Summary
Changes in oocyte quality can have great impact on the developmental potential of early embryos. Here we test whether nuclear genome transfer from a developmentally incompetent to a developmentally competent oocyte can restore developmental potential. Using in vitro oocyte aging as a model system we performed nuclear transfer in mouse oocytes at metaphase II or at the first interphase, and observed that development to the blastocyst stage and to term was as efficient as in control embryos. The increased developmental potential is explained primarily by correction of abnormal cytokinesis at anaphase of meiosis and mitosis, by a reduction in chromosome segregation errors, and by normalization of the localization of chromosome passenger complex components survivin and cyclin B1. These observations demonstrate that developmental decline is primarily due to abnormal function of cytoplasmic factors involved in cytokinesis, while the genome remains developmentally fully competent.
Keywords: genome exchange, oocyte aging, mouse, fragmentation
Graphical Abstract
Highlights
-
•
In vitro aging as a model of oocyte fragmentation and poor developmental potential
-
•
Nuclear transfer restores normal cytokinesis of in vitro aged mouse oocytes
-
•
Nuclear transfer restores development to term of in vitro aged oocytes
-
•
Developmental potential is primarily limited by the cytoplasm, not the nucleus
In this article, Egli and colleagues show that nuclear transfer can prevent abnormal cytokinesis and fragmentation of fertilized eggs at mitosis, and restore developmental potential to mouse eggs aged in vitro.
Introduction
A decline in developmental potential occurs when oocytes remain unfertilized for prolonged periods in vitro: the timing for optimal fertilization and development in mice is less than 12 hr post ovulation (Morton et al., 1997, Prietal et al., 2001, Wakayama et al., 2004), and in human within 4–12 hr after ovulation/oocyte retrieval (Chen and Kattera, 2003, Morton et al., 1997). Delayed fertilization affects development at various stages, during preimplantation development (Nagy et al., 1993), implantation (Chen and Kattera, 2003, Yuzpe et al., 2000), and development to term (Chen and Kattera, 2003).
In vitro postovulatory aging of oocytes occurs during prolonged in vitro culture of oocytes before fertilization, and its development capacity can be impaired in terms of preimplantation development and implantation rate. The decreased developmental rate is associated with an increase in chromosomal abnormalities, fragmentation, and abnormal configuration of metaphase spindles is often found in mouse postovulatory aged oocytes (Tatone et al., 2011). Bai et al. (2006) transferred oocyte genomes from 25–26 hr post oocyte retrieval into oocyte cytoplasm of 2–3 hr post oocyte retrieval, and found that postovulatory aged oocytes cannot develop beyond the two-cell stage; however, high blastocyst formation rate (86.2%) and offspring were obtained from reconstructed oocytes from 25- to 26-hr post oocyte retrieval nucleus and 2- to 3-hr post oocyte retrieval cytoplasm. Development to term was also obtained, although the efficiency was not compared with manipulated controls and thus did not suggest enough evidence for the availability of genome exchange to restore oocyte aging. The underlying molecular mechanisms of these defects remain poorly understood. Therefore, a better understanding of the molecular events that lead to a reduced developmental potential, and the development of methods to rescue abnormal cytokinesis, may have applications for reproductive treatment.
Here we use postovulatory aged mouse oocytes as a model of developmentally compromised oocytes to determine the utility of genome transfer to rescue developmental potential. Using a total of 50 nuclear transfer experiments on 1,645 oocytes, we found that abnormal localization of survivin and cyclin B1 is associated with abnormalities in cytokinesis, resulting in developmental failure of postovulatory aged oocytes. Genome transfer at either metaphase or interphase restored these defects, resulting in efficient development.
Results
Postovulatory Aged Oocytes Show Poor Preimplantation Development
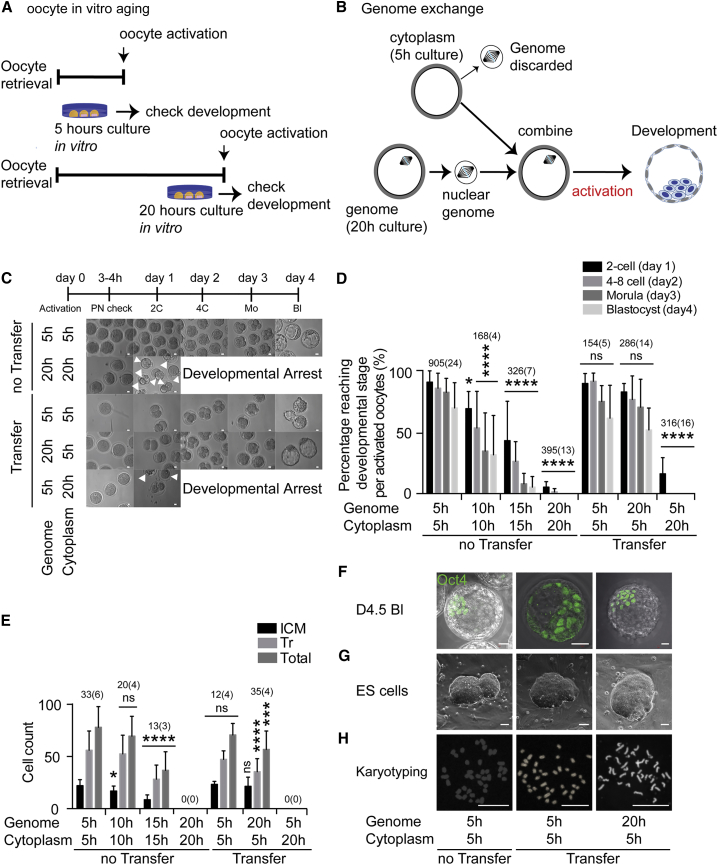
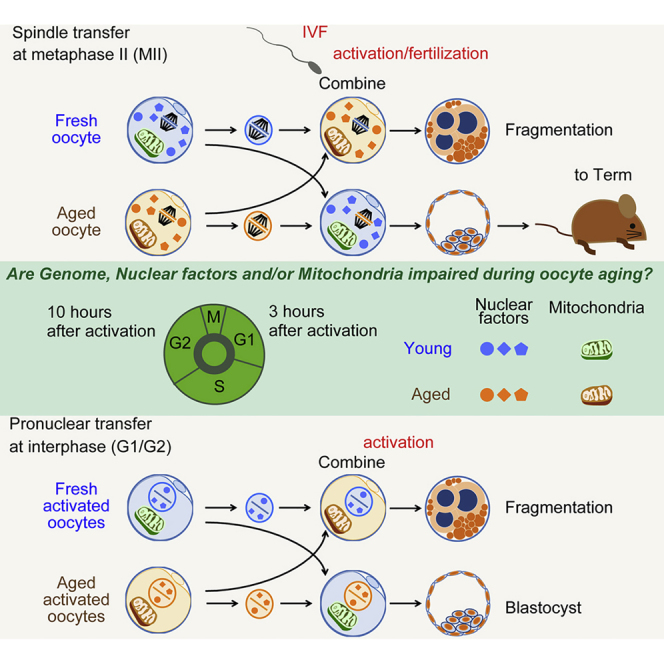
We used a model system for failed fertilization in which oocytes remain unfertilized for prolonged periods in vitro, resulting in developmental arrest at the cleavage stages (Figure 1A). To determine the timing of developmental decline of postovulatory oocytes in mice, we used parthenogenetic activation of oocytes instead of fertilization because it avoids the possibility that the contribution of the sperm genome can complement for potential defects in the oocyte genome. Oocytes were activated with ionomycin, 6-dimethylaminopurine (DMAP), puromycin, and cytochalasin B to prevent polar body extrusion, which results in diploid parthenotes. Control oocytes activated at 5 hr post oocyte retrieval developed efficiently to the blastocyst stage. Oocytes aged for 10 hr and 15 hr post retrieval showed greatly impaired preimplantation development compared with 5-hr incubated oocytes (Figures 1C and 1D). In addition, the cell number of both inner cell mass and trophectoderm of the few blastocysts derived from both 10- and 15-hr aged oocytes showed a lower cell number (Figure 1E), an indication for compromised development. Furthermore, no oocytes activated at 20 hr post oocyte retrieval could reach the blastocyst stage, and frequently failed at the first cleavage with extensive fragmentation (Figure 1C, arrowheads).
Figure 1.
Decline in Preimplantation Developmental Competence during Postovulatory Aging and Rescue after Genome Transfer at MII
(A) Strategy for oocyte retrieval and the timing of in vitro culture and oocyte activation.
(B) Genome exchange between 5-hr and 20-hr incubated oocytes. The genome of 5-hr incubated oocytes is transferred into enucleated, 20-hr incubated oocytes (5G20C). After manipulation, they are artificially activated and observed for preimplantation development. A reciprocal exchange is also conducted.
(C) Preimplantation development of activated oocytes. Arrowheads indicate fragmentation.
(D) Genome exchange rescues preimplantation development of 20-hr incubated oocytes. The number of oocytes and the number of experiments (in parentheses) is indicated above each column. The error bars show mean ± SEM. ∗p < 0.05, ∗∗∗∗p < 0.0001; ns, not significant.
(E) Cell numbers in blastocyst from genome exchanged oocytes. The total number of blastocysts and the number of experiments (in parentheses) is indicated above each column. The error bars show mean ± SEM. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
(F–H) Oct4 immunostaining at blastocyst stage (F), ESC derivation (G), and karyotype of ESCs (H). Scale bars represent 20 μm.
Genome Exchange at Meiosis Rescues Development of Postovulatory Aged Oocytes
To determine whether the poor developmental capacity of postovulatory aged oocytes was caused by cytoplasmic deterioration or chromosomal damage, or both, we performed nuclear genome exchange between 5-hr post oocyte retrieval oocytes and 20-hr post oocyte retrieval oocytes (Figure 1B). To reveal defects in the cytoplasm, we transferred the genome of 5-hr post oocyte retrieval oocytes into enucleated, 20-hr post oocyte retrieval oocytes (5G20C oocytes). Since spindle and chromosomes extracted from 5-hr post oocyte retrieval oocytes are normal, the development of 5G20C eggs would reveal cytoplasmic defects. As a control for these manipulations, we transferred nuclear genomes between 5-hr post oocyte retrieval oocytes (5G5C oocytes). Moreover, the genome from 20-hr post retrieval oocytes was transferred into 5-hr post oocyte retrieval oocytes (20G5C oocytes) to reveal damage to the DNA of postovulatory aged oocytes. The fusion rate of the auto- and hetero-transferred karyoplast-cytoplast pairs was 98.3% (980 of 996 oocytes observed), with no significant differences among the different groups. More than 97% of oocytes (1,605/1,645) survived the manipulations. After activation, 60.0% ± 7.4% of 20G5C oocytes developed to the blastocyst stage, but none of the 5G20C oocytes developed (Figure 1D). The cell number of the inner cell mass in 20G5C blastocysts was identical that of the blastocysts developing from nonmanipulated 5-hr post oocyte retrieval oocytes (Figure 1E). Twenty-three blastocysts obtained from 20G5C oocytes were used for embryonic stem cell (ESC) derivation and gave rise to six cell lines. Karyotyping analysis of the cell lines showed that its chromosomal number was normal, 40, XX (Figure 1H). Therefore, the cytoplasm was primarily affected by postovulatory aging, while the genome remained competent to support preimplantation development.
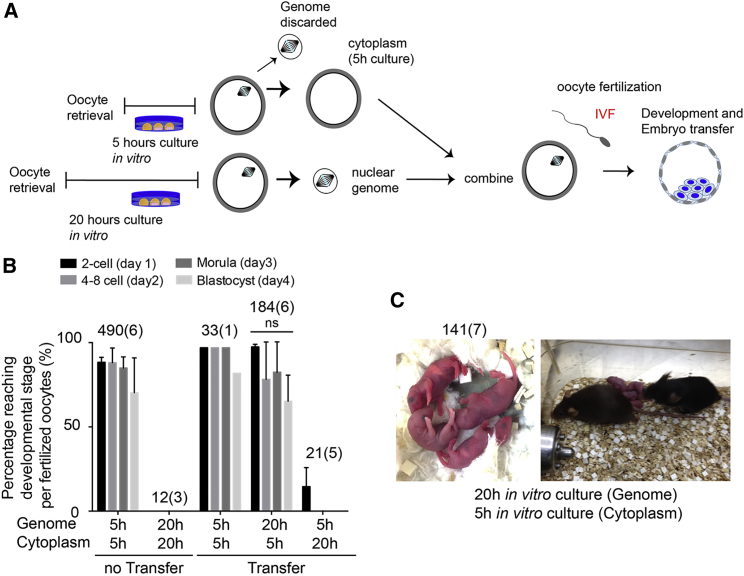
Development to Term after Fertilization of Reconstructed Oocytes
To determine whether nuclear genome transfer from postovulatory aged oocytes to newly ovulated oocytes could allow development to term, we fertilized 20G5C oocytes (Figures 2A and 2B). As a control, we used 5-hr and 20-hr post oocyte retrieval oocytes. Fertilization of both 20G5C and 5-hr oocytes was efficient (184/230 [80.0%], 570/626 [91.0%] fertilized) (Figure S1), while fertilization of unmanipulated 20C oocytes was inefficient (12/145 [8.2%] fertilized). After in vitro fertilization (IVF), high blastocyst formation rates (77.5% and 63.0%) were observed in the unmanipulated 5-hr oocytes and 20G5C oocytes. Unmanipulated 20-hr oocytes and 5G20C oocytes did not develop beyond the 2-cell stage (Figure 2B). 20G5C oocytes developed in vitro to the blastocyst stage, and 141 embryos were transferred into seven pseudopregnant foster females, which resulted in the birth of 12 pups, 4 males and 8 females (Figure 2C). This is comparable with nonmanipulated 5-hr oocytes (8/100, 3 males and 5 females) and 5G5C oocytes (2/27, 1 male and 1 female). In addition, 10 of 12 pups derived from 20G5C embryos were fertile (10/12) for 10 months after delivery. One male and one female of 12 pups derived from 20G5C, as well as one male and one female of eight pups derived from nonmanipulated 5-hr oocytes, did not produce any offspring, suggesting that infertility was not related to the manipulation. Thus we found that genome exchange could rescue postovulatory aged oocytes and allow normal development to term. Consistent with our previous conclusion using parthenogenesis, developmentally competent cytoplasm was sufficient to fully restore developmental competence to a postovulatory aged oocyte nucleus.
Figure 2.
Genome Exchange at MII Rescues Full-Term Development of Postovulatory Aged Oocytes
(A) Strategy for oocyte manipulation. The genome of 5-hr incubated oocytes is transferred into enucleated, 20-hr incubated oocytes (5G20C); oocytes are fertilized in vitro and allowed to develop.
(B) Preimplantation development after in vitro fertilization. The total number of 2PN oocytes and the number of experiments (in parentheses) is indicated above each column. The error bars show mean ± SEM. ns, not significant.
(C) Pups born after genome exchange (left) and fertile adults (right). The total number of transferred embryos and the number of experiments (in parentheses) is indicated.
Interphase Genome Transfer Is Also Effective at Restoring Developmental Potential
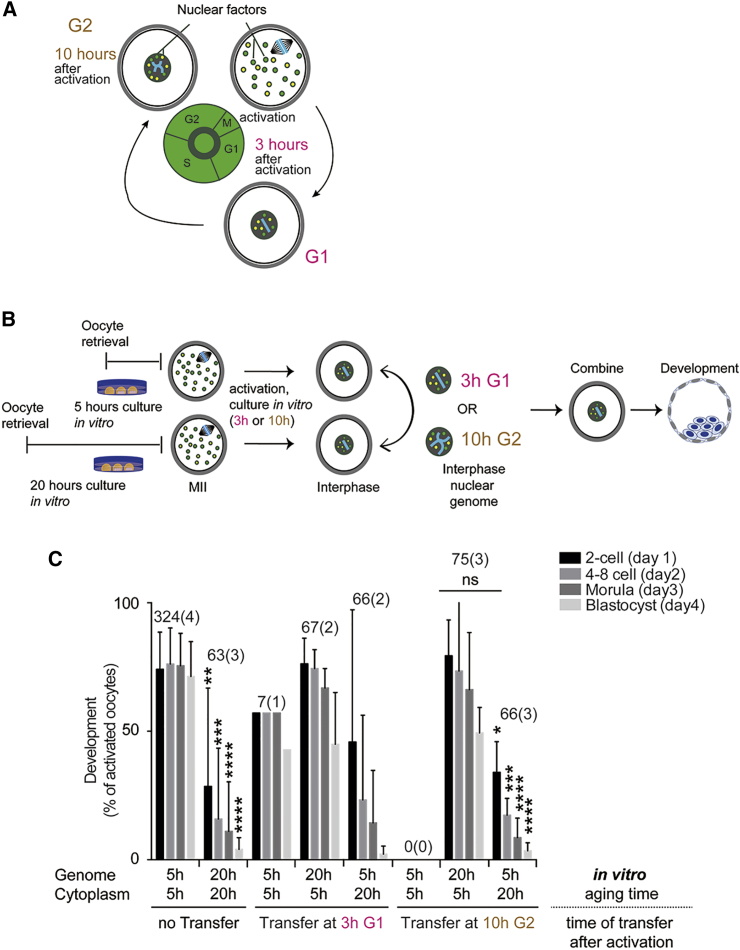
The cytoplasm of oocytes at metaphase contains nuclear factors involved in DNA replication and gene expression during embryonic development (for review see Egli et al., 2008). At interphase these factors localize to the nucleus, while mitochondria and components of the translation machinery localize to the cytoplasm throughout the cell cycle (Figure 3A). If factors involved in nuclear processes are affected by postovulatory aging, nuclear transfer at interphase might be unable to restore developmental potential because it would deplete factors from the developmentally competent oocyte and replace these with compromised nuclear proteins. For instance, replication in the presence of compromised DNA replication machinery might result in irreversible damage to the DNA. Therefore, transfer at interphase at different time points, at G1 or G2, can identify the cellular process/cellular compartment that is most affected by postovulatory aging.
Figure 3.
Pronuclear Transfer Restores Developmental Competence
(A) Schematic for the localization of nuclear factors. At metaphase, nuclear factors (colored dots) disperse throughout the cytoplasm. At interphase, nuclear factors, such as those required for DNA replication, localize within the nuclear envelope.
(B) Schematic for genome exchange between 5-hr and 20-hr incubated oocytes at interphase. Both 5-hr and 20-hr incubated oocytes are artificially activated while suppressing polar body extrusion and potentially abnormal cytokinesis of 20-hr oocytes. At interphase (3 hr and 10 hr after activation, respectively), reciprocal exchange is conducted and preimplantation development is observed.
(C) Preimplantation development after pronuclear transfer at 3 hr and at 10 hr after activation. The total number of oocytes and the number of experiments (in parentheses) is indicated above each column. The error bars show mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
We performed reciprocal pronuclear genome transfer between eggs activated 20 hr or 5 hr after oocyte retrieval. Transfer was performed at 3 hr after activation when cells were in G1 of the cell cycle, and preimplantation development was observed (Figure 3B). The transfer of nuclear genomes at interphase from eggs activated 20 hr after retrieval to eggs activated 5 hr post retrieval showed 53.7% (36/67 oocytes [two experiments]) development to the blastocyst stage (Figure 3C). On the contrary, only 3.0% (2/66 oocytes [two experiments]) of transfers from eggs activated at 5 hr to eggs activated at 20 hr post retrieval developed to the blastocyst stage. To determine whether nuclear DNA replicated normally in 20-hr oocytes, we also performed nuclear transfer at 10 hr after activation, corresponding to the G2 phase of the cell cycle (Figure 3B). These reconstructed eggs developed to the blastocyst stage at a similar efficiency as when transfer was performed at G1 (48.0%, 36/75 oocytes [three experiments]) (Figure 3C). Thus, the genome remains developmentally competent despite replicating in a developmentally compromised oocyte. On the contrary, only 3.0% (2/66 oocytes [three experiments]) transfers of nuclei from eggs activated at 5 hr post retrieval to the cytoplasm of eggs activated at 20 hr post retrieval developed to the blastocyst stage. Therefore, factors that remain cytoplasmic throughout the cell cycle are primarily responsible for the developmental decline of postovulatory aged oocytes. Nevertheless, a minor difference was seen between transfer at metaphase and interphase. A small number of 5- to 20-hr interphase transfer oocytes (3%) reached the blastocyst stage, while none reached the blastocyst stage when transfer was performed at metaphase (Figure 1D). Also, while interphase transfer rescued 48% of postovulatory aged oocytes, transfer at metaphase rescued 60%. This suggests that nuclear factors also contribute to the developmental decline of postovulatory aged oocytes, but to a much smaller extent than factors that remain cytoplasmic throughout the cell cycle.
Genome Exchange at Meiosis Rescues Chromosome Alignment and Spindle Function
In contrast to transfer at interphase, the transfer of a genome at meiosis also involves the transfer of the meiotic spindle. The replacement of spindle components could in principle affect developmental potential. Therefore, we investigated the molecular integrity and functionality of the spindle apparatus after postovulatory aging and after genome transfer.
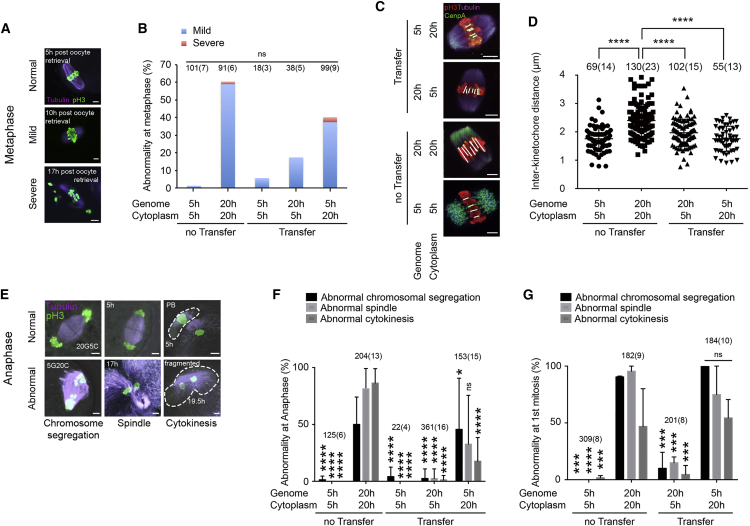
We first investigated spindle morphology and chromosome alignment in 20-hr oocytes. Alignment defects were ranked from none, to mild, to severe (Figure 4A); chromosomes in the no chromosomal defect group aligned tightly on the metaphase plate, those in the mild chromosomal defect group showed misalignment on the metaphase plate, whereas chromosomes in the severe group were dispersed. The proportion of oocytes showing mild chromosomal alignment defects increased during postovulatory aging, and reached 56% at 20 hr (Figures 4B and S2A). Severe abnormalities were only observed after more than 17 hr of postovulatory aging. To better understand chromosome alignment defects in postovulatory aged oocytes, we stained for kinetochores/centrosomes. Greater distances between sister kinetochores (interkinetochore) are thought to have a causal relationship with deterioration in chromosome cohesion and resulting mis-segregation in meiosis I of both young and reproductively aged oocytes (Lister et al., 2010, Merriman et al., 2012). We measured the distance between kinetochores marked by the centromeric protein Cenp A. The interkinetochore distance was increased in unmanipulated 20-hr aged eggs (2.34 ± 0.05 μm, mean ± SEM, n = 23 eggs, 130 kinetochore pairs) compared with unmanipulated 5-hr eggs (1.75 ± 0.05 μm, n = 14 eggs, 69 kinetochore pairs; p < 0.0001) (Figures 4C and 4D). Increased interkinetochore distances were found in aged oocytes with both normal or mild metaphase II (MII) alignment defects.
Figure 4.
Genome Exchange Restores Spindle Morphology and Function
(A) Chromosome alignment in MII oocytes. Examples of defects in postovulatory aged oocytes are shown.
(B) Frequency of abnormal MII alignment of nonmanipulated and manipulated oocytes. The total number of oocytes and the number of experiments (in parentheses) is indicated above each column. ns, not significant.
(C) Immunofluorescence images of kinetochores, spindles, and chromosomes stained with Cenp A (green), p-H3 (red), and tubulin (purple) at MII. Scale bars represent 5 μm.
(D) Quantification of interkinetochore distance for all (discernible) sister kinetochore pairs. Each dot shows one pair. The number of kinetochores and the number of oocytes (in parentheses) are indicated. ∗∗∗∗p < 0.0001.
(E) Chromosome segregation at anaphase of the second meiosis after artificial activation. Examples of chromosomal defects, spindle abnormality, and cytokinetic abnormality are shown. The areas bound by dashed lines present the second polar body (PB) (top) and fragmentation (bottom). Time cultured after oocyte retrieval before activation is indicated.
(F) Frequency of abnormalities of nonmanipulated and manipulated oocytes at anaphase of meiosis. The error bars show mean ± SEM. ∗p < 0.05, ∗∗∗∗p < 0.0001; ns, not significant.
(G) Frequency of abnormal first mitosis in nonmanipulated and manipulated oocytes at anaphase of first mitosis. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
To determine whether genome exchange can correct chromosome alignment defects, we performed genome exchange between 5-hr and 20-hr post retrieval oocytes. A reduction of mild severe chromosome alignment defects, though not statistically significant because of high variability, was seen when 20-hr genomes were transferred into 5-hr cytoplasm (Figure 4B). Conversely, the transfer of 5-hr genomes to 20-hr cytoplasm resulted in a deterioration of chromosome alignment, though not to the extent seen in 20-hr oocytes (Figure 4B). To determine whether transfer restored normal kinetochore distance, we transferred genomes from postovulatory aged to newly ovulated oocytes (20G5C oocytes). Oocytes were fixed 1 hr after manipulation. Interkinetochore distance was largely normalized (1.92 ± 0.04 μm, n = 15 eggs, 102 kinetochore pairs; p < 0.0001) (Figures 4C and 4D). On the contrary, interkinetochore distance of 5G20C oocytes (1.71 ± 0.05 μm, n = 13 eggs, 55 kinetochore pairs) did not change compared with 5-hr oocytes (1.75 ± 0.05 μm, n = 14 eggs, 69 kinetochore pairs; not significant).
To examine functionality of the spindle after postovulatory aging and transfer, we activated oocytes and observed them at anaphase of the second meiosis. The majority of oocytes aged for 17 hr or more post retrieval failed to form a normal anaphase spindle and underwent cytokinesis with extensive fragmentation (Figures 4E and 4F). At 20 hr post retrieval, 75% showed abnormalities (153/204). Remarkably, transfer of the genome from 20-hr aged to 5-hr oocytes led to a significant and consistent improvement of spindle function at anaphase of MII. Ninety-seven percent of 20G5C oocytes (353/361) showed a normal anaphase spindle formation and cytokinesis (p < 0.0001) (Figure 4F). A small number of abnormal segregation events (2.5%) remained (Figure 4F), corresponding to the percentage of severe abnormalities at metaphase. Conversely, when genomes from 5-hr oocytes were transferred to 20-hr cytoplasm, anaphase spindle morphology and chromosome segregation deteriorated dramatically. Only 45% of 20-hr oocytes (92/204) showed normal segregation (Figure 4F).
To determine the ability of postovulatory aged and transferred oocytes to cleave at the first mitosis, we suppressed the cytokinesis at the second meiosis using cytochalasin B. Oocytes formed two nuclei, but no polar body, and then entered mitosis. Fifty-five percent of 20-hr oocytes (101/182) fragmented (Figure 4G). On the contrary, 94% of 20G5C oocytes (190/201) showed normal cytokinesis (p < 0.001). The improvement was seen independent of the cell-cycle timing of genome transfer: 190 of 201 cleaved normally after metaphase transfer at the second meiosis (Figure 4G), 62 of 67 after G1 transfer (Figure 3C), and 67 of 75 after G2 transfer at the first mitosis (Figure 3C).
Therefore, while cytoplasm from newly ovulated oocytes can restore normal spindle function and reduce the formation of errors at anaphase of meiosis or mitosis, postovulatory aged cytoplasm induces the defects in spindles that are normal at transfer. This is consistent with the conclusion that components of the cytoplasm are primarily affected by postovulatory aging.
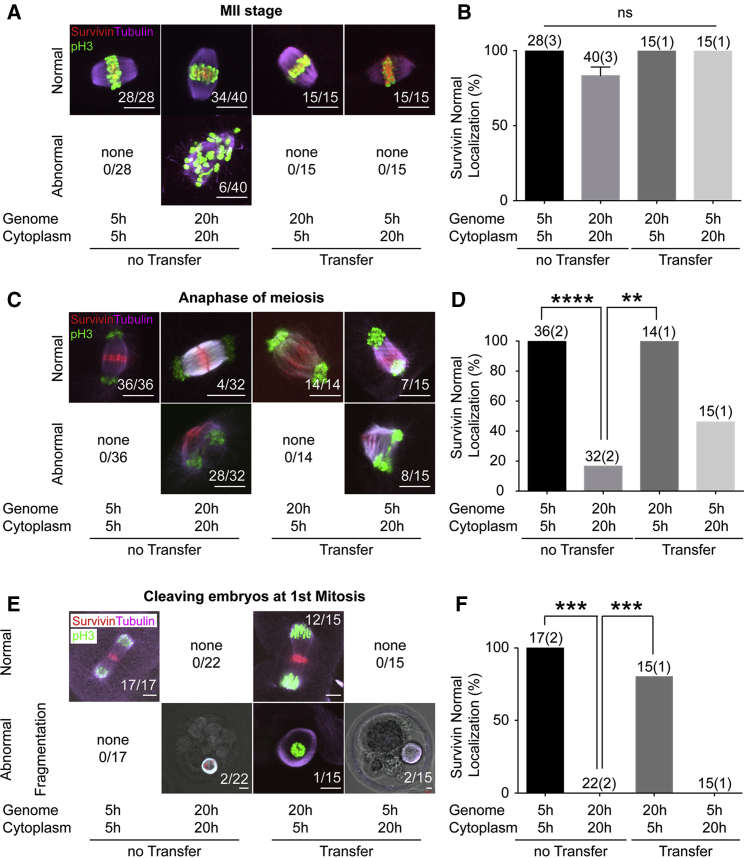
Survivin and Cyclin B1 Mislocalize in Aged Spindles and Are Restored after Transfer
To identify the molecular determinants affecting cell division in postovulatory aged oocytes, we performed immunochemistry for key spindle components. Components of the chromosome passenger complex are critical components of chromosome segregation at anaphase and cytokinesis (reviewed in van der Waal et al., 2012). Survivin is known to be a key component of the chromosome passenger complex in the formation of the midbody and localization of the cleavage furrow (Temme et al., 2003). We found that survivin localized to the center of the spindle of all 5-hr oocytes, but 15% of 20-hr oocytes (6/40) showed an abnormal survivin localization pattern, such as in the cytoplasm apart from spindle or throughout the spindle randomly at metaphase (not significant) (Figures 5A and 5B). At anaphase of meiosis, survivin localized to the midbody in all 5-hr oocytes, but localized on the longitudinal axis of the spindle in most 20-hr oocytes (p < 0.0001) (Figures 5C and 5D). A similar difference was seen at anaphase of the first mitosis: while 5-hr oocytes showed survivin localization to the midbody, 20-hr oocytes fragmented without proper chromosome segregation or distinct survivin localization (p < 0.001) (Figures 5E and 5F). When we transferred genomes from postovulatory aged to newly ovulated oocytes (20G5C oocytes), the localization of survivin was normalized at all stages, at metaphase and anaphase of meiosis, and at anaphase of the first mitosis (Figures 5A–5F). In contrast, in 5G20C oocytes, survivin localization was abnormal at anaphase of meiosis and at anaphase of the first mitosis. These results suggest that aged cytoplasm adversely affects survivin localization and cytokinesis, while new cytoplasm ensures normal survivin localization.
Figure 5.
Genome Exchange Restores Survivin Localization
(A, C, and E) Immunofluorescence images of spindles and chromosomes stained with p-H3 (green), tubulin (purple), and survivin (red) at MII (A), anaphase of meiosis II (C), and anaphase of the first mitosis (E). Examples of normal and abnormal survivin localization are shown. All images are projections of confocal z series. Scale bars represent 5 μm.
(B, D, and F) Quantification of localization patterns at MII (B), anaphase (D), and the first mitosis (F). The total number of oocytes and the number of experiments (in parentheses) is indicated above each column. The error bars show mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant.
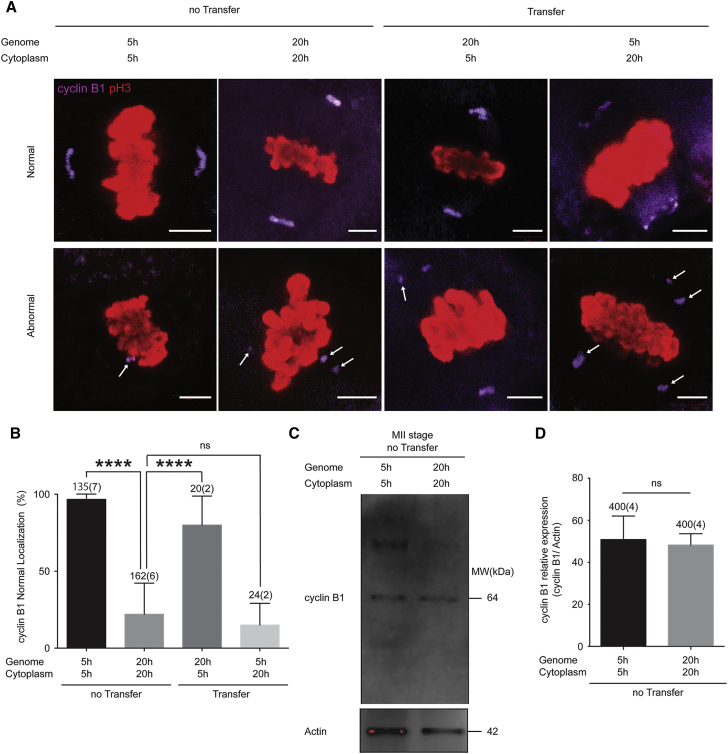
We also determined the localization of cyclin B1, a kinase localized to both the cytoplasm and the spindle poles (Hoffmann et al., 2012). In 94% of 5-hr oocytes (128/135), cyclin B1 localized at the spindle poles (Figure 6A), but in 22% of 20-hr oocytes (35/162) multiple ectopic clusters of cyclin B1 were found in the cytoplasm (indicated by arrows in Figure 6A). Eighty-five percent of the 20G5C oocytes (17/20) showed normal localization of cyclin B1. On the contrary, only 8.3% of 5G20C oocytes (2/24) showed normal localization of cyclin B1 (Figure 6B). Amounts of cyclin B1 expressed in unmanipulated oocytes were also measured by immunoblotting (Figure 6C), but there was no significant difference between 5-hr and 20-hr oocytes (four experiments) (Figure 6D). Therefore, nuclear transfer restores localization and function of critical spindle components.
Figure 6.
Genome Exchange Restores Cyclin B1 Localization at Metaphase
(A) Immunofluorescence at MII stage for p-H3 (red) and cyclin B1 (purple). Examples for normal and abnormal localization of cyclin B1 are shown. All images are projections of confocal z series. Arrows indicate ectopic clusters of cyclinB1 in the cytoplasm. Scale bars represent 5 μm.
(B) Quantification of cyclin B1 localization patterns. The error bars show mean ± SEM. ∗∗∗∗p < 0.0001; ns, not significant.
(C and D) Western blotting analysis (C) and quantification (D) of cyclin B1 at MII and actin as a loading control. A representative result is shown from four independent experiments. The error bars show mean ± SEM. ns, not significant.
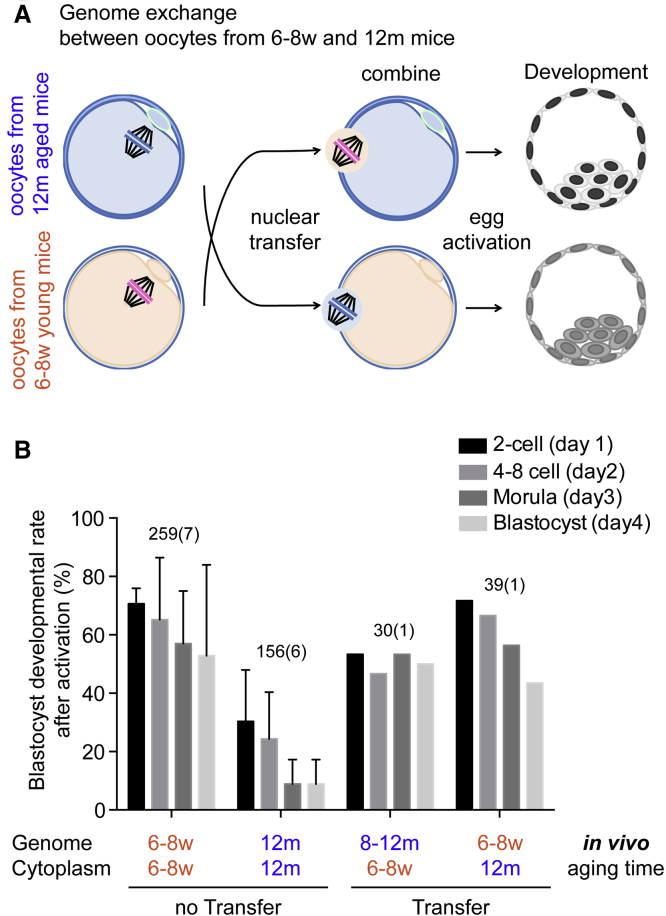
Genome Exchange at Meiosis Improves Development of Reproductively Aged Oocytes
To determine whether genome exchange can rescue reproductively aged oocytes, we transferred the genome of oocytes retrieved from aged mice (12 months) into enucleated oocytes retrieved from young mice (6–8 weeks) (12m8w oocytes) and observed blastocyst development after artificial activation (Figures 7A and 7B). We found that the transfer of the nuclear genome into young cytoplasm improved developmental competence (Figure 7B). The transfer of a young nucleus to an old cytoplasm also resulted in a modest improvement in developmental potential (Figure 7B). These results are consistent with the results of postovulatory aging. Further studies will focus on the molecular mechanism underlying how and why nuclear transfer restores developmental potential of reproductively aged oocytes.
Figure 7.
Genome Exchange at MII Increases Preimplantation Development of In Vivo Aged Oocytes
(A) Genome exchange between oocytes retrieved from young mice (6–8 weeks) and those from reproductively aged mice (12 months). The genome of reproductively aged oocytes is transferred into enucleated young oocytes (12m8w). After manipulation, they are artificially activated and observed for preimplantation development. A reciprocal exchange (8w12m) is also conducted.
(B) Preimplantation development of activated oocytes. Preimplantation development was observed after activation for 4 days and counted every day.
Discussion
The results presented here provide proof of principle that genome transfer can rescue developmentally incompetent oocytes. We reconstructed oocytes from 5-hr post oocyte retrieval cytoplasm and genomes extracted from oocytes left unfertilized for 20 hr post retrieval, and observed cleavage and development to term after IVF. The resulting offspring was normal and fertile. These results demonstrate although the genome remains largely unaffected by postovulatory aging, the ability of the cytoplasm to support development rapidly deteriorates. Postovulatory aged oocytes showed spindle abnormalities, increased kinetochore distance at metaphase, fragmentation at cleavage, and abnormal localization of cyclin B1 at metaphase, and of the chromosome passenger complex component survivin at anaphase. Remarkably, nuclear genome transfer into cytoplasm of newly ovulated oocytes could largely correct these defects. The converse experiment, the transfer of genomes from newly ovulated oocytes into aged cytoplasm, showed that these abnormalities are induced by the postovulatory aged cytoplasm.
Human embryos frequently fail to develop to high-quality blastocysts, a significant obstacle to IVF treatments. It is possible that genome transfer, as done for preventing transmission of mitochondrial disease (Craven et al., 2010, Paull et al., 2012, Tachibana et al., 2013), might also be useful to improve fertility treatments. Although such experiments on human oocytes are yet to be conducted, developmental potential can be improved after transfer from aged to young mouse oocytes. We transferred oocyte genomes from mice aged 10–12 months (reproductive aging) into oocytes of young mice aged 6–8 weeks, and found that 50% (15/30) of 12m8w oocytes and 43.5% (17/39) of 8w12m oocytes reached blastocyst stage, whereas 10.2% (16/156) of reproductively aged oocytes developed. These results suggest that reproductive aging affects both genome and the cytoplasm. both genome and cytoplasm are damaged during reproductive aging. Consistent with our results, Mitsui et al. (2009) transferred oocyte genomes from mice aged 10–12 months into oocytes of young mice aged 3–5 months, and found that development to term increased from 6.3% for in vivo aged oocytes to 27.1% for the reconstructed oocytes.
Although the mechanisms of in vitro postovulatory aging and in vivo reproductive aging differ, there are some remarkable cytological similarities relevant to human reproduction. It has been reported that aging of human and mouse oocytes can result in disorganization of the spindle and defects in chromosome alignment (Battaglia et al., 1996), including increased kinetochore distance (Merriman et al., 2012). Moreover, the frequent fragmentation seen in our postovulatory aged oocytes is reminiscent of the frequent fragmentation observed in human preimplantation embryos. Fragmentation is a common feature of human embryos (Antczak and Van Blerkom, 1999, Hardy, 1999), and is reported to correlate with low implantation rate (Alikani et al., 1999) and low live-birth rate (Luke et al., 2014, Racowsky et al., 2011), as well as with chromosomal abnormalities (Chavez et al., 2012) and apoptosis (Hardy, 1999). The molecular mechanisms leading to fragmentation are unclear; our data show that ectopic localization of cyclin B1 to cytoplasmic clusters is likely responsible for the frequent fragmentation of postovulatory aged oocytes. Thus genome transfer may be useful to restore developmental potential of oocytes for patients with high embryo fragmentation. Additional experiments are required to address these questions in human oocytes.
Experimental Procedures
Collection and Manipulation of Oocytes
Female B6D2F1 mice were superovulated at 6–8 weeks and 12 months by injection with 5 IU of pregnant mare serum gonadotropin (Sigma-Aldrich), followed 46–48 hr later by 5 IU of human chorionic gonadotropin (hCG, Sigma-Aldrich). Following the removal of cumulus cells by incubation in GMOPSplus (Vitrolife) containing 300 μg/mL hyaluronidase (Sigma-Aldrich), oocytes were washed thoroughly in synthetic oviductal medium enriched with potassium (EmbryoMax KSOM medium [1×] with 1/2 amino acids; Millipore) and then selected for collection based on morphology. The postovulatory aged oocytes were then incubated in KSOMaa at 37°C in an atmosphere of 95% air/5% CO2 for 15 hr and were used for subsequent experiments.
Oocyte Activation
Oocytes were activated using 3 μM ionomycin in CZB medium for 5 min at 37°C, followed by culture in 10 μM puromycin, 2 μM 6-DMAP, and 5 μg/mL cytochalasin B for 3–4 hr. Activated constructs were cultured in KSOMaa medium. Oocytes at 20 hr post ovulation, 5G20C, 12-month aged oocytes, and 8w12m oocytes were placed into ionomycin containing medium twice, 1 hr apart, because one-time ionomycin treatment could activate only 25% of 20-hr oocytes (14/56 oocytes [four experiments]) or 37% of 5G20C oocytes (25/68 oocytes [six experiments]). After activation, 27% of 20-hr post oocyte retrieval oocytes (128/474) and 39% of 5G20C oocytes (128/327) did not form a pronucleus. On the contrary, 99% of 5-hr post oocyte retrieval oocytes (1,027/1,036) and 93% of 20-hr nucleus to 5-hr cytoplasm (299/321) oocytes (20G5C oocytes) formed a pronucleus, and thus were successfully activated. Culture was performed in an MINC incubator fed with gas containing 6% CO2, 5% oxygen, and 89% nitrogen at a flow rate of 20 mL/min.
Nuclear Transfer of Oocytes
Oocytes were placed in separate drops of GMOPSplus containing 5 mg/mL cytochalasin B (Sigma) covered with mineral oil (Irvine Scientific). Karyoplasts were aspirated into a pipette with a diameter of 20 mm (Humagen) after incubation in medium containing cytochalasin B for 3–5 min. If the karyoplast contained a larger amount of cytoplasm, the extra cytoplasm was removed by pressing the cytoplasm against the zona pellucida. Karyoplasts were inserted below the zona pellucida of another enucleated oocyte, and fused using either inactivated Sendai virus HVJ-E (GenomeOne, Cosmo Bio), diluted with fusion buffer 1:40, or direct injection, performed in cell fusion medium (0.26 M mannitol, 0.1 mM MgSO4, 0.05% BSA, and 0.5 mM HEPES) using two to eight fusion pulses of 20 μs width and 1.3 V cm2 strength (LF201, NEPA Gene) (Paull et al., 2012). All manipulations were performed on a 37°C heated stage (Tokai Hit) of a Nikon TE 2000U inverted microscope, using Narishige micromanipulators.
In Vitro Fertilization, Embryo Culture, and Embryo Transfer
IVF was performed as described by Nagy et al. (2003). In brief, oocytes were collected from hormone-stimulated females 13 hr after injection with hCG. Sperm was obtained from the cauda epididymis of DBA2 and incubated at 37°C for 15–30 min in HTF medium (Kitazato) supplemented with 5% fetal bovine serum (FBS) before the addition of the oocytes. During the IVF procedure, to eliminate the negative effect of zona pellucida hardening in the aging process on the fertilization (Wolf and Hamada, 1976) and also considering that manipulation itself introduces a hole into the zona pellucida, we opened all of the zona pellucida of oocytes for fertilization using a Xyclone laser. The presence of pronuclei was scored 8 hr after the initiation of the IVF reaction, and zygotes with two pronuclei were selected for uterine transfer. Fertilized eggs were cultured in KSOMaa medium for 4 days, and examined and photographed daily. Blastocysts were transferred to the uterus of day-2.5 pseudopregnant ICR females. Cesarean section was performed on embryonic day 19.5. Surviving pups were fostered to an ICR foster mother that had given birth either on the same day or 1–4 days earlier.
Blastocyst Outgrowth and Embryonic Stem Cell Isolation
For the derivation of mouse ESCs, blastocysts were plated on irradiated mouse embryonic fibroblast feeder layers in mouse ES medium containing the mitogen-activated protein kinase inhibitor PD98059 (Cell Signaling) and LIF (Chemicon) according to a standard procedure (Nagy et al., 2003). Zona pellucidae of blastocysts at day 4 post activation were removed using acidic Tyrode’s solution (Sigma). The blastocysts were cultured individually in the ES medium on gelatinized chamber slides at 37°C in an atmosphere of 5% CO2.
ESC Chromosomal Counting
ESCs were treated with trypsin and incubated in 0.56% (w/v) KCl, stained with Hoechst 33342, and fixed with a 1:3 mixture of glacial acetic acid and methanol. Chromosomal spreads of ESCs were stained with DRAQ5 (1:2,000) and counted for each cell line.
Ethical Approval for the Use of Animals
All experiments were conducted in accordance with the Laboratory Animal Care and Use Committee of Columbia University. Animal work was approved by the Columbia Institutional Animal Care and Use Committee.
Immunocytochemistry of Oocytes
Oocytes were fixed in 3% paraformaldehyde (PFA) and 0.125% Triton X-100 for survivin or cyclin B1 staining, or in 2% PFA and 0.25% Triton X-100 for centromere staining for 10 min at room temperature, permeabilized in PBS with 0.5% Triton X-100 for 30 min, and blocked in blocking solution consisting of PBS with 10% FBS and 0.05% sodium azide for 1 hr at room temperature. Immunocytochemical staining was performed by incubating in primary antibody at 4°C in blocking solution overnight, followed by secondary conjugated antibodies in blocking buffer at room temperature for 1 hr. The cellular DNA was stained with Hoechst 33342 for 5–10 min and used for confocal imaging. β-Tubulin antibody (Millipore, 05-661; Sigma, 118K4843), phospho-histone 3 antibody (Millipore, MABE76), survivin antibody (Cell Signaling, 71G4b7), and cyclin B1 (Abcam, ab72) at a concentration of 1:1,000, and anti-centromere protein antibody (Cell Signaling, 15–235) at a concentration of 1:100 were used. The appropriate secondary antibodies (immunoglobulin G [IgG]) (Molecular Probes/Invitrogen) were diluted at 1:1,000; goat anti-human (A-11013) or donkey anti-rat (A-20218) IgG conjugated with Alexa Fluor 488, donkey anti-rabbit IgG conjugated with Alexa Fluor 555 (A-31572), donkey anti-rat IgG conjugated with Alexa Fluor 594 (A-21209), and donkey anti-mouse IgG conjugated with Alexa Fluor 647 (A-31571). The cells were washed and viewed by laser confocal microscopy (LSM510, Carl Zeiss). All samples were processed simultaneously.
Immunoblot Analysis
An amount of extracted protein corresponding to 100 oocytes was solubilized in sample buffer solution without 2-mercaptoethanol (Nacalai Tesque), loaded per lane of the NuPAGE Novex Tris-Acetate Mini Gels (Invitrogen), and transferred to Immobilon-P transfer membrane (Millipore). The membrane was soaked in skim milk containing TBST (Tris-buffered saline with 0.1% Tween 20) for 30 min at room temperature before an overnight incubation at 4°C with primary antibody, also diluted in blocking solution. The anti-cyclin B1 antibody (BD Pharmingen, 554,79) and anti-actin antibody (Abcam, ab8227) were used at 1:300 dilution. The membrane was then washed three times with TBST, incubated with a horseradish peroxidase-conjugated secondary anti-mouse (Cell Signaling Technology, 7076S) or anti-rabbit (7074S) antibody at 1:1,000 dilution directed against the primary antibody for 60 min, and washed three times with TBST. The signal was detected by enhanced chemiluminescence (SuperSignal West Dura Extended Duration Substrate, Thermo Scientific) following the manufacturer’s recommendations.
Statistical Analysis
Either a chi-square test or a one-way ANOVA with Bonferroni’s multiple comparison testing was used for statistical analysis. Statistical analyses were performed using GraphPad Prism software (GraphPad). p < 0.05 was considered to indicate statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 in the figures).
Author Contributions
Nuclear transfer was performed by D.E. Oocyte activation, microscopic observation, immunostaining, immunoblot, data collection, and statistical analysis were performed by M.Y. D.E. and M.Y. designed experiments, interpreted data, and wrote the paper.
Acknowledgments
This research was supported the NYSCF-Robertson award to D.E., and laboratory startup funds to D.E. from Columbia University. M.Y. is supported by the New York Stem Cell Foundation and in part by the Uehara Memorial Foundation and the Takeda Science Foundation, Japan. D.E. is an NYSCF-Robertson Investigator. The authors would like to thank Chuang-Chen Lin for carrying out embryo transfer, and Lauren Bauer Vensand, Carmen Dusenberry, Travis Kroeker, and Brandon Pearl for expert laboratory support.
Published: February 23, 2017
Footnotes
Supplemental Information includes two figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.01.020.
Supplemental Information
References
- Alikani M., Cohen J., Tomkin G., Garrisi G.J., Mack C., Scott R.T. Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil. Steril. 1999;71:836–842. doi: 10.1016/s0015-0282(99)00092-8. [DOI] [PubMed] [Google Scholar]
- Antczak M., Van Blerkom J. Temporal and spatial aspects of fragmentation in early human embryos: possible effects on developmental competence and association with the differential elimination of regulatory proteins from polarized domains. Hum. Reprod. 1999;14:429–447. doi: 10.1093/humrep/14.2.429. [DOI] [PubMed] [Google Scholar]
- Bai Z.D., Liu K., Wang X.Y. Developmental potential of aged oocyte rescued by nuclear transfer following parthenogenetic activation and in vitro fertilization. Mol. Reprod. Dev. 2006;73:1448–1453. doi: 10.1002/mrd.20538. [DOI] [PubMed] [Google Scholar]
- Battaglia D.E., Goodwin P., Klein N.A., Soules M.R. Influence of maternal age on meiotic spindle assembly in oocytes from naturally cycling women. Hum. Reprod. 1996;11:2217–2222. doi: 10.1093/oxfordjournals.humrep.a019080. [DOI] [PubMed] [Google Scholar]
- Chavez S.L., Loewke K.E., Han J., Moussavi F., Colls P., Munne S., Behr B., Reijo Pera R.A. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat. Commun. 2012;3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Kattera S. Rescue ICSI of oocytes that failed to extrude the second polar body 6 h post-insemination in conventional IVF. Hum. Reprod. 2003;18:2118–2121. doi: 10.1093/humrep/deg325. [DOI] [PubMed] [Google Scholar]
- Craven L., Tuppen H.A., Greggains G.D., Harbottle S.J., Murphy J.L., Cree L.M., Murdoch A.P., Chinnery P.F., Taylor R.W., Lightowlers R.N. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Birkhoff G., Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat. Rev. Mol. Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy K. Apoptosis in the human embryo. Rev. Reprod. 1999;4:125–134. doi: 10.1530/ror.0.0040125. [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Krol M., Polanski Z. Spindle assembly checkpoint-related meiotic defect in oocytes from LT/Sv mice has cytoplasmic origin and diminishes in older females. Reproduction. 2012;144:331–338. doi: 10.1530/REP-11-0362. [DOI] [PubMed] [Google Scholar]
- Lister L.M., Kouznetsova A., Hyslop L.A., Kalleas D., Pace S.L., Barel J.C., Nathan A., Floros V., Adelfalk C., Watanabe Y. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr. Biol. 2010;20:1511–1521. doi: 10.1016/j.cub.2010.08.023. [DOI] [PubMed] [Google Scholar]
- Luke B., Brown M.B., Stern J.E., Jindal S.K., Racowsky C., Ball G.D. Using the Society for Assisted Reproductive Technology Clinic Outcome System morphological measures to predict live birth after assisted reproductive technology. Fertil. Steril. 2014;102:1338–1344. doi: 10.1016/j.fertnstert.2014.07.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriman J.A., Jennings P.C., McLaughlin E.A., Jones K.T. Effect of aging on superovulation efficiency, aneuploidy rates, and sister chromatid cohesion in mice aged up to 15 months. Biol. Reprod. 2012;86:49. doi: 10.1095/biolreprod.111.095711. [DOI] [PubMed] [Google Scholar]
- Mitsui A., Yoshizawa M., Matsumoto H., Fukui E. Improvement of embryonic development and production of offspring by transferring meiosis-II chromosomes of senescent mouse oocytes into cytoplasts of young mouse oocytes. J. Assist. Reprod. Genet. 2009;26:35–39. doi: 10.1007/s10815-008-9282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton P.C., Yoder C.S., Tucker M.J., Wright G., Brockman W.D., Kort H.I. Reinsemination by intracytoplasmic sperm injection of 1-day-old oocytes after complete conventional fertilization failure. Fertil. Steril. 1997;68:488–491. doi: 10.1016/s0015-0282(97)00223-9. [DOI] [PubMed] [Google Scholar]
- Nagy Z.P., Joris H., Liu J., Staessen C., Devroey P., Van Steirteghem A.C. Intracytoplasmic single sperm injection of 1-day-old unfertilized human oocytes. Hum. Reprod. 1993;8:2180–2184. doi: 10.1093/oxfordjournals.humrep.a138000. [DOI] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. Third Edition. Cold Spring Harbor Laboratory; 2003. Manipulating the Mouse Embryo: A Laboratory Manual. [Google Scholar]
- Paull D., Emmanuele V., Weiss K.A., Treff N., Stewart L., Hua H., Zimmer M., Kahler D.J., Goland R.S., Noggle S.A. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature. 2012;493:632–637. doi: 10.1038/nature11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prietl G., van der Ven H., Krebs D. Artificial insemination: noninvasive management of subfertile couples. In: Rabe H., Diedrich K., Strowitzki T., editors. Manual on Assisted Reproduction. Second Edition. Springer; 2001. [Google Scholar]
- Racowsky C., Stern J.E., Gibbons W.E., Behr B., Pomeroy K.O., Biggers J.D. National collection of embryo morphology data into Society for Assisted Reproductive Technology Clinic Outcomes Reporting System: associations among day 3 cell number, fragmentation and blastomere asymmetry, and live birth rate. Fertil. Steril. 2011;95:1985–1989. doi: 10.1016/j.fertnstert.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Tachibana M., Amato P., Sparman M., Woodward J., Sanchis D.M., Ma H., Gutierrez N.M., Tippner-Hedges R., Kang E., Lee H.S. Towards germline gene therapy of inherited mitochondrial diseases. Nature. 2013;493:627–631. doi: 10.1038/nature11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatone C., Di Emidio G., Barbaro R., Vento M., Ciriminna R., Artini P.G. Effects of reproductive aging and postovulatory aging on the maintenance of biological competence after oocyte vitrification: insights from the mouse model. Theriogenology. 2011;76:864–873. doi: 10.1016/j.theriogenology.2011.04.017. [DOI] [PubMed] [Google Scholar]
- Temme A., Rieger M., Reber F., Lindemann D., Weigle B., Diestelkoetter-Bachert P., Ehninger G., Tatsuka M., Terada Y., Rieber E.P. Localization, dynamics, and function of survivin revealed by expression of functional survivinDsRed fusion proteins in the living cell. Mol. Biol. Cell. 2003;14:78–92. doi: 10.1091/mbc.E02-04-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waal M.S., Hengeveld R.C., van der Horst A., Lens S.M. Cell division control by the chromosomal passenger complex. Exp. Cell Res. 2012;318:1407–1420. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Wakayama S., Thuan N.V., Kishigami S., Ohta H., Mizutani E., Hikichi T., Miyake M., Wakayama T. Production of offspring from one-day-old oocytes stored at room temperature. J. Reprod. Dev. 2004;50:627–637. doi: 10.1262/jrd.50.627. [DOI] [PubMed] [Google Scholar]
- Wolf D.P., Hamada M. Age-dependent losses in the penetrability of mouse eggs. J. Reprod. Fertil. 1976;48:213–214. doi: 10.1530/jrf.0.0480213. [DOI] [PubMed] [Google Scholar]
- Yuzpe A.A., Liu Z., Fluker M.R. Rescue intracytoplasmic sperm injection (ICSI)-salvaging in vitro fertilization (IVF) cycles after total or near-total fertilization failure. Fertil. Steril. 2000;73:1115–1119. doi: 10.1016/s0015-0282(00)00522-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.