Abstract
microRNA (miRNA) is abnormally expressed in numerous diseases, and it was intimately associated with cell proliferation and apoptosis. However, the mechanism by which miRNAs control cataractogenesis remains unclear. In the current study, it was demonstrated that miR-34a was highly expressed in the cataractous lens by stem-loop reverse transcription-quantitative polymerase chain reaction. Trying to investigate the role of miR-34a in human lens epithelial cells, miR-34a mimics were transfected into SRA01/04 cells, and this suppressed proliferation and induced apoptosis. Subsequently, E2F3 was confirmed as a direct target of miR-34a. Downregulation of E2F3 by small interfering (si) RNA siE2F3 resulted in proliferation inhibition and apoptosis of SRA01/04 cells. Furthermore, it was demonstrated that miR-34a and siE2F3 downregulated E2F3 expression at a protein level. In summary, the current study demonstrated that miR-34a suppressed the proliferation and induced apoptosis of SRA01/04 cells by downregulating E2F3. These observations provide novel insights with potential therapeutic applications for the treatment of cataracts.
Keywords: cataract, E2F3, miR-34a
Introduction
Cataracts are the leading cause of blindness. Although cataract surgery has a high success rate, it cannot meet the great need for treatment, particularly in developing countries. At present, no effective drugs to cure cataracts are available, thus the identification and development of novel drugs to cure or relieve cataract-related vision loss is required. MicroRNAs (miRNAs) are endogenous, small, non-coding, regulatory RNAs approximately 22 nucleotides in size, which partially or completely bind to complementary recognition sequences of mRNA, resulting in translational repression or cleavage and degradation of mRNA, thus regulating the expression of mRNA targets (1). A previous study using miRNA chips demonstrated increased expression of miRNA 34a (miR-34a) in cataracts compared with transparent lenses (2). Previous studies have identified that miR-34a is involved in apoptosis and senescence (3–5), and E2F3 is known to be a regulatory factor of the cell cycle, which can induce quiescent lens cells to re-enter the cell cycle. Inappropriate cell cycle re-entry can be accompanied by programmed cell death (6–8) and can also occur in human lens epithelial cells, which is associated with cataractogenesis (9). Various cataractogenesis models, including those based on UV (10,11), selenite (12), hypergalactosemia (13–15), N-methyl-N-nitrosourea (15), naphthalene (16), CPT-11 (17) and muscarinic receptor antagonists (18), indicate that the apoptosis of lens epithelial cells largely accounts for cataract formation. Therefore, the current study investigated the effect of miR-34a on human lens epithelial cells and hypothesized that the effect takes place through the E2F3 pathway.
Materials and methods
Capsule sample
Three transparent lens capsule samples were collected from eyes that were obtained 8 to 24 h postmortem from the Eye Bank of Zhongshan Ophthalmic Center (Guangzhou, China) and 3 cataractous capsule samples were collected from age-associated cataract patients during surgery subsequent to obtaining written informed consent from each patient. All lens specimens were centered anterior capsules with a ~5–6mm diameter obtained using anterior continuous curvilinear capsulorhexis. These samples were randomly divided into 3 groups for testing miR-34a expression, with each group consisting of one pair of individual transparent and cataractous lens capsules. The research was performed in accordance with the Declaration of Helsinki for Research Involving Human Tissue and with the approval of the Sun Yat-Sen University-Zhongshan Ophthalmic Center-Institutional Review Board (Guangzhou, China) (SYSU-ZOC-IRB).
RNA extraction and quantification of miR-34a by stem-loop reverse transcription-quantiative polymerase chain reaction (RT-qPCR)
All tissue samples were homogenized in TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the total RNA was isolated according to the manufacturer's instructions. A total of 1 µg RNA was reverse transcribed into first strand cDNA using the PrimeScript RT Reagent kit (RR037A; Takara Biotechnology Co., Ltd., Dalian, China) in a 20 µl reaction mixture containing the following: 5X PrimeScript buffer (4 µl), PrimeScript RT Enzyme Mix I (1 µl), Oligo Dt Primer (1 µl), stem-loop R (1 µl) and total RNA (1 µg), made up to 20 µl with RNase-free dH2O. The RT reaction was performed as follows: 37°C for 15 min, 85°C for 5 sec, then terminated at 4°C. RNA concentration was determined by absorption at 260 nm. Quantification of miR-34a by stem-loop RT-qPCR was performed as described previously (2). Primers were as follows: miR-34a, FACACTCCAGCTGGGTGGCAGTGTCTTAGCT, stem-loop RCTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAACCAG and general reverse TGGTGTCGTGGAGTCG; U6, FCTCGCTTCGGCAGCACAandRAACGCTTCACGAATTTGCGT.
miRNA target prediction
The candidate targets for miR-34a were identified using the miRWalk 1.0 database algorithm for miRNA target prediction (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html), selecting miRanda, miRWalk, PicTar5 and TargetScan as the prediction programs.
Cell culture
The 293T cells were obtained from the Human Virus Institute of the Medical College of Sun Yat-Sen University. The 293T cells were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM; Gibco® Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco® Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). The cells were incubated in a humidified 37°C incubator containing 5% CO2. The SRA01/04 cells were obtained from the from the Cancer Institute of Chinese Academy of Medical Sciences (Beijing, China). The SRA01/04 cells were cultured in low glucose DMEM (Gibco® Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin. The cells were incubated in a humidified 37°C incubator containing 5% CO2.
Clone and miRNA
The potential binding sites of miR-34a on the 3′-untranslated region (UTR) of human E2F3 were cloned into a dual luciferase vector psiCHECK2 (Promega Corporation, Madison, WI, USA) named psiCHECK-E2F3. A mutant 3′-UTR fragment of E2F3 with mutations in miR-34a seed binding sites was generated, called psiCHECK-E2F3 mut. The miRNA mimics/small interfering (si)E2F3 were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China), and were transfected at a concentration of 50 nM using RNAimax (Invitrogen; Thermo Fisher Scientific, Inc.).
Luciferase assay
The 293T cells were plated onto the 24-well plate. The cells were transfected with psiCHECK-E2F3 or psiCHECK-E2F3 mut, in the presence miR-34a or the mimic control (Guangzhou RiboBio Co., Ltd.). After 48 h, the firefly and Renilla luciferase activities were assayed using the Dual-Glo Luciferase Assay System (Promega Corporation) in a Tecan Safire Microplate Reader II (Tecan Group, Ltd., Männedorf, Switzerland). The ratio of the luminescent signals from Renilla vs. firefly represents the target specificity of miR-34a. All experiments were performed in triplicate.
Flow cytometry
The SRA01/04 cells were plated into a 12-well plate; subsequent to transfection for 120 h, the SRA01/04 cells were collected and then subjected to an apoptosis assay. Apoptosis was determined by Annexin V-fluorescein isothiocyanate/propidium iodide staining with the apoptosis detection kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Subsequent to treatment according to the manufacturer's instructions, the specimens were assessed by flow cytometry (BD Biosciences, San Jose, CA, USA).
Cell proliferation assay
Cell proliferation was determined by the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to the manufacturer's instructions. Briefly, SRA1/04 cells were seeded into a 96-well flat-bottomed plate, grown at 37°C for 8 h, and transfected with (50 nM) miRNA mimics. Subsequently 10 µl CCK-8 dye was added to each well, cells were incubated at 37°C for 0.5 h and the absorbance was measured at 450 nm in the microplate reader.
Nuclear staining
The nuclei were stained with DAPI (Beyotime Institute of Biotechnology, Inc., Jiangsu, China) according the manufacturer's instructions. Fluorescence images were captured using a confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany).
TUNEL
The TUNEL (Roche Diagnostics, Basel, Switzerland) method was used to label the 3′ end of apoptotic cell fragmented DNA. SRA01/04 cells were plated onto chamber slides in 24-well plates and were treated as described above. Subsequent to transfection for 120 h, cells were washed with phosphate-buffered saline (PBS) and were fixed with 4% paraformaldehyde for 30 min at room temperature, and were treated with 0.1% Triton X-100 for 2 min on ice. Chamber slides were rinsed in PBS and incubated for 60 min at 37°C with 50 µl TUNEL reaction mixture. Subsequent to washing with PBS, the glass coverslips were analyzed by confocal microscopy.
Immunofluorescence
SRA01/04 cells were plated onto chamber slides in 24-well plates and were treated as described above. Subsequent to transfection for 120 h, the cells were washed twice with ice cold PBS. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature, then 1% bovine serum albumin solution (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) was added as the blocking buffer for 1 h at 37°C. Subsequent to washing with PBS twice, cells were incubated overnight at 4°C with the anti-active caspase 3 (Asp175) antibody (#9661; 1:400; Cell Signaling Technology, Inc., Danvers, MA, USA). Subsequent to washing in PBS, the Cy3-conjugated secondary antibody (111-165-046; 1:500; Jackson ImmunoResearch, Inc., PA, USA) was added. Following incubation with the antibodies, the nuclei were stained with DAPI (Beyotime Institute of Biotechnology and the images were captured using the confocal microscope.
Western blotting
SDS-PAGE and western blot analysis were performed according to standard procedures. The cell pellets were collected at 72 h post-transfection for protein extraction. The cells were extracted with lysis buffer containing 150 mM NaCl, 1% NP-40, 0.1% SDS, 2 mg/ml aprotinin and 1 mM PMSF for 30 min at 4°C. The protein extracts were separated on 10% SDS-PAGE gels and transferred onto PVDF membranes. Subsequent to blocking in Tris-buffered saline with Tween-20 (TBST) containing 25 mmol/l Tris-HCl, pH 7.5, 137 mmol/l NaCl, 2.7 mmol/l KCl and 0.05% Tween-20 with 5% nonfat milk for 1 h at 37°C, the membranes were incubated with the primary antibody against E2F3 (sc-878; 1:200; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or GAPDH (ab9485; 1:2,500; Abcam, Cambridge, MA, USA) in TBST with 5% nonfat milk at 4°C overnight. The membranes were extensively washed three times with TBST and incubated with the goat anti-rabbit IgG secondary antibody conjugated with horseradish peroxidase (ab205718; 1:2,000; Abcam) at room temperature for 1 h. Subsequent to additional washes with TBST, the proteins were visualized with an ECL kit (Beyotime Institute of Biotechnology).
Statistical analysis
Statistical comparisons were made using Student's t-test for unpaired data or using one-way analysis of variance with the Bonferroni post-hoc test for multiple comparisons (GraphPad Prism, version 5.01; GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-34a is upregulated in the cataractous lens
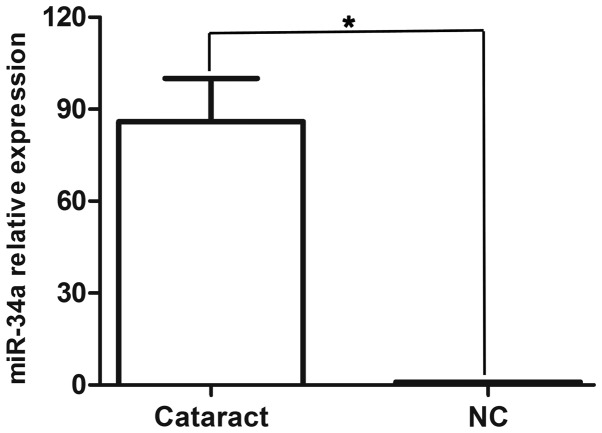
Previous miRNA chip data indicated higher expression of miR-34a in cataractous lenses when compared with transparent lenses (2), and these microarray results were confirmed by stem-loop RT-qPCR. The results indicated that miR-34a was expressed greater than 90-fold in cataractous lenses compared with transparent lenses (Fig. 1).
Figure 1.
miR-34a was highly expressed in cataractous lens. The expression of miR-34a was upregulated in fresh specimens of anterior lens capsules of age-related cataracts using stem loop reverse transcription-quantitative polymerase chain reaction, compared with transparent lenses (NC), which indicated that miR-34a expression was greater than 90-fold in cataracts compared with the transparent lens. Data are presented as the mean ± standard deviation; *P<0.05. miR, microRNA; NC, normal control.
miR-34a suppresses SRA01/04 proliferation and induces apoptosis
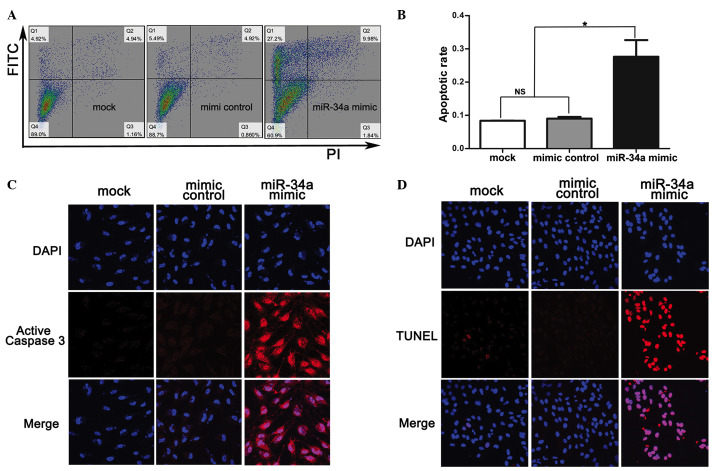
To investigate the effect of miR-34a on SRA01/04 human lens epithelium cell proliferation and apoptosis, miR-34a mimics were transfected into SRA01/04 cells and it was identified that cell apoptosis and slow growth occurred subsequent to transfection (Fig. 2A). The CCK-8 assay detected the growth conditions of SRA01/04, the absorbance at 450 nm was observed to be reduced in the SRA01/04 cells transfected with miR-34a mimics compared with the mimic control at 72, 96, 120 and 144 h following transfection with miRNA mimics (Fig. 2B). The apoptotic rate measured by flow cytometry was significantly increased in SRA01/04 cells transfected with miR-34a mimics (27.63±5%) compared with the mimic control (9±0.5%) and mock (8.37±0.06%) (Fig. 3A and B). In addition, apoptotic indicators TUNEL and active caspase 3 were observed to be highly expressed in SRA01/04 cells transfected with miR-34a mimics compared with the mimic control and mock (Fig. 3C and D).
Figure 2.
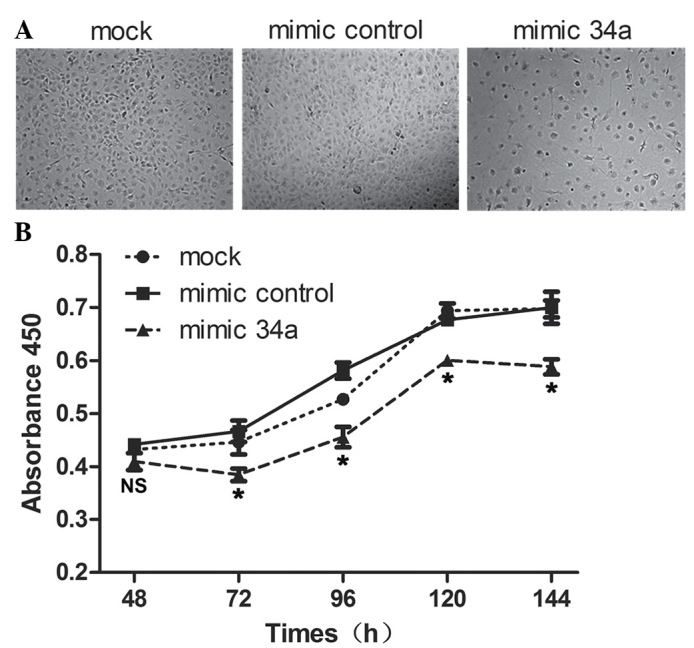
miR-34a inhibited the proliferation of SRA01/04. (A) The cell density was reduced in SRA01/04 cells transfected with the miR-34a mimic (magnification, ×50). (B) SRA01/04 cells were transfected with the miR-34a mimic, mock or mimic control, and a cell viability assay (Cell Counting Kit-8) was performed at 48, 72, 96, 120 and 144 h subsequent to transfection with microRNA mimics. Data are presented as the mean ± standard deviation, (miR-34a mimic vs. mimic control and mock; *P<0.05). miR, microRNA; NS, non-significant.
Figure 3.
miR-34a induced the apoptosis of SRA01/04. (A and B) SRA01/04 transfected with mock, mimic control, miR-34a mimic and Annexin V-FITC and PI staining for the detection of apoptosis by flow cytometry; (C) In situ analysis of apoptosis in SRA01/04 by TUNEL. Red color indicates TUNEL-positive cells. (D) Immunofluorescence microscopic analysis of active caspase 3 protein expression in SRA01/04 cells. It was indicated that SRA01/04 cells transfected with miR-34a mimic exhibited increased apoptosis compared with mock and mimic control. Magnification, ×40. Data are presented as the mean ± standard deviation; *P<0.05. miR, microRNA; FITC, fluorescein isothiocyanate; PI, propidium iodide; NS, non-significant.
Identification of candidate target genes for miR-34a
To determine the potential targets of miR 34a in SRA01/04 cells, the miRNA target prediction tool miRWalk (selecting miRanda, miRWalk, PicTar5 and TargetScan) was used, and this identified 20 putative target messenger RNAs of miR-34a (TOPORS, PGRMC2, UHRF2, CPEB2, CTNND2, CAMSAP1, EML5, DBC1, DCX, JAG1, E2F3, E2F5, ACSL1, RAB21, POGZ, RTF1, CAMTA1, SATB2, ZDHHC17 and ZNF281). Of these candidates, E2F3 was selected due to its involvement in cell apoptosis and the cell cycle (6–8), and possible association with cataractogenesis (9).
miR-34a targets the 3′-UTR of E2F3 mRNA
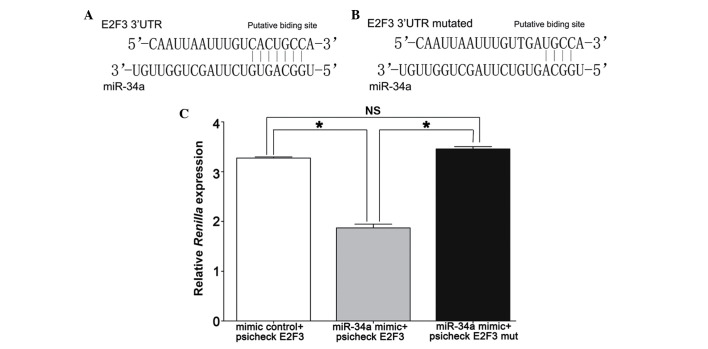
Using TargetScan to analyze the 3′-UTR of E2F3 to identify potential binding sites for miR-34a, a single recognition sequence containing a conserved 8-mer exact seed match at positions 2730–2737 bp (Fig. 4A and B) was identified in the E2F3 3′UTR. This indicated that miR-34a may directly bind to E2F3 3′UTR to regulate E2F3 expression at the transcriptional level.
Figure 4.
E2F3 was a direct target of miR-34a. (A and B) Schematic representation of WT and mut putative miR-34a-binding sites in the 3′-UTR of E2F3. (C) The 293T cells were transfected with miR-34a mimic and psiCHECK2 containing either the WT or mut putative binding sites for miR-34a, and 48 h later the luciferase assay was performed. The data indicated that miR-34a mimic reduced the Renilla activity in the reporter construct containing the E2F3 site (1.87±0.13), whereas no effect was observed with a construct containing a mut E2F3 seed site (3.46±0.09) and mimic control (3.27±0.04). Data are presented as the mean ± standard deviation; *P<0.05. miR, microRNA; WT, wild-type; mut, mutant; UTR, untranslated region; NS, non-significant.
To validate E2F3 as a direct target of miR-34a, luciferase reporter constructs (psiCHECK E2F3) were generated, in which the E2F3 3′-UTR containing the miR-34a seed target was placed behind the Renilla luciferase gene of the plasmid psiCHECK2. miR-34a mimics inhibited Renilla luciferase activity (1.87±0.13), whereas no effect was observed when the miR-34a target site was mutated (3.46±0.09), which was similar to that of the mimic control (3.27±0.04) (Fig. 4C).
E2F3 siRNA (siE2F3) suppresses SRA01/04 proliferation and induces apoptosis
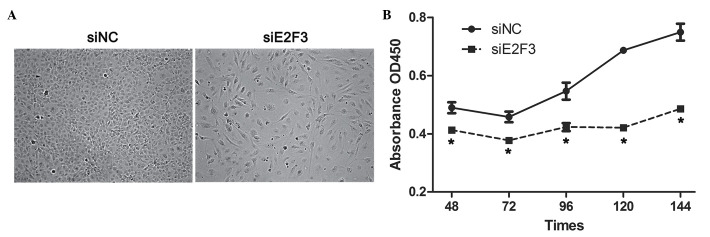
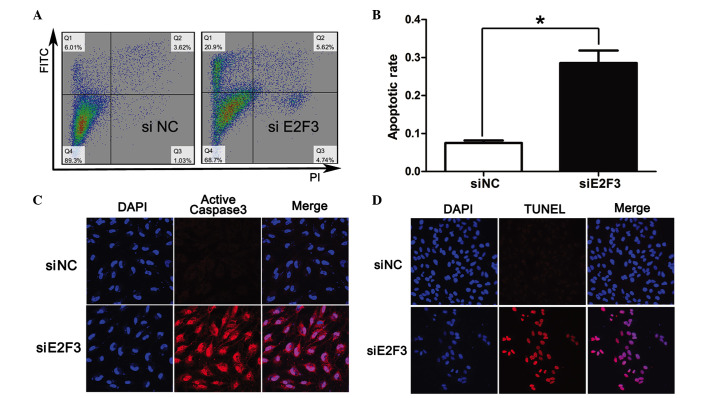
In order to determine the role of E2F3 in proliferation and apoptosis of SRA01/04 cells, siE2F3 were transfected into SRA01/04 cells and this was observed to significantly suppress proliferation of SRA01/04 cells at 48, 72, 96, 120 and 144 h subsequent to siE2F3 transfection compared with the negative control (siNC) (Fig. 5). In addition, the results indicated a higher apoptotic rate in SRA01/04 cells transfected with siE2F3 (28.5±1.9%) compared with siNC (7.5±0.4%) (Fig. 6A and B). In addition, apoptotic indicators TUNEL and active caspase 3 were highly expressed in SRA01/04 cells transfected with siE2F3 when compared with siNC (Fig. 6C and D).
Figure 5.
siE2F3 inhibited the proliferation of SRA01/04 cells. (A) The cell density was reduced in SRA01/04 transfected with siRNA siE2F3 (magnification, ×50); (B) SRA01/04 cells were transfected with siE2F3 or siNC, and a cell viability assay (Cell Counting Kit-8) was performed at 48, 72, 96, 120 and 144 h subsequent to siRNA transfection. Data are presented as the mean ± standard deviation; *P<0.05. si, small interfering; NC, negative control; OD, optical density.
Figure 6.
siE2F3 induced the apoptosis of SRA01/04 cells. (A and B) SRA01/04 cells transfected with siE2F3 or siNC, and Annexin V-FITC and PI staining for the detection of apoptosis by flow cytometry. (C) Immunofluorescence microscopic analysis of active caspase 3 protein expression of SRA01/04 cells. (D) The apoptosis of SRA01/04 cells observed by the TUNEL assay. Red color indicated TUNEL-positive cells. Those results indicated that SRA01/04 transfected with miR-34a exhibited higher apoptosis compared with the mock and mimic control. Magnification, ×40. Data are presented as the mean ± standard deviation; *P<0.05. si, small interfering; NC, negative control; FITC, fluorescein isothiocyanate; PI, propidium iodide.
miR-34a and siE2F3 reduce E2F3 expression
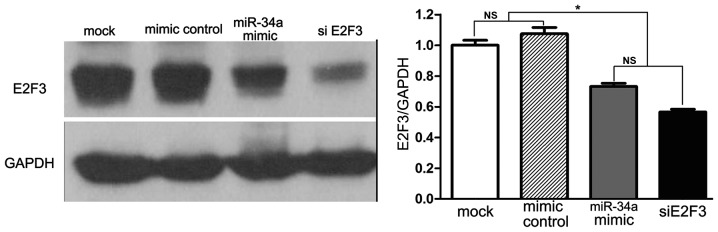
Subsequent to verifying that miR-34a and siE2F3 inhibit proliferation and induce apoptosis of SRA01/04 cells, it was investgated whether miR-34a and siE2F3 reduce the expression of E2F3. Results indicate that miR-34a mimics and siE2F3 significantly inhibited E2F3 protein expression (Fig. 7).
Figure 7.
miR-34a and siE2F3 reduce E2F3 expression. SRA01/04 cells transfected with miR-34a mimic or siE2F3 exhibited reduced E2F3 expression compared with the mock and mimic control. Data are presented as the mean ± standard deviation; *P<0.05. miR, microRNA; si, small interfering; NS, non-significant.
Discussion
miRNAs are endogenous and non-coding RNAs, approximately 22 nucleotides in length, which perform important regulatory roles by targeting mRNAs for cleavage or translational repression (1). Greater than one third of human genes appear to be conserved miRNA targets (19). Numerous diseases have been indicated to involve changes in the miRNA expression profile. As miRNAs are involved in the regulation of cellular proliferation, differentiation and apoptosis, it is suggested that they take part in complex, multifactorial and environmentally influenced cellular processes, leading to the aging of cells and organisms, and human disease (20).
According to a previous study (2), miR-34a is markedly increased in cataracts compared with transparent lenses. This led to the hypothesis of the current study, that miR-34a may participate in cataractogenesis and its mechanisms. Tazawa et al (21) and Welch et al (22) previously demonstrated that introducing miR-34a inhibited cancer cell proliferation and induced apoptosis, accompanying downregulated E2F3 expression. In addition, E2F3 is associated with cell proliferation, differentiation and apoptosis (23). Therefore, using the miRNA prediction software, it was identified that miR-34a may take part in cataractogenesis by inducing lens epithelial cell apoptosis by downregulating E2F3.
miR-34a is located in the 1p36.23 chromosome region, and it is abnormally expressed in various tumor types including human neuroblastoma (22,24), prostate cancer, colon cancer and pancreatic cancer (25), and it is highly expressed in age-associated diseases including cardiac aging (26), atherosclerotic cardiovascular disease (27) and cataracts (28). Ectopic overexpression of miR-34a in numerous tumor cell lines leads to the reactivation of the apoptotic pathway, which suggests that miR-34a may be a potent tumor suppressor gene (29). In addition, miR-34a has been reported to be involved in senescence. In a premature senescence model, induction by hydrogen peroxide resulted in significant upregulation of miR-34a (30), however, antisense inhibition of miR-34a blocked the onset of replicative senescence (31). Chien et al (28) demonstrated positive correlations between miR-34a levels and lens opacity severity, which suggested that miR-34a may serve a role in lens senescence. Ito et al (27) identified that miR-34a expression increased in senescent human umbilical cord vein endothelial cells and in the heart and spleen of older mice. In addition, Zhao et al (32) identified that the overexpression of miR-34a in bone-marrow-derived endothelial progenitor cells from rats led to the induction of senescence by suppressing the sirtuin 1 gene.
The results indicated that miR-34a was markedly increased in cataracts compared with transparent lenses, which was in agreement with the results of Chien et al (28). Cataractogenesis has been reported to be closely associated with lens epithelial cell apoptosis, and it is widely accepted that the normal lens epithelial cells are essential for maintenance of metabolic homeostasis and transparency of the lens (33). If the lens epithelial cells lose function, then the microenvironment of the lens gets disturbed and the lens becomes opaque (34). In order to explain the association between lens epithelial cell apoptosis and cataractogenesis, there is direct evidence that depletion or stress-induced apoptotic death of lens epithelial cells can initiate cataract formation in adult lenses (9). In addition, in vitro and in vivo studies have demonstrated that cataract formation as a result of treatment of adult lenses with stress factors, such as calcimycin and H2O2, induced the apoptosis of lens epithelial cells (35,36). A previous study (37) implicated that normal lens epithelial cells exhibited reduced apoptosis compared with that of cataracts, which suggests that lens epithelial cell apoptosis may be a common cellular basis for the initiation of non-congenital cataract formation. Guggenmoos-Holtzmann et al (38) and Balaram et al (39) identified that the number of lens epithelial cells gradually reduced in line with aging. Therefore, it is suggested that through the different stages of growth and differentiation in the adult lens, various stress conditions can induce apoptosis of the lens epithelial cells, resulting in eventual non-congenital cataractogenesis (40), including age-associated cataracts.
The E2F transcription factor family that contains 8 members (E2F1-8) serves a crucial role in the regulation of cellular proliferation, differentiation and apoptosis (23). The ablation of E2F3 in mouse embryonic fibroblasts (MEFs) markedly reduce E2F target gene expression (41) and significantly compromise the proliferation of MEFs (42), which suggests that E2F3 serves a critical role in the control of cell proliferation. In the current study, the suppression of E2F3 by miR-34a and siE2F3 suppressed SRA01/04 cell proliferation and induced cell apoptosis, which suggested that miR-34a acted via through E2F3.
In summary, the results of the present study indicated that miR-34a was highly expressed in cataractous lens epithelial cells, and miR-34a suppresses proliferation and induces apoptosis of SRA01/04 cells in a similar way to siE2F3. In addition, miR-34a and siE2F3 significantly reduced E2F3 protein expression. All of these results demonstrated that miR-34a induced SRA01/04 human lens epithelial cell apoptosis through E2F3, and may be closely involved in the pathogenesis of cataracts. These observations may aid in the development of novel therapeutic strategies.
Acknowledgements
The current study was supported by the National Natural Science Foundation of China (grant nos. 81270980 and 81000389), the Pearl River Science and Technology New Star Project of Guangzhou City (grant no. 2014J2200060), the Guangdong Provincial Natural Science Foundation for Distinguished Young Scholars of China (grant no. 2014A030306030) and the Science and Technology Program of Guangdong Province (grant no. 2013B021800054).
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu C, Lin H, Wang Q, Chen W, Luo H, Chen W, Zhang H. Discrepant expression of microRNAs in transparent and cataractous human lenses. Invest Ophthalmol Vis Sci. 2012;53:3906–3912. doi: 10.1167/iovs.11-9178. [DOI] [PubMed] [Google Scholar]
- 3.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 4.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hermeking H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 6.Wenzel PL, Chong JL, Sáenz-Robles MT, Ferrey A, Hagan JP, Gomez YM, Rajmohan R, Sharma N, Chen HZ, Pipas JM, et al. Cell proliferation in the absence of E2F1-3. Dev Biol. 2011;351:35–45. doi: 10.1016/j.ydbio.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, Liang D, Yang T, Leone G, Overbeek PA. Distinct capacities of individual E2Fs to induce cell cycle re-entry in postmitotic lens fiber cells of transgenic mice. Dev Neurosci. 2004;26:435–445. doi: 10.1159/000082285. [DOI] [PubMed] [Google Scholar]
- 8.Chong JL, Tsai SY, Sharma N, Opavsky R, Price R, Wu L, Fernandez SA, Leone G. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol Cell Biol. 2009;29:414–424. doi: 10.1128/MCB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Q, Liu JP, Li DW. Apoptosis in lens development and pathology. Differentiation. 2006;74:195–211. doi: 10.1111/j.1432-0436.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- 10.Ayala M, Strid H, Jacobsson U, Söderberg PG. p53 expression and apoptosis in the lens after ultraviolet radiation exposure. Invest Ophthalmol Vis Sci. 2007;48:4187–4191. doi: 10.1167/iovs.06-0660. [DOI] [PubMed] [Google Scholar]
- 11.Galichanin K, Löfgren S, Bergmanson J, Söderberg P. Evolution of damage in the lens after in vivo close to threshold exposure to UV-B radiation: Cytomorphological study of apoptosis. Exp Eye Res. 2010;91:369–377. doi: 10.1016/j.exer.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Tamada Y, Fukiage C, Nakamura Y, Azuma M, Kim YH, Shearer TR. Evidence for apoptosis in the selenite rat model of cataract. Biochem Biophys Res Commun. 2000;275:300–306. doi: 10.1006/bbrc.2000.3298. [DOI] [PubMed] [Google Scholar]
- 13.Murata M, Ohta N, Sakurai S, Alam S, Tsai J, Kador PF, Sato S. The role of aldose reductase in sugar cataract formation: Aldose reductase plays a key role in lens epithelial cell death (apoptosis) Chem Biol Interact 130–132. 2001:617–625. doi: 10.1016/S0009-2797(00)00289-1. [DOI] [PubMed] [Google Scholar]
- 14.Takamura Y, Kubo E, Tsuzuki S, Akagi Y. Apoptotic cell death in the lens epithelium of rat sugar cataract. Exp Eye Res. 2003;77:51–57. doi: 10.1016/S0014-4835(03)00083-6. [DOI] [PubMed] [Google Scholar]
- 15.Yoshizawa K, Oishi Y, Nambu H, Yamamoto D, Yang J, Senzaki H, Miki H, Tsubura A. Cataractogenesis in neonatal Sprague-Dawley rats by N-methyl-N-nitrosourea. Toxicol Pathol. 2000;28:555–564. doi: 10.1177/019262330002800408. [DOI] [PubMed] [Google Scholar]
- 16.Pandya U, Saini MK, Jin GF, Awasthi S, Godley BF, Awasthi YC. Dietary curcumin prevents ocular toxicity of naphthalene in rats. Toxicol Lett. 2000;115:195–204. doi: 10.1016/S0378-4274(00)00191-0. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima Y, Nakamura T, Enomoto T, Murata Y. Loss of one allele of the p53 gene in the lens epithelial tumor in transgenic mice suppresses apoptosis induced by a topoisomerase I inhibitor (CPT-11) Cancer Lett. 2002;179:165–173. doi: 10.1016/S0304-3835(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 18.Durand G, Hubert MF, Kuno H, Cook WO, Boussiquet-Leroux C, Owen R, Fujimaki Y, Kemi M, Virat M, van Zwieten MJ. Muscarinic receptor antagonist-induced lenticular opacity in rats. Toxicol Sci. 2002;66:166–172. doi: 10.1093/toxsci/66.1.166. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Harries LW. MicroRNAs as mediators of the ageing process. Genes (Basel) 2014;5:656–670. doi: 10.3390/genes5030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 23.Maiti B, Li J, de Bruin A, Gordon F, Timmers C, Opavsky R, Patil K, Tuttle J, Cleghorn W, Leone G. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem. 2005;280:18211–18220. doi: 10.1074/jbc.M501410200. [DOI] [PubMed] [Google Scholar]
- 24.Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 25.Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Tréguer K, Carmona G, Bonauer A, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398:735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Chien KH, Chen SJ, Liu JH, Chang HM, Woung LC, Liang CM, Chen JT, Lin TJ, Chiou SH, Peng CH. Correlation between microRNA-34a levels and lens opacity severity in age-related cataracts. Eye (Lond) 2013;27:883–888. doi: 10.1038/eye.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 30.Maes OC, Sarojini H, Wang E. Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J Cell Physiol. 2009;221:109–119. doi: 10.1002/jcp.21834. [DOI] [PubMed] [Google Scholar]
- 31.Fujita K, Mondal AM, Horikawa I, Nguyen GH, Kumamoto K, Sohn JJ, Bowman ED, Mathe EA, Schetter AJ, Pine SR, et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11:1135–1142. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299:E110–E116. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinoshita JH. Mechanisms initiating cataract formation. Proctor Lecture. Invest Ophthalmol. 1974;13:713–724. [PubMed] [Google Scholar]
- 34.Piatigorsky J. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation. 1981;19:134–153. doi: 10.1111/j.1432-0436.1981.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 35.Li WC, Kuszak JR, Wang GM, Wu ZQ, Spector A. Calcimycin-induced lens epithelial cell apoptosis contributes to cataract formation. Exp Eye Res. 1995;61:91–98. doi: 10.1016/S0014-4835(95)80062-X. [DOI] [PubMed] [Google Scholar]
- 36.Li DW, Spector A. Hydrogen peroxide-induced expression of the proto-oncogenes, c-jun, c-fos and c-myc in rabbit lens epithelial cells. Mol Cell Biochem. 1997;173:59–69. doi: 10.1023/A:1006828402225. [DOI] [PubMed] [Google Scholar]
- 37.Li WC, Kuszak JR, Dunn K, Wang RR, Ma W, Wang GM, Spector A, Leib M, Cotliar AM, Weiss M, et al. Lens epithelial cell apoptosis appears to be a common cellular basis for non-congenital cataract development in humans and animals. J Cell Biol. 1995;130:169–181. doi: 10.1083/jcb.130.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guggenmoos-Holzmann I, Engel B, Henke V, Naumann GO. Cell density of human lens epithelium in women higher than in men. Invest Ophthalmol Vis Sci. 1989;30:330–332. [PubMed] [Google Scholar]
- 39.Balaram M, Tung WH, Kuszak JR, Ayaki M, Shinohara T, Chylack LJ., Jr Noncontact specular microscopy of human lens epithelium. Invest Ophthalmol Vis Sci. 2000;41:474–481. [PubMed] [Google Scholar]
- 40.Zhang L, Yan Q, Liu JP, Zou LJ, Liu J, Sun S, Deng M, Gong L, Ji WK, Li DW. Apoptosis: Its functions and control in the ocular lens. Curr Mol Med. 2010;10:864–875. doi: 10.2174/156652410793937741. [DOI] [PubMed] [Google Scholar]
- 41.Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]