Summary
Genome editing in induced pluripotent stem cells is currently hampered by the laborious and expensive nature of identifying homology-directed repair (HDR)-modified cells. We present an approach where isolation of cells bearing a selectable, HDR-mediated editing event at one locus enriches for HDR-mediated edits at additional loci. This strategy, called co-targeting with selection, improves the probability of isolating cells bearing HDR-mediated variants and accelerates the production of disease models.
Graphical Abstract
Highlights
-
•
Increases the efficiency of genome editing in human iPSCs
-
•
Enhances detectability of variants of interest derived by homology-directed repair
-
•
Is a simple, scalable, and adaptable strategy for knocking in variants of interest
The potential of CRISPR/Cas9-based genome editing in iPSCs is currently hampered by the laborious nature of identifying cells modified by the lesser-used homology-directed repair (HDR) pathway. Geurts, McDermott-Roe, and colleagues present a straightforward approach, co-targeting with selection, based on co-modification that enriches for cells undergoing HDR thereby improving targeting efficiencies.
Introduction
Programmable nucleases are seeing widespread application in the genome engineering field on account of their ability to permit precise genetic modifications in cell cultures and whole organisms. The CRISPR/Cas9 (CRISPR-associated) system (Bhaya et al., 2011) has attracted particular attention on account of its flexibility, ease of use, and cost-effectiveness compared with alternative nucleases (e.g., zinc-finger nucleases [ZFNs] and transcription activator-like effector nucleases [TALENs]). Approaches utilizing ZFNs, TALENs, and, increasingly, CRISPR/Cas9 for the creation of genetically modified induced pluripotent stem cell (iPSC) lines that can be converted into pertinent somatic cell types for exploration of contextually relevant pathophysiological states have become a go-to strategy for delineating variant/disease association (reviewed in Hockemeyer and Jaenisch, 2016). Precision genome editing typically involves incorporation of an exogenously supplied DNA donor with the desired variant, often containing one or more additional sequence incorporations to prevent nuclease re-cutting (Long et al., 2014), into the genome of the host cell via the homology-directed repair (HDR) pathway following a nuclease-mediated, double-strand break. Despite enhancements in the efficiency with which donor DNA can be incorporated into the genome, HDR-based editing in iPSCs using either vector- or single-stranded oligodeoxynucleotide (ssODN)-based donors occurs infrequently, often less than 1% (Soldner et al., 2011, Wang et al., 2014). Consequently, identifying a cell that bears a mutation of interest, which can entail extended maintenance, expansion, and analysis of hundreds of clonal populations, is laborious, expensive, and not readily scalable.
Increasing evidence suggests that HDR, which represents the lesser-used method of genome repair, is dependent on various cell-autonomous factors. Mitotic manipulation, temporal regulation of Cas9 expression, and suppression of the non-homologous end-joining (NHEJ) pathway have all been shown to enhance HDR editing in vitro to varying degrees (Gutschner et al., 2016, Lin et al., 2014, Maruyama et al., 2015, Yu et al., 2015). Selecting cells based on reagent delivery and integration of Cas9 into the genome have also been shown to enhance the frequency of HDR (Ding et al., 2013a, Ding et al., 2013b, Gonzalez et al., 2014). Elegant strategies have been devised to isolate precision-modified cells, including knockin of excisable selectable cassettes and serial enrichment of positive subfractions (Miyaoka et al., 2014, Yusa et al., 2011). Here we implemented a simple and adaptable method that obviates chemical perturbation, avoids stable Cas9 expression, maintains inherent DNA repair competence, does not require additional time or equipment, and is applicable to mismatch repair-proficient cell systems. We envisaged a potential HDR-competence spectrum across any population of transfected iPSCs, whereby a small subpopulation would naturally be more receptive to the incorporation of donor DNA via HDR while other cells remain refractory. In such a receptive cell, multiple independent HDR events could occur simultaneously meaning, in theory, an HDR-based primary editing event to incorporate a selectable marker at one locus could be accompanied by one or more independent user-specified HDR-mediated edits at other loci. Hence, isolation of cells based on the primary, selectable modification would enrich for the secondary, passenger modification(s). The methodology outlined herein is conceptually analogous to strategies previously described for the selection of cells harboring NHEJ-mediated gene-disruption events (Liao et al., 2015, Moriarity et al., 2014) although we extend the principle for isolation of cells bearing HDR-mediated precision editing.
Results and Discussion
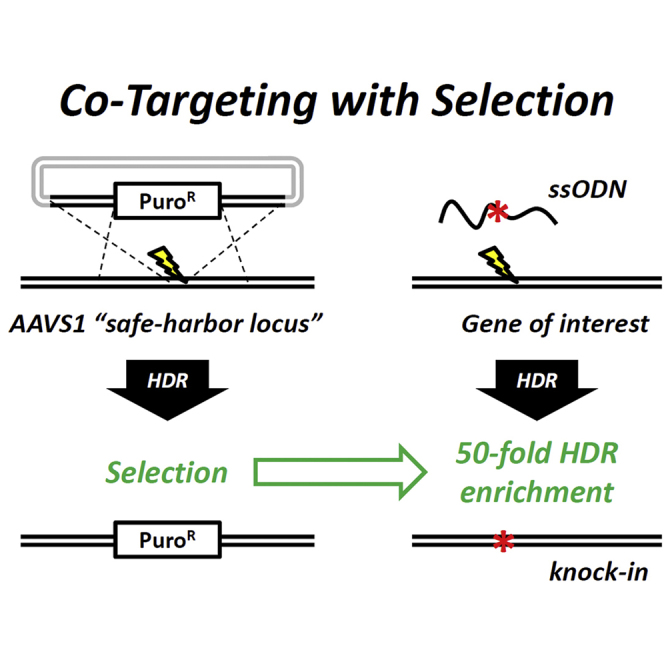
To test our hypothesis, we devised and implemented a strategy that we refer to as co-targeting with selection (CTS). CTS involves simultaneous transfection of human iPSCs with (1) a nuclease and donor plasmid designed to incorporate, via HDR, an antibiotic-resistance cassette into the AAVS1 safe harbor site (Sadelain et al., 2012) on chromosome 19, and (2) CRISPR/Cas9-based reagents and a cognate ssODN designed to introduce a variant of interest at a second locus, followed by maintenance in antibiotic-containing medium for approximately 10 days to select for resistant (and theoretically HDR-competent) clones (Figures 1A and 1B). Antibiotic-resistant colonies are then isolated, clonally expanded, and screened for knockin of the variant of interest. CTS does not alter the duration from transfection to isolation and analysis, but based on our experience and data reported herein, markedly enhances the representation of cells bearing passenger modifications (knockin alleles at the gene of interest) in the final population.
Figure 1.
Rationale for CTS and Proof of Feasibility
(A) Cells are edited simultaneously with gene-specific CRISPR/Cas9 and ssODN containing a variant of interest (red asterisk) as well as plasmids expressing AAVS1-specific TALENs and a puromycin resistant (PuroR) donor cassette.
(B) Components from (A) are transfected into iPSCs where HDR-receptive cells (green) are more likely to incorporate donor DNA than HDR-refractory cells (red).
(C) Precision-edited versus indel-containing alleles, detected via direct Sanger sequencing of relevant PCR products in hB53 hiPS6 iPSCs following CTS (n = 39) compared with no selection (n = 46) or transient exposure to puromycin (n = 48) (left panel), where n is the number of individual iPSC clones analyzed. Percent of clones bearing the CRYAB:c.325G>C variant (middle panel) and the number of heterozygous/homozygous clones (right panel).
(D) Representative chromatograms showing local CRYAB sequence of a wild-type (WT) clone (top) and that of a clone bearing a CRYAB:c.325G>C (homozygous) knockin allele (black arrow, c.325G>C variant; gray arrow, Cas9-blocking silent variant). See also Figures S1, S2, S4 and Table S1.
We first applied the CTS method to a single gene (CRYAB) in hB53 hiPS6 iPSCs (Riedel et al., 2014). Following transfection of a pre-validated CRYAB-specific single guide RNA (sgRNA)-expressing pX330 vector and ssODN donor template (for incorporation of the passenger modification) as well as a commercially available AAVS1-specific TALEN pair and puromycin N-acetyltransferase (pac)-containing donor vector driven by a constitutive promoter (for incorporation of the selectable modification), cells were treated with puromycin per our CTS protocol (to enrich for cells that underwent HDR, Figure S1), for 48 hr (to eliminate untransfected cells), or not at all (to mimic a selection-free system) (Figure S2). Clones were then harvested and analyzed via Sanger sequencing for modification at the gene of interest. The total number of editing events (NHEJ and HDR) was >7-fold greater in CTS cells relative to unselected cells and, crucially, the HDR/NHEJ ratio was >4-fold greater (Figure 1C, left panel, and Table S1). This corresponded to a ∼40% likelihood of picking a precision-modified clone, both heterozygous and homozygous, from the final culture with CTS compared with 2% (no treatment) or 4% (transient puromycin treatment) (Figures 1C, middle and right panel, and 1D). Interestingly, we did not observe enhancement in donor incorporation following isolation of transiently transfected cells in this experiment, which may be due to the pac cassette presence on the AAVS1 donor-targeting construct and not on the gene-specific nuclease (pX330) plasmid. Therefore, direct comparisons with published methods where selection is performed for transfection of the nuclease containing plasmid (Ding et al., 2013a, Ding et al., 2013b) and CTS were not performed in this study. These data suggested that selecting for HDR-receptive cells via a selectable modification significantly enriched for cells bearing passenger modifications (precision edit events) at the site of interest.
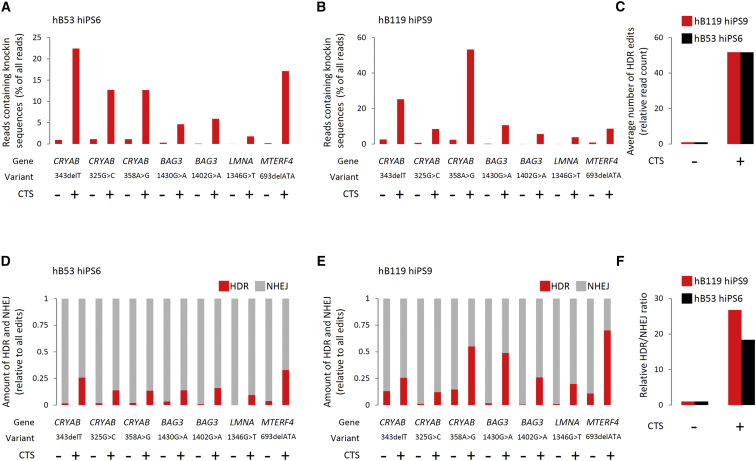
To benchmark the impact of CTS on HDR representation in a more quantitative manner and across multiple loci, we applied the workflow to a total of seven disease-associated variants across four different genes (CRYAB, BAG3, LMNA, and MTERF4) in two separate iPSC lines (hB53 hiPS6 and hB119 hiPS9) and analyzed editing outcomes via deep-sequencing. Following CTS, cells were pooled and deep-sequenced using the Illumina MiSeq platform. Average sequencing coverage following read trimming and quality filtering across all experiments was >150,000×, and the error rate was estimated to be less than 0.1%. We observed an average 3.7-fold (hB53 hiPS6) and 3.3-fold (hB119 hiPS9) increase in the total number of edits (HDR and NHEJ) across all loci with CTS compared with those without CTS (Table S2). Focusing on precision editing events and considering all seven variants, we observed an average 50-fold increase in HDR with CTS compared with those without CTS in both cell lines (Figures 2A–2C; Table S2). In support of our hypothesis that CTS enriches for HDR, we observed an overt shift in the balance between the two modes of repair such that the HDR/NHEJ ratio was enhanced on average 18-fold (hB53 hiPS6) and 27-fold (hB119 hiPS9) with CTS compared with those without CTS (Figures 2D–2F; Table S2). Considering all loci and both cell lines, the HDR rate following CTS was ∼14%, which corresponds to >1 in 10 clones bearing a precision edit.
Figure 2.
Quantitative Analysis of CTS-Enabled Precision Editing Across Multiple Genes
(A and B) Representation of precision (HDR only) editing events, based on read sequence and normalized to total read count, with (+) and without (−) CTS at multiple loci in hB53 hiPS6 (A) and hB119 hiPS9 (B) iPSC lines (−) CTS indicates cells handled in the same way as (+) CTS except for the addition of puromycin.
(C) Average fold change in HDR-mediated knockin with CTS relative to that without CTS, considering all loci in hB53 hiPS6 and hB119 hiPS9 iPSC lines.
(D and E) Relative proportions of reads bearing NHEJ- and HDR-based edits with (+) and without (−) CTS at multiple loci in hB53 hiPS6 (D) and hB119 hiPS9 (E) iPSC lines.
(F) Average increase in the HDR/NHEJ ratio in hB53 hiPS6 and hB119 hiPS9 iPSC lines with CTS relative to without CTS. See also Figure S3 and Table S2.
While CTS led to a gross increase in incorporation of the intended passenger modifications, deep-sequencing data revealed significant variation in locus targetability. For example, representation of the CRYAB:c.343delT variant was 22% (hB53 hiPS6) and 25% (hB119 hiPS9) following CTS (1% [hB53 hiPS6] and 3% [hB119 hiPS9] without CTS), whereas representation of the LMNA:c.1346G>T variant was 1% (hB53 hiPS6) and 4% (hB119 hiPS9) HDR following CTS (<0.05% in both cell lines without CTS). Notwithstanding, the average >100-fold increase in HDR at LMNA with CTS means that isolating a precision-edited clone is feasible (∼1 cell in 40 with CTS compared with ∼1 cell in ∼3,800 without, assuming heterozygosity) and suggests that loci which are inherently refractory to precision editing may be amenable via CTS. We also observed that the extent of ssODN incorporation was seemingly independent of its orientation relative to the sgRNA target strand (Table S3).
We next assessed how reflective the HDR editing rates calculated via deep-sequencing were of actual editing rates. Hence, the CTS protocol was repeated for all seven variants independently, and approximately 250 clonal populations were then picked and analyzed via Sanger sequencing. We successfully isolated cell lines for all seven mutations and observed a concordance between the quantities of HDR editing events determined via deep-sequencing and direct sequencing (Figures 3 and S3; Tables S2 and S3). We note that pooled deep-sequencing measures allelic representation at a specific time point and does not reflect zygosity or account for differing cellular growth rates from which the pool was derived. Extent of heterozygosity seemed to correlate, at least for the CRYAB mutations, with increasing distance from the CRISPR/Cas9 cut site (i.e., increased observation of heterozygous knockin clones with CRYAB:c.325G>C), but not in the case of BAG3:c.1430G>A, where we failed to isolate any mutant homozygous lines even though the targeted nucleotide was immediately adjacent to the CRISPR/Cas9 cut site. We speculate, as others have, that the nature and extent of zygosity is dictated by the distance between the CRISPR/Cas9 cut site and the targeted nucleotide as well as locus-dependent factors, such as chromatin organization (Paquet et al., 2016, Ward, 2015, Yang et al., 2013).
Figure 3.
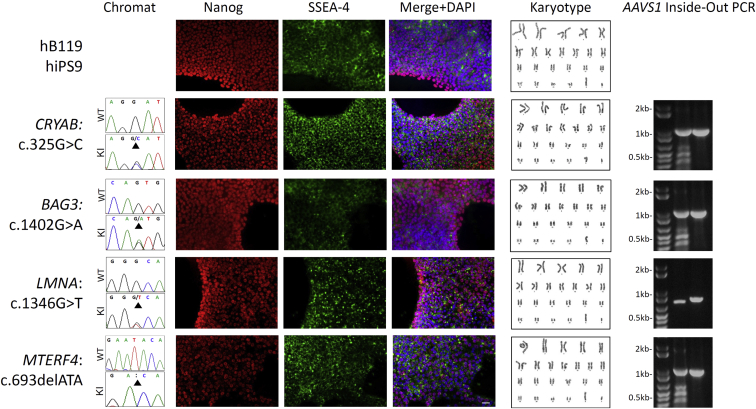
Validation of Knockin Cell Lines Generated with CTS
Knockin clones were generated for each disease-associated variant of interest shown in Table S3, with representative clones harboring variants in each gene shown here. Chromatograms (chromats) showing Sanger sequencing results of original cell line (WT) and knockin line (KI) with variants indicated by black arrows. Immunocytochemistry showing pluripotency markers Nanog and SSEA-4 for each knockin cell line harboring the respective variant of interest. Images were merged and counterstained with DAPI. Scale bar, 100 μm. Representative karyotypes for each cell line. Agarose gel showing AAVS1 inside-out PCR for both the 5′ (middle lane) and 3′ (right lane) integration sites (Experimental Procedures), which demonstrates site-specific integration of the selection construct via HDR. See also Figure S3 and Table S3.
We analyzed the zygosity and specificity of pac cassette knockin at the AAVS1 locus by Southern blotting and an integration-specific PCR assay (Figures S4A and S4B). We found that 50% of the clones (24/48) were correctly targeted without additional random integration. Of these, 21/24 were heterozygous and 3/24 were homozygous (Figure S4C). We observed concordance between results from Southern blotting and the PCR-based assay. With the PCR-based assay, representative cell lines harbored the pac cassette at AAVS1 (Figure 3) and none displayed evidence of additional aspecific integration events (data not shown). In addition, all lines analyzed exhibited the typical pluripotent cell morphology and karyotypic stability as well as expression of pluripotency markers (Figure 3), and maintained a capacity to form high-representation cardiomyocyte cultures (data not shown).
Despite improvements in sgRNA design (Doench et al., 2016), we nevertheless analyzed the top potential off-target sites (Experimental Procedures) in multiple mutant cell lines via Sanger sequencing and detected no signs of aspecific cleavage (data not shown). Furthermore, deep-sequencing of potential off-target sites (Experimental Procedures) revealed that CTS did not enrich for aberrant cutting compared with unselected cells (data not shown). The TALENs targeting AAVS1 have been previously demonstrated to have minimal off-target cleavage (Hockemeyer et al., 2011). Collectively, these data suggest negligible reagent promiscuity and that while CTS enriches for cells bearing HDR-edited alleles, it does not enrich for off-target mutations. We note that GUIDE-seq (Tsai et al., 2015) or similar would be required for whole-genome examination of off-target cutting.
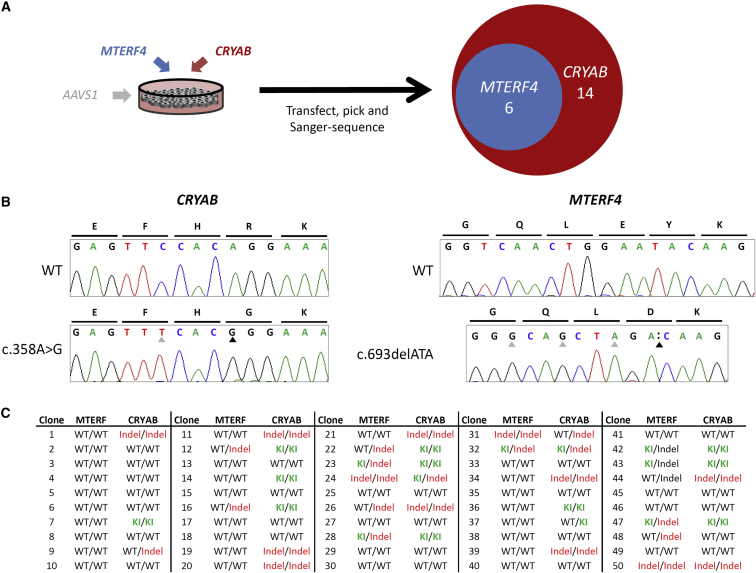
CTS is an inexpensive, rapid, straightforward, and readily scalable method, conceptually analogous to other marker-assisted enrichment strategies (Arribere et al., 2014), which increases the likelihood of isolating cells bearing knockin alleles by providing a non-integrating reporter of the HDR pathway activity akin to that devised by Flemr and Buhler (2015). While the precise cellular mechanisms of ssODN-mediated double-strand break repair remain to be elucidated, the observed enrichment by CTS via canonical homologous recombination of the double-stranded pac cassette donor into AAVS1 suggests a transient state of HDR permissiveness and a potential overlap in these repair mechanisms in iPSCs. We encountered significant variability in inter-locus targetability and suspect that local sequence composition and chromatin organization likely influence repair preference. Given the multifactorial nature of complex diseases and especially the role of modifier loci, we envisage CTS being of potential utility for simultaneous recapitulation of multiple candidate variants. Indeed, using CTS, we concurrently delivered editing reagents designed to incorporate passenger mutations at two different loci and isolated multiple clones bearing both edits (Figure 4). It will be interesting to determine whether application of CTS in conjunction with polycistronic sgRNA delivery systems (Cong et al., 2013) will permit highly parallelized HDR-based genome editing.
Figure 4.
Simultaneous Dual Modification using CTS
(A) Dual modification was attempted on hB53 hiPS6 using components for targeting MTERF4:c.693delATA and CRYAB:c.358A>G. Following CTS, 50 clones were sequenced and, of those, 14 had knockin at CRYAB (depicted in the red circle) and 6 had knockin at MTERF4 (depicted in the blue circle). All 6 of the cells with MTERF4 knockin also had knockin at CRYAB. Given individual editing rates, the likelihood of co-occurrence assuming random distribution of events is 5%. We observed a disproportionate co-occurrence of dual modification with a FET < 0.001.
(B) Representative chromatograms of dual-targeted clones at each loci and WT sequence. Black arrows indicate variant of interest position. Silent, engineered blocking mutations that prevent re-targeting by Cas9 are indicated by gray arrows.
(C) Table including genotypes of individual clones at both (MTERF4 and CRYAB) loci. WT, unmodified; KI, knockin; Indel, insertion or deletion. See also Table S3.
Operationally, CTS provides a marked improvement in the efficiency of isolating precision-modified iPSC lines compared with direct cloning-based methods, without extended hands-on time or a requirement for additional instrumentation. Following methodological refinements and legislating for effect range, we conservatively estimate that a single well-trained technician could generate ten precision-edited cell lines in 1 month. CTS is generalizable in that, while the system described uses antibiotic resistance as the selectable modification, alternative HDR-based reporters (e.g., insertion of a GFP cassette) could theoretically be employed. In our experiments, half of the analyzed clones were correctly targeted at AAVS1 with no additional random integration events, consistent with data reported previously (Hockemeyer et al., 2011), and this would likely be further improved with a gene trap approach. The site of the selectable modification could also be adapted depending on context and user requirements. For example, knockin of a GFP tag into a cardiac transcription factor such as NKX2-5 (Elliott et al., 2011) would yield iPSCs that harbor a variant of interest in tandem with a reporter which assists cardiomyocyte isolation. Furthermore, compared with alternative knockin strategies facilitated by targeted insertion of a selectable marker, CTS does not require the production of gene-specific custom targeting vectors, making it a readily scalable strategy. In addition, while we observed no adverse effects of AAVS1 targeting or carriage of the pac cassette on cell behavior or differentiation potential (although appropriate isogenic control cell lines harboring only the pac cassette should be utilized for phenotypic evaluation), removal of the pac cassette could be performed through transfection of cells with piggyBac transposase, although excision/re-integration rates would need to be empirically determined. Removal of the pac cassette would be required for any subsequent modification of generated cell lines with the same CTS strategy. Finally, a detailed mechanistic examination of the processes (e.g., engagement of HDR proteins, chromatin reorganization) that distinguish HDR-responsive from HDR-refractory cells and which contribute to the reported observations will be necessary and will undoubtedly catalyze discovery of additional factors which augment precision genome editing in all cell systems.
Experimental Procedures
Detailed Experimental Procedures are available in the Supplemental Information.
Targeting Reagents
CRISPR target sites proximal to the SNP of interest were identified using ZiFiT Targeter Version 4.2 and were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (a gift from Feng Zhang, Addgene plasmid no. 42230) as described previously (Cong et al., 2013). Cleavage efficacy of designed vectors was validated using the Cel-1 Surveyor assay as described previously (Geurts et al., 2009, Miller et al., 2007). Cognate, variant-specific ssODNs were designed and include silent mutations to prevent re-cutting of Cas9 following HDR. The AAVS1 Safe Harbor TALE-Nuclease Kit was purchased from System Biosciences, including pAAVS1 Dual Promoter Donor Vector (GE602A-1) and the TALE-Nuclease Vectors, pZT-AAVS1 L1 TALE-N Vector (GE601A-1) and pZT-AAVS1 R1 TALE-N Vector (GE601A-1) previously shown to have minimal off-target cleavage (Hockemeyer et al., 2011). A second AAVS1 Safe Harbor Kit was purchased from Transposagen, the Puro-TK with XTN TALEN (catalog no. KSH-004).
iPSC Lines and Culture
All human subject research was approved by the Medical College of Wisconsin and University of Utah institutional review boards. The human iPSC lines used in this study are hB53 hiPS6 (Riedel et al., 2014) and hB119 hiPS9, derived as described previously (Riedel et al., 2014). Informed consent was obtained for this procedure. iPSCs were cultured as described previously (Mitzelfelt et al., 2016).
iPSC Transfection
Transfection of relevant components was performed in iPSCs using a 4D-Nucleofector (Lonza). Following transfection, cells underwent the CTS protocol outlined in Figure S1 and detailed in the Supplemental Experimental Procedures. For deep-sequencing analysis of pooled populations (Figure 2), iPSCs were transfected and cultured as described, except that 2-day post transfection cells were separated into two groups, –CTS and +CTS, with corresponding –CTS and +CTS samples derived from the same initial transfection. +CTS conditions were as shown in Figure S1; –CTS conditions were identical, except that puromycin was omitted. Cells were pooled and genomic DNA was analyzed as described below.
Genotyping PCR and Sanger Sequencing for Clones
Genomic DNA was isolated and PCR was carried out using gene-specific primers. Resulting amplicons were Sanger sequenced using amplification primers.
Illumina MiSeq Library Preparation
Genomic DNA was isolated from pooled populations and samples were prepared and analyzed with the Illumina MiSeq, as described previously (Kistler et al., 2015).
Illumina MiSeq Analysis Methods
Reads were quality filtered using Trimmomatic (Bolger et al., 2014). Sorted/indexed BAM files were aligned to reference sequences (obtained from Ensembl release 84) with Bowtie2. Extent of HDR-mediated donor integration was quantified by interrogating FASTQ files for informative segments of the donor sequence (typically ∼50 nucleotides, spanning the targeted nucleotide and CRISPR cut site) via in-house code, manual inspection, and third-party software.
PCR-Based Analysis of Integration of the AAVS1 Donor Vector
HDR was confirmed at the AAVS1 locus using inside-out PCR with one primer falling inside the exogenous sequence and one primer outside the homology arm (Figure S4B).
Immunocytochemistry and Karyotyping
Immunocytochemistry and karyotyping were performed as described previously (Mitzelfelt et al., 2016).
Author Contributions
Conceptualization, A.M.G., K.A.M., and C.M.D.R.; Methodology, A.M.G., K.A.M., and C.M.D.R.; Software and Formal Analysis, C.M.D.R.; Investigation, K.A.M., C.M.D.R., M.N.G., M.M., C.T.K., M.J.C., K.D.K., M.R., and S.L.; Resources, D.P.D., D.H., C.J.J., and M.T.F.; Writing – Original Draft, C.M.D.R.; Writing – Review & Editing, C.M.D.R., K.A.M., A.M.G., and I.J.B.; Visualization, C.M.D.R. and K.A.M.; Supervision, A.M.G., C.M.D.R., and K.A.M.; Funding Acquisition, A.M.G., I.J.B., K.A.M., C.M.D.R., and J.W.V.
Acknowledgments
We thank Shirng-Wern Tsaih and Michael Tschannen (Human Molecular Genetics Center, Medical College of Wisconsin) for advice and technical support. This work was supported by NIH New Innovator Award 1DP2OD008396 (A.M.G.) and NIH Director's Pioneer Award Grant 8DP1HL17650-04 (I.J.B.), the Steven Cullen Healthy Heart Award (A.M.G.), made possible by the Research and Education Program Fund, a component of the Advancing a Healthier Wisconsin endowment at the Medical College of Wisconsin, a W.M. Keck Foundation Medical Research Grant (J.W.V.), and a Ruth L. Kirschstein National Research Service Award F31 Individual Fellowship 1F31AR067618-01A1 (K.A.M.).
Published: February 23, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.01.021.
Contributor Information
Chris McDermott-Roe, Email: mcdec@mail.med.upenn.edu.
Aron M. Geurts, Email: ageurts@mcw.edu.
Supplemental Information
References
- Arribere J.A., Bell R.T., Fu B.X.H., Artiles K.L., Hartman P.S., Fire A.Z. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198:837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Lee Y.-K., Schaefer E.A.K., Peters D.T., Veres A., Kim K., Kuperwasser N., Motola D.L., Meissner T.B., Hendriks W.T. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., Regan S.N., Xia Y., Oostrom L.A., Cowan C.A., Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotech. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.A., Braam S.R., Koutsis K., Ng E.S., Jenny R., Lagerqvist E.L., Biben C., Hatzistavrou T., Hirst C.E., Yu Q.C. NKX2-5eGFP/w hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Flemr M., Buhler M. Single-step generation of conditional knockout mouse embryonic stem cells. Cell Rep. 2015;12:709–716. doi: 10.1016/j.celrep.2015.06.051. [DOI] [PubMed] [Google Scholar]
- Geurts A.M., Cost G.J., Freyvert Y., Zeitler B., Miller J.C., Choi V.M., Jenkins S.S., Wood A., Cui X., Meng X. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Zhu Z., Shi Z.D., Lelli K., Verma N., Li Q.V., Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Haemmerle M., Genovese G., Draetta Giulio F., Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016;14:1555–1566. doi: 10.1016/j.celrep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler K.E., Vosshall L.B., Matthews B.J. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11:51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Tammaro M., Yan H. Enriching CRISPR-Cas9 targeted cells by co-targeting the HPRT gene. Nucleic Acids Res. 2015;43:e134. doi: 10.1093/nar/gkv675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotech. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.-L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotech. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Mitzelfelt K.A., Limphong P., Choi M.J., Kondrat F.D., Lai S., Kolander K.D., Kwok W.M., Dai Q., Grzybowski M.N., Zhang H. Human 343delT HSPB5 chaperone associated with early-onset skeletal myopathy causes defects in protein solubility. J. Biol. Chem. 2016;291:14939–14953. doi: 10.1074/jbc.M116.730481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y., Chan A.H., Judge L.M., Yoo J., Huang M., Nguyen T.D., Lizarraga P.P., So P.-L., Conklin B.R. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods. 2014;11:291–293. doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarity B.S., Rahrmann E.P., Beckmann D.A., Conboy C.B., Watson A.L., Carlson D.F., Olson E.R., Hyland K.A., Fahrenkrug S.C., McIvor R.S. Simple and efficient methods for enrichment and isolation of endonuclease modified cells. PLoS One. 2014;9:e96114. doi: 10.1371/journal.pone.0096114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- Riedel M., Jou C.J., Lai S., Lux R.L., Moreno A.P., Spitzer K.W., Christians E., Tristani-Firouzi M., Benjamin I.J. Functional and pharmacological analysis of cardiomyocytes differentiated from human peripheral blood mononuclear-derived pluripotent stem cells. Stem Cell Rep. 2014;3:131–141. doi: 10.1016/j.stemcr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2012;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- Soldner F., Laganière J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L.I., Myers R.H., Lindquist S. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotech. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.D. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ. Repair Genet. 2015;199:363–377. doi: 10.1534/genetics.114.172361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Guell M., Byrne S., Yang J.L., De Los Angeles A., Mali P., Aach J., Kim-Kiselak C., Briggs A.W., Rios X. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Rashid S.T., Strick-Marchand H., Varela I., Liu P.-Q., Paschon D.E., Miranda E., Ordonez A., Hannan N.R.F., Rouhani F.J. Targeted gene correction of [agr]1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.