Abstract
Patients with unresectable, chemo-refractory colorectal cancer liver metastases (CRCLM) have limited local treatment options. We report our institutional experience on the efficacy of resin-based yttrium-90 (90Y) radioembolization for the treatment of CRCLM and our findings on associated circulating cell-free DNA (ccfDNA) studies. A total of 58 patients treated with 90Y for CRCLM at the Medstar Georgetown University Hospital had a median survival of 6 months [95% confidence interval (CI), 4.55–7.45 months] after treatment, with a 12-month survival rate of 33%. The median survival from treatment stratified by mutational status was longer in the wild-type (WT) as compared to the KRAS mutant patients at 7 vs. 5 months, but did not achieve statistical significance (p=0.059). Median tumor local control duration after 90Y treatment was 2 months (95% CI, 0.34–3.66 months) for the entire cohort and was longer in the WT vs. the mutant patients (2 vs. 1 month, respectively, p=0.088). Plasma was prospectively collected from a subset of 9 patients both before and after single lobe treatment, and ccfDNA concentration and fragmentation index (FI) were measured using quantitative PCR and atomic-force microscopy (AFM). In the WT and KRAS mutant patients, DNA FI was reduced from a median of 0.73–0.65 after treatment. A reduction in DNA FI after single lobe treatment was associated with an improved overall survival (p=0.046). Analysis by AFM of paired pre- and post-treatment samples from KRAS mutant and WT patients revealed a larger average decrease in fragment size in the WT patients (p=0.013). 90Y radioembolization extends local control for CRCLM, however, KRAS mutant tumors may be more radio-resistant to treatment. Changes in the FI of patients following treatment were noted and may be evaluated in a larger study for relevance as a biomarker of response.
Keywords: yttrium-90, radioembolization, KRAS, colorectal cancer, liver metastases, atomic-force microscopy, ccfDNA, PCR
Introduction
Colorectal cancer (CRC) is the third most frequent type of cancer diagnosed and the second leading cause of cancer-related deaths in the United States, with an estimated 132,700 new diagnoses and 49,700 deaths in 2015 (1). Approximately 25% of patients present with metastatic disease, while another 25–35% of patients develop metastatic disease during or after treatment, with the liver being the most common site of metastases (2). Surgical resection has been shown to be the only potentially curative option for colon cancer metastasis to the liver (3), resulting in a 25–40% overall survival at 5 years compared to 0–5% in those not having a liver resection. However, only about 15% of patients are considered to be resectable at presentation (2). While improvements in chemotherapy regimens and increasingly aggressive surgical approaches have resulted in improvements in survival rates for metastatic patients (4), patients with unresectable, chemo-refractory colorectal cancer liver metastases (CRCLM) have limited local treatment options. In addition to being prognostic for overall survival, the presence of hepatic disease is the major contributor to the cause of death in approximately half of the metastatic CRC patients (5), indicating that local control of the liver is an important aspect of disease management and outcomes.
For unresectable liver metastases, local therapies include stereotactic body radiation therapy (SBRT), hepatic arterial infusion of chemotherapy, arterially directed embolic therapy, radiofrequency ablation, and liver directed radiation. Selection of these different modalities often depends on the extent, size, and location of the disease (6). Yttrium-90 (90Y) radioembolization exploits the physiological difference in the blood flow between tumors and the normal liver to preferentially deliver radiation to the metastases, thereby shrinking the tumors while sparing the healthy liver (7). Liver directed radiation has shown promising results in a variety of institutional studies, demonstrating promising survival outcomes and low toxicities (8–10). A recent randomized phase III trial, SIRFLOX, which compared first line chemotherapy vs. the same chemotherapy with selective internal radiation, demonstrated a significant delay of disease progression in the liver (11).
CRC is a heterogeneous disease composed of multiple disease subtypes (12). While about 25% of CRCs are associated with a family history, suggesting a role for inherited genetic mutations, only a small percentage of family historylinked cancers are associated with highly penetrant mutations in major genes. The majority of CRCs occur sporadically, developing in a multi-step process, involving an accumulation of mutations in tumor-suppressor genes and oncogenes (13). Mutations in KRAS have been associated with 33–48% of CRCs. The KRAS protein is a downstream effector of EGFR receptor tyrosine kinase activity, which activates intracellular signaling cascades mediated by the RAF/MEK, MAPK, AKT and PI3K pathways (14). Mutations in KRAS have been linked to CRC progression, with KRAS mutants having worse progression-free survival and overall survival compared to wild-type (WT) patients (13,15–17). The impact of KRAS mutation on radiation response is less clear, with one group reporting a greater survival benefit after 90Y radioembolization in WT vs. KRAS mutant patients. However, as there was no evaluation of local control after therapy, the differential assessment of radiation response in the two groups remains unclear (18).
The presence of abnormally high levels of circulating cell-free DNA (ccfDNA) in the plasma of cancer patients has been well established and is thought to result from a variety of different mechanisms including apoptosis, necrosis, and circulating tumor cell lysis (19,20). In CRC patients, an elevated plasma DNA concentration has been shown to be diagnostic for benign vs. malignant disease and to correlate with worse progression-free survival and overall survival after chemotherapy (21–23). DNA fragmentation index (FI), which is measured as the ratio of longer to shorter DNA fragments, has been shown to be higher in patients with advanced stages of breast cancer and with post-operative recurrence (24). In addition, recently, the level of ccfDNA fragmentation has also been shown to correlate with survival in metastatic colon cancer patients (22). However, changes in ccfDNA concentration and FI after 90Y radioembolization have not been assessed.
Here we report on our institutional experience on the efficacy of resin-based 90Y radioembolization for the treatment of CRCLM and the changes in ccfDNA levels and FI with liver directed internal radiation, investigating whether gene mutations in KRAS could serve as prognostic factors for 90Y treatment of CRCLM.
Materials and methods
Patient selection
This retrospective review and prospective blood evaluation was approved by the Institutional Review Board (IRB 2008-344) of Georgetown University. From May 2011 to March 2015, a total of 58 patients underwent 90Y treatment for CRCLM at our institution. Patients eligible for study inclusion had histologically-confirmed CRC with liver metastases. Patients were included irrespective of prior treatment, including prior chemotherapy, surgery, or radiation therapy. All patients were not considered candidates for surgical resection and many of the patients were considered refractory to chemotherapy. Lesions were considered for treatment in any location within the liver. Patients generally were expected to have a life expectancy of at least 6 months and adequate hepatic function.
Pre-treatment imaging, planning, and treatment
All patients underwent pre-procedural imaging with computed tomography (CT), FDG PET-CT, and/or MRI to determine liver tumor location, size, and volume. Three dimensional volumetric software was utilized to evaluate the total tumor volume, total liver volume, and total liver tumor burden (ratio of TV/LV). A pre-procedure planning angiogram and embolization of extrahepatic arteries at risk of nontarget perfusion of 90Y microspheres was performed. Hepatopulmonary shunt estimation was performed using Tc-99m-MAA, and this was judged to be adequate if <20%. Therapy with 90Y microspheres was performed within 2 weeks of initial planning angiography. 90Y microsphere infusion followed the manufacturer's guidelines. Following delivery, a SPECT of the abdomen was performed to assess the distribution of the microspheres. The patients were discharged home the same day.
Depending on extent of disease within the liver at presentation, patients received either bilobar (right and left lobe) or unilobar treatment. For bilobar treatment, the first lobe treatment was followed by the second lobe treatment approximately 30 days later. Infusion of 90Y microspheres was performed by both the interventional radiologist and radiation oncologist. Patients were treated to a mean dose of 43.87 Gy and mean activity of 24.28 mCi.
Plasma isolation, ccfDNA extraction, qPCR, and AFM preparation
Sample handling was in accordance with published NCI biospecimen best practices. A 10 ml sample of blood was collected in a K2-EDTA tube. Blood samples were processed within 30–60 min with an initial spin at 800 × g for 10 min at 4°C. The plasma was transferred to another tube and centrifuged at 13,000 × g for 10 min to remove the remaining blood cells. The supernatant plasma samples were then aliquoted into 2 ml/tube and either immediately processed for DNA extraction or stored at −80°C. ccfDNA was extracted from 5 ml of plasma using the QIAamp DNA Mini blood kit (Qiagen, Valencia, CA, USA) according to the ‘Blood and Body Fluid Protocol’. ccfDNA samples were kept at −20°C until use.
Plasma was prospectively collected from 9 patients both pre- and post-treatment, and ccfDNA concentrations and FI values were determined using quantitative PCR as previously described (25). Briefly, qPCR amplifications were performed in duplicate in a 20-µl PCR mix, including the amplification primer and DNA extract. Thermal cycling consisted of three repeated steps: a 3-min Hot Start polymerase activation-denaturation step at 95°C, followed by 40 repeated cycles at 95°C for 10 sec and 60°C for 30 sec. Melting curves were obtained by increasing the temperature from 55 to 90°C with a plate reading every 0.2°C. Serial dilutions of genomic DNA were used as a standard for quantification. The quantity of ccfDNA fragments of different sizes was assessed using an integrated PCR system that targeted intronic sequences within the same regions. The degree of ccfDNA FI was assessed by calculating the ratio of the concentration determined by using the primer set amplifying a large target (>250 bp) to the concentration determined by using the primer set amplifying a short target (<100 bp). These values were normalized, and the ratio of large fragments (~ 250 bp) to small fragments (<100 bp) was recorded as the DNA FI. The ratio of DNA FI values before and after treatment (pre-treatment FI/post-treatment FI) were stratified by values less than or greater than 1.
Samples for atomic-force microscopy (AFM) analysis were prepared as previously described (26). Briefly, a 2 µl ccfDNA sample in a Tris and EDTA solution was deposited onto APS-mica. The sample was then rinsed with deionized water and dried with nitrogen gas. It was mounted on a Nanoscope IIIa AFM for imaging (Veeco/Digital Instruments, Santa Barbara, CA, USA). Images were acquired in tapping mode in air using silicon tapping mode probes. Ten images were acquired for each ccfDNA sample. Acquired AFM images were analyzed using commercially available software, FemtoScan Online (Advanced Technologies Center, Moscow, Russia), for measurements of ccfDNA fragment lengths. Measured DNA fragments were sorted by size, and the fragment size distribution was obtained by dividing the number of fragments in each bin by the total number of fragments. DNA fragment size conversions were made on the assumption that 1 bp was equivalent to a length of 0.34 nm. AFM was performed on 6 patients, 4 KRAS WT and 2 KRAS mutants.
Follow-up and statistical analysis
A retrospective review was conducted for clinical outcomes, demographic information, and tumor mutational status. In 47 patients, interval imaging, including CT, PET-CT, and/or MRI, was available for tumor response assessment using RECIST criteria as a score of complete response, partial response, stable disease, or progressive disease (27). Because use of 90Y microspheres is a liver-directed therapy, tumor response was limited to an assessment of hepatic lesions in the liver lobe or lobes treated, with whole liver local control assessed for bilobar treatments and treated liver lobe local control assessed for unilobar treatments. Tumor mutational status was available for 49 patients based on the review of documented pathology reports. Actuarial survival and progression-free survival were evaluated by the Kaplan-Meier method. Univariate analysis was performed using Kaplan-Meier, and multivariate analysis was assessed using Cox regression analysis. A two-tailed t-test was used to assess changes in fragment size with AFM in WT and KRAS mutant patients before and after single lobe 90Y treatment.
Results
Patient characteristics
Patient characteristics are noted in Table I. Our patient population consisted of 58 patients diagnosed with colon cancer at a median age of 56 years (range, 31–85 years). Half of the patients were male, 86% had a performance status ≤1, and 67% of the patients had metastatic disease outside the liver. Patients received 90Y treatment for a median of 20 months (range, 5–52 months) after diagnosis with hepatic metastases. As summarized in Table II, our patients received a median of two chemotherapy treatment lines prior to 90Y treatment, ranging from 1 to 5. Every patient had exposure to either 5-FU or capecitabine prior to treatment.
Table I.
Patient characteristics, n=58.
| Patient characteristics | n (%) |
|---|---|
| Age at 90Y treatment (years) | |
| Median (range) | 56 (31–85) |
| ≤40 | 3 (5) |
| >40 to ≤50 | 13 (22) |
| >50 to ≤60 | 16 (28) |
| >60 to ≤70 | 16 (28) |
| >70 | 10 (17) |
| Race | |
| White | 36 (62) |
| Black | 12 (21) |
| Other | 10 (17) |
| Gender | |
| Male | 29 (50) |
| Female | 29 (50) |
| Performance status (ECOG) | |
| 0 | 14 (24) |
| 1 | 36 (62) |
| ≥2 | 7 (12) |
| Unknown | 1 (2) |
| Location of lesion treated | |
| Right lobe | 50 (86) |
| Left lobe | 40 (69) |
| Both lobes | 32 (55) |
| Presence of extrahepatic disease | |
| Absent | 19 (33) |
| Present | 39 (67) |
90Y, yttrium-90.
Table II.
Chemotherapy characteristics, n=58.
| Chemotherapy characteristics | n (%) |
|---|---|
| No. of previous chemotherapy lines | |
| 1 | 12 (21) |
| 2 | 29 (50) |
| ≥3 | 17 (29) |
| Exposure to 5-fluoruracil or capecitabine | |
| No | 0 (0) |
| Yes | 58 (100) |
| Exposure to leucovorin | |
| No | 9 (16) |
| Yes | 49 (84) |
| Exposure to oxaliplatin | |
| No | 5 (9) |
| Yes | 53 (91) |
| Exposure to irinotecan | |
| No | 27 (47) |
| Yes | 31 (53) |
| Exposure to bevacizumab | |
| No | 14 (24) |
| Yes | 44 (76) |
| Exposure to cetuximab or panitumumab | |
| No | 52 (90) |
| Yes | 6 (10) |
| Exposure to nilotinib or sunitinib | |
| No | 54 (93) |
| Yes | 4 (7) |
| Exposure to other agentsa | |
| No | 44 (76) |
| Yes | 14 (24) |
Includes PARP inhibitors, anti-PDL1, docetaxel, C-met inhibitors, and tezacitabine.
As summarized in Table III, tumor mutation information was available for 49 patients, with 27 (55%) being KRAS mutant, 21 (43%) being WT, and 1 (2%) being Her2/neu overexpressing. A larger proportion of KRAS WT patients were treated with anti-EGFR-targeted agents before 90Y radioembolization, but the overall exposure to EGFR inhibitors was low (10%, Table II).
Table III.
Mutation status, n=49.
| Mutation status | n (%) | Median age (years) |
|---|---|---|
| KRAS WT | 21 (43) | 53 |
| KRAS mutated | 27 (55) | 60 |
| Her2/neu | 1 (2) | 38 |
WT, wild-type.
90Y radioembolization procedure
A total of 90 treatments were delivered to 58 patients (Table I). Eight patients received treatment to the left lobe alone, 18 received treatment to the right lobe alone, and 32 received treatment to both liver lobes spaced approximately a month apart. The delivered activity during 90Y treatment ranged from 8.9 to 59.4 mCi.
Imaging response and liver progression-free survival
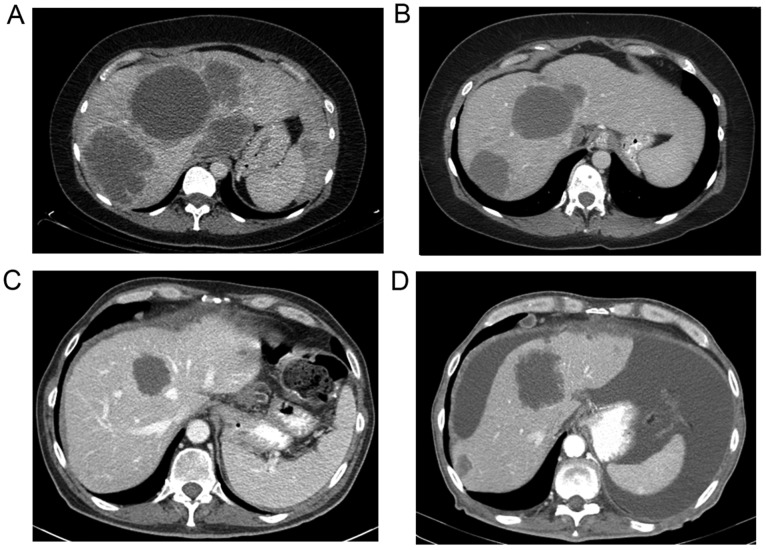
In 47 patients, interval imaging, including CT, PET/CT and/or MRI, was available for tumor response assessment. Fig. 1 shows representative CT images before (Fig. 1A and C) and after treatment (Fig. 1B and D) in a WT (Fig. 1A and B) and KRAS mutant (Fig. 1C and D) patient. The median time of imaging after the procedure was 2 months following treatment. RECIST criteria were utilized to measure the response in each liver lobe treated (28). Imaging assessment showed a partial response in 9 patients (19%), stable disease in 20 (43%), and progressive disease in 18 (38%).
Figure 1.
Representative CT images of DNA in (A and B) wild-type and (C and D) KRAS mutant patients before (A and C) and after (B and D) 90Y treatment.
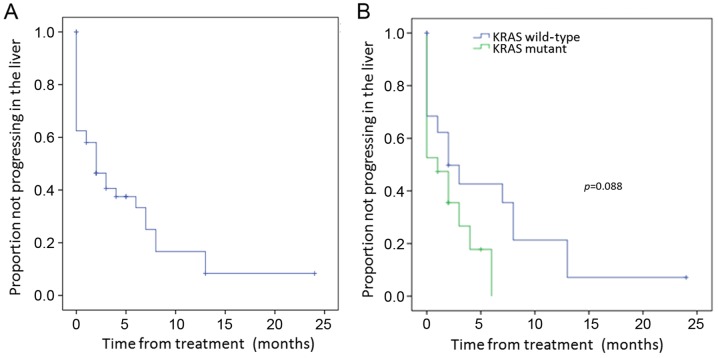
Median liver lobe progression-free survival duration after 90Y treatment was 2 months [95% confidence interval (CI), 0.34–3.66 months] for the entire cohort (Fig. 2A). The median local control duration assessed according to tumor mutation was longer in the WT vs. KRAS mutant patients of 2 vs. 1 month, respectively (p=0.088) (Fig. 2B). While this was not significant with univariate analysis, it was significant with multivariate analysis (p=0.031) (Table IV). Progression-free survival in the liver was not significantly different by univariate or multivariate analysis in patients stratified by prior treatment number of chemotherapy lines (Table IV). However, patients with prior exposure to nilotinib or sunitinib had worse progression-free survival in the liver with univariate analysis (5.6±1.3 vs. 0.5±0.5 months, p=0.034), but this was not significant with multivariate analysis (Table V).
Figure 2.
Progression-free survival in the liver: (A) all patients; (B) patients by mutational status.
Table IV.
Univariate and multivariate analyses for progression-free survival and overall survival.
| Progression-free survival in liver | Overall survival treatment | Overall survival diagnosis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | Univariate | Multivariate | |||||
| Variables | Reference | P-value | HR (95% CI) | P-value | P-value | HR (95% CI) | P-value | P-value | HR (95% CI) | P-value |
| Age (years) | <60 | 0.230 | 2.12 (0.41–10.99) | 0.370 | 0.044 | 0.82 (0.22–3.09) | 0.766 | 0.598 | 1.92 (0.47–7.95) | 0.366 |
| Gender | Female | 0.065 | 1.55 (0.60–3.98) | 0.364 | 0.442 | 0.87 (0.38–1.98) | 0.739 | 0.458 | 0.82 (0.34–1.94) | 0.644 |
| Performance status | ≤1 vs. ≥2 | 0.395 | 0.63 (0.13–3.08) | 0.568 | 0.001 | 0.24 (0.09–0.61) | 0.003 | 0.017 | 0.21 (0.07–0.62) | 0.005 |
| Smoking | No | 0.742 | 1.24 (0.36–4.27) | 0.731 | 0.444 | 1.38 (0.51–3.76) | 0.524 | 0.013 | 2.03 (0.62–6.68) | 0.245 |
| Presence of extrahepatic disease | No | 0.445 | 0.83 (0.30–2.30) | 0.723 | 0.775 | 0.90 (0.40–2.01) | 0.801 | 0.722 | 1.36 (0.58–3.18) | 0.475 |
| Mutation status | KRAS WT | 0.088 | 0.29 (0.09–0.90) | 0.031 | 0.059 | 0.58 (0.26–1.33) | 0.200 | 0.322 | 0.69 (0.28–1.74) | 0.435 |
| FI | <1 | 0.716 | NA | 0.170 | NA | 0.040 | NA | |||
| No. of previous chemotherapy lines | Continuous | 3.87 (0.47–32.16) | 0.211 | 1.11 (0.17–7.18) | 0.911 | 0.2 (0.05-.89) | 0.035 | |||
| 1 vs. ≥2 | 0.387 | 2.96 (0.23–38.73) | 0.409 | 0.652 | 1.60 (0.19–13.59) | 0.667 | 0.278 | 0.21 (0.03–1.47) | 0.115 | |
| ≤2 vs. ≥3 | 0.703 | 5.07 (0.35–73.60) | 0.235 | 0.845 | 1.00 (0.11–9.28) | 0.998 | 0.801 | 0.24 (0.04–1.50) | 0.128 | |
FI, fragmentation index; WT, wild-type; NA, not applicable.
Table V.
Univariate and multivariate analyses for progression-free survival and overall survival with chemotherapy exposure.
| Progression-free survival in liver | Overall survival treatment | Overall survival diagnosis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | Univariate | Multivariate | ||||
| Variables | P-value | HR (95% CI) | P-value | P-value | HR (95% CI) | P-value | P-value | HR (95% CI) | P-value |
| Oxaliplatin | 0.5 | 0.61 (0.19–2.0) | 0.413 | 0.009 | 0.12 (0.02–0.90) | 0.039 | 0.039 | 0.174 (0.02–1.33) | 0.092 |
| Leucovorin | 0.464 | 1.35 (0.51–3.55) | 0.545 | 0.063 | 1.89 (0.84–4.28) | 0.127 | 0.391 | 0.94 (0.41–2.15) | 0.886 |
| Irinotecan | 0.514 | 0.79 (0.34–1.81) | 0.569 | 0.689 | 0.71 (0.38–1.34) | 0.291 | 0.114 | 1.28 (0.64–2.56) | 0.479 |
| Bevacizumab | 0.073 | 0.39 (0.12–1.32) | 0.132 | 0.426 | 1.36 (0.68–2.75) | 0.385 | 0.858 | 1.0 (0.50–2.0) | 0.988 |
| Cetuximab or panitumab | 0.799 | 0.56 (0.17–1.84) | 0.338 | 0.708 | 1.34 (0.46–3.87) | 0.59 | 0.332 | 1.65 (0.55–4.93) | 0.372 |
| Nilotinib or sunitinib | 0.034 | 0.45 (0.15–1.34) | 0.15 | 0.01 | 0.32 (0.11–0.97) | 0.045 | 0.1 | 0.47 (0.16–1.41) | 0.176 |
Survival outcomes
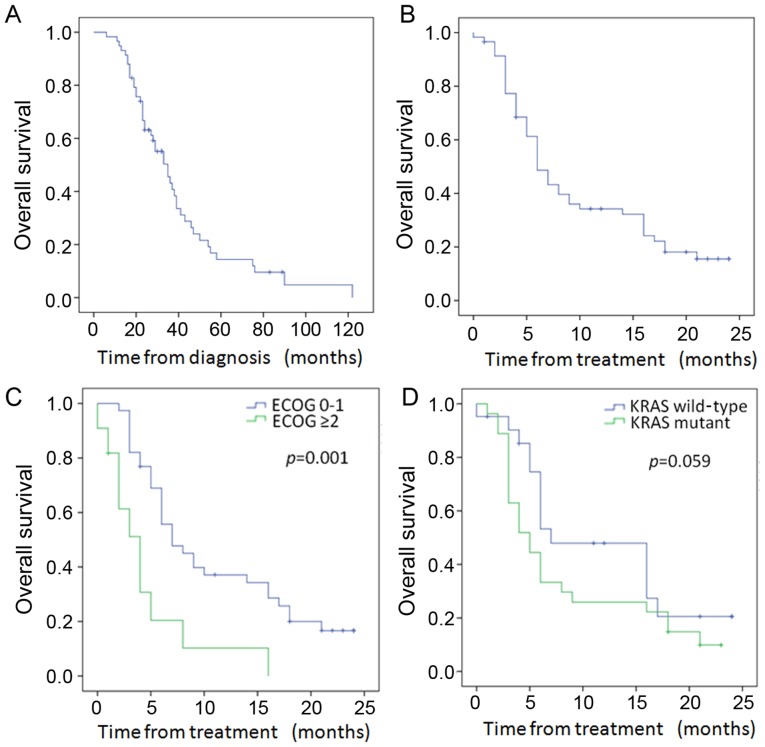
Of the 58 patients, 46 died during follow-up. As shown in Fig. 3A, the median survival from diagnosis was 35 months (95% CI, 26.9–43.1 months). The median survival after 90Y was 6 months (95% CI, 4.55–7.45 months) in the entire cohort (Fig. 3B), with a 12-month survival of 33%. Patients with ECOG performance status ≤1 had significantly improved median survival over those with ECOG ≥2 (7 vs. 4 months, p=0.001) (Fig. 3C and Table IV). In addition, with univariate analysis, patients younger than 60 also had improved survival after treatment (p=0.044) (Table IV). The median survival from treatment stratified by mutational status (Fig. 3D and Table IV) was longer in the WT as compared to the KRAS mutant patients at 7 months (95% CI, 1.8–12.2) vs. 5 months (95% CI, 3.0–7.0), but did not achieve statistical significance (p=0.059). While overall survival from treatment stratified by the number of prior chemotherapy lines was not significant with either univariate or multivariate analysis (Table IV), prior exposure to oxaliplatin was associated with improved survival from treatment (22.5±1.1 vs. 9.0±1.0 months, p=0.009) and prior exposure to nilotinib/sunitinib was associated with a worse survival from treatment (10.5±1.1 vs. 3.8±0.48 months, p=0.01) (Table V). Futhermore, with multivariate analysis, survival after treatment was significantly associated with performance status [p=0.003; HR, 0.24 (CI, 0.09–0.61); Table IV] and prior exposure to oxaliplatin and nilotinib/sunitinib (p=0.039 and p=0.045, respectively; Table V).
Figure 3.
Overall survival: (A) all patients from diagnosis; (B) overall survival from treatment all patients; (C) overall survival of patients by performance status from treatment; (D) overall survival of patients by mutation status from treatment.
Survival from diagnosis was improved in those patients with ECOG performance status ≤1 (42.3±4.5 vs. 27.3±6.5 months, p=0.17; Table IV). With multivariate analysis, both the performance status and the number of previous chemotherapy lines were significant for survival from cancer diagnosis (Table IV). Prior exposure to oxaliplatin was statistically significant for overall survival from diagnosis (70.5±8.8 vs. 37.4±3.8 months, p=0.039) with univariate analysis, but did not maintain significance with multivariate analysis (Table V).
ccfDNA analysis
ccfDNA, which was collected immediately prior to 90Y administration and 2–4 weeks after single lobe treatment, was detected in all of the 9 analyzed samples by PCR. In the WT and KRAS mutant patients, DNA FI was decreased from a median of 0.73–0.65 after treatment. As shown in Fig. 4, a decrease in the ratio of DNA FI (pre-treatment FI/post-treatment FI), or an increase in the fraction of small DNA fragments, after single lobe treatment was associated with improved outcomes. While overall survival from treatment did not meet statistical significance in patients with a DNA FI ratio pre/post-treatment of >1 vs. <1 (Fig. 4A, p=0.17), overall survival from diagnosis was significantly improved in those patients (Fig. 4B, p=0.04).
Figure 4.
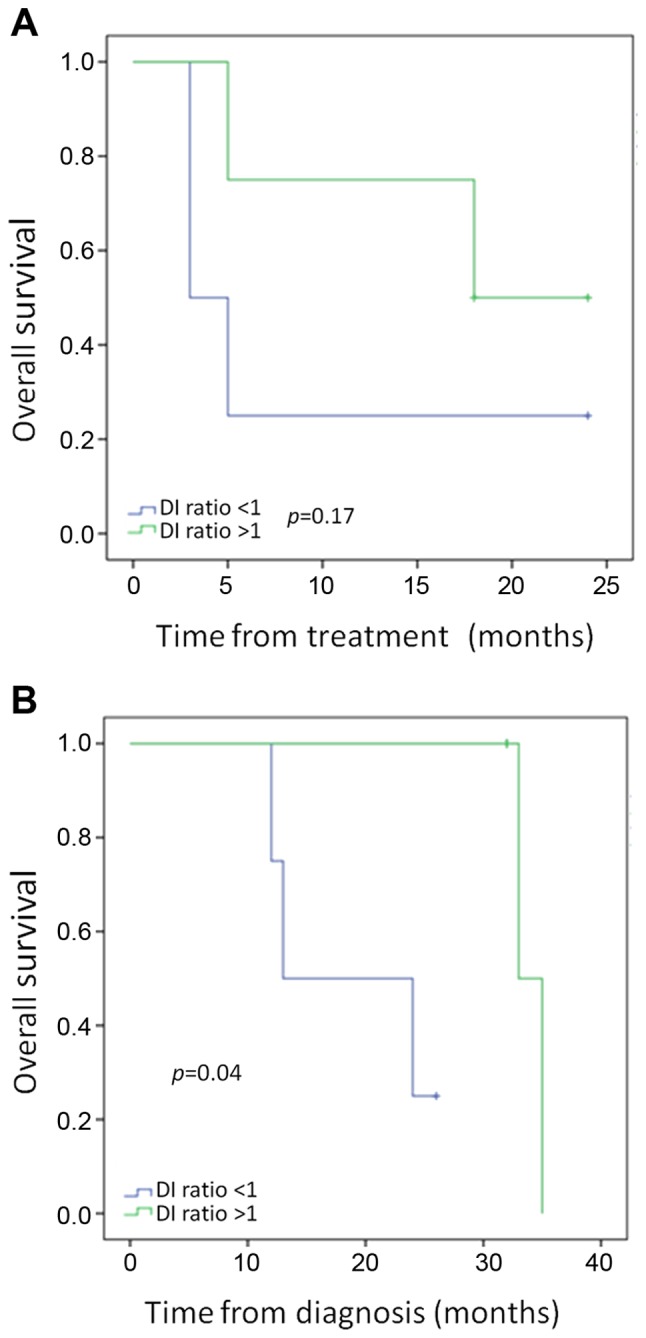
Evaluation of KRAS mutant and wild-type patients for changes in circulating cell-free DNA. (A) Overall survival from treatment according to DNA fragmentation index (FI) changes; (B) overall survival from diagnosis according to DNA FI changes.
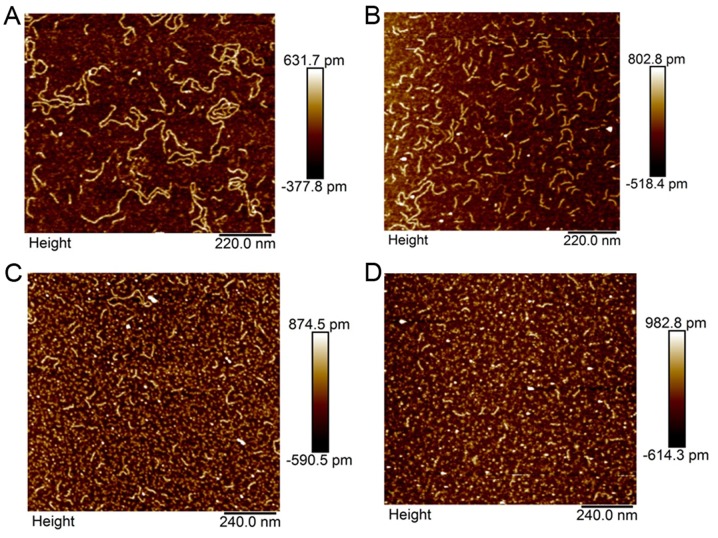
AFM analysis
Analysis by AFM of paired pre- and post-treatment samples was performed in 4 WT and 2 KRAS mutant patients. An illustrative example is shown in Fig. 5 of sample AFM images of WT (Fig. 5A and B) and KRAS mutant (Fig. 5C and D) DNA before (Fig. 5A and C) and after (Fig. 5B and D) single liver lobe exposure to 90Y. This figure demonstrates the capability of AFM in measuring individual DNA fragments and illustrates the treatment related differences in WT and KRAS mutant patient ccfDNA.
Figure 5.
Representative atomic-force microscopy images of DNA in (A and B) wild-type (WT) and (C and D) KRAS mutant patients before (A and C) and after (B and D) yttrium-90 (90Y) treatment. 90Y treatment appeared to induce significant fragmentation in circulating cell-free DNA for KRAS WT but not in KRAS mutant patients.
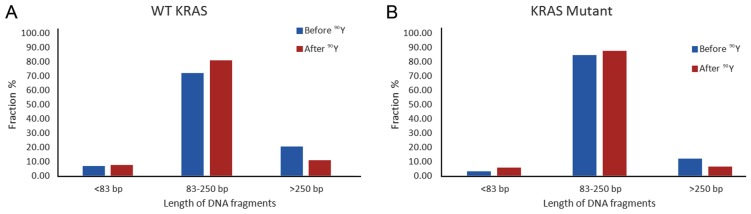
In a pooled analysis of 4 WT and 2 KRAS mutant patients, the average size of ccfDNA bp change after single lobe treatment was significantly larger in the WT patients, with the WT ccfDNA decreasing from 251.86 to 154.65 bp as compared to the KRAS mutant ccfDNA decreasing from 177.98 to 155.09 bp (p=0.013). Quantification of changes in different fragment size groups before and after treatment is shown in Fig. 6. While both WT and KRAS mutant patient ccfDNA had decreased percentage of >250 bp size range and higher proportion of 85–350 bp ccfDNA after single lobe treatment, the magnitude of change was larger in the WT patients, with the 85–350 bp size fragments increasing by 12.3% in the WT (Fig. 6A) and 3.6% in the KRAS mutant (Fig. 6B).
Figure 6.
Comparison of the fractional fragment size distribution of circulating cell-free DNA (ccfDNA) from (A) wild-type (WT) and (B) KRAS mutant samples before and after single lobe treatment with yttrium-90 (90Y). The estimated amount of each ccfDNA fraction expressed as the percentage of the total ccfDNA estimated as the sum of the ccfDNA amount of the three size fractions in (A) WT and (B) KRAS mutant patients before (blue) and after (red) treatment in (A) four pooled WT samples and (B) two pooled KRAS mutant samples.
Discussion
In this study we demonstrated that KRAS mutant patients had less radiation-related changes in ccfDNA after treatment than WT patients and that radiation treatment changes in ccfDNA, as measured by DNA FI, were predictive for patient overall survival. While several studies have previously described the predictive potential of ccfDNA levels in overall survival after chemotherapy for metastatic CRC patients (23,29), this is the first study to demonstrate the treatment FI ratio as a potential predictive marker for survival.
Molecular biomarker research has revealed CRC to be a heterogeneous disease with multiple molecular disease subtypes (12). CRC biomarker research has led to the development of new chemotherapy regimens and novel monoclonal antibody therapies targeting critical molecules in cell signaling pathways, and these treatment advances have resulted in improvements in overall survival for metastatic CRC (4). There is evidence that tumor response to chemotherapy is predictive for overall survival. In a study by Adam et al, response to chemotherapy correlated with improved overall survival in patients who received neoadjuvant chemotherapy prior to curative liver metastatic tumor resection (30). Mechanisms of resistance to targeted therapies are abundant, with the majority of KRAS WT tumors not responding to anti-EGFR therapy (31). Anti-EGFR therapy resistance has been associated with other gene mutations, epigenetic silencing, and augmented expression of other receptor tyrosine kinases, resulting in a complex tumor microenvironment that may be difficult to predict treatment response (12).
Multiple clinical trials have confirmed the lack of clinical benefit of anti-EGFR therapy against RAS mutant tumors (32,33). The meta-analysis from Sorich et al in 2015 demonstrated that tumors without RAS mutations exhibited an improved clinical response compared to RAS mutant tumors, regardless of the specific targeted therapies or partner chemotherapy used (34). In addition, KRAS mutant tumors have been associated with poor survival outcomes and treatment responses in the absence of targeted agents (16,35,36), which may indicate that these tumors are less responsive to systemic therapies. The impact of KRAS mutation status on radiation treatment response is less clear, but is an area of investigation. Lahti et al reported KRAS mutation status to be predictive for overall survival after 90Y radioembolization for unresectable CRC liver metastases (18). While overall survival after 90Y treatment in KRAS mutant vs. WT patients did not quite meet statistical significance in our study, there was a clear separation in the curves. Notably, mutation status was significant on multivariate analysis for liver progression-free survival in our study. Intriguingly, it is possible that the same differential chemotherapy responses of WT and KRAS mutant tumors that have been well described are also true for radiation treatment.
El Messaoudi et al recently demonstrated that higher ccfDNA concentrations, higher levels of mutant ccfDNA, and the level of ccfDNA fragmentation correlated with shorter overall survival in metastatic CRC patients (22). While there was no difference in overall survival from treatment as a function of DNA integrity index in our study, analysis of survival from diagnosis was statistically longer in those patients with a decrease in their fragment ccfDNA size after treatment. The discrepancy in survival may be a function of the small number of patients in our study. However, it may also be a function of the limitations of 90Y in treating metastatic CRC, which have been previously outlined in the recently published SIRFLOX study (11), with the overall aim of 90Y treatment being to control liver disease and not to control systemic burden. Radiation treatment-related DNA FI changes are likely reflective of tumor response capability. Indeed, DNA FI may reflect tumoricidal changes and could serve as a biomarker to assess treatment response.
Our study sample with molecular data is small. While our findings are hypothesis driving, definitive conclusions are limited until a more robust analysis can be performed on a larger number of patients. In addition, while all of our patients received chemotherapy prior to 90Y treatment and many were considered chemotherapy refractory, our patient population was diverse in their overall level of health and in the natural history of their disease. It is not surprising that overall survival was improved in patients with better performance statuses. While no overall survival differences were observed among patients with or without extrahepatic disease, the overall survival differences seen based on the number and types of chemotherapy exposures likely reveals variation in patient performance status and disease course. Further molecular studies on a larger group of patients are necessary to validate our observational findings.
In conclusion, 90Y radioembolization is an effective treatment for CRCLM in extending local control for liver dominant metastatic disease. However, KRAS mutant tumors may be more radio-resistant to treatment. In this study, changes in ccfDNA FI were correlated with overall survival, likely indicating that these changes are reflective of treatment response. Measurements of FI may have the potential to be another molecular biomarker that could predict treatment response to therapy.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Nordlinger B, Adam R, Köhne CH, Pozzo C, Poston G, Ychou M, Rougier P. European Colorectal Metastases Treatment Group: Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42:2212–2221. doi: 10.1016/j.ejca.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Stangl R, Altendorf-Hofmann A, Charnley RM, Scheele J. Factors influencing the natural history of colorectal liver metastases. Lancet. 1994;343:1405–1410. doi: 10.1016/S0140-6736(94)92529-1. [DOI] [PubMed] [Google Scholar]
- 4.Gallagher DJ, Kemeny N. Metastatic colorectal cancer: From improved survival to potential cure. Oncology. 2010;78:237–248. doi: 10.1159/000315730. [DOI] [PubMed] [Google Scholar]
- 5.Helling TS, Martin M. Cause of death from liver metastases in colorectal cancer. Ann Surg Oncol. 2014;21:501–506. doi: 10.1245/s10434-013-3297-7. [DOI] [PubMed] [Google Scholar]
- 6.Xing M, Kooby DA, El-Rayes BF, Kokabi N, Camacho JC, Kim HS. Locoregional therapies for metastatic colorectal carcinoma to the liver - an evidence-based review. J Surg Oncol. 2014;110:182–196. doi: 10.1002/jso.23619. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy A, Coldwell D, Sangro B, Wasan H, Salem R. Radioembolization for the treatment of liver tumors general principles. Am J Clin Oncol. 2012;35:91–99. doi: 10.1097/COC.0b013e3181f47583. [DOI] [PubMed] [Google Scholar]
- 8.Sato KT, Lewandowski RJ, Mulcahy MF, Atassi B, Ryu RK, Gates VL, Nemcek AA, Jr, Barakat O, Benson A, III, Mandal R, et al. Unresectable chemorefractory liver metastases: Radioembolization with 90Y microspheres - safety, efficacy, and survival. Radiology. 2008;247:507–515. doi: 10.1148/radiol.2472062029. [DOI] [PubMed] [Google Scholar]
- 9.Hickey R, Lewandowski RJ, Prudhomme T, Ehrenwald E, Baigorri B, Critchfield J, Kallini J, Gabr A, Gorodetski B, Geschwind JF, et al. 90Y Radioembolization of colorectal hepatic metastases using glass microspheres: Safety and survival outcomes from a 531-patient multicenter study. J Nucl Med. 2016;57:665–671. doi: 10.2967/jnumed.115.166082. [DOI] [PubMed] [Google Scholar]
- 10.Maleux G, Deroose C, Laenen A, Verslype C, Heye S, Haustermans K, De Hertogh G, Sagaert X, Topal B, Aerts R, et al. Yttrium-90 radioembolization for the treatment of chemorefractory colorectal liver metastases: Technical results, clinical outcome and factors potentially influencing survival. Acta Oncol. 2016;55:486–495. doi: 10.3109/0284186X.2015.1101151. [DOI] [PubMed] [Google Scholar]
- 11.van Hazel GA, Heinemann V, Sharma NK, Findlay MP, Ricke J, Peeters M, Perez D, Robinson BA, Strickland AH, Ferguson T, et al. SIRFLOX: Randomized phase III trial comparing first-line mFOLFOX6 (plus or minus bevacizumab) versus mFOLFOX6 (plus or minus bevacizumab) plus selective internal radiation therapy in patients with metastatic colorectal cancer. J Clin Oncol. 2016;34:1723–1731. doi: 10.1200/JCO.2015.66.1181. [DOI] [PubMed] [Google Scholar]
- 12.Linnekamp JF, Wang X, Medema JP, Vermeulen L. Colorectal cancer heterogeneity and targeted therapy: A case for molecular disease subtypes. Cancer Res. 2015;75:245–249. doi: 10.1158/0008-5472.CAN-14-2240. [DOI] [PubMed] [Google Scholar]
- 13.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waring P, Tie J, Maru D, Karapetis CS. RAS mutations as predictive biomarkers in clinical management of metastatic colorectal cancer. Clin Colorectal Cancer. 2016;15:95–103. doi: 10.1016/j.clcc.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Osumi H, Shinozaki E, Suenaga M, Matsusaka S, Konishi T, Akiyoshi T, Fujimoto Y, Nagayama S, Fukunaga Y, Ueno M, et al. RAS mutation is a prognostic biomarker in colorectal cancer patients with metastasectomy. Int J Cancer. 2016;139:803–811. doi: 10.1002/ijc.30106. [DOI] [PubMed] [Google Scholar]
- 16.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: The multicenter ‘RASCAL’ study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 17.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 18.Lahti SJ, Xing M, Zhang D, Lee JJ, Magnetta MJ, Kim HS. KRAS status as an independent prognostic factor for survival after Yttrium-90 radioembolization therapy for unresectable colorectal cancer liver metastases. J Vasc Interv Radiol. 2015;26:1102–1111. doi: 10.1016/j.jvir.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 20.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spindler KL, Appelt AL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Cell-free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. Int J Cancer. 2014;135:2984–2991. doi: 10.1002/ijc.28946. [DOI] [PubMed] [Google Scholar]
- 22.El Messaoudi S, Mouliere F, Du Manoir S, Bascoul-Mollevi C, Gillet B, Nouaille M, Fiess C, Crapez E, Bibeau F, Theillet C, et al. Circulating DNA as a strong multi-marker prognostic tool for metastatic colorectal cancer patient management care. Clin Cancer Res. 2016;22:3067–3077. doi: 10.1158/1078-0432.CCR-15-0297. [DOI] [PubMed] [Google Scholar]
- 23.Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18:1177–1185. doi: 10.1158/1078-0432.CCR-11-0564. [DOI] [PubMed] [Google Scholar]
- 24.Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, Hoon DS. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–4276. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 25.Mouliere F, El Messaoudi S, Pang D, Dritschilo A, Thierry AR. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol. 2014;8:927–941. doi: 10.1016/j.molonc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pang D, Winters TA, Jung M, Purkayastha S, Cavalli LR, Chasovkikh S, Haddad BR, Dritschilo A. Radiation-generated short DNA fragments may perturb non-homologous end-joining and induce genomic instability. J Radiat Res (Tokyo) 2011;52:309–319. doi: 10.1269/jrr.10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 29.Spindler KL, Pallisgaard N, Andersen RF, Ploen J, Jakobsen A. Gemcitabine and capecitabine for heavily pre-treated metastatic colorectal cancer patients - a phase II and translational research study. Anticancer Res. 2014;34:845–850. [PubMed] [Google Scholar]
- 30.Adam R, Pascal G, Castaing D, Azoulay D, Delvart V, Paule B, Levi F, Bismuth H. Tumor progression while on chemotherapy: A contraindication to liver resection for multiple colorectal metastases? Ann Surg. 2004;240:1052–1064. doi: 10.1097/01.sla.0000145964.08365.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 32.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 33.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 34.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: A meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 35.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, et al. Kirsten ras mutations in patients with colorectal cancer: The ‘RASCAL II’ study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: Results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]