in the original purification process of plasma proteins) (Guilbert and Iscove, 1976) for in vitro HSC culture. BSA-based serum-free cultures have been well established for pluripotent stem cells. However, stable in vitro expansion of HSCs remains difficult and non-reproducible. This is at least in part due to the use of different batches (lots) of BSA-FV by different laboratories.
To address these issues, we tested 15 different lots of commercially available BSA-FV; each exhibited different abilities to maintain HSCs and unique protein profiles. To identify the best molecular candidates for HSC maintenance in BSA-FV, we developed a fully defined culture system using all-recombinant proteins. Using this approach, we provide evidence that HSC maintenance is strongly supported by two factors in BSA-FV, interleukin-1α (IL-1α) and hemopexin (HPX). Further investigation found that HPX reduced HSC intracellular reactive oxygen species (ROS) levels and that HPX was present on non-myelinating Schwann cells, a constituent of HSC niche in BM. These findings highlight the utility of all-recombinant protein-based systematic analysis for ex vivo HSC self-renewal and differentiation, and identification of bona fide growth factors that contribute to these cell-fate decisions.
Results
HSC Maintenance Depends on Varying Levels of Unidentified Proteins Contained in BSA-FV
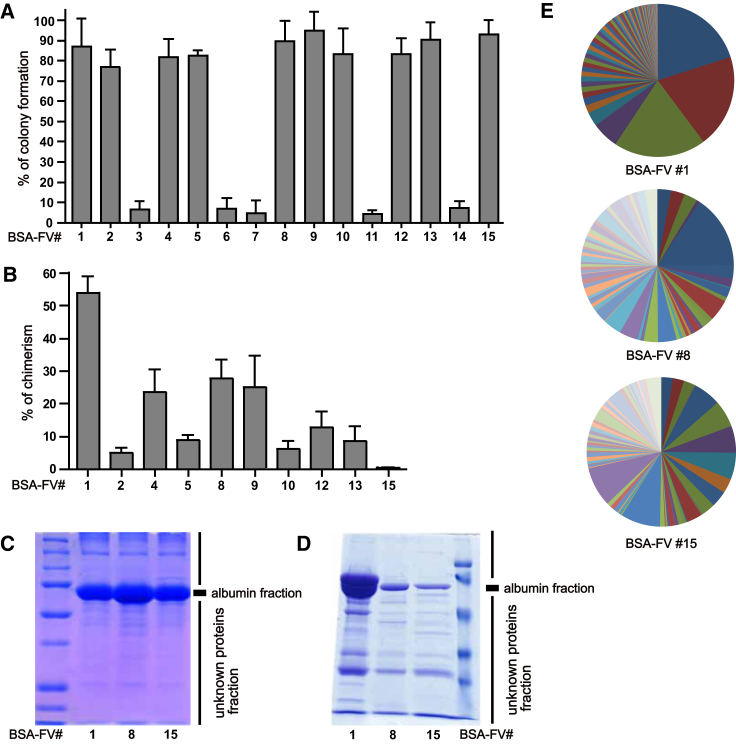
During our attempts at in vitro maintenance of HSCs, we realized that different BSA-FV lots varied tremendously in their colony-forming ability. We therefore evaluated 15 different lots of BSA-FV based on the ability to support colony formation from single CD34- KSL HSCs (Osawa et al., 1996). After 11 days in culture with each BSA-FV lot, we confirmed that 5 out of 15 lots showed very poor colony-formation ability (Figure 1A). We next cultured HSCs with the remaining ten lots of BSA-FV for 1 week and examined their blood system repopulating potential using competitive repopulation assays. To our surprise, different lots of BSA-FV varied in the ability to accelerate HSC-mediated blood system reconstitution (Figure 1B), and did not correlate with colony formation. Culture in BSA-FV #1 resulted in peripheral blood (PB) chimerism (at 12 weeks after the transplantation) of more than 50%, while BSA-FV #15 cultured cells were essentially undetectable. We hypothesized that these results were due to different compositions of unknown factors in each lot of BSA-FV.
Figure 1.
Heterogeneity in BSA-FV Lots Affects HSC Maintenance
(A) Percentage of single CD34− HSCs forming colonies after in vitro culture for 11 days with cytokines and various BSA-FV lots. Mean ± SEM from three independent experiments (n = 30 per BSA-FV culture condition).
(B) Percentage peripheral blood (PB) chimerism from 40 CD34−KSL cells, cultured for 1 week with cytokines and various BSA-FV lots and transplanted into lethally irradiated recipients together with 106 bone marrow (BM) competitor cells. PB chimerism 12 weeks after transplantation. Mean ± SD of three independent experiments (n = 10 mice per BSA-FV culture condition).
(C and D) Protein analysis of total BSA-FV (C) and albumin-depleted BSA-FV (D) by SDS-PAGE.
(E) The ratio of proteins identified by HPLC-MS analyses in different lots of BSA-FV (BSA-FV #1, #8, #15), excluding albumin.
To test this hypothesis, we attempted to detect the presence of unknown factors in BSA-FV using SDS-PAGE analysis. In addition to the major albumin protein, a number of unidentified bands were detected in BSA-FV #1, #8, and #15 (Figures 1C and 1D). Interestingly, in agreement with lot-to-lot variability in band patterns, further high-performance liquid chromatography-mass spectrometry (HPLC-MS) analysis revealed that different lots of BSA-FV exhibit distinct protein profiles (Figure 1E and Table S1). These results suggest that differences in HSC maintenance can be ascribed to varying levels of unidentified proteins contained in BSA-FV.
HSC Maintenance Varies Depending on Different Lots of BSA-FV
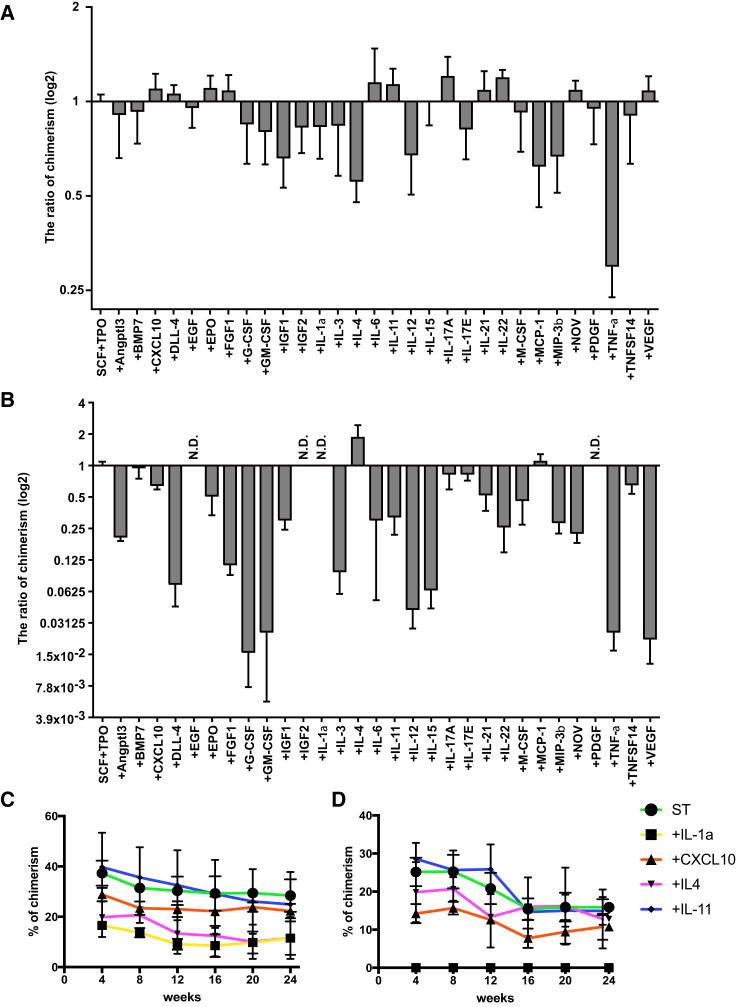
To assess the ability of BSA-FV to augment cytokine-mediated HSC maintenance, we cultured 40 CD34−KSL cells in the presence of 1% BSA-FV, SCF, TPO, and 30 different individual ligands. These 30 ligands were selected based on the expression of their receptors on CD34− HSCs, identified from a previously published DNA microarray expression dataset. After 1 week, cultured cells were then transplanted into lethally irradiated recipient mice along with competitor cells and the ratio of PB chimerism determined 12 weeks after transplantation. To our surprise, the effect of each cytokine on the maintenance of HSCs changed dramatically when different BSA-FV lots were used. For instance, although IL-11 was reported to promote the growth of primitive hematopoietic progenitors in cooperation with IL-4 (Jacobsen et al., 1995), the ratio of chimerism induced by IL-11 was higher than that of control when BSA-FV #1 was used (Figure 2A), but the opposite result was obtained for BSA-FV #8 (Figure 2B). In addition, PB chimerism was detected in the presence of epidermal growth factor, insulin-like growth factor 2, or platelet-derived growth factor (PDGF) when BSA-FV #1 was used, whereas we could not observe any PB chimerism under the same conditions when BSA-FV #8 was used. From the viewpoint of reproducibility, these results suggest that simple addition of BSA-FV to cell culture should be reconsidered for efficient in vitro maintenance of HSCs. To further confirm these results, we repeated the same experiment focusing on IL-1α, CXCL10, IL-4, and IL-11. The results clearly demonstrated that, as was the case for IL-1α, the effect of the cytokine on the ratio of PB chimerism depended on the BSA-FV lot (Figures 2C and 2D).
Figure 2.
The Ratio of Peripheral Blood Chimerism Depends on BSA-FV Lot
(A and B) Percentage PB chimerism from 40 CD34−KSL HSCs cultured for 1 week in the presence of 30 different cytokines, SCF, TPO, and BSA-FV #1 (A) or #8 (B), and transplanted into lethally irradiated recipient mice together with 106 BM competitor cells. PB chimerism at 12 weeks after transplantation (n = 5 mice per BSA-FV culture condition). Mean ± SD from two independent experiments, calculated as a ratio relative to SCF + TPO only conditions.
(C and D) As above, but displayed as percentage PB chimerism for selected cytokines (IL-1α, CXCL10, IL-4, IL-11) cultured with 1% BSA-FV #1 (C) or #8 (D) between 4 and 24 weeks post transplantation.
Recombinant Protein-Based Culture Is a Promising Standard Culture System for HSC Maintenance and Expansion
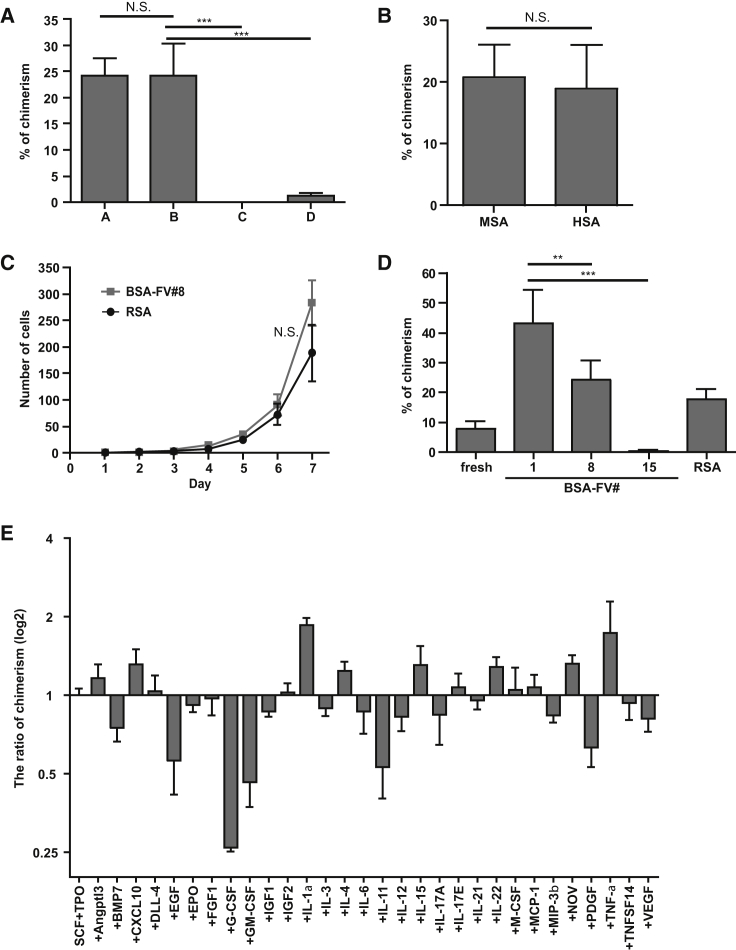
To improve reproducibility in HSC proliferation, we established a standard BSA-FV free HSC culture system. After trial and error, we focused on a purified recombinant serum albumin (RSA). We first analyzed the effect of four different RSAs on HSC maintenance and the ratio of chimerism (Figure 3A). Although RSA produced by rice (#C and #D) did not show any effect, RSA produced by yeast (#A and #B) exhibited repopulation ability and multi-lineage output (Figure S1A), regardless of lot or manufacturer. Furthermore, human RSA (HSA) and mouse RSA (MSA) produced by yeast equally maintained HSC reconstitution ability and multi-lineage output (Figures 3B and S1B). These results indicate that culture conditions containing RSA could be a promising candidate for recombinant protein-based culture systems.
Figure 3.
Recombinant Serum Albumin Can Replace BSA-FV for Ex Vivo Culture of HSCs
(A) Percentage PB chimerism from 40 CD34−KSL cells, following 1 week of culture with cytokines and 1% RSA produced by either yeast (A and B) and rice (C and D). PB chimerism 12 weeks after transplantation into lethally irradiated recipients together with 106 BM competitor cells (n = 8 mice per RSA culture condition). Mean ± SD from two independent experiments.
(B) As in (A) but cultured with 1% MSA or HSA. Mean ± SD from three independent experiments (n = 8 mice per RSA culture condition).
(C) Single CD34−KSL HSCs were cultured for 1 week in 96-well microtiter plates in S-Clone SF-03 supplemented with SCF, TPO, and 1% BSA-FV #8 (gray line) or RSA (black line). Cell numbers were counted every 24 hr under a microscope. Mean ± SEM (n = 40 per RSA culture condition).
(D) As in (A) but for either freshly isolated HSCs cultured for 7 days with 1% BSA-FV #1, #8, #15, or RSA. Mean ± SD from two independent experiments (n = 10 mice per RSA culture condition).
(E) Percentage PB chimerism of 40 CD34−KSL HSCs (cultured for 1 week in the presence of different cytokines, SCF, TPO, and RSA) 12 weeks after transplantation into lethally irradiated recipient mice together with 106 BM competitor cells. Mean ± SD from two independent experiments (n = 5 mice per cytokine culture condition), calculated as a ratio relative to SCF + TPO only conditions.
Statistical significance denoted by ∗∗p < 0.05, ∗∗∗p < 0.005, or N.S. (not significant) as determined by unpaired t test.
To assess the ability of HSC to proliferate in RSA cultures, we compared single HSC growth in RSA- and BSA-FV #8-based cultures. RSA-cultured HSCs exhibited growth kinetics equivalent to those cultured in BSA-FV #8 (Figure 3C). HSCs were also cultured with RSA for 1 week, and tested for their potential to reconstitute the blood system. We found that the ratio of PB chimerism after the transplantation was lower than that of BSA-FV #1, but similar to that of BSA-FV #8 (Figure 3D). RSA-, as well as BSA-FV #1- and #8-cultured HSCs, all displayed increased PB chimerism following transplantation over freshly isolated CD34−KSL HSCs (Figure 3D). A 7-day culture with RSA increased PB chimerism by approximately 2-fold.
Using the above recombinant protein-based culture system, we reassessed the effect of the 30 ligands to maintain HSCs ex vivo. These experiments identified several factors that increased the ratio of PB chimerism at 12 weeks after transplantation (Figure 3E). These results were distinct from the BSA-FV-based cultures, highlighting the BSA-mediated augmentation of ligand effects. Among these 30 ligands we focused on IL-1α, which exhibited highly elevated reconstitution ability when added to the RSA-based culture. IL-1α has been previously reported to suppress colony formation of mouse KSL cells (Yonemura et al., 1996). However, competitive repopulation assays after in vitro culture showed a significantly higher ratio of PB chimerism compared with controls (Figures S2A and S2B). In addition, 16 weeks after transplantation we performed secondary transplantation and found that IL-1α exhibits much higher levels of chimerism than controls (Figure S2C). As IL-1α did not alter the proliferation kinetics of HSCs in vitro (Figure S2D), these findings strongly suggest that IL-1α is a bone fide HSC maintenance factor.
Hemopexin Contained in BSA-FV Is Responsible for HSC Maintenance
HSC maintenance factors may not necessarily be ligands of receptors expressed on HSCs. As shown in Figure 1E, HPLC-MS analysis of BSA-FV #1, #8, and #15 revealed distinct protein profiles. Integrating these protein profiles with the HSC repopulation assay data allowed us to shortlist putative HSC maintenance factors for investigation with a recombinant protein-based culture system. To determine the functional significance of these factors, we prepared recombinant proteins and assessed their effect on HSC repopulating ability by competitive repopulation assays following 1 week of culture with or without each recombinant protein.
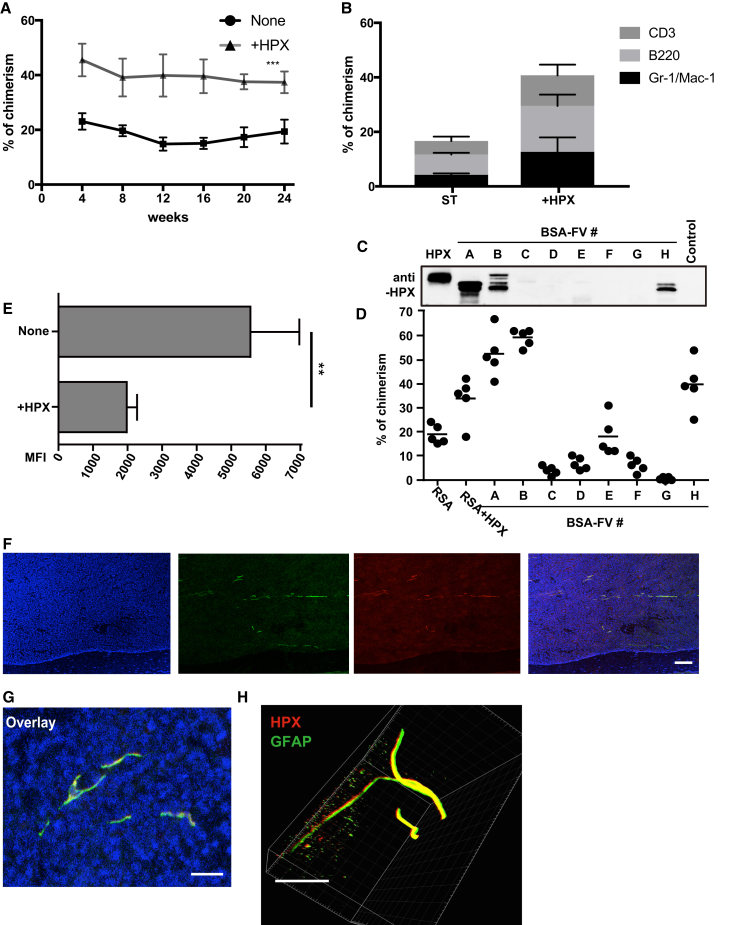
One factor tested by this system, HPX, significantly increased levels of PB chimerism when added to the in vitro HSC culture in both primary and secondary transplantation recipients (Figures 4A, 4B, and S3A). Further studies demonstrated that as little as 5 ng/mL HPX could induce this increase in HSC repopulating ability (Figure S3B). To assess whether HPX is present in other lots of BSA-FV, we analyzed HPX levels in different lots of BSA-FV by western blotting, and found that three out of eight BSA-FV lots (#A, #B, and #H) contained a significant amount of HPX (Figure 4C). Consistent with this observation, competitive repopulation assays clearly demonstrated that HSCs cultured with HPX-positive BSA-FV exhibited higher chimerism than HPX-negative controls (Figure 4D).
Figure 4.
The All-Recombinant Protein-Based Culture System Contributes to Identification of HSC Maintenance Factors Contained in BSA-FV
(A and B) The percentage PB chimerism from 40 CD34−KSL HSCs following culture for 1 week in the presence SCF, TPO, RSA, and with or without HPX (10 ng/mL) before transplantation into lethally irradiated recipient mice together with 106 BM competitor cells. PB chimerism measured over 24 weeks post transplantation (n = 4 or 5 mice per culture condition) and displayed as total PB chimerism (A) and blood-lineage chimerism (B) within the myeloid (Gr-1/Mac-1+), B cell (B220+), and T cell (CD3+) lineages at 12 weeks. Mean ± SD from two independent experiments.
(C) Immunoblotting for HPX of albumin-depleted BSA-FV.
(D) Lethally irradiated recipient mice were transplanted with 106 BM competitor cells and 40 CD34−KSL HSCs cultured for 1 week with 1% BSA-FV or 1% RSA with or without HPX (all in the presence of SCF and TPO). Percentage PB chimerism at 12 weeks post transplantation (n = 5 mice per BSA-FV culture condition). Mean ± SD of three independent experiments.
(E) Effect of HPX on ROS levels of in vitro cultured HSCs, as measured by flow cytometric analysis of HySOx staining. Mean fluorescence intensity (MFI) values ± SD (n = 5 per culture condition).
(F and G) Fluorescence imaging of BM sections co-stained with anti-HPX (red), anti-GFAP (green), and DAPI (blue) antibodies. Scale bars: (F) 100 μm; (G) 4 μm.
(H) 3D fluorescence image of a representative tibia BM, stained with anti-HPX (red) and anti-GFAP (green). Scale bar, 150 μm.
Statistical significance denoted by ∗∗p < 0.05 and ∗∗∗p < 0.005 as determined by unpaired t test.
HPX is a heme-binding plasma glycoprotein (Muller-Eberhard, 1988) that prevents heme-mediated oxidative stress (Tolosano and Altruda, 2002). Given the described negative effects of ROS on HSC function, we hypothesized that HPX enhances HSC function through inhibiting oxidative stress. We therefore evaluated the in vitro effect of HPX on ROS production in HSCs (Figure S3C). Consistent with our hypothesis, we observed a significant reduction of ROS activity in HSCs cultured with HPX, compared with control cultured HSCs (Figure 4E). HPX did not alter HSC proliferation kinetics in vitro (Figure S3D), highlighting that HPX enhances HSC maintenance rather than cell proliferation.
HPX expression has previously been reported in liver, CNS, retina, and peripheral nerves (Chen et al., 1998, Swerts et al., 1992, Tolosano et al., 1996). Given that peripheral nerves include non-myelinating Schwann cells, which are also a constituent of HSC niche (Yamazaki et al., 2011), we were interested in whether this BM cell type also expressed HPX. We therefore investigated HPX expression by immunostaining BM sections along with glial fibrillary acidic protein (GFAP) expression, and confirmed overlapping expression of HPX and GFAP in all sections (Figures 4F and 4G). Furthermore, using a recently developed whole-body imaging by tissue decolorization technique, we performed 3D immunohistochemistry of whole BM and further confirmed co-localization of HPX and GFAP (Figure 4H and Movie S1). While the majority of HPX co-localized with GFAP, HPX staining was also seen elsewhere within the BM, suggesting that other BM niche cells express HPX. These results raise the intriguing possibility that HPX protects HSCs from oxidative stress in specialized BM niches, although further work is necessary to define the role of HPX in vivo.
Finally, we attempted to quantitate the effects of HPX and IL-1α on in vitro HSC maintenance and expansion using limiting dilution analysis by transplanting 1,000, 100, and 10 cells (following a 7-day culture) alongside 2 × 105 BM competitors. Surprisingly, all mice displayed significant donor cell chimerism (>90% in some mice) within the PB after 4 weeks (Figure S4). While these results precluded quantitation of HSC frequency, they do highlight the ability of RSA-based cultures to expand blood system reconstituting cells in vitro. After all, single HSCs on average give rise to approximately 200 progenies during the 7-day RSA culture (Figure 3C), yet substantial PB chimerism was detected from aliquots of 100 and 10 HSC progenies (1/2 and 1/20 HSC equivalents, respectively).
Discussion
Reproducibility is of central importance in experimental science. Unfortunately, in the case of in vitro maintenance of HSCs, reproducible results are often not obtained. Several factors are likely responsible, including the variability of culture reagents, insufficient culture conditions, and (im)purity of phenotypic HSCs. Data presented here highlight the wide variety of in vitro HSC maintenance capabilities of different BSA-FV preparations. To overcome these limitations, we have developed a fully defined all-recombinant protein-based system for the in vitro culture of murine HSCs. Using this system, we have demonstrated that IL-1α and HPX, often contained within BSA-FV, significantly enhance in vitro maintenance of HSCs. This system therefore provides a basis on which to develop a reproducible in vitro HSC maintenance and expansion culture system.
Development of stable in vitro expansion conditions for HSCs would have important implications for both basic research and clinical use. In particular, stable ex vivo expansion of HSCs would reduce current shortages in donors for clinical BM transplantation and could afford ex vivo gene correction approaches for inherited hematological disorders. Furthermore, recent efforts to generate HSCs in vitro by directed differentiation and reprogramming are currently limited by our inability to maintain any HSCs generated in vitro. Here, we propose an all-recombinant protein-based system with which to develop such culture conditions, and systematic approaches by which to develop optimal conditions.
Based on DNA microarray data, we tested the functions of 30 different candidate factors by adding them one by one to HSC culture medium. Consequently, we were able to identify two HSC maintenance factors, IL-1α and HPX. In previous studies, IL-1α was reported to support the proliferation of mouse HSCs in cooperation with IL-3 (Jubinsky and Stanley, 1985, Mochizuki et al., 1987), while another group reported that the same factor reduces the colony-forming and reconstitution ability of HSCs (Yonemura et al., 1996). These contradictory results highlight the problems of using variable reagents such as conditioned medium or FBS. In the present study, we were able to reveal that IL-1α in fact contributes to the PB reconstitution potential of HSCs (Figures S1A and S1B). However, we note that high concentrations of IL-1α (100 ng/mL) inhibited colony formation (data not shown).
Furthermore, we were able to identify HPX as an HSC maintenance factor contained within BSA-FV. HPX is thought to inhibit heme-induced ROS generation. While hematopoietic defects in HPX knockout mice have not been described (Tolosano et al., 1999), our results demonstrated that HPX reduced ROS level of cultured HSCs and improved their reconstitution capability. HPX is unlikely to be the only HSC maintenance factor in BSA-FV, and we believe that protein profiling of “good” BSA-FV batches, as described here, will allow further identification of novel HSC maintenance and expansion factors.
In conclusion, we believe that the all-recombinant protein-based system described here is an exciting research tool for generating highly robust and reproducible data, and one that will provide important contributions in our efforts to identify growth and differentiation factors for HSCs, as well as many other different stem cell populations.
Experimental Procedures
HSC Purification
Mouse CD34−KSL HSCs were purified from BM cells of 8- to 10-week-old mice. See Supplemental Experimental Procedures for a detailed description.
HSC Culture
CD34−KSL HSCs were deposited into 96-well microtiter plates containing 200 μL of serum-free medium S-Clone SF-03 (Sanko Junyaku) supplemented with 1% BSA (Sigma and Wako), MSA, or HSA (Albumin Bioscience, Sigma, Bioverde) and cytokines (50 ng/mL mouse SCF, 50 ng/mL human TPO).
Competitive Repopulation Assays
Competitive repopulation assays were performed using the Ly5 congenic mouse system. Forty CD34−KSL cells from B6-Ly5.1 and the BM competitor cells (1 × 106) from B6-F1 mice were transplanted into B6-Ly5.2 mice irradiated at a dose of 9.8 Gy. See Supplemental Experimental Procedures for a detailed description.
Sample Purification for Mass Spectrometry
Depletion of albumin from BSA-FV was performed using Melon Gel IgG Spin Purification Kit (Thermo Scientific) according to the manufacturer's instructions.
BM Immunofluorescence and 3D Imaging
Frozen BM sections were prepared and immunostained according to the Kawamoto method (Kawamoto, 2003). 3D imaging was performed as previously described (Susaki et al., 2015).
Author Contributions
A.I., I.R., T.K., and S.Y. planned and performed the experiments and wrote the manuscript. M.M. helped perform experiments. A.W., K.S., T.N., J.O., Y.T., C.L., M.O., Y.N., H.E., and H.N. provided scientific discussion and technical support. H.E. provided scientific discussion. S.Y., A.W., and H.N. directed the study and wrote the manuscript.
Acknowledgments
We thank Drs. R. Yamamoto, Ishii, Y. Yamazaki, and A Tojo for technical help and advice, and Dr. M. Kasai for critical reading of the manuscript. This work was supported by grants from Japan Science and Technology Agency (JST) (grant no. 50625580), the Ministry of Education, Culture, Sport, Science, and Technology (Japan), California Institute of Regenerative Medicine (grant no. LA1-06917), Siebel Foundation, and Ludwig Foundation.
Published: February 23, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, one table, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.01.015.
Contributor Information
Hiromitsu Nakauchi, Email: nakauchi@ims.u-tokyo.ac.jp.
Satoshi Yamazaki, Email: y-sato4@ims.u-tokyo.ac.jp.
Supplemental Information
References
- Barker J.E. Sl/Sld hematopoietic progenitors are deficient in situ. Exp. Hematol. 1994;22:174–177. [PubMed] [Google Scholar]
- Calvi L.M., Adams G.B., Weibrecht K.W., Weber J.M., Olson D.P., Knight M.C., Martin R.P., Schipani E., Divieti P., Bringhurst F.R. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Chen W., Lu H., Dutt K., Smith A., Hunt D.M., Hunt R.C. Expression of the protective proteins hemopexin and haptoglobin by cells of the neural retina. Exp. Eye Res. 1998;67:83–93. doi: 10.1006/exer.1998.0494. [DOI] [PubMed] [Google Scholar]
- Eaves C.J. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 2015;125:2605–2613. doi: 10.1182/blood-2014-12-570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbert L.J., Iscove N.N. Partial replacement of serum by selenite, transferrin, albumin and lecithin in haemopoietic cell cultures. Nature. 1976;263:594–595. doi: 10.1038/263594a0. [DOI] [PubMed] [Google Scholar]
- Iwama A., Oguro H., Negishi M., Kato Y., Morita Y., Tsukui H., Ema H., Kamijo T., Katoh-Fukui Y., Koseki H. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Jacobsen F.W., Keller J.R., Ruscetti F.W., Veiby O.P., Jacobsen S.E. Direct synergistic effects of IL-4 and IL-11 on proliferation of primitive hematopoietic progenitor cells. Exp. Hematol. 1995;23:990–995. [PubMed] [Google Scholar]
- Jubinsky P.T., Stanley E.R. Purification of hemopoietin 1: a multilineage hemopoietic growth factor. Proc. Natl. Acad. Sci. USA. 1985;82:2764–2768. doi: 10.1073/pnas.82.9.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch. Histol. Cytol. 2003;66:123–143. doi: 10.1679/aohc.66.123. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Morrison S.J. Uncertainty in the niches that maintain haematopoietic stem cells. Nat. Rev. Immunol. 2008;8:290–301. doi: 10.1038/nri2279. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Terhorst C., Morrison S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Ku H., Yonemura Y., Kaushansky K., Ogawa M. Thrombopoietin, the ligand for the Mpl receptor, synergizes with steel factor and other early acting cytokines in supporting proliferation of primitive hematopoietic progenitors of mice. Blood. 1996;87:4544–4551. [PubMed] [Google Scholar]
- Mendez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma'ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki D.Y., Eisenman J.R., Conlon P.J., Larsen A.D., Tushinski R.J. Interleukin 1 regulates hematopoietic activity, a role previously ascribed to hemopoietin 1. Proc. Natl. Acad. Sci. USA. 1987;84:5267–5271. doi: 10.1073/pnas.84.15.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Eberhard U. Hemopexin. Methods Enzymol. 1988;163:536–565. doi: 10.1016/0076-6879(88)63049-7. [DOI] [PubMed] [Google Scholar]
- Osawa M., Hanada K., Hamada H., Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Sauvageau G., Thorsteinsdottir U., Eaves C.J., Lawrence H.J., Largman C., Lansdorp P.M., Humphries R.K. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Susaki E.A., Tainaka K., Perrin D., Yukinaga H., Kuno A., Ueda H.R. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 2015;10:1709–1727. doi: 10.1038/nprot.2015.085. [DOI] [PubMed] [Google Scholar]
- Swerts J.P., Soula C., Sagot Y., Guinaudy M.J., Guillemot J.C., Ferrara P., Duprat A.M., Cochard P. Hemopexin is synthesized in peripheral nerves but not in central nervous system and accumulates after axotomy. J. Biol. Chem. 1992;267:10596–10600. [PubMed] [Google Scholar]
- Tolosano E., Altruda F. Hemopexin: structure, function, and regulation. DNA Cell Biol. 2002;21:297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- Tolosano E., Cutufia M.A., Hirsch E., Silengo L., Altruda F. Specific expression in brain and liver driven by the hemopexin promoter in transgenic mice. Biochem. Biophys. Res. Commun. 1996;218:694–703. doi: 10.1006/bbrc.1996.0124. [DOI] [PubMed] [Google Scholar]
- Tolosano E., Hirsch E., Patrucco E., Camaschella C., Navone R., Silengo L., Altruda F. Defective recovery and severe renal damage after acute hemolysis in hemopexin-deficient mice. Blood. 1999;94:3906–3914. [PubMed] [Google Scholar]
- Walasek M.A., van Os R., de Haan G. Hematopoietic stem cell expansion: challenges and opportunities. Ann. N. Y. Acad. Sci. 2012;1266:138–150. doi: 10.1111/j.1749-6632.2012.06549.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Iwama A., Takayanagi S., Morita Y., Eto K., Ema H., Nakauchi H. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 2006;25:3515–3523. doi: 10.1038/sj.emboj.7601236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S., Ema H., Karlsson G., Yamaguchi T., Miyoshi H., Shioda S., Taketo M.M., Karlsson S., Iwama A., Nakauchi H. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- Yonemura Y., Ku H., Hirayama F., Souza L.M., Ogawa M. Interleukin 3 or interleukin 1 abrogates the reconstituting ability of hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 1996;93:4040–4044. doi: 10.1073/pnas.93.9.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Niu C., Ye L., Huang H., He X., Tong W.G., Ross J., Haug J., Johnson T., Feng J.Q. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.