Abstract
Dermal papilla cells (DPCs) are located at the base of hair follicles, and are known to induce hair follicle regeneration. Platelet-rich plasma (PRP) functions in hair follicle regeneration. To investigate the influence of PRP on DPCs, the present study analyzed RNA-seq data of human hair dermal papilla cells (HHDPCs) that were treated or untreated by PRP. The data included in the RNA-seq were from two normal and two treated HHDPC samples. Following identification by Cuffdiff software, differentially expressed genes (DEGs) underwent enrichment analyses, and protein-protein interaction networks were constructed using Cytoscape software. Additionally, transcription factor (TF)-DEG and TF-long non-coding RNA (lncRNA) regulatory networks were constructed. A total of 178 differentially expressed lncRNA were screened, 365 were upregulated and 142 were downregulated. Notably, upregulated cyclin dependent kinase 1 (CDK1) (degree=76), polo-like kinase 1 (PLK1) (degree=65), cell division cycle 20 (degree=50), cyclin B1 (degree=49), aurora kinase B (degree=47), cyclin dependent kinase 2 (degree=46) and downregulated v-myc avian myelocytomatosis viral oncogene homolog (MYC) (degree=12) had higher degrees in networks. In addition, CCAAT/enhancer binding protein β, E2F transcription factor 1 (E2F1), early growth response 1 and MYC may be key TFs for their target genes, and were enriched in pathways associated with the cell cycle. They may also be involved in cell proliferation via various interactions with other genes, for example CDK1-PLK1 and E2F1→CDK1. These dysregulated genes induced by PRP may affect proliferation of HHDPCs.
Keywords: human dermal papilla cells, platelet-rich plasma, differentially expressed genes, protein-protein interaction network, regulatory network
Introduction
As a type of dermal cell located in the base of the hair follicle, dermal papilla cells (DPCs) have the ability to induce hair follicle regeneration and hair growth (1). Platelet-rich plasma (PRP) is plasma enriched with a higher proportion of platelets, compared with that with that which is usually found in whole blood. α-granules in platelets contain growth factors, indicating that PRP has the ability to promote cell proliferation and differentiation (2). Previous studies have reported that PRP may induce mesenchymal stem cell proliferation and chondrogenic differentiation in vitro (3–5). It has been reported that PRP is essential in hair follicle regeneration (6). Thus, it was hypothesized that PRP may contribute to hair follicle regeneration via changes in DPC proliferation levels. Hair follicle regeneration is currently known to have an impact in treating alopecia and dermal wounds (7,8). The initial generation of a hair follicle is intimately linked with signal exchange between mesenchymal and epithelial cells via the formation of hair placodes (9,10). Alopecia is defined as a loss of hair from the body or head, and may be induced by nutritional deficiencies, fungal infection and traumatic damage (11). Increased chronic ulcers, skin disease, trauma caused by burns and accidental skin defects mean the restoration of dermal wounds is becoming an important medical concern (12). Thus, it is important to investigate the influence of PRP on DPC and develop therapeutic strategies for hair follicle regeneration.
Previous studies have investigated mechanisms of hair follicle regeneration. For example, S100 calcium binding protein A4 and S100 calcium binding protein A6 may be important in activating stem cells at the beginning of follicle regeneration (13,14). Wingless-type mouse mammary tumor virus integration site family member 10b promotes hair follicle growth and regeneration via activation of the canonical Wnt signaling pathway and, thus, may be used as therapeutic target in the treatment of hair follicle-associated diseases (15,16). Via the Gpr44 receptor, prostaglandin D2 may function in inhibiting hair follicle regeneration (17). As a crucial ATPase of the BAF chromatin-remodeling complex, brahma-related gene 1 may regulate the processes of epidermal repair and hair regeneration in bulge stem cells (18).
Preliminary studies demonstrated that PRP at a concentration of 5% should be used to treat human hair DPCs (HHDPCs) in the present study (data not shown). Using RNA-seq data of HHDPCs from normal samples and samples treated with 5% PRP, the present study aimed to identify differentially expressed genes (DEGs) and predict their possible function using Gene Ontology (GO) and pathway enrichment analyses. The interactions and associations between the DEGs were investigated using protein-protein interaction (PPI) networks. Furthermore, regulatory networks were constructed to screen key genes and transcription factors (TFs).
Materials and methods
PRP preparation
Samples of whole blood (10 ml) were taken from the median cubital vein of each of the 8 male, healthy participants (mean age, 24.9 years) and mixed with 3.2% sodium citrate (vol/vol=10:1). PRP was prepared using a two-step centrifugation method. Firstly, the whole blood was centrifuged at 400 × g for 10 min at room temperature, allowing separation of blood into three layers, the topmost platelet-poor plasma layer, an intermediate PRP layer and the bottommost red blood cell layer. Subsequently, the upper two layers were centrifuged again at 3,800 × g for 10 min at room temperature. The platelets in PRP were activated by 0.2 ml 10% CaCl2 and 1,000 U bovine thrombin. After a 10 min incubation, PRP was centrifuged at 1,500 × g for 5 min at room temperature, and the supernatant was stored at −80°C. All participants provided informed consent, and the present study was approved by the ethics committee of the Hangzhou First People's Hospital (Hangzhou, China).
HHDPCs cultivation
At 37°C in a humidified 5% CO2 incubator (Thermo Fisher Scientific, Inc., Waltham, MA, USA), the HHDPCs (Shanghai Hu Zheng Industrial Co., Ltd., Shanghai, China) were cultivated in a medium consisting of 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) and 1% double antibody (Gibco; Thermo Fisher Scientific, Inc.). HHDPCs were passaged at 80–90% confluence and pancreatin was used for digestion (Gibco; Thermo Fisher Scientific, Inc.). Subsequently, the cells were centrifuged at 300 × g for 5 min at room temperature and the supernatant was removed. HHDPCs were preserved in a frozen stock solution which consisted of 10% dimethyl sulfoxide, 40% FBS and 50% RPMI-1640.
RNA extraction and RNA-seq library construction
Following cell counting, HHDPCs were spread on 6-well plates (Applied Biosystems; Thermo Fisher Scientific, Inc.; 2×105 cells/well) and starved for 24 h. HHDPCs in the treatment group were treated with 5% PRP whilst the HHDPCs in the control group were treated with RPMI-1640 medium containing 10% FBS and 1% penicillin-streptomycin solution. Each experiment was repeated twice. Total RNA from HHDPC treatment samples and normal HHDPC samples was extracted using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. The purity and integrity of total RNA were checked by a spectrophotometer (Merinton Instrument, Ltd., Beijing, China) and 2% agarose gel electrophoresis. Following this, the RNA-seq library was prepared using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs, Inc. Ipswich, MA, USA) according to the manufacturer's protocols. Briefly, mRNAs were isolated and divided into ~200 nt fragments. Subsequently, double-stranded cDNAs were synthesized and modified, and DNA cluster amplification was performed using a Phusion® Human Specimen Direct Polymerase Chain Reaction kit (Thermo Fisher Scientific, Inc.). The reaction mixture was subjected to the following cycling conditions: An initial denaturation step at 94°C for 30 sec, followed by 11 cycles of denaturation at 98°C for 10 sec, annealing at 65°C for 30 sec and extension at 72°C for 30 sec, and a final extension step at 72°C for 5 min. Using Illumina HiSeq 2500 v4 100PE (Illumina, Inc., San Diego, CA, USA), high-throughput sequencing was conducted for the RNA-seq library.
DEGs screening
RNA-seq data were preprocessed by the Next Generation Sequencing Quality Control Toolkit (19). Any sequences containing >20% bases with a quality value <20 were filtered out. Using TopHat2 software (ccb.jhu.edu/software/tophat/index.shtml) (20), RNA-seq data was aligned to human genome hg19, which was downloaded from the University of California, Santa Cruz website (genome.ucsc.edu) (21). The parameter was set as no-mixed and other parameters used default settings. Cuffdiff software (cufflinks.cbcb.umd.edu/) (22) was used to identify the DEGs between RNA-seq data of normal HHDPCs and treated HHDPCs. The adjusted P<0.05 and |log2fold change (FC)|>1 served as the cut-off criteria.
Functional and pathway enrichment analysis
GO analysis aims to describe subcellular location, molecular function and the biological processes of gene products (23). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database introduces functions of molecules or genes (24). Using the GOFunction (25) of bioconductor (www.bioconductor.org/), as well as annotation files GO.db (26) and org.Hs.eg.db (27), GO and KEGG pathway enrichment analyses were performed for the DEGs. P<0.05 and ≥2 genes enriched in one pathway served as the cut-off criteria.
PPI network construction
The STRING online software (28) was applied to screen interactions of proteins encoded by DEGs, and the Cytoscape software (www.cytoscape.org/) (29) was used to visualize the PPI network. A combined score >0.7 was used as the cut-off criterion.
Regulatory network construction
Using information provided by the ENCODE website (genome.ucsc.edu/ENCODE/encode.hg17.html) regarding TF binding sites, (30), in addition to genetic and long non-coding RNAs (lncRNAs) location information on the genome, differentially expressed TF-DEG pairs and differentially expressed TF-differentially expressed lncRNA pairs were screened. A certain amount of overlapping occurred between TF binding sites and the cut-off criterion was defined as 1,000 bp upstream to 500 bp downstream of the transcription start site.
Results
DEGs analysis
Compared with normal HHDPC samples, there were 365 upregulated and 142 downregulated genes screened in the treated HHDPC samples. Furthermore, 178 differentially expressed (including 131 upregulated and 47 downregulated) lncRNAs were identified in the treated HHDPC samples.
Functional and pathway enrichment analysis
The enriched KEGG pathways for upregulated genes were listed in Table I, including cell cycle (P<0.0001), progesterone-mediated oocyte maturation (P<0.0001) and DNA replication (P=0.000614). The enriched KEGG pathways for downregulated genes included p53 signaling pathway (P<0.0001), mitogen-activated protein kinase signaling pathway (P=0.00709) and cell cycle (P=0.020687; Table II). In addition, no GO functions were enriched for the DEGs.
Table I.
Pathways enriched for upregulated genes.
| Category | Term | Description | Gene number | Gene symbol | P-value |
|---|---|---|---|---|---|
| KEGG | 4110 | Cell cycle | 25 | BUB1, BUB1B | <0.0001 |
| KEGG | 4914 | Progesterone-mediated oocyte maturation | 12 | CCNA2, CCNB1 | 8.13×10−8 |
| KEGG | 4114 | Oocyte meiosis | 13 | AURKA, BUB1 | 2.12×10−7 |
| KEGG | 5130 | Pathogenic Escherichia coli infection | 8 | ACTB, ACTG1 | 1.10×10−5 |
| KEGG | 4115 | p53 signaling pathway | 8 | CCNB1, CCNB2 | 4.72×10−5 |
| KEGG | 4145 | Phagosome | 11 | ACTB, ACTG1 | 0.000182 |
| KEGG | 4540 | Gap junction | 8 | CDK1, PDGFRB | 0.000346 |
| KEGG | 3030 | DNA replication | 5 | FEN1, MCM3 | 0.000614 |
| KEGG | 4974 | Protein digestion and absorption | 7 | COL18A1, COL2A1 | 0.000971 |
| KEGG | 4510 | Focal adhesion | 11 | ACTB, ACTG1 | 0.001748 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; BUB1, BUB1 mitotic checkpoint serine/threonine kinase; BUB1B, BUB1 mitotic checkpoint serine/threonine kinase B; CCNA2, cyclin A2; CCNB1, cyclin B1; AURKA, aurora kinase A; ACTB, β-actin; ACTG1, γ-actin 1; CCNB2, cyclin B2; CDK1, cyclin dependent kinase 1; PDGFRB, platelet derived growth factor receptor β; FEN1, flap structure-specific endonuclease 1; MCM3, mini chromosome maintenance complex component 3; COL18A1, collagen type XVIII α1 chain; COL2A1, collagen type II α 1 chain.
Table II.
Pathways enriched for downregulated genes.
| Category | Term | Description | Gene number | Gene symbol | P-value |
|---|---|---|---|---|---|
| KEGG | 4115 | p53 signaling pathway | 6 | BBC3, CCND2 | <0.0001 |
| KEGG | 250 | Alanine, aspartate and glutamate metabolism | 3 | ASNS, GLS2, GOT1 | 0.002425 |
| KEGG | 5219 | Bladder cancer | 3 | CDKN1A, MYC, VEGFA | 0.005288 |
| KEGG | 4010 | MAPK signaling pathway | 7 | DDIT3, DUSP2 | 0.00709 |
| KEGG | 330 | Arginine and proline metabolism | 3 | GLS2, GOT1, SAT1 | 0.010639 |
| KEGG | 910 | Nitrogen metabolism | 2 | ASNS, GLS2 | 0.016056 |
| KEGG | 4964 | Proximal tubule bicarbonate reclamation | 2 | GLS2, PCK2 | 0.016056 |
| KEGG | 4110 | Cell cycle | 4 | CCND2, CDKN1A | 0.020687 |
| KEGG | 4612 | Antigen processing and presentation | 3 | CIITA, HLA-DQA1, HLA-DQA2 | 0.026471 |
| KEGG | 5310 | Asthma | 2 | HLA-DQA1, HLA-DQA2 | 0.026583 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; BBC3, BCL2 binding component 3; CCND2, cyclin D2; ASNS, asparagine synthetase (glutamine-hydrolyzing); GLS2, glutaminase 2; GOT1, glutamic-oxaloacetic transaminase 1; CDKN1A, cyclin dependent kinase inhibitor 1A; MYC, v-myc avian myelocytomatosis viral oncogene homolog; VEGFA, vascular endothelial growth factor A; DDIT3, DNA damage inducible transcript 3; DUSP2, dual specificity phosphatase 2; SAT1, spermidine/spermine N1-acetyltransferase 1; PCK2, phosphoenolpyruvate carboxykinase, mitochondrial; CIITA, class II major histocompatibility complex transactivator; HLA-DQA1, major histocompatibility complex, class II, DQ α 1; HLA-DQA2, major histocompatibility complex, class II, DQ α 2; MAPK, mitogen-activated protein kinase.
PPI network analysis
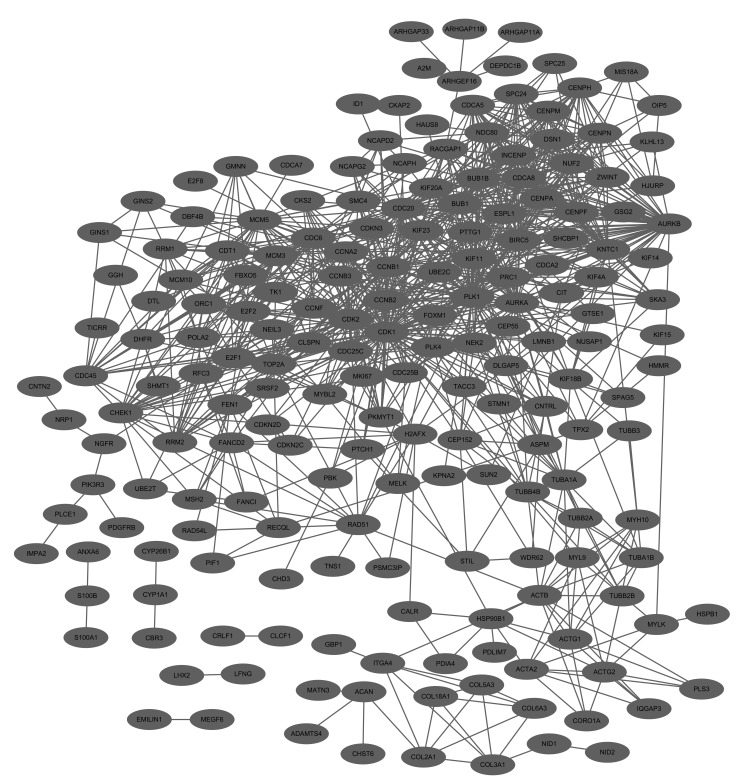
The PPI network for upregulated genes had 192 nodes and 1,017 interactions (Fig. 1). In this network, cyclin-dependent kinase 1 (CDK1, degree=76), polo-like kinase 1 (PLK1, degree=65), cell division cycle 20 (CDC20, degree=50), cyclin B1 (CCNB1, degree=49), aurora kinase B (AURKB, degree=47) and cyclin-dependent kinase 2 (CDK2, degree=46) demonstrated higher degrees. In addition, these genes interacted with each other for example, CDK1-PLK1, CDC20-CCNB1, AURKB-CDK2 and CDK1-AURKB in the PPI network.
Figure 1.
Protein-protein interaction network for upregulated genes. The gray nodes represent the upregulated genes.
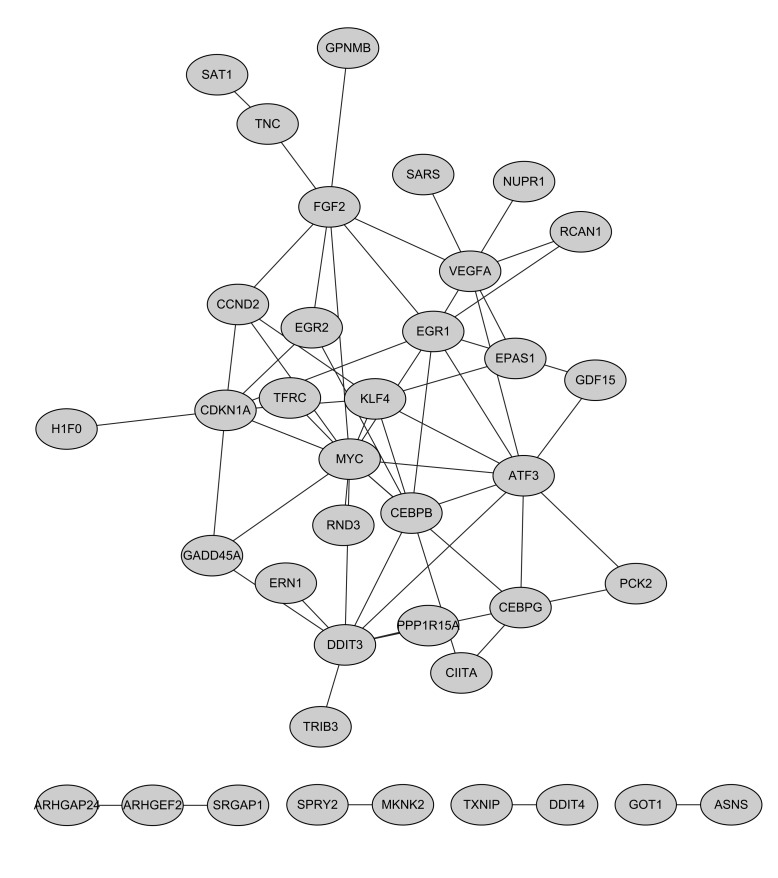
The PPI network for downregulated genes had 38 nodes and 58 interactions (Fig. 2). In this network, c-myc (MYC, degree=12), activating transcription factor 3 (degree=9), DNA-damage-inducible transcript 3 (degree=8) and early growth response 1 (EGR1, degree=8) indicated higher degrees.
Figure 2.
Protein-protein interaction network for downregulated genes. The gray nodes indicate the downregulated genes.
Regulatory network analysis
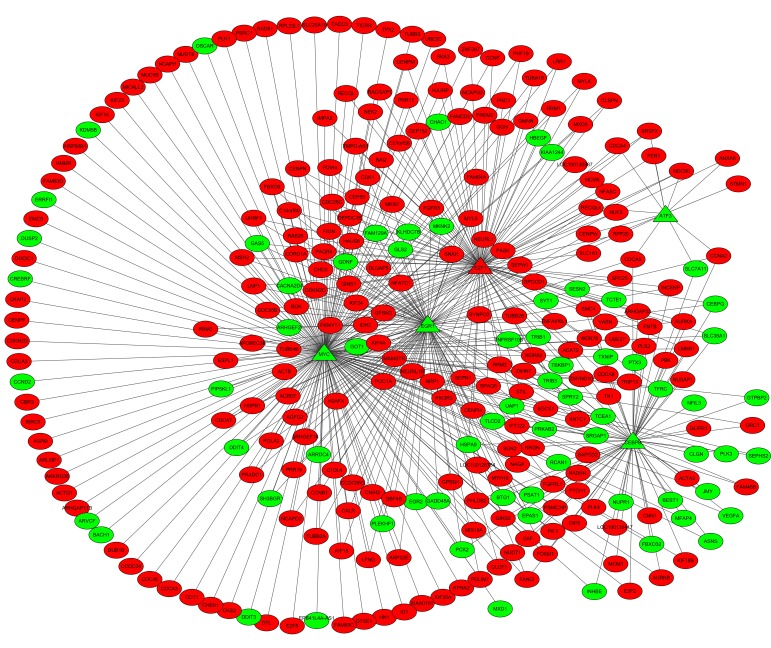
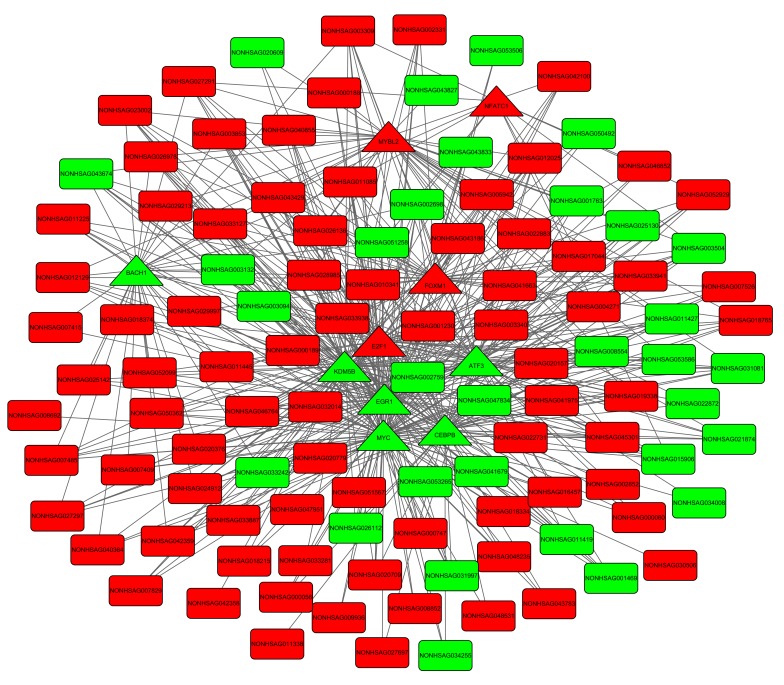
A total of 453 TF-DEG pairs and 530 TF-lncRNA pairs were identified. The TF-DEG and TF-lncRNA regulatory networks are presented in Figs 3 and 4. Notably, the target genes of TFs CCAAT/enhancer binding protein-β (C/EBP-β), E2F transcription factor (E2F) 1, EGR1 and MYC were enriched in pathways associated with the cell cycle. In the TF-DEG regulatory network, these TFs targeted CDK1, PLK1, CCNB1 and AURKB (for example, MYC→PLK1, MYC→CCNB1, MYC→AURKB, CEBPB→AURKB, E2F1→CDK1 and E2F1→CCNB1).
Figure 3.
TF-DEG regulatory network. The red and green nodes represent upregulation and downregulation, respectively. The circles and triangles indicate for DEGs and TFs, respectively. TF, transcription factor; DEG, differentially expressed gene.
Figure 4.
TF-lncRNA regulatory network. The red and green nodes represent upregulation and downregulation, respectively. The circles and rectangles indicate TFs and lncRNAs, respectively. TF, transcription factor; lncRNA, long non-coding RNA.
Discussion
The present study screened 178 differentially expressed lncRNA, 365 upregulated genes and 142 downregulated genes in treated HHDPC samples compared with normal HHDPC samples. Only selected DEGs and lncRNAs have been presented in the present study. DEGs enriched in pathways have been listed in Tables I and II. Pathway enrichment indicated that these upregulated and downregulated genes were enriched in proliferation-associated pathways, including those involved in the cell cycle and DNA replication, indicating that PRP may affect cell proliferation of HHDPCs. The DEGs and lncRNAs that were key nodes in the networks have been mentioned. In the PPI networks, upregulated CDK1 (degree=76), PLK1 (degree=65), CDC20 (degree=50), CCNB1 (degree=49), AURKB (degree= 47), CDK2 (degree=46) and downregulated MYC (degree=12) had higher degrees. Furthermore, CEBPB, E2F1, EGR1 and MYC, which may be key TFs for their target genes, were enriched in pathways associated with the cell cycle.
CDK1 activity is continuously required to ensure continuing end resection and to maintain a double-strand break-induced checkpoint. Furthermore, CDK1 is involved in later stages of homologous recombination (31). It has been demonstrated that CDK1 is the only crucial cell cycle CDK and is able to drive cell division alone (32). As an important moderator of cell division (33), PLK1 is essential for mitotic entry at the appropriate time (34). PLK1 appears to be required for centrosome-mediated microtubule activities and spindle assembly. In addition, siRNAs targeted against PLK1 may be potential anti-proliferative agents that inhibit neoplastic cells (35,36). This suggests that CDK1 and PLK1 may be associated with cell proliferation.
CDC20 can activate anaphase-promoting complex (APC/C) in metaphase of the cell cycle, and its C terminus contains a WD40 repeat domain that regulates protein-protein interactions (37,38). Suppression of CCNB1, which is a cyclin controlling mitotic entry, induces a marked arrest of G2/M phase in HCT116 and SW480 cells, preventing the expression of CDK1 and cell division cycle 25C (39,40). AURKB functions in chromosome bi-orientation, spindle assembly and cytokinesis, and may be degraded by APC/C bound to cadherin 1 in G1 and in late mitosis (41). In animal cells, CDK2/cyclin E can trigger initiation of centrosome duplication by targeting substrate nucleophosmin/B23 (42–44). Thus, the expression levels of CDC20, CCNB1, AURKB and CDK2 may be associated with cell proliferation. In the PPI network for upregulated DEGs, it was observed that numerous DEGs demonstrated a certain level of interaction with each other, for example, CDK1-PLK1, CDC20-CCNB1, AURKB-CDK2 and CDK1-AURKB. Therefore, it may be other genes, including PLK1 and CCNB, which had interaction with CDC20, CCNB1, AURKB and CDK2, may also function in cell proliferation.
The proto-oncogene MYC encodes transcription factor c-myc which is associated with regulation of cell proliferation and differentiation (45). Conditional activation or constitutive expression of wild-type CEBPB may promote differentiation and suppress proliferation of 32D-BCR/ABL cells, which is contrary to a DNA binding-deficient CEBPB mutant (46). It is reported that E2F1, E2F2 and E2F3 function as transcriptional activators in the process of the G1/S transition (47). As a member of the Egr transcription factor family, EGR1 is involved in cell growth, proliferation and stress responses in various types of tissues (48). This may suggest the expression levels of CEBPB, E2F1, EGR1 and MYC were associated with cell proliferation. In the TF-DEG regulatory network, it was observed that TFs could target CDK1, PLK1, CCNB1 and AURKB, for example MYC→PLK1, MYC→CCNB1, MYC→AURKB, CEBPB→AURKB, E2F1→CDK1 and E2F1→CCNB1, suggesting that TFs may be involved in cell proliferation via regulation of CDK1, PLK1, CCNB1 and AURKB.
In conclusion, the present study conducted a comprehensive bioinformatics analysis of genes that may be associated with cell proliferation. A total of 178 differentially expressed lncRNA were identified, and additionally, 365 upregulated and 142 downregulated genes were screened. It was indicated that CDK1, PLK1, CDC20, CCNB1, AURKB, CDK2, CEBPB, E2F1, EGR1 and MYC may be associated with cell proliferation of HHDPCs. Thus, PRP may be important in HHDPCs. Further investigation is required in order to elucidate the functional mechanisms of these genes in HHDPCs. In addition, the proliferative ability of HHDPCs treated with PRP will be investigated in future work.
Acknowledgements
The present study was supported by the Science and Technology Projects Fund of Hangzhou City (grant no. 20130633B04).
References
- 1.Jahoda C, Horne K, Oliver R. Induction of hair growth by implantation of cultured dermal papilla cells. Nature. 1984;311(1984):560–562. doi: 10.1038/311560a0. [DOI] [PubMed] [Google Scholar]
- 2.Eppley BL, Woodell JE, Higgins J. Platelet quantification and growth factor analysis from platelet-rich plasma: Implications for wound healing. Plast Reconstr Surg. 2004;114:1502–1508. doi: 10.1097/01.prs.0000138251.07040.51. [DOI] [PubMed] [Google Scholar]
- 3.Kocaoemer A, Kern S, Klueter H, Bieback K. Human AB serum and thrombin-activated platelet-rich plasma are suitable alternatives to fetal calf serum for the expansion of mesenchymal stem cells from adipose tissue. Stem Cells. 2007;25:1270–1278. doi: 10.1634/stemcells.2006-0627. [DOI] [PubMed] [Google Scholar]
- 4.Vogel JP, Szalay K, Geiger F, Kramer M, Richter W, Kasten P. Platelet-rich plasma improves expansion of human mesenchymal stem cells and retains differentiation capacity and in vivo bone formation in calcium phosphate ceramics. Platelets. 2006;17:462–469. doi: 10.1080/09537100600758867. [DOI] [PubMed] [Google Scholar]
- 5.Mishra A, Tummala P, King A, Lee B, Kraus M, Tse V, Jacobs CR. Buffered platelet-rich plasma enhances mesenchymal stem cell proliferation and chondrogenic differentiation. Tissue Engineering Part C Methods. 2009;15:431–435. doi: 10.1089/ten.tec.2008.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reese RJ. Autologous platelet rich plasma (PRP): What do we know? Important concepts relevant to hair restoration surgery. Hair Transplant Forum Int. 2010;2010:14–17. [Google Scholar]
- 7.Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, Cotsarelis G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 8.Gilhar A, Kalish RS. Alopecia areata: A tissue specific autoimmune disease of the hair follicle. Autoimmun Rev. 2006;5:64–69. doi: 10.1016/j.autrev.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Barsh G. Of ancient tales and hairless tails. Nat Genet. 1999;22:315–316. doi: 10.1038/11876. [DOI] [PubMed] [Google Scholar]
- 10.Oro AE, Scott MP. Splitting hairs: Dissecting roles of signaling systems in epidermal development. Cell. 1998;95:575–578. doi: 10.1016/S0092-8674(00)81624-4. [DOI] [PubMed] [Google Scholar]
- 11.Kulick D, Hark L, Deen D. The bariatric surgery patient: A growing role for registered dietitians. J Am Diet Assoc. 2010;110:593–599. doi: 10.1016/j.jada.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Oshima H, Inoue H, Matsuzaki K, Tanabe M, Kumagai N. Permanent restoration of human skin treated with cultured epithelium grafting-wound healing by stem cell based tissue engineering. Hum Cell. 2002;15:118–128. doi: 10.1111/j.1749-0774.2002.tb00106.x. [DOI] [PubMed] [Google Scholar]
- 13.Ito M, Kizawa K. Expression of calcium-binding S100 proteins A4 and A6 in regions of the epithelial sac associated with the onset of hair follicle regeneration. J Investig Dermatol. 2001;116:956–963. doi: 10.1046/j.0022-202x.2001.01369.x. [DOI] [PubMed] [Google Scholar]
- 14.Kizawa K, Toyoda M, Ito M, Morohashi M. Aberrantly differentiated cells in benign pilomatrixoma reflect the normal hair follicle: Immunohistochemical analysis of Ca-binding S100A2, S100A3 and S100A6 proteins. Br J Dermatol. 2005;152:314–320. doi: 10.1111/j.1365-2133.2004.06391.x. [DOI] [PubMed] [Google Scholar]
- 15.Li YH, Zhang K, Yang K, Ye JX, Xing YZ, Guo HY, Deng F, Lian XH, Yang T. Adenovirus-mediated Wnt10b overexpression induces hair follicle regeneration. J Investig Dermatol. 2013;133:42–48. doi: 10.1038/jid.2012.235. [DOI] [PubMed] [Google Scholar]
- 16.Li YH, Zhang K, Ye JX, Lian XH, Yang T. Wnt10b promotes growth of hair follicles via a canonical Wnt signalling pathway. Clin Exp Dermatol. 2011;36:534–540. doi: 10.1111/j.1365-2230.2011.04019.x. [DOI] [PubMed] [Google Scholar]
- 17.Nelson AM, Loy DE, Lawson JA, Katseff AS, FitzGerald GA, Garza LA. Prostaglandin D2 inhibits wound-induced hair follicle neogenesis through the receptor, Gpr44. J Investig Dermatol. 2013;133:881–889. doi: 10.1038/jid.2012.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong Y, Li W, Shang C, Chen RM, Han P, Yang J, Stankunas K, Wu B, Pan M, Zhou B, et al. Brg1 governs a positive feedback circuit in the hair follicle for tissue regeneration and repair. Dev Cell. 2013;25:169–181. doi: 10.1016/j.devcel.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Patel RK, Jain M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: Enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Zhou X, Zhu J, Gu Y, Zhao W, Zou J, Guo Z. GO-function: Deriving biologically relevant functions from statistically significant functions. Brief Bioinform. 2012;13:216–227. doi: 10.1093/bib/bbr041. [DOI] [PubMed] [Google Scholar]
- 26.Carlson M, Falcon S, Pages H, Li N. A set of annotation maps describing the entire Gene Ontology. Version 2.4.5. 2007 [Google Scholar]
- 27.Carlson M, Falcon S, Pages H, Li N. org. Hs. eg. db: Genome wide annotation for Human. Version 3.4.0. 2013 [Google Scholar]
- 28.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas DJ, Rosenbloom KR, Clawson H, Hinrichs AS, Trumbower H, Raney BJ, Karolchik D, Barber GP, Harte RA, Hillman-Jackson J, et al. The ENCODE project at UC santa cruz. Nucleic Acids Res. 2007;35:D663–D667. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 33.Petronczki M, Lénárt P, Peters JM. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Sumara I, Giménez-Abián JF, Gerlich D, Hirota T, Kraft C, de la Torre C, Ellenberg J, Peters JM. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr Biol. 2004;14:1712–1722. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 35.Spänkuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, Strebhardt K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst. 2002;94:1863–1877. doi: 10.1093/jnci/94.24.1863. [DOI] [PubMed] [Google Scholar]
- 36.Gumireddy K, Reddy MR, Cosenza SC, Boominathan R, Baker SJ, Papathi N, Jiang J, Holland J, Reddy EP. ON01910, a non-ATP-competitive small molecule inhibitor of Plk1, is a potent anticancer agent. Cancer Cell. 2005;7:275–286. doi: 10.1016/j.ccr.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Yu H. Cdc20: A WD40 activator for a cell cycle degradation machine. Mol Cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang Y, Yu H, Liang X, Xu J, Cai X. Chk1-induced CCNB1 overexpression promotes cell proliferation and tumor growth in human colorectal cancer. Cancer Biol Ther. 2014;15:1268–1279. doi: 10.4161/cbt.29691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abel D, Abdul-Hamid O, Dijk M, Oudejans CB. Transcription factor STOX1A promotes mitotic entry by binding to the CCNB1 promotor. PLoS One. 2012;7:e29769. doi: 10.1371/journal.pone.0029769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- 42.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/S0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 43.Tarapore P, Okuda M, Fukasawa K. A mammalian in vitro centriole duplication system: Evidence for involvement of CDK2/cyclin E and nucleophosmin/B23 in centrosome duplication. Cell Cycle. 2002;1:75–78. [PubMed] [Google Scholar]
- 44.Tokuyama Y, Horn HF, Kawamura K, Tarapore P, Fukasawa K. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J Biol Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- 45.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 46.Guerzoni C, Bardini M, Mariani SA, Ferrari-Amorotti G, Neviani P, Panno ML, Zhang Y, Martinez R, Perrotti D, Calabretta B. Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood. 2006;107:4080–4089. doi: 10.1182/blood-2005-08-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 48.Min IM, Pietramaggiori G, Kim FS, Passegué E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell stem cell. 2008;2:380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]