Abstract
The androgen receptor (AR) is frequently expressed in breast cancer; however, its prognostic value remains unclear. AR expression in breast cancer has been associated with improved outcomes in estrogen receptor (ER)-positive breast cancer compared with ER-negative disease. Eliminating AR function in breast cancer is critically important for breast cancer progression. However, the mechanism underlying AR regulation remains poorly understood. The study of microRNAs (miRNAs) has provided important insights into the pathogenesis of hormone-dependent cancer. To determine whether miRNAs function in the AR regulation of breast cancer, the present study performed miRNA expression profiling in AR-positive and -negative breast cancer cell lines. A total of 153 miRNAs were differentially expressed in AR-positive compared with AR-negative breast cancer cells; 52 were upregulated and 101 were downregulated. A number of these have been extensively associated with breast cancer cell functions, including proliferation, invasion and drug-resistance. Furthermore, through pathway enrichment analysis, signaling pathways associated with the prediction targets of the miRNAs were characterized, including the vascular endothelial growth factor and mammalian target of rapamycin signaling pathways. In conclusion, the results of the present study indicated that the expression of miRNAs may be involved in the mechanism underlying AR regulation of breast cancer, and may improve understanding of the role of AR in breast cancer.
Keywords: microRNA, profiling, differential, androgen receptor, breast cancer
Introduction
Breast cancer remains a major health problem for women worldwide; accounting for one in three newly diagnosed cases of cancer in women. Targeting the estrogen receptor (ER) has achieved marked progress in the treatment of patients with ER-positive breast cancer. Although the ER is critical for breast cancer carcinogenesis, the most commonly expressed hormone receptor in invasive breast cancer is the androgen receptor (AR), which is expressed in 72.9% of primary breast cancer cases (1). Furthermore, the AR has been characterized in terms of its role as a predictive factor, and the clinical significance of its expression in breast cancer. The AR is inhibitory in breast cancer, counteracting the oncogenic effect of the ER. Due to this crosstalk, AR expression is associated with improved prognosis in ER-positive breast cancer (2,3). However, the prognostic value of AR expression in triple negative breast cancer (TNBC) remains unclear, with certain studies suggesting reduced mortality (4), and others indicating poor prognosis (5). A recent systematic review of 19 separate studies investigated the association between AR expression, overall survival and disease free survival, and revealed that the expression of AR is a marker of good prognosis, with an approximate doubling of overall survival at 3 or 5 years (6). Therefore, further examination of the role of the AR in breast cancer is essential, and may contribute to the development of novel therapies for AR-positive tumors, particularly ER−, progesterone receptor (PR)−, AR+ breast cancer patients who derive limited benefit from current chemotherapeutic strategies.
MicroRNAs (miRNAs) are small non-coding RNAs, ~22 nucleotides in length, which regulate the expression of multiple genes by degrading messenger RNA (mRNA) or interrupting the translation process (7). Aberrant expression of miRNAs contributes to cancer pathogenesis by inducing oncogenes, inhibiting tumor suppressor genes or disrupting important signaling pathways. Dysregulation of miRNAs occurs in breast cancer pathogenesis. Sets of genes have been identified to be dysregulated in breast cancer using miRNA expression profiling studies (8,9). Furthermore, functional studies have confirmed miRNAs as tumor suppressors and oncogenes in breast cancer (10).
miRNAs impact the pathobiology of hormone-regulated cancers. The potential association between AR expression and miRNA signature in prostate cancer has been examined. A miRNA profiling study of 6 prostate cancer (PCa) cell lines, 9 prostate cancer xenograft models and 9 prostate carcinoma samples identified a significant link between miRNAs and AR expression (11), and revealed a potential correlation for further investigation. Similarly, in 2007, Shi et al (12) observed differential expression of miR-125b in androgen-dependent and -independent PCa cells, as well as in benign and malignant prostate tissues. This study suggested that androgen-AR signaling may regulate the differential expression of miR-125b. Furthermore, a study in 2011 described 71 miRNAs that influenced the expression levels of AR in human PCa cells, and 13 miRNAs were validated to regulate the long AR 3′untranslated region (UTR) (13). Taken together, these findings indicate a potential link between AR expression and miRNAs. In breast cancer, miRNAs have been studied primarily with regard to ER and human epidermal growth receptor 2. AR-associated miRNAs in breast cancer have been less well investigated.
To reveal the association between miRNAs and AR in breast cancer, miRNA expression profiling was performed in breast cancer cell lines representative of various AR expressions. Further target prediction was conducted on the significantly dysregulated miRNAs. Target genes were classified into different pathways according to their biological functions, as determined by the Gene Ontology (GO) system. Vascular endothelial growth factor (VEGF) and mammalian target of rapamycin (mTOR) signaling pathways were demonstrated to correlate with the significantly dysregulated miRNAs. The results of the present study revealed a correlation between differential miRNA expression and AR expression levels in breast cancer, and described a putative interaction between the AR, VEGF and mTOR signaling pathways. These results may improve understanding of the role of the AR in breast cancer.
Materials and methods
Cell culture
The Hs578T, MDA-MB-231, MCF-7 and SK-BR-3 human breast cancer cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). Hs578T and MCF-7 cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin/streptomycin and 4 mg/ml insulin, whereas MDA-MB-231 and SK-BR-3 cells were cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS and 1% penicillin/streptomycin. All cells were cultured at 37°C, in an atmosphere of 5% CO2.
RNA preparation
Samples were harvested from Hs578T, MDA-MB-231, MCF-7 and SK-BR-3 human breast cancer cells. Total RNA was prepared using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), and the quality and quantity of the RNA were assessed using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.). Optical density (OD) 260/280 ≥1.6 and OD 260/230 ≥1 indicated acceptable RNA purity, whereas acceptable RNA integrity (RNA integrity number ≥5) was determined using an Agilent RNA 6000 Nano assay (Agilent Technologies, Inc., Santa Clara, CA, USA). Gel electrophoresis was used to evaluate genomic DNA contamination.
miRNAs and gene expression analysis
RNA samples were subjected to Human OneArray® version 6 (Phalanx Biotech Group, Hsinchu, Taiwan). Data were analyzed with Rosetta Resolver System software (Rosetta Biosoftware, Seattle, WA, USA). Standard selection criteria were used to identify differentially expressed genes: i) Absolute log2 fold change ≥0.585; absolute fold change ≥1.5 and ii) false discovery rate <0.05, which were subsequently categorized into up and downregulated genes for AR-positive vs. -negative breast cancer cells.
miRNA target prediction and miRNA-gene interaction analysis
The miRNAs whose expression was significantly dysregulated between AR-positive and -negative breast cancer cells were selected for target prediction. Predicted target genes were identified by at least three of the seven well-established databases: DIANA (diana.imis.athena-innovation.gr/DianaTools/index.php), miRanda (www.microrna.org/microrna/home.do), miBridge (sitemaker.umich.edu/mibridge/home), PicTar (pictar.mdc-berlin.de/), PITA (genie.weizmann.ac.il/pubs/mir07/mir07_data.html), RNA22 (cm.jefferson.edu/rna22/) and TargetScan (www.targetscan.org/vert_71/). Pathway enrichment analysis was performed using the GO system to gain insight into the molecular networks and canonical pathways associated with differentially expressed miRNA. Of the enriched pathways with a P-value of <0.05, the associations between the differentially expressed miRNAs and their candidate target genes were visualized with an interaction network, for the VEGF and mTOR signaling pathways.
Results
miRNAs are differentially expressed in AR-positive and -negative breast cancer cell lines
Previous reports have indicated that AR protein expression levels may be affected by a set of miRNAs in PCa cells (13); another 25 miRNAs have been reported to be differentially expressed in PCa compared with matched benign tissues (14). This evidence suggests that there is an association between miRNA expression and the AR signaling pathway. However, the mechanism underlying AR-mediated regulation of miRNAs in PCa and TNBC remains unclear. To further identify differentially expressed miRNAs in breast cancer cell lines with varying AR expression, miRNA expression profiling was performed in breast cancer cell lines representative of AR-negative (Hs578T: ER−/PR−/HER2−/AR−) and AR-positive (MDA-MB-231: ER−/PR−/HER2−/AR+; MCF-7: ER+/PR+/HER2−/AR+; SK-BR-3: ER−/PR−/HER2+/AR+) molecular subtypes. A total of 153 miRNAs were identified as significantly altered in AR-positive vs. AR-negative cell lines, with at least a threefold change in expression (P<0.05; Fig. 1). Of the 52 upregulated miRNAs, miR-933 and miR-5793 had the most significant increases, with miR-933 expression undergoing the greatest upregulation (2.83-fold; P=0.026). Of the 101 downregulated miRNAs, miR-4792 expression underwent the greatest decrease (5.93-fold; P=0.002).
Figure 1.
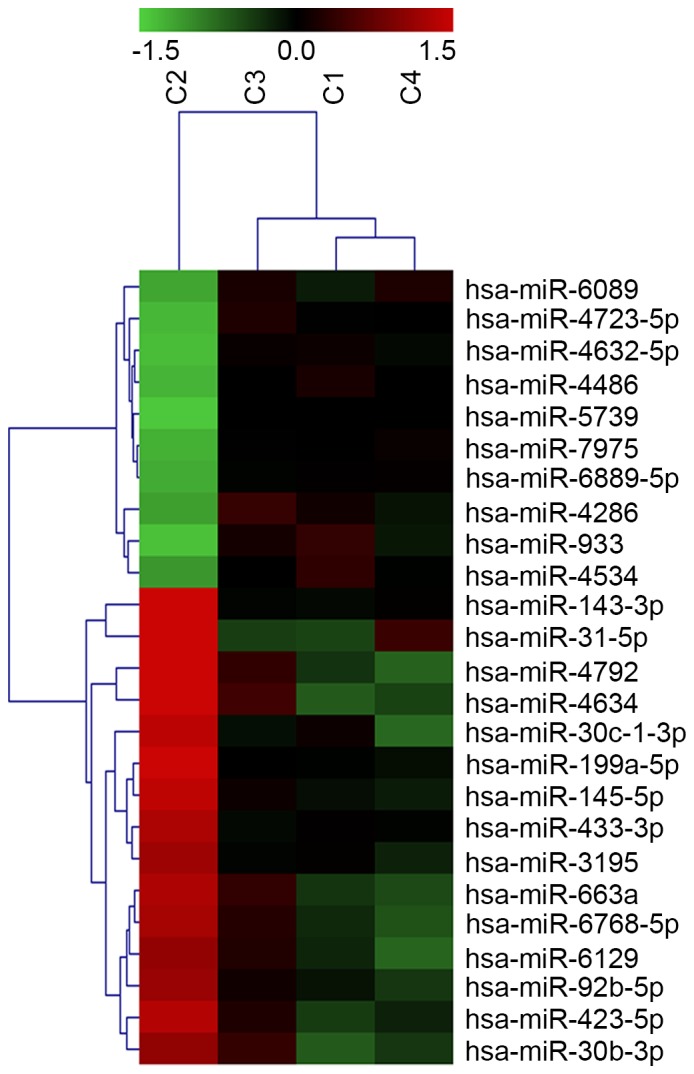
Differential expression of miRNAs. Of the 153 differentially expressed miRNAs in AR-positive vs. -negative breast cancer cell lines, hierarchical clustering of the 25 most variably expressed are presented. Rows, individual microRNAs; columns, individual breast cancer cell lines (C1, MDA-MB-231; C2, Hs578T; C3, MCF-7; C4, SK-BR-3). The colors in the heatmap represent the normalized expression values with downregulation represented by shades of green and upregulation by shades of red. miRNA, microRNA.
Predicted targets of the differentially expressed miRNAs and their involvement in signaling pathways
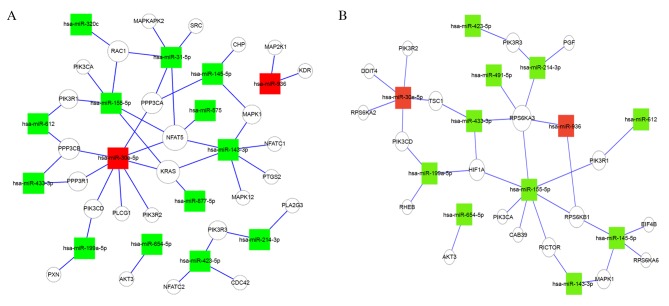
As a single miRNA may target multiple mRNA transcripts, the dysregulation of sets of miRNAs may impact significantly on cancerogenesis via the regulation of multiple signaling pathways. To determine the probable biological functions of the differentially expressed miRNAs, the potential targets of the 153 differently expressed miRNAs were predicted in seven online databases: DIANA, miRanda, miRBridge, PicTar, PITA, RNA22 and TargetScan. A total of 5,576 target genes were predicted, and classified into different pathways according to their basic biological functions by the GO system. Multiple effectors of pathways involved in tumor cell proliferation and invasion were characterized, for example in the VEGF and mTOR signaling pathways (Fig. 2). Due to the importance of the VEGF and mTOR signaling pathways in breast cancer, the VEGF-miRNA-target gene network (Fig. 3A) and mTOR-miRNA-target gene network (Fig. 3B) were constructed to reveal the association between AR-associated miRNAs and their target effectors in the two signaling pathways. The present study revealed a putative role of AR in the VEGF and mTOR signaling pathways, suggesting that AR may regulate the pathways through up or downregulating miRNAs. Further work is required to improve the understanding of the role of AR in these signaling pathways.
Figure 2.
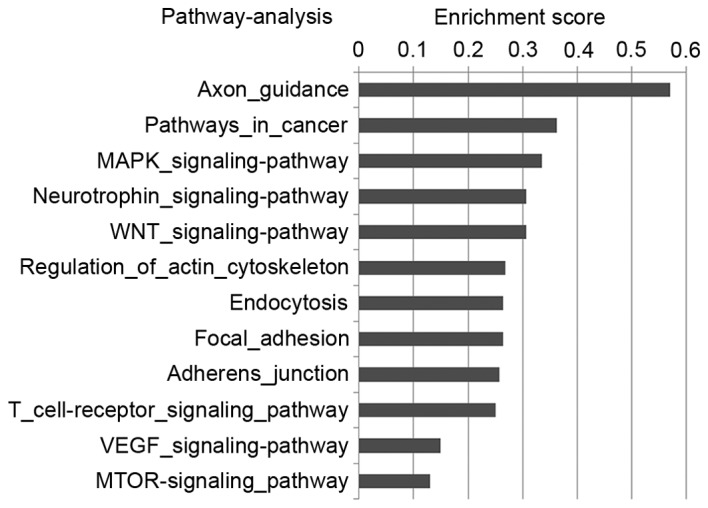
Signaling pathways of the target genes of differentially expressed miRNAs. The target genes of the differentially expressed miRNAs were classified into various signaling pathways. The vertical axis represents the pathway category, and the horizontal axis the enrichment score. miRNA, microRNA.
Figure 3.
Signaling pathway networks. (A) VEGF-miRNA-target genes network. (B) mTOR-miRNA-target genes network. The red/green square nodes represent upregulated/downregulated miRNAs in AR-positive vs. -negative breast cancer cell lines. The hollow circles represent predicted target genes. VEGF, vascular endothelial growth factor; miRNA, microRNA; mTOR, mammalian target of rapamycin; AR, androgen receptor.
Discussion
The present study performed a screening to identify AR-associated miRNAs in four breast cancer cell lines. By focusing on miRNAs that were associated with AR in three separate cell lines, cell-dependent bias was reduced. A total of 153 miRNAs were identified as differentially expressed. Of these, 52 were upregulated and 101 were downregulated. By predicting the targets of differentially expressed miRNAs, interactions between AR, mTOR and VEGF signaling pathways were indicated, suggesting that the AR may interact with miRNAs and contribute to breast cancer carcinogenesis via VEGF or mTOR signaling pathways. To date, studies validating the association between the AR and miRNA expression have primarily been conducted in PCa. Few studies have reported the role of AR-associated miRNAs in breast cancer. Thus, the present study increases the understanding of the role of AR in breast cancer.
miRNAs have previously been implicated in the regulation of AR signaling in prostate cancer. Chromatin immunoprecipitation analysis confirmed AR binding to putative androgen responsive elements (AREs) within the promoter regions of these miRNAs, thereby regulating miRNA expression (15). Porkka et al (11) in 2007 were the first to reveal an association between the AR and miRNAs. Following miRNA expression profiling of PCa cell lines and carcinoma samples, hierarchical clustering of the aberrantly expressed miRNAs accurately separated the carcinomas according to AR status, indicating that the AR may regulate the expression of miRNAs. Furthermore, a number of studies have described the roles of miRNAs as regulators of AR activity. Östling et al (13) demonstrated that the majority of the miRNAs regulating the AR targeted an extended 6 kb 3′UTR. A total of 71 unique miRNAs were identified to influence the level of AR expression in PCa cell lines. Therefore, there exists a potential link between AR signaling and miRNAs, with implications for the development of strategies to inhibit AR function via miRNAs-associated pathways.
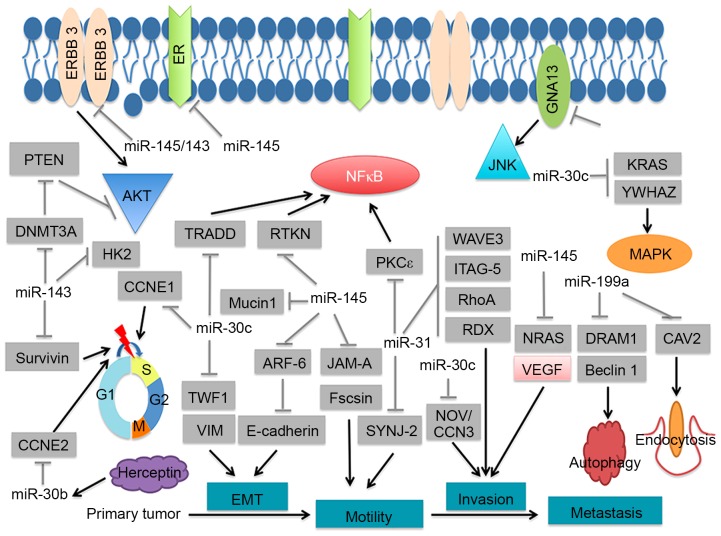
A differential miRNA expression pattern, involving 153 miRNAs, was detected in the present study, 52 upregulated and 101 downregulated, thus identifying those miRNAs that may be significant in breast cancer development. Certain significantly dysregulated miRNAs in the upregulated and downregulated groups have been previously associated with breast cancer progression. Notably, certain of the significantly downregulated miRNAs have been associated with breast cancer cell functions, including proliferation, invasion and drug-resistance (Table I; Fig. 4). For example, miR-143 and miR-145, components of the miR-143/145 cluster, were significantly downregulated. miR-143 and miR-145 have been demonstrated to possess anti-tumorigenic activity (Table I). Furthermore, expression of the miR-143/145 cluster was revealed to function as a tumor suppressor in breast cancer via the inhibition of ERBB3 (17). Stimulation of EGF pathways, including ERBB3, leads to enhanced AR stability and promotes AR binding to AREs. Therefore, the miR-143/145 cluster may be involved in the underlying mechanism of the AR in breast cancer. Furthermore, miR-31, an miRNA that was significantly downregulated in the present study, has been identified to directly target and destabilize the AR through its coding sequence. The mutual regulation between miR-31 and AR maintains prostate cellular homeostasis, and the loss of miR-31 contributes to PCa progression through unregulated AR expression. In breast cancer, miR-31 serves as a tumor suppressor via inhibition of guanine nucleotide binding protein alpha 13, WAS protein family member 3, β1-Integrin and protein kinase C ε (Table I). miR-181, another dysregulated miRNA in the present study, has been demonstrated to indirectly target AR to function as a growth suppressive miRNA in prostate cancer (40). Taken together, the dysregulated miRNAs identified by the present study may regulate the expression of AR in breast cancer. It may be useful to determine whether the dysregulated pattern is implicated in the role of the AR in breast cancer. The majority of the upregulated miRNAs identified in the present study have not been widely reported as aberrantly expressed in other cancers, which suggests that AR-associated breast cancer may have a specific signature. Further studies are required to confirm the expression of the dysregulated miRNAs by polymerase chain reaction anaylsis.
Table I.
Significantly downregulated miRNAs in the present study are associated with breast cancer.
| Author, year | miRNA | Cancer | Target gene | Clinical specimen/animal model/cell line | Function | Ref. |
|---|---|---|---|---|---|---|
| Ng, 2014 | miR-143 | Breast | DNMT3A | MDA-MB-231, T47D | Inhibit cell proliferation | 16 |
| Yan, 2014 | ERBB3 | Breast cancer tissues, MDA-MB-231, MCF-7, xenograft mouse model | Inhibit cell proliferation and invasion | 17 | ||
| Jiang, 2012 | HK2 | MDA-MB-231, ZR-75-30, xenograft mouse model | Inhibit cell proliferation and invasion | 18 | ||
| Yu, 2012 | Survivin | MCF-7 | Inhibit cell proliferation | 19 | ||
| Rasheed, 2015 | miR-31 | Breast | GNA13 | Breast cancer tissues, MDA-MB-231, MCF-10a | Inhibit cell invasion | 20 |
| Augoff, 2011 | β1-Integrin | MDA-MB-231 | Inhibit cell invasion | 21 | ||
| Sossey-Alaoui, 2011 | Wave3 | Breast cancer tissues, MDA-MB-231 | Inhibit cell invasion | 22 | ||
| Körner, 2013 | PKCε | MDA-MB-231, MCF-10A | Inhibit cell invasion | 23 | ||
| Valastyan, 2010 | RDX ITGA5 RhoA | MDA-MB-231, xenograft mouse model | Inhibit cell invasion | 24 | ||
| Ben-Chetrit, 2015 | SYNJ2 | Breast cancer tissues, MD-MB-231, MCF-10a, xenograft mouse model | Inhibit cell invasion | 25 | ||
| Dobson, 2014 | miR-30c | Breast | NOV/CCN3 | MDA-MB-231 | Promote cell invasion | 26 |
| Shukla, 2015 | TRADD CCNE1 | Breast cancer tissues, MDA-MB-231 | Inhibit cell proliferation and invasion | 27 | ||
| Tanic, 2012 | KRAS | MDA-MB-436 | Inhibit cell growth | 28 | ||
| Bockhorn, 2013 | TWF1 VIM | MDA-MB-231, xenograft mouse model | Inhibit cell invasion | 10 | ||
| Bockhorn, 2013 | TWF1 | Breast cancer tissues, MDA-MB-231, xenograft mouse model | Inhibit chemo-resistance | 29 | ||
| Fang, 2014 | YWHAZ | MCF-7, xenograft mouse model | Inhibit chemo-resistance | 30 | ||
| Yi, 2013 | miR-199a | Breast | DRAM1 Beclin 1 | MDA-MB-231, MCF-7 | Regulate apoptosis | 31 |
| Shatseva, 2011 | CAV-2 | MDA-MB-231 | Inhibit cell proliferation | 32 | ||
| Yan, 2014 | miR-145 | Breast | ERBB3 | Breast cancer tissues, MDA-MB-231, MCF-7, xenograft mouse model | Inhibit cell proliferation and invasion | 17 |
| Eades, 2015 | ARF6 | Breast cancer tissues, MDA-MB-231 | Inhibit cell invasion | 33 | ||
| Zou, 2012 | N-RAS | Breast cancer tissues, MDA-MB-231, MCF-7, | Inhibit cell growth, Invasion, and | 34 | ||
| VEGF-A | xenograft mouse model | angiogenesis | ||||
| Wang, 2009 | RTKN | MFC-7 | Inhibit cell growth | 35 | ||
| Spizzo, 2010 | ER-α | MDA-MB-231, MCF-7 | Inhibit cell growth | 36 | ||
| Gotte, 2010 | JAM-A Fascin | MB-MDA-231, MCF-7, MDA-MB-468, SK-BR-3 | Inhibit cell invasion and motility | 37 | ||
| Liu, 2014 | Muc-1 | Breast cancer tissues, MDA-MB-231, LM2-4142 | Inhibit cell growth and invasion | 38 | ||
| Ichikawa, 2012 | miR-30b-3p | Breast | CCNE2 | SKBR3; BT474 | Inhibit cell growth | 39 |
miRNA, microRNA.
Figure 4.
Involvement of the significantly dysregulated microRNAs in breast cancer signaling pathways.
However, studies on AR-associated miRNAs in breast cancer are limited. To the best of our knowledge, only two studies have been performed to date. Nakano et al (41) identified miRNAs induced by dihydrotestosterone (DHT) in an AR-positive breast cancer cell line. A total of 13 miRNAs were upregulated and 28 miRNAs were downregulated significantly. Of the aberrantly expressed miRNAs, miR-363, with the fold-change (8.15-fold) targets IQ motif- and WD repeats-containing 1 to influence the mechanism underlying the action of androgens. Furthermore, to identify critical miRNAs associated the androgen-induced AR signaling pathway in breast cancer, Lyu et al (42) examined global miRNA expression in DHT-treated MCF-7 and MDA-MB-453 cells. A total of 10 differentially expressed miRNAs were identified in MDA-MB-453 cells. Of these, 4 upregulated miRNAs (let-7a, b, c and d) were detected. Androgen-induced AR activating signal was revealed to directly upregulate let-7a expression, downregulate CMYC and KRAS expression and inhibit cell proliferation in ER−, PR−, AR+ breast cancer cells. miR-30b, another critical miRNA associated with the androgen-induced AR activating signal in breast cancer (42), was aberrantly expressed in the present study, which screened the AR 3′UTR for potential miRNA target sites. miR-30b was predicted by miRanda to target the 3′UTR of the AR. Taken together, these data suggested a potential link between AR signaling and miRNAs in breast cancer. To investigate the role of AR-associated miRNAs in breast cancer, further investigations are required. The present study has identified a distinct miRNA expression pattern in AR-positive breast cancer cell lines, which indicates a critical role for miRNAs in AR-positive and -negative breast cancer.
Pathway enrichment analysis suggested that the differentially expressed miRNAs collectively targeted various signaling pathways associated with cell proliferation and invasion. Notably, VEGF and mTOR pathways, the key pathways of breast tumorigenesis, were identified as associated with the deregulated miRNAs of the present study (43). Previous studies have revealed that the involvement of miRNAs was critical for VEGF-induced angiogenesis and mTOR-associated tumor proliferation (44,45). In the present study, a putative correlation was revealed between AR, VEGF and mTOR signaling pathways. The prognostic value of AR expression in TNBC remains elusive, with certain studies suggesting reduced mortality (4), and others indicating poor prognosis (5). Furthermore, our previous study identified a role for AR in tumorigenesis role and the inhibitory effect of AR antagonist in AR-positive mesenchymal stem-like TNBC in vitro and in vivo, suggesting that AR inhibition may be a potential therapeutic approach for AR-positive TNBC patients (46). AR may therefore be a tumorigenic factor in breast cancer. AR-associated miRNAs may be involved in the VEGF and mTOR signaling pathways to promote tumor cell proliferation.
In conclusion, the results of the present study indicated that there is an imbalance of miRNAs levels in AR-positive breast cancer cells compared with AR-negative cells, suggesting an important role for miRNAs in the function of the AR in breast cancer. Following classification of the miRNAs targets into different pathways, a potential interaction between AR, VEGF and mTOR signaling pathways was revealed. These results suggested a potential underlying mechanism of AR in breast cancer via the dysregulation of the expression pattern of miRNAs.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81272252) and the Foundation for Clinical Medicine, Science and Technology Special Project of Jiangsu Province (grant no. BL2014071) to X. G.
References
- 1.Park S, Koo J, Park HS, Kim JH, Choi SY, Lee JH, Park BW, Lee KS. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2010;21:488–492. doi: 10.1093/annonc/mdp510. [DOI] [PubMed] [Google Scholar]
- 2.Tsang JY, Ni YB, Chan SK, Shao MM, Law BK, Tan PH, Tse GM. Androgen receptor expression shows distinctive significance in ER positive and negative breast cancers. Ann Surg Oncol. 2014;21:2218–2228. doi: 10.1245/s10434-014-3629-2. [DOI] [PubMed] [Google Scholar]
- 3.Loibl S, Müller BM, von Minckwitz G, Schwabe M, Roller M, Darb-Esfahani S, Ataseven B, du Bois A, Fissler-Eckhoff A, Gerber B, et al. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast Cancer Res Treat. 2011;130:477–487. doi: 10.1007/s10549-011-1715-8. [DOI] [PubMed] [Google Scholar]
- 4.He J, Peng R, Yuan Z, Wang S, Peng J, Lin G, Jiang X, Qin T. Prognostic value of androgen receptor expression in operable triple-negative breast cancer: A retrospective analysis based on a tissue microarray. Med Oncol. 2012;29:406–410. doi: 10.1007/s12032-011-9832-0. [DOI] [PubMed] [Google Scholar]
- 5.Hu R, Dawood S, Holmes MD, Collins LC, Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA, Tamimi RM. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res. 2011;17:1867–1874. doi: 10.1158/1078-0432.CCR-10-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vera-Badillo FE, Templeton AJ, de Gouveia P, Diaz-Padilla I, Bedard PL, Al-Mubarak M, Seruga B, Tannock IF, Ocana A, Amir E. Androgen receptor expression and outcomes in early breast cancer: A systematic review and meta-analysis. J Natl Cancer Inst. 2014;106:djt319. doi: 10.1093/jnci/djt319. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dvinge H, Git A, Gräf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 9.Mulrane L, McGee SF, Gallagher WM, O'Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 10.Bockhorn J, Yee K, Chang YF, Prat A, Huo D, Nwachukwu C, Dalton R, Huang S, Swanson KE, Perou CM, et al. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat. 2013;137:373–382. doi: 10.1007/s10549-012-2346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 12.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, de Vere White RW. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Östling P, Leivonen SK, Aakula A, Kohonen P, Mäkelä R, Hagman Z, Edsjö A, Kangaspeska S, Edgren H, Nicorici D, et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- 14.Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, Giannopoulou E, Alves P, Tewari AK, Gerstein MB, Beltran H, et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013;73:1232–1244. doi: 10.1158/0008-5472.CAN-12-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, et al. miR-21: An androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng EK, Li R, Shin VY, Siu JM, Ma ES, Kwong A. MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumor Biol. 2014;35:2591–2598. doi: 10.1007/s13277-013-1341-7. [DOI] [PubMed] [Google Scholar]
- 17.Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13:220. doi: 10.1186/1476-4598-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S, Li B, Li Y, Li D, Wang ED, Liu MF. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31:1985–1998. doi: 10.1038/emboj.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, Zhang X, Dhakal IB, Beggs M, Kadlubar S, Luo D. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer. 2012;12:29. doi: 10.1186/1471-2407-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasheed SA, Teo CR, Beillard EJ, Voorhoeve PM, Zhou W, Ghosh S, Casey PJ. MicroRNA-31 controls G protein alpha-13 (GNA13) expression and cell invasion in breast cancer cells. Mol Cancer. 2015;14:67. doi: 10.1186/s12943-015-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 is a broad regulator of β1-integrin expression and function in cancer cells. Mol Cancer Res. 2011;9:1500–1508. doi: 10.1158/1541-7786.MCR-11-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sossey-Alaoui K, Downs-Kelly E, Das M, Izem L, Tubbs R, Plow EF. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int J Cancer. 2011;129:1331–1343. doi: 10.1002/ijc.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Körner C, Keklikoglou I, Bender C, Wörner A, Münstermann E, Wiemann S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon) J Biol Chem. 2013;288:8750–8761. doi: 10.1074/jbc.M112.414128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010;70:5147–5154. doi: 10.1158/0008-5472.CAN-10-0410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Ben-Chetrit N, Chetrit D, Russell R, Körner C, Mancini M, Abdul-Hai A, Itkin T, Carvalho S, Cohen-Dvashi H, Koestler WJ, et al. Synaptojanin 2 is a druggable mediator of metastasis and the gene is overexpressed and amplified in breast cancer. Sci Signal. 2015;8:ra7. doi: 10.1126/scisignal.2005537. [DOI] [PubMed] [Google Scholar]
- 26.Dobson JR, Taipaleenmäki H, Hu YJ, Hong D, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Pratap J. hsa-mir-30c promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/CCN3. Cancer Cell Int. 2014;14:73. doi: 10.1186/s12935-014-0073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla K, Sharma AK, Ward A, Will R, Hielscher T, Balwierz A, Breunig C, Münstermann E, König R, Keklikoglou I, Wiemann S. MicroRNA-30c-2-3p negatively regulates NF-κB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol Oncol. 2015;9:1106–1119. doi: 10.1016/j.molonc.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanic M, Yanowsky K, Rodriguez-Antona C, Andrés R, Márquez-Rodas I, Osorio A, Benitez J, Martinez-Delgado B. Deregulated miRNAs in hereditary breast cancer revealed a role for miR-30c in regulating KRAS oncogene. PLoS One. 2012;7:e38847. doi: 10.1371/journal.pone.0038847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Y, Shen H, Cao Y, Li H, Qin R, Chen Q, Long L, Zhu XL, Xie CJ, Xu WL. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz J Med Biol Res. 2014;47:60–69. doi: 10.1590/1414-431X20133324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi H, Liang B, Jia J, Liang N, Xu H, Ju G, Ma S, Liu X. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett. 2013;587:436–443. doi: 10.1016/j.febslet.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 32.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–2836. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 33.Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou C, Xu Q, Mao F, Li D, Bian C, Liu LZ, Jiang Y, Chen X, Qi Y, Zhang X, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11:2137–2145. doi: 10.4161/cc.20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, Zhou H, Zhao RC. miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol. 2009;34:1461–1466. [PubMed] [Google Scholar]
- 36.Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2010;17:246–254. doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Götte M, Mohr C, Koo CY, Stock C, Vaske AK, Viola M, Ibrahim SA, Peddibhotla S, Teng YH, Low JY, et al. miR-145-dependent targeting of Junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene. 2010;29:6569–6580. doi: 10.1038/onc.2010.386. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Wang X, Yang X, Liu Y, Shi Y, Ren J, Guleng B. miRNA423-5p regulates cell proliferation and invasion by targeting trefoil factor 1 in gastric cancer cells. Cancer Lett. 2014;347:98–104. doi: 10.1016/j.canlet.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 39.Ichikawa T, Sato F, Terasawa K, Tsuchiya S, Toi M, Tsujimoto G, Shimizu K. Trastuzumab produces therapeutic actions by upregulating miR-26a and miR-30b in breast cancer cells. PLoS One. 2012;7:e31422. doi: 10.1371/journal.pone.0031422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong SJ, Liu J, Wang X, Qu LX. microRNA-181 promotes prostate cancer cell proliferation by regulating DAX-1 expression. Exp Ther Med. 2014;8:1296–1300. doi: 10.3892/etm.2014.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano K, Miki Y, Hata S, Ebata A, Takagi K, McNamara KM, Sakurai M, Masuda M, Hirakawa H, Ishida T, et al. Identification of androgen-responsive microRNAs and androgen-related genes in breast cancer. Anticancer Res. 2013;33:4811–4819. [PubMed] [Google Scholar]
- 42.Lyu S, Yu Q, Ying G, Wang S, Wang Y, Zhang J, Niu Y. Androgen receptor decreases CMYC and KRAS expression by upregulating let-7a expression in ER−, PR−, AR+ breast cancer. Int J Oncol. 2014;44:229–237. doi: 10.3892/ijo.2013.2151. [DOI] [PubMed] [Google Scholar]
- 43.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 44.Chang SH, Lu YC, Li X, Hsieh WY, Xiong Y, Ghosh M, Evans T, Elemento O, Hla T. Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. J Biol Chem. 2013;288:4908–4921. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oneyama C, Okada M. MicroRNAs as the fine-tuners of Src oncogenic signalling. J Biochem. 2015;157:431–438. doi: 10.1093/jb/mvv036. [DOI] [PubMed] [Google Scholar]
- 46.Zhu A, Li Y, Song W, Xu Y, Yang F, Zhang W, Yin Y, Guan X. Antiproliferative effect of androgen receptor inhibition in Mesenchymal stem-like triple-negative breast cancer. Cell Physiol Biochem. 2016;38:1003–1014. doi: 10.1159/000443052. [DOI] [PubMed] [Google Scholar]