Abstract
Squamous cell carcinoma is the most common cancer in the oral cavity. We previously demonstrated that transforming growth factor-β1 (TGF-β1) promotes the epithelial-mesenchymal transition (EMT) of human oral squamous cell carcinoma (hOSCC) cells; however, it remains to be clarified whether the TGF-β superfamily member bone morphogenetic protein (BMP) affects this process in hOSCC cells. Here, we examined the independent and collective effects of TGF-β1 and BMP-2 on EMT and mesenchymal-epithelial transition (MET) in a panel of four hOSCC cell lines. Notably, we found that HSC-4 cells were the most responsive to BMP-2 stimulation, which resulted in the upregulation of Smad1/5/9 target genes such as the MET inducers ID1 and cytokeratin 9 (CK9). Furthermore, BMP-2 downregulated the mesenchymal marker N-cadherin and the EMT inducer Snail, but upregulated epithelial CK9 expression, indicating that BMP-2 prefers to induce MET rather than EMT. Moreover, TGF-β1 dampened BMP-2-induced epithelial gene expression by inhibiting Smad1/5/9 expression and phosphorylation. Functional analysis revealed that TGF-β1 and BMP-2 significantly enhanced HSC-4 cell migration and proliferation, respectively. Collectively, these data suggest that TGF-β positively regulates hOSCC invasion in the primary tumor, whereas BMP-2 facilitates cancer cell colonization at secondary metastatic sites. Thus, the invasive and metastatic characteristics of hOSCC appear to be reciprocally regulated by BMP and TGF-β.
Keywords: bone morphogenetic protein-2, epithelial-mesenchymal transition, ID1, mesenchymal-epithelial transition, metastasis, Smad, squamous cell carcinoma, transforming growth factor-β
Introduction
Epithelial-mesenchymal transition (EMT) is characterized by the loss of epithelial markers with a reciprocal gain in mesenchymal phenotype and migratory potential (1). While EMT is essential for embryonic development and adult tissue maintenance (2,3), it is also necessary for desmoplasia and cancer cell migration (4). Conversely, mesenchymal-epithelial transition (MET) is a physiological and embryological phenomenon induced by cytokines (5,6). During EMT, transcription factors, such as Snail (7) and Slug (8) are upregulated and induce mesenchymal gene expression and suppress that of epithelial genes (9).
Transforming growth factor-β (TGF-β) is an important inducer of EMT (10,11). Bone morphogenetic proteins (BMPs) are members of the TGF-β superfamily, of which 20 have been discovered in humans to date (12). BMPs were first identified for their pro-osteogenic effects, but recent studies have revealed their additional significance as tissue morphogenetic factors (13), particularly for BMP-2, −4 and −7 (14). In particular, BMP-2 is a cytokine used to treat bone defects and is being investigated in regenerative studies (15,16).
BMP signaling is induced when a heterodimeric membrane kinase binds BMP and subsequently triggers Smad protein phosphorylation, similar to the mechanism of TGF-β pathway activation. However, downstream BMP-induced signals are mediated by Smad1/5/9, whereas TGF-β signaling is mediated by Smad2/3. These receptor-regulated Smad complexes (Smad1/5/9 and Smad2/3) bind the common mediator Smad4, and collectively translocate into the nucleus. The Smad complex then binds the DNA promoter region upstream of target genes to induce their expression (17). The effects of BMP on cancer cells are relatively uncharacterized when compared to those of TGF-β, but generally thought to promote EMT (18,19). However, BMP-2 was recently reported to suppress EMT in the presence of TGF-β (20). Thus, the overall effects of BMP on EMT likely depend on the cellular context.
Squamous cell carcinoma is the most common cancer of the oral cavity in humans (21,22). Although BMPs are thought to be involved in cancer metastasis, the mechanisms underlying BMP-induced EMT/MET have not yet been clarified at the molecular level (12). Nevertheless, a few studies have demonstrated the positive effects of BMP on cancer progression using human oral squamous cell carcinoma (hOSCC) cell lines and carcinoma tissue (23–26). Moreover, while bone is a major target for hOSCC metastasis (26,27), the cellular and molecular mechanisms facilitating this process remain to be elucidated.
Previous studies by our group revealed that TGF-β1 responsiveness was correlated with EMT-related gene expression in six hOSCC cell lines. Notably, TGF-β1 enhanced the migration of HSC-4 hOSCC cells via the Slug/Wnt-5b/MMP-10 and integrin α3β1/FAK signaling axes (28,29). Since cells are subjected to multiple simultaneous signals from extracellular ligands and must then integrate and interpret them, this study investigated the independent and collective effects of TGF-β1 and BMP-2 on EMT and MET in HSC-4 cells. In addition, we evaluated how TGF-β1 affects the BMP-2-induced MET in HSC-4 cells at the molecular level.
Materials and methods
Materials
Cell lines were obtained from the Japanese Collection of Research Bioresources (Osaka, Japan). Recombinant human TGF-β1 and BMP-2 were purchased from Peprotech, Inc. (Rocky Hill, NJ, USA). The TGF-βR1 kinase inhibitor SB431542 was provided by Merck-Millipore (Frankfurt, Germany). The BMP type I receptor kinase inhibitor LDN-193189 was purchased from Selleck Chemical (Houston, TX, USA). The proteasome inhibitor MG132 was obtained from Merck Millipore. The protease inhibitor cocktail and phosphatase inhibitor cocktail 1 and 2 were purchased from Sigma-Aldrich (St. Louis, MO, USA). All of the other reagents were of analytical grade.
Cell culture
All of the cell lines were grown at 37°C in 5% CO2. Human HSC-2 and HSC-4 squamous cell carcinoma cells were cultured in MEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA). SAS cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% FBS. HSC-3 cells were cultured in DMEM (Sigma-Aldrich) containing 10% FBS. The culture medium was replaced with serum-free medium 24 h prior to cytokine-stimulated experiments. In the majority of experiments, 2.0×105 hOSCC cells were cultured in 500 µl serum-free media containing 10 ng/ml TGF-β1 or 20 ng/ml BMP-2 for the indicated time-periods in 24-well tissue culture plates.
Western blot analysis
For Smad phosphorylation analysis, 3.0×106 cells were lysed in RIPA buffer (Sigma-Aldrich) containing a protease and phosphatase inhibitor cocktail (Sigma-Aldrich) and the protein content was assessed with a BCA reagent (Thermo Fisher Scientific, Waltham, MA, USA). To examine marker protein expression, 1.0×106 cells were cultured in a 6-well plate in serum-free MEM with or without 10 ng/ml TGF-β1 for the indicated time-points. Harvested cells were homogenized in SDS sample buffer containing a protease and phosphatase inhibitor cocktail (Sigma-Aldrich). Proteins were separated by 12.5% SDS-PAGE (ATTO Co., Tokyo, Japan) and subsequently transferred onto PVDF membranes (Merck Millipore). The membranes were probed with the primary antibodies, including rabbit anti-Smad1 (#6944) and rabbit anti-Smad5 (#9517; both from Cell Signaling Technology, Beverly, MA, USA), rabbit anti-Smad9 [Smad8 (R-64); Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA] and rabbit anti-phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad9 (Ser465/467) (#13820; Cell Signaling Technology). Mouse anti-β-actin (clone C4; Santa Cruz Biotechnology, Inc.) was used as a loading control in the Smad phosphorylation experiments. The blots were then incubated with alkaline phosphatase-conjugated secondary antibody and signals were visualized using an alkaline phosphatase substrate kit (BCIP/NBT substrate kit; Vector Laboratories, Inc., Burlingame, CA, USA).
Reverse transcriptase quantitative-PCR (RT-qPCR)
For total RNA preparation, 2.0×105 cells were cultured in 24-well tissue culture plates. Total RNA was isolated using the Isogen reagent (Nippon Gene, Co., Ltd., Toyama, Japan) according to the manufacturer's instructions. RNA was reverse transcribed into first-strand cDNA with an RT-PCR system kit (Takara Bio, Inc., Shiga, Japan). qPCR was performed on a Thermal Cycler Dice Real-Time System (Takara Bio, Inc.) using SYBR® Premix Ex Taq™ II (Takara Bio, Inc.) with human gene-specific primers (Table I). Target gene expression was normalized to an internal β-actin reference and expressed in terms of fold-change relative to the control sample (30).
Table I.
Primer sequences for the qPCR analysis.
| Genes | Primer sequences | |
|---|---|---|
| BMP-2 | F | 5′-AAGATTCCTAAGGCATGCTGTGTC-3′ |
| R | 5′-TCGTCAAGGTACAGCATCGAGA-3′ | |
| Cytokeratin 9 | F | 5′-TCAGCTGACTGGGCAGAACA-3′ |
| R | 5′-ACCTCATGCTCGATCTGGGTTA-3′ | |
| Cytokeratin 18 | F | 5′-AGGAGTATGAGGCCCTGCTGAA-3′ |
| R | 5′-TTGCATGGAGTTGCTGCTGTC-3′ | |
| E-cadherin | F | 5′-TACACTGCCCAGGAGCCAGA-3′ |
| R | 5′-TGGCACCAGTGTCCGGATTA-3′ | |
| ID1 | F | 5′-CGGAATCTGAGGGAGAACAAG-3′ |
| R | 5′-CTGAGAAGCACCAAACGTGA-3′ | |
| N-cadherin | F | 5′-CGAATGGATGAAAGACCCATCC-3′ |
| R | 5′-TCGTCAAGGTACAGCATCGAGA-3′ | |
| NEDD4 | F | 5′-GATTTGTAAACCGAATCCAGAAGCA-3′ |
| R | 5′-CCAGTCATTCACATCAACATCTCC-3′ | |
| NEDD4L | F | 5′-CCAATGGGTCAGAAATAATGGTCA-3′ |
| R | 5′-AAGGCGTTCATCTGCTTCTGG-3′ | |
| Smad1 | F | 5′-ACAGTCTGTGAACCATGGATTTGA-3′ |
| R | 5′-TGAGGTGAACCCATTTGAGTAAGAA-3′ | |
| Samd5 | F | 5′-GCTTTCATCCCACCACTGTCTGTA-3′ |
| R | 5′-CCTGCCGGTGATATTCTGCTC-3′ | |
| Smad6 | F | 5′-GAGCTGAGCCGAGAGAAAGA-3′ |
| R | 5′-AGATGCACTTGGAGCGAGTT-3′ | |
| Smad7 | F | 5′-TGCAACCCCCATCACCTTAG-3′ |
| R | 5′-TCGTCAAGGTACAGCATCGAGA-3′ | |
| Smad9 | F | 5′-TGGCCCAGTCAGTTCACCAC-3′ |
| R | 5′-CATGAAGATGAATCTCAATCCAGCA-3′ | |
| Smurf1 | F | 5′-CCGCATCGAAGTGTCCAGAG-3′ |
| R | 5′-CCCACGGAATTTCACCATCAG-3′ | |
| Smurf2 | F | 5′-TGCACTAACAACCTGCCGAAAG-3′ |
| R | 5′-CTTGTCATTCCACAGCAAATCCAC-3′ | |
| Snail | F | 5′-GACCACTATGCCGCGCTCTT-3′ |
| R | 5′-TCGCTGTAGTTAGGCTTCCGATT-3′ | |
| β-actin | F | 5′-GGAGATTACTGCCCTGGCTCCTA-3′ |
| R | 5′-GACTCATCGTACTCCTGCTTGCTG-3′ | |
F, forward; R, reverse.
Proliferation assay
HSC-4 cell proliferation was evaluated by alamarBlue® assay (AbD Serotec, Oxon, UK) according to the manufacturer's instructions. Briefly, 1.0×104 cells/well were subcultured in 96-well plates in MEM supplemented with 10% FBS for 24 h. The culture medium was then replaced with MEM supplemented without FBS with or without TGF-β1 (10 ng/ml) or BMP-2 (20 ng/ml) for 2 days. The cells were washed once with PBS and then incubated with 100 µl alamarBlue® (10% alamarBlue® in Ham's F-12) at 37°C in 5% CO2 for 1.5 h. The absorbances at 570 and 600 nm were used to measure the reduced and oxidized forms of reagent, respectively, using a microplate reader. Assays were performed independently at least 3 times (n=6).
Cell migration assays
Boyden chamber migration assays were performed using cells transfected with Slug siRNA as aforementioned. Then, cells were treated with 10 ng/ml TGF-β1 or 20 ng/ml BMP-2 under serum-free conditions for 72 h and subsequently plated at a density of 1.0×105 cells in the upper chamber of a Boyden apparatus in serum-free media, and were allowed to migrate into the lower chamber containing medium supplemented with 10% FBS for 24 h at 37°C. The chamber filter was fixed in 4% paraformaldehyde and stained with DAPI (1:500; Sigma-Aldrich) for 10 min. Migrating cells were counted in 9 fields on the membrane. Values indicate the mean number of migrating cells compared to the controls.
Statistical analysis
All experiments were performed at least 3 times independently. Results are expressed as the means ± standard deviation (SD). Data were analyzed using two-tailed, independent Student's t-tests. P<0.05 was considered statistically significant.
Results
BMP-2 responsiveness in the hOSCC cell lines
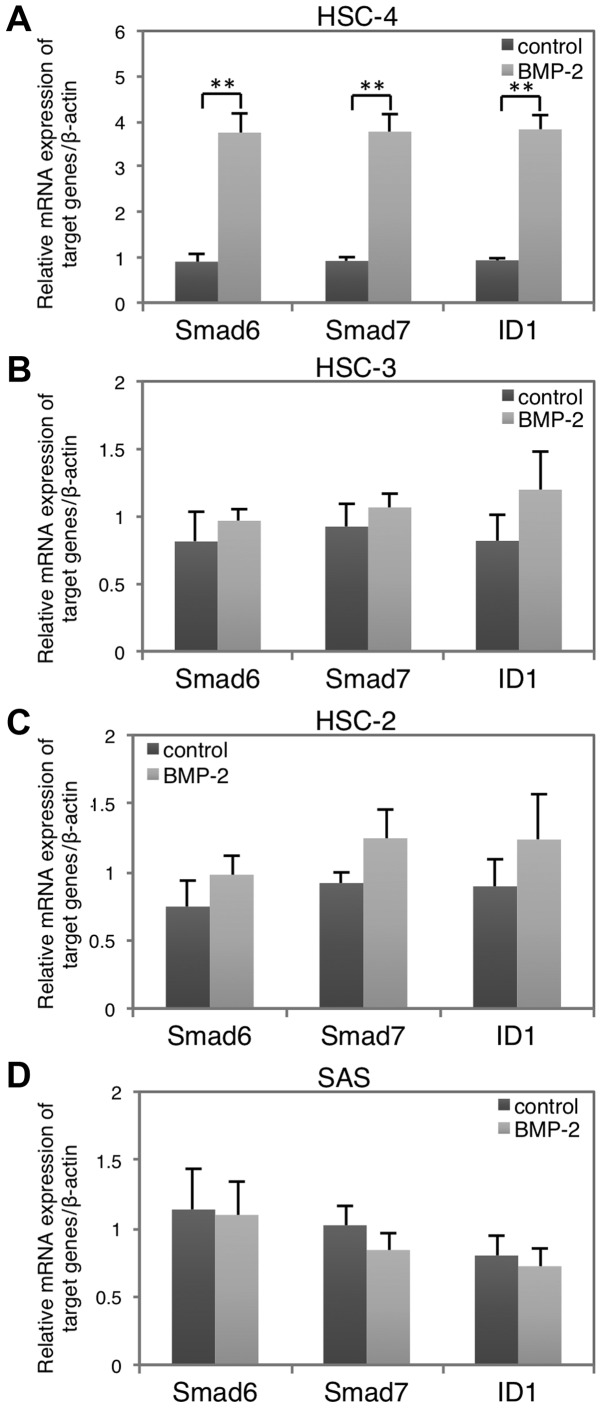
We first studied the cellular response to BMP-2 in four hOSCC cell lines: HSC-2, HSC-3, HSC-4, and SAS. Smad6, Smad7 and ID1 are targets of BMP signaling (31). Expression analysis revealed that BMP-2 induced a significant upregulation of all three target genes in the HSC-4 cells (Fig. 1A), but no marked changes were observed in the other cell lines (Fig. 1B-D). Thus, we used HSC-4 cells to investigate the effects of BMP-2 on MET in the following experiments. In addition, HSC-4 cells are also responsive to TGF-β1 stimulation based on our previous examinations (28).
Figure 1.
Difference in BMP-2 responsiveness in the hOSCC cell lines. Expression of BMP-2 target genes Smad6, Smad7 and ID1 was evaluated in (A) HSC-4, (B) HSC-3, (C) HSC-2 and (D) SAS hOSCC cell lines following treatment with 10 ng/ml BMP-2 for 3 h (light gray bars) vs. untreated controls (dark gray bars). Data represent the mean ± SD from triplicate experiments (**P<0.01). hOSCC, human oral squamous cell carcinoma; BMP, bone morphogenetic protein.
BMP-2-induced response in HSC-4 cells
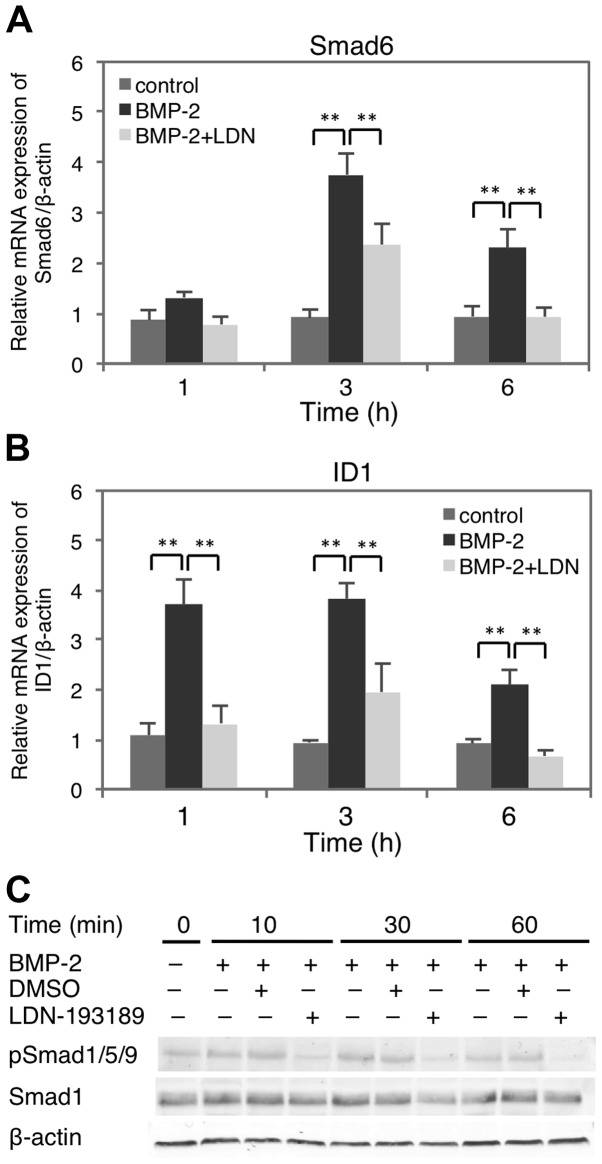
To confirm BMP-2 signaling in HSC-4 cells, the time course of Smad6 and ID1 gene expression was investigated in the presence or absence of LDN-193189, a selective inhibitor of activin receptor-like kinase (ALK)-2/3 (Fig. 2A and B). The results showed that both genes were upregulated 1 h after BMP-2 treatment, peaking at 3 h post-stimulation, which was significantly inhibited by LDN-193189. Moreover, Smad1/5/9 phosphorylation was increased in the BMP-2-treated HSC-4 cells, but inhibited in the presence of LDN-193189 (Fig. 2C). Thus, these results indicated that BMP-2 elicits Smad1/5/9 activation and target gene expression in HSC-4 cells.
Figure 2.
Time-points of the BMP-2-induced response in HSC-4 cells. (A) Smad6 and (B) ID1 gene expression was examined in cells treated with BMP-2 (dark-grey bars) or controls (medium-gray bars) for the indicated times. Some samples were pre-treated with 50 nM LDN-193189 for 30 min before BMP-2 treatment (light-gray bars). Data represent the mean ± SD from triplicate experiments (**P<0.01). (C) Smad1/5/9 phosphorylation and Smad1 protein levels were monitored in BMP-2 stimulated cells for up to 60 min. β-actin served as a loading control; BMP, bone morphogenetic protein.
Effects of BMP-2 or TGF-β1 on epithelial and mesenchymal status in HSC-4 cells
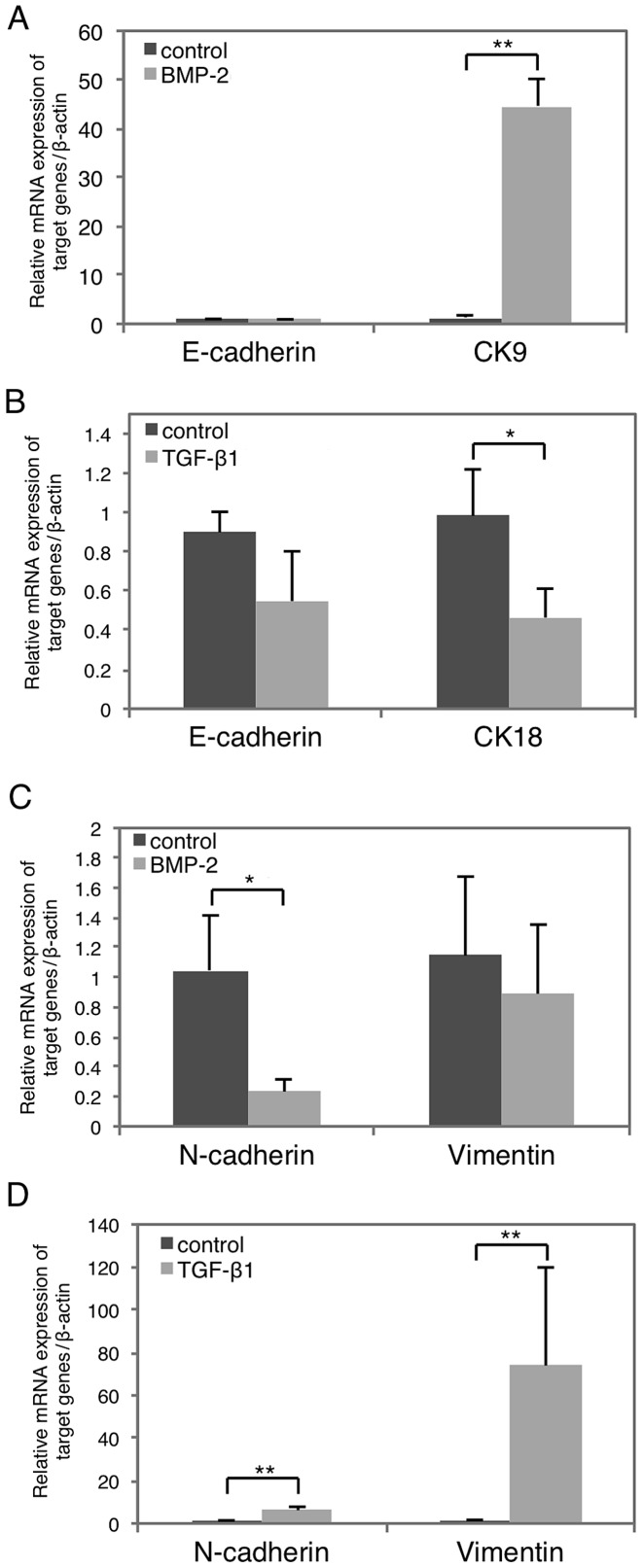
Next, we examined how BMP-2 alters the epithelial and mesenchymal characteristics of HSC-4 cells by examining marker expression with RT-qPCR (Fig. 3). Notably, the epithelial marker cytokeratin 9 (CK9) was clearly upregulated following BMP-2-stimulation (Fig. 3A, right), whereas that of N-cadherin was significantly suppressed (Fig. 3C, left). Conversely, TGF-β1 stimulation resulted in the significant suppression of epithelial cytokeratin 18 (CK18) marker (Fig. 3B, right), as well as the induction of the mesenchymal markers N-cadherin and vimentin (Fig. 3D). However, the expression of epithelial marker E-cadherin was not affected by either BMP-2 or TGF-β1 stimulation (Fig. 3A and B, left), and mesenchymal vimentin was not significantly affected by BMP-2 (Fig. 3C, right). Moreover, the morphology of BMP-2-treated HSC-4 cells was not different from that of control cells (data not shown). Collectively, these data indicated that TGF-β1 suppresses epithelial marker expression and promotes that of mesenchymal markers, resulting in EMT. In contrast, BMP-2 has the opposite effect promoting MET.
Figure 3.
Effects of BMP-2 or TGF-β1 on epithelial and mesenchymal marker expression in HSC-4 cells. Cells were stimulated with (light gray bars) or without (dark grey bars) 10 ng/ml BMP-2 or TGF-β1 for 48 h. (A and B) E-cadherin, (A) CK9 and (B) CK18 were examined as epithelial markers. (C and D) N-cadherin and vimentin were analyzed as mesenchymal markers. Data represent the mean ± SD from triplicate experiments (*P<0.05; **P<0.01). BMP, bone morphogenetic protein; TGF-β1, transforming growth factor-β1.
TGF-β1 abrogates BMP-2-mediated effects on MET/EMT gene expression in a dose-dependent manner
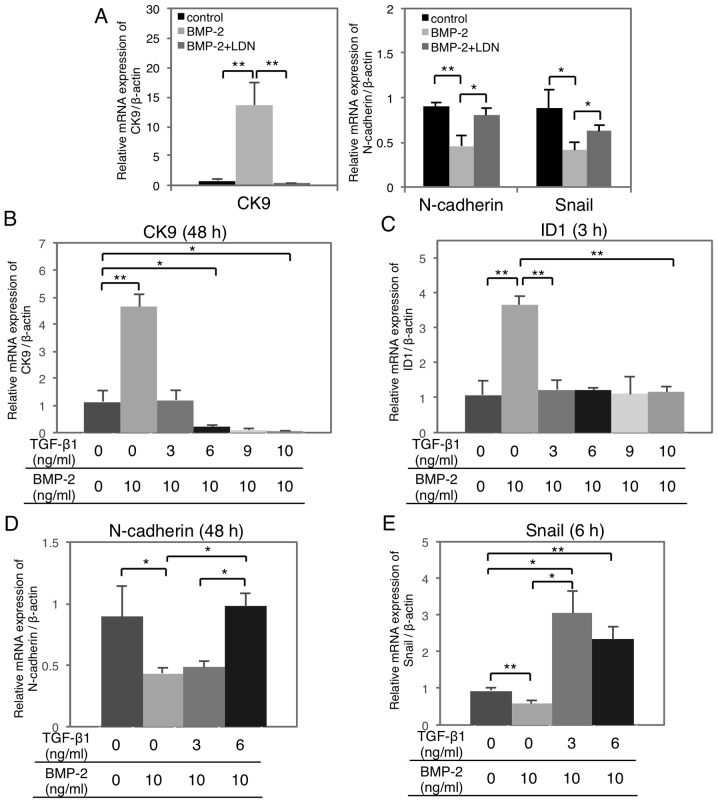
To examine the function of TGF-β1 on BMP-2-induced MET, HSC-4 cells were treated with increasing doses of TGF-β1 with or without BMP-2 stimulation. BMP-2-induced epithelial CK9 (Fig. 4A, left) and ID1 upregulation (Fig. 2B), whereas the N-cadherin and Snail downregulation (Fig. 4A, right) was significantly suppressed by LDN-193189. Moreover, BMP-2-induced CK9 and ID1 upregulation was significantly suppressed by TGF-β1 in a dose-dependent manner (Fig. 4B and C). Similarly, BMP-2-induced N-cadherin and Snail downregulation were significantly suppressed by TGF-β1 stimulation in a dose-dependent manner (Fig. 4D and E).
Figure 4.
TGF-β1 abrogates the BMP-2-mediated effects on gene expression in a dose-dependent manner. (A) HSC-4 cells were cultured with or without 10 ng/ml BMP-2 for 48 h (CK9 and N-cadherin) or 6 h (Snail). Some cells were pre-treated with LDN-193189 (50 nM) for 30 min before BMP-2 stimulation. The expression of CK9, N-cadherin and Snail was analyzed by RT-qPCR. (B-E) Cells were co-stimulated with increasing doses of TGF-β1 as indicated for (B and D) 48 h, (C) 3 h and (E) 6h with 10 ng/ml BMP-2 simultaneously. (B) CK9, (C) ID1, (D) N-cadherin and (E) Snail were analyzed by RT-qPCR. Data represent the mean ± SD from triplicate experiments (*P<0.05; **P<0.01). TGF-β1, transforming growth factor-β1; BMP, bone morphogenetic protein.
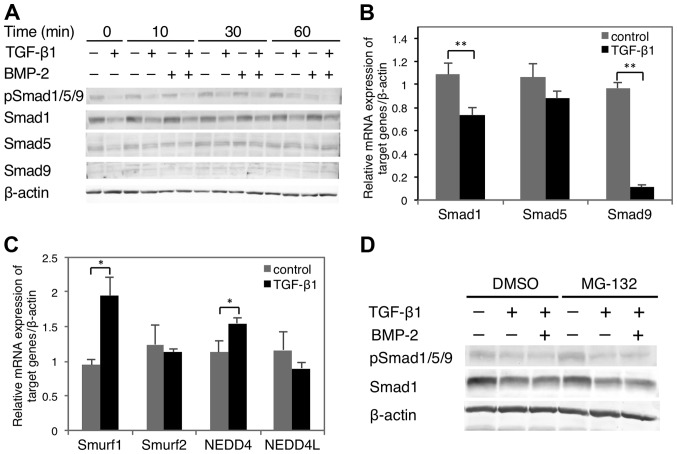
Effect of TGF-β1 on Smad1/5/9 expression and phosphorylation
Western blot analysis showed that TGF-β1 hindered Smad1/5/9 phosphorylation in HSC-4 cells in the presence or absence of BMP-2 (Fig. 5A). In addition, while TGF-β1 suppressed Smad1 expression independently of BMP-2, it had no marked effect on Smad5 expression. Smad9 expression was undetectable regardless of exogenous stimulation. With respect to RNA expression, both Smad1 and 9 were significantly decreased by TGF-β1 stimulation, whereas that of Smad5 was unaffected (Fig. 5B). TGF-β1 stimulation alters the levels of phosphorylated (p)Smad1/5 and pSmad2 by regulating the E3 ligase activities of Smurf1 (32,33), NEDD4 (34) Smurf2 (35) and NEDD4L (36), respectively. Notably, Smurf1 and NEDD4 were significantly upregulated by TGF-β1 stimulation, whereas Smurf2 and NEDD4L were mostly unchanged (Fig. 5C). However, the TGF-β1-mediated degradation of BMP-2-induced pSmad1/5/9 remained intact after treatment with MG132 proteasome inhibitor. This suggested that BMP signal attenuation by TGF-β1 occurs in a proteasome-independent manner (Fig. 5D), possibly through the Smad1/9 downregulation (Fig. 5B). Thus, the effect of TGF-β1 on BMP-2 signaling likely results from the suppression of Smad1/9 expression rather than phosphorylation.
Figure 5.
Effect of TGF-β1 on BMP-2-induced Smad1/5/9 protein expression and phosphorylation in HSC-4 cells. (A) HSC-4 cells were cultured with or without 10 ng/ml TGF-β1 for 48 h, and subsequently treated with or without BMP-2 (20 ng/ml) for the indicated times. Smad1/5/9 expression and phosphorylation were then examined by western blot analysis. (B) Smad1, Smad5 and Smad9 mRNA expression levels following treatment with (black bars) or without (gray bars) TGF-β1 (10 ng/ml) for 48 h. (C) Smurfl, Smurf2, NEDD4 and NEDD4L mRNA expression levels were examined after treatment with (black bars) or without (gray bars) TGF-β1 for 48 h. Data represent the mean ± SD from triplicate experiments (**P<0.01). (D) HSC-4 cells were pre-treated with an MG-132 proteasome inhibitor (0.5 µM) or DMSO (vehicle) for 30 min and then treated with TGF-β1 and/or BMP-2 as described in (A). Smad1/5/9 expression and phosphorylation were then examined by western blot analysis. TGF-β1, transforming growth factor-β1; BMP, bone morphogenetic protein.
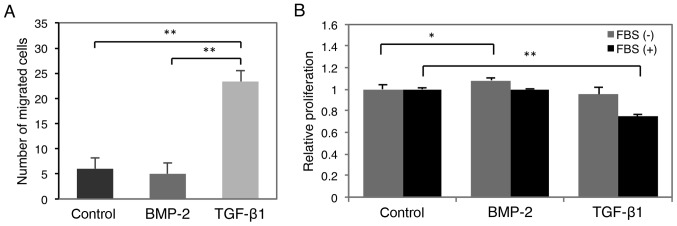
Effect of BMP-2 and TGF-β1 on HSC-4 cell migration and proliferation
The effect of BMP-2 on cell migration and proliferation was investigated to assess its potential effect on hOSCC progression (Fig. 6). This analysis revealed that TGF-β1 significantly enhanced the migratory capacity of HSC-4 cells 72 h after TGF-β1 stimulation, whereas BMP-2 did not (Fig. 6A). Alternatively, BMP-2 facilitated cell proliferation 48 h after stimulation, but was significantly suppressed in the presence of TGF-β1 (Fig. 6B).
Figure 6.
Effect of BMP-2 or TGF-β1 on HSC-4 cell migration and proliferation. (A) Cell migration was evaluated in cells stimulated with BMP-2 (10 ng/ml; dark gray bar), TGF-β1 (10 ng/ml; light gray bar), or left untreated (black bar) using a Boyden chamber assay. Data represent the mean ± SD from triplicate experiments (*P<0.01). (B) Proliferation was monitored in cells stimulated with or without 10 ng/ml BMP-2 or TGF-β1 in serum-free (gray bar) or 10% FBS-supplemented (black bar) media by alamarBlue® assay. Data represent the mean ± SD of 6 wells (*P<0.05, **P<0.01). BMP, bone morphogenetic protein; TGF-β1, transforming growth factor-β1.
Discussion
In order to detect BMP-2-responsive hOSCC, Smad6 and ID1 expression (Fig. 1) and Smad1/5/9 phosphorylation (Fig. 2) were examined in four hOSCC cell lines after BMP-2 stimulation. HSC-4 cells were the most responsive to both BMP-2 and TGF-β1 and thus selected for further analysis. Results from the present study demonstrate that BMP-2 promoted the expression of epithelial CK9, but suppressed that of the mesenchymal markers N-cadherin and vimentin (Fig. 3C), suggesting that it promotes MET. Moreover, these BMP-2-inducible effects were significantly inhibited by TGF-β1 stimulation in HSC-4 cells in a dose-dependent manner (Fig. 4B-E). Collectively, these results strongly suggest that TGF-β1 inhibits BMP-2-induced MET in hOSCC.
Further analysis revealed that TGF-β1 suppressed BMP-2-induced Smad1/5/9 phosphorylation (Fig. 5A) and Smad1/9 expression (Fig. 5B) and promoted the expression of E3 protein ligases that target BMP-2 pathway effectors in HSC-4 cells (Fig. 5C). As such, TGF-β1 is likely a key regulator of Smad1/5/9 pathway suppression downstream of BMP-2 in hOSCC cells. ID (inhibitor of DNA binding) proteins are a family of four transcriptional regulators, including ID1 (31). ID1 expression is reportedly enhanced by BMP-2-mediated Smad1/5 pathway activity (Figs. 1A, 2B and 2C) (37,38). ID proteins have significant implications in cancer progression as Stankic et al reported that ID1 induced MET during metastatic breast cancer cell colonization (39). Moreover, Del Pozo Martin et al (40) reported that metastatic colonization is induced by the interaction between mesenchymal cancer cells and stromal fibroblasts, which secrete factors to induce MET via BMP/Smad1/5 signaling. Our study showed that ID1 protein expression was increased when cultured in activated fibroblast-conditioned media, but was blocked by LDN-193189 treatment. Based on these data, TGF-β1 may suppress MET by disrupting BMP-2-mediated Smad1/5/9 signaling, resulting in ID1 downregulation in HSC-4 cells. On the other hand, Snail is upregulated during EMT and generates a positive feedback loop (10). Notably, Snail expression was significantly suppressed by BMP-2 in HSC-4 cells (Fig. 4A, right and 4E); however, whether BMP-2-induced Smad1/5/9 signaling plays an important role in Snail suppression in HSC-4 cells remains unclear.
Cancer metastasis is the result of cancer cell MET, as well as their proliferative burst after homing to these metastatic sites (12,40). As shown in Fig. 6B, BMP-2 significantly induced HSC-4 cell proliferation, which was not observed following TGF-β1 treatment. If hOSCC cells are susceptible to BMP-2 stimulation at metastatic sites, they likely retain a high proliferative capacity to promote secondary tumor formation. Collectively, this evidence supports that BMP-2 positively regulates metastatic colonization in hOSCC. On the other hand, TGF-β1 induces EMT (28) and increases cell migration (Fig. 6A) and invasion (29) in primary hOSCC tumors. In addition, TGF-β1 may inhibit tumor progression by attenuating BMP-2-induced MET at metastatic sites.
Yang et al (20) reported that BMP-2 suppresses EMT in TGF-β1-induced renal interstitial fibrosis. Interestingly, BMP-2 attenuated TGF-β1-induced EMT of NRK-49F kidney fibroblasts downregulating Snail expression. Alternatively, we found that the BMP-2-induced Snail downregulation was significantly inhibited by TGF-β1 stimulation in a dose-dependent manner (Fig 4E), suggesting that TGF-β1 suppresses the BMP-2-induced MET by disrupting the induction of Snail in hOSCC cells. Recently, it was reported that BMP-4 may inhibit TGF-β1-induced EMT in primary retinal pigment epithelium cells through the Smad2/3 pathway (41). Therefore, it will be necessary to determine whether TGF-β1-induced EMT is inhibited by BMP stimulation in hOSCC cells in the future.
Our findings partly clarify the molecular mechanisms underlying EMT and MET in hOSCC and may facilitate the discovery of molecular drug targets to attenuate hOSCC progression.
Acknowledgements
We would like to thank Editage (www.editage.jp) for English language editing. This study was supported in part by a Grant-in-aid for Scientific Research (no. 90118274 to S.K., no. 26293426 to T.S. and no. 2667052 and 16H05534 to A.I.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; Grant-in-aid for the Strategic Medical Science Research Center from the Ministry of Education, Culture, Sports, Science and Technology of Japan, 2010–2014.
Glossary
Abbreviations
- BMP
bone morphogenetic protein
- CK9
cytokeratin 9
- CK18
cytokeratin 18
- EMT
epithelial-mesenchymal transition
- hOSCC
human oral squamous cell carcinoma
- MET
mesenchymal-epithelial transition
- TGF-β
transforming growth factor-β
References
- 1.Smith A, Teknos TN, Pan Q. Epithelial to mesenchymal transition in head and neck squamous cell carcinoma. Oral Oncol. 2013;49:287–292. doi: 10.1016/j.oraloncology.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial- mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Weber CE, Li NY, Wai PY, Kuo PC. Epithelial-mesenchymal transition, TGF-β, and osteopontin in wound healing and tissue remodeling after injury. J Burn Care Res. 2012;33:311–318. doi: 10.1097/BCR.0b013e318240541e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He J, Xu Y, Koya D, Kanasaki K. Role of the endothelial-to-mesenchymal transition in renal fibrosis of chronic kidney disease. Clin Exp Nephrol. 2013;17:488–497. doi: 10.1007/s10157-013-0781-0. [DOI] [PubMed] [Google Scholar]
- 5.Fabregat I, Malfettone A, Soukupova J. New insights into the crossroads between EMT and stemness in the context of cancer. J Clin Med. 2016;5:37. doi: 10.3390/jcm5030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moustakas A, Heldin P. TGFβ and matrix-regulated epithelial to mesenchymal transition. Biochim Biophys Acta. 2014;1840:2621–2634. doi: 10.1016/j.bbagen.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, De Herreros A García. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 8.Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial-mesenchymal transition through β-catenin-T-cell factor-4-dependent expression of transforming growth factor-β3. Mol Biol Cell. 2008;19:4875–4887. doi: 10.1091/mbc.E08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 11.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wakefield LM, Hill CS. Beyond TGFβ: Roles of other TGFβ superfamily members in cancer. Nat Rev Cancer. 2013;13:328–341. doi: 10.1038/nrc3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simic P, Vukicevic S. Bone morphogenetic proteins in development and homeostasis of kidney. Cytokine Growth Factor Rev. 2005;16:299–308. doi: 10.1016/j.cytogfr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: A critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Wu M, Chen G, Li YP. TGF-β and BMP signaling in osteoblast, skeletal development and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopf J, Paarmann P, Hiepen C, Horbelt D, Knaus P. BMP growth factor signaling in a biomechanical context. Biofactors. 2014;40:171–187. doi: 10.1002/biof.1137. [DOI] [PubMed] [Google Scholar]
- 17.Chen G, Deng C, Li YP. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8:272–288. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormack N, O'Dea S. Regulation of epithelial to mesenchymal transition by bone morphogenetic proteins. Cell Signal. 2013;25:2856–2862. doi: 10.1016/j.cellsig.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Kang MH, Kim JS, Seo JE, Oh SC, Yoo YA. BMP2 accelerates the motility and invasiveness of gastric cancer cells via activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway. Exp Cell Res. 2010;316:24–37. doi: 10.1016/j.yexcr.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Yang YL, Ju HZ, Liu SF, Lee TC, Shih YW, Chuang LY, Guh JY, Yang YY, Liao TN, Hung TJ, et al. BMP-2 suppresses renal interstitial fibrosis by regulating epithelial-mesenchymal transition. J Cell Biochem. 2011;112:2558–2565. doi: 10.1002/jcb.23180. [DOI] [PubMed] [Google Scholar]
- 21.Graves CA, Abboodi FF, Tomar S, Wells J, Pirisi L. The translational significance of epithelial-mesenchymal transition in head and neck cancer. Clin Transl Med. 2014;3:60. doi: 10.1186/s40169-014-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert R, Sauvaget C, de Camargo Cancela M, Sankaranarayanan R. Epidemiology of cancer from the oral cavity and oropharynx. Eur J Gastroenterol Hepatol. 2011;23:633–641. doi: 10.1097/MEG.0b013e3283484795. [DOI] [PubMed] [Google Scholar]
- 23.Kokorina NA, Lewis JS, Jr, Zakharkin SO, Krebsbach PH, Nussenbaum B. rhBMP-2 has adverse effects on human oral carcinoma cell lines in vivo. Laryngoscope. 2012;122:95–102. doi: 10.1002/lary.22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kokorina NA, Zakharkin SO, Krebsbach PH, Nussenbaum B. Treatment effects of rhBMP-2 on invasiveness of oral carcinoma cell lines. Laryngoscope. 2011;121:1876–1880. doi: 10.1002/lary.21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sand JP, Kokorina NA, Zakharkin SO, Lewis JS, Jr, Nussenbaum B. BMP-2 expression correlates with local failure in head and neck squamous cell carcinoma. Otolaryngol Head Neck Surg. 2014;150:245–250. doi: 10.1177/0194599813513003. [DOI] [PubMed] [Google Scholar]
- 26.Kejner AE, Burch MB, Sweeny L, Rosenthal EL. Bone morphogenetic protein 6 expression in oral cavity squamous cell cancer is associated with bone invasion. Laryngoscope. 2013;123:3061–3065. doi: 10.1002/lary.24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan J, Johnson NW, Zhou G, Parsons PG, Boyle GM, Gao J. Potential molecular targets for inhibiting bone invasion by oral squamous cell carcinoma: A review of mechanisms. Cancer Metastasis Rev. 2012;31:209–219. doi: 10.1007/s10555-011-9335-7. [DOI] [PubMed] [Google Scholar]
- 28.Saito D, Kyakumoto S, Chosa N, Ibi M, Takahashi N, Okubo N, Sawada S, Ishisaki A, Kamo M. Transforming growth factor-β1 induces epithelial-mesenchymal transition and integrin α3β1-mediated cell migration of HSC-4 human squamous cell carcinoma cells through Slug. J Biochem. 2013;153:303–315. doi: 10.1093/jb/mvs144. [DOI] [PubMed] [Google Scholar]
- 29.Hino M, Kamo M, Saito D, Kyakumoto S, Shibata T, Mizuki H, Ishisaki A. Transforming growth factor-β1 induces invasion ability of HSC-4 human oral squamous cell carcinoma cells through the Slug/Wnt-5b/MMP-10 signalling axis. J Biochem. 2016;159:631–640. doi: 10.1093/jb/mvw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Lasorella A, Benezra R, Iavarone A. The ID proteins: Master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 32.Shi W, Chen H, Sun J, Chen C, Zhao J, Wang YL, Anderson KD, Warburton D. Overexpression of Smurf1 negatively regulates mouse embryonic lung branching morphogenesis by specifically reducing Smad1 and Smad5 proteins. Am J Physiol Lung Cell Mol Physiol. 2004;286:L293–L300. doi: 10.1152/ajplung.00228.2003. [DOI] [PubMed] [Google Scholar]
- 33.Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14:2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim BG, Lee JH, Yasuda J, Ryoo HM, Cho JY. Phospho-Smad1 modulation by nedd4 e3 ligase in BMP/TGF-β signaling. J Bone Miner Res. 2011;26:1411–1424. doi: 10.1002/jbmr.348. [DOI] [PubMed] [Google Scholar]
- 35.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 36.Gao S, Alarcón C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massagué J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogata T, Wozney JM, Benezra R, Noda M. Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc Natl Acad Sci USA. 1993;90:9219–9222. doi: 10.1073/pnas.90.19.9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langenfeld EM, Kong Y, Langenfeld J. Bone morphogenetic protein 2 stimulation of tumor growth involves the activation of Smad-1/5. Oncogene. 2006;25:685–692. doi: 10.1038/sj.onc.1209110. [DOI] [PubMed] [Google Scholar]
- 39.Stankic M, Pavlovic S, Chin Y, Brogi E, Padua D, Norton L, Massagué J, Benezra R. TGF-β-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell Reports. 2013;5:1228–1242. doi: 10.1016/j.celrep.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Pozo Martin Y, Park D, Ramachandran A, Ombrato L, Calvo F, Chakravarty P, Spencer-Dene B, Derzsi S, Hill CS, Sahai E, et al. Mesenchymal cancer cell-stroma crosstalk promotes niche activation, epithelial reversion, and metastatic colonization. Cell Reports. 2015;13:2456–2469. doi: 10.1016/j.celrep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao H, Li H, Yang S, Li M, Zhao C, Zhang J, Xu G, Wang F. Inhibitory effect of bone morphogenetic protein 4 in retinal pigment epithelial-mesenchymal transition. Sci Rep. 2016;6:32182. doi: 10.1038/srep32182. [DOI] [PMC free article] [PubMed] [Google Scholar]