Abstract
Gene expression data of hepatocellular carcinoma (HCC) was compared with that of cirrhosis (C) to identify critical genes in HCC. A total of five gene expression data sets were downloaded from Gene Expression Omnibus. HCC and healthy samples were combined as dataset HCC, whereas cirrhosis samples were included in dataset C. A network was constructed for dataset HCC with the package R for performing Weighted Gene Co-expression Network Analysis. Modules were identified by cluster analysis with the packages flashClust and dynamicTreeCut. Hub genes were screened out by calculating connectivity. Functional annotations were assigned to the hub genes using the Database for Annotation, Visualization and Integration Discovery, and functional annotation networks were visualized with Cytoscape. Following the exclusion of outlier samples, 394 HCC samples and 47 healthy samples were included in dataset HCC and 233 cirrhosis samples were included in dataset C. A total of 6 modules were identified in the weighted gene co-expression network of dataset HCC (blue, brown, turquoise, green, red and yellow). Modules blue, brown and turquoise had high preservation whereas module yellow exhibited the lowest preservation. These modules were associated with transcription, mitosis, cation transportation, cation homeostasis, secretion and regulation of cyclase activity. Various hub genes of module yellow were cytokines, including chemokine (C-C motif) ligand 22 and interleukin-19, which may be important in the development of HCC. Gene expression profiles of HCC were compared with those of cirrhosis and numerous critical genes were identified, which may contribute to the progression of HCC. Further studies on these genes may improve the understanding of HCC pathogenesis.
Keywords: hepatocellular carcinoma, cirrhosis, gene expression data, weighted gene co-expression network analysis, modules, hub genes
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer. It is the fifth most frequently occurring cancer worldwide and the third most common cause of cancer-associated mortality (1). Numerous advances have been made in the understanding of the pathogenesis of HCC. Various signal transduction pathways are implicated in HCC, including the Wnt/β-catenin (2), pRb (3,4), mitogen-activated protein kinase (5,6) and Ras signaling pathways (7), and these are being extensively studied to identify potential biomarkers.
HCC is predominantly associated with cirrhosis, which is associated with alcohol, hepatitis B, hepatitis C and non-alcoholic fatty liver disease. Liang et al (8) conducted a meta-analysis and demonstrated that primary biliary cirrhosis is associated with a greater risk of HCC. The regulation/dysregulation of apoptosis of (pre) neoplastic cells as well as proliferation may be important in the process of hepatocarcinogenesis (9). Laouirem et al (10) reported that progression from cirrhosis to cancer is associated with early ubiquitin post-translational modifications. Inhibition of inflammation and mitosis may delay the development of HCC from hepatitis virus-associated cirrhosis (11). Geranylgeranyl diphosphate synthase 1 may be important during the development of HCC from cirrhosis and is of clinical significance for the biological characterization of HCC (12). However, the molecular mechanisms underlying the progression of cirrhosis to HCC remain to be fully elucidated.
Gene expression profiling has been widely used to detect hepatocarcinogenesis and to identify its underlying molecular mechanisms. Nam et al (13) performed gene expression profiling to investigate molecular changes from dysplastic nodules to HCC. Villanueva et al (14) combined clinical, pathological and gene expression data to predict recurrence of HCC. Jia et al (15) identified potential biomarkers of HCC using gene expression profiling.
Reanalyzing existing gene expression data with bioinformatics tools may provide novel insights into the pathogenesis of HCC. The present study therefore analyzed gene expression data of HCC and cirrhosis samples to identify critical genes involved in the development of cirrhosis to HCC. The findings of the present study may improve the understanding of the molecular mechanisms underlying HCC.
Materials and methods
Gene expression data
A total of five gene expression datasets (GSE63898, GSE17548, GSE14323, GSE60502 and GSE62232) were downloaded from Gene Expression Omnibus (ncbi.nlm.nih.gov/geo/; Table I). Cirrhosis HCC samples from GSE14323 were excluded. In addition, nine sets of gene expression data were collected with HG-U133A whereas others were collected with HG-U133A_2 from Gene Expression Omnibus (GEO). To reduce systematic errors, the 9 samples (GSM358103, GSM358104, GSM358105, GSM358106, GSM358107, GSM358108, GSM358109, GSM358110, GSM358111) were removed. Finally, 441 HCC and healthy samples were included in dataset HCC, and 234 cirrhosis samples were included in dataset C.
Table I.
Summary of five gene expression datasets.
| Accession | Sample groups | No. of samples | Platform |
|---|---|---|---|
| GSE63898 | Cirrhosis/HCC | 396 | HG-U219 |
| GSE17548 | Cirrhosis/HCC | 37 | HG-U133_Plus_2 |
| GSE14323 | Healthy/cirrhosis/HCC/cirrhosisHCC | 124 | HG-U133A/HG-U133A_2 |
| GSE60502 | Healthy/HCC | 36 | HG-U133A |
| GSE62232 | Healthy/HCC | 91 | HG-U133_Plus_2 |
HCC, hepatocellular carcinoma.
Pretreatment of raw data was conducted with package affy (http://www.bioconductor.org/packages/release/bioc/html/affy.html) (16) and probes were mapped into genes. Raw data was acquired using four different platforms and a total of 12,012 common genes were selected for the following analysis. Normalization was performed using the quantiles method (16). Gene expression data were divided into two datasets: HCC (HCC and healthy samples) and C (cirrhosis samples).
The comparability of the HCC and C datasets was evaluated by correlations of gene expression levels and connectivity. A high correlation indicated high comparability. Connectivity was calculated using the softConnectivity function from package Weighted Gene Co-expression Network Analysis (WGCNA; labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/) (17) with 5,000 randomly selected genes.
Cluster analysis was conducted with hclust (http://www.r-tutor.com/gpu-computing/clustering/hierarchical-cluster-analysis) to exclude outliers. Finally, 394 HCC samples and 47 healthy samples were included in dataset HCC, and 233 cirrhosis samples were included in dataset C.
Construction of weighted gene co-expression networks. A weighted gene co-expression network was constructed for dataset HCC with package WGCNA (2). Cluster analysis was performed with the flashClust function from package flashClust (labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/Rpackages/flashClust/) and modules were identified with the cutreeHybrid function from package dynamicTreeCut (labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/BranchCutting/). The preservation of modules in dataset C was examined with the modulePreservation function from package WGCNA.
Screening of hub genes
Module Eigengene (ME) was calculated with the moduleEigengenes function from package WGCNA. Connectivity between each gene and ME was calculated with signedKME, which indicated the module membership (MM) of the genes in the module. Genes with high MM values were regarded as hub genes.
Functional annotations
Genes with MM >0.6 were regarded as module-specific genes. All module-specific genes were identified and functional enrichment analysis was performed with the Database for Annotation, Visualization and Integration Discovery (david.abcc.ncifcrf.gov/) (18), including Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (19). Functional networks were constructed with the Enrichment Map (20) in Cytoscape (www.cytoscape.org) (21).
Results
HCC and C datasets
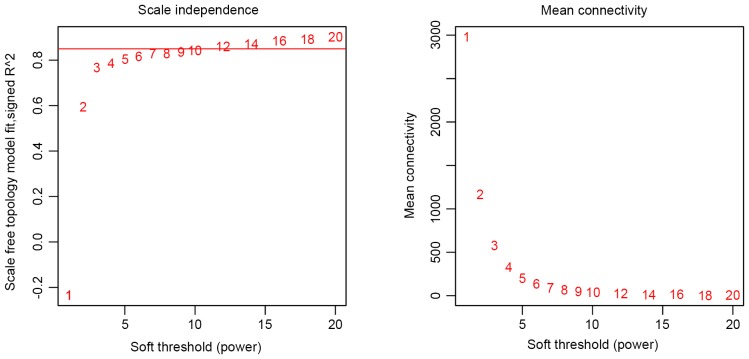
The comparability between the HCC and C datasets was examined with correlations of gene expression levels and connectivity. To calculate connectivity, weighted gene co-expression networks were constructed with 5,000 genes. As described previously (22) the optimal soft threshold power was set as 7 (Fig. 1).
Figure 1.
Optimizing soft-threshold power using network topology as a function of soft threshold power. The left panel shows the scale-free fit index (y-axis) as a function of the soft-threshold power (x-axis). The values of the corresponding soft threshold power are labeled. A guidance line is shown at the value of 0.85 for fit index. When the fit index reaches a plateau (a relatively high value of 0.85), the minimal value of soft threshold power should be 7. The right panel displays the mean connectivity (degree, y-axis) as a function of the soft-threshold power (x-axis). When power is 7, the mean connectivity reaches a steady value.
As presented in Fig. 2, the comparability between dataset HCC and dataset C was good. The correlation of gene expression was 0.97 and the connectivity value was 0.69.
Figure 2.
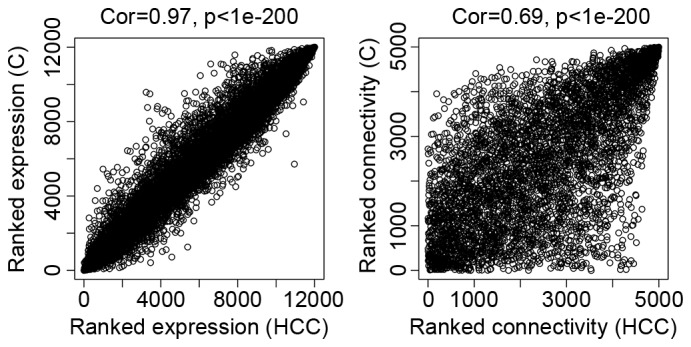
The comparability of datasets HCC and C. The left panel reveals the correlation of expression ranks of genes between the two datasets. The correlation is 0.97 and P<1e-200. The right panel reveals the correlation of connectivity ranks of 5,000 random genes between the two data sets. The correlation is 0.69 and P<1e-200. HCC, hepatocellular carcinoma; C, cirrhosis; Cor, correlation.
The cluster analysis result presented in Fig. 3 revealed that no outlier sample was observed in dataset HCC, whereas one outlier sample was detected in dataset C. Therefore, 394 HCC and 47 healthy samples were included in dataset HCC and 233 cirrhosis samples were included in dataset C.
Figure 3.
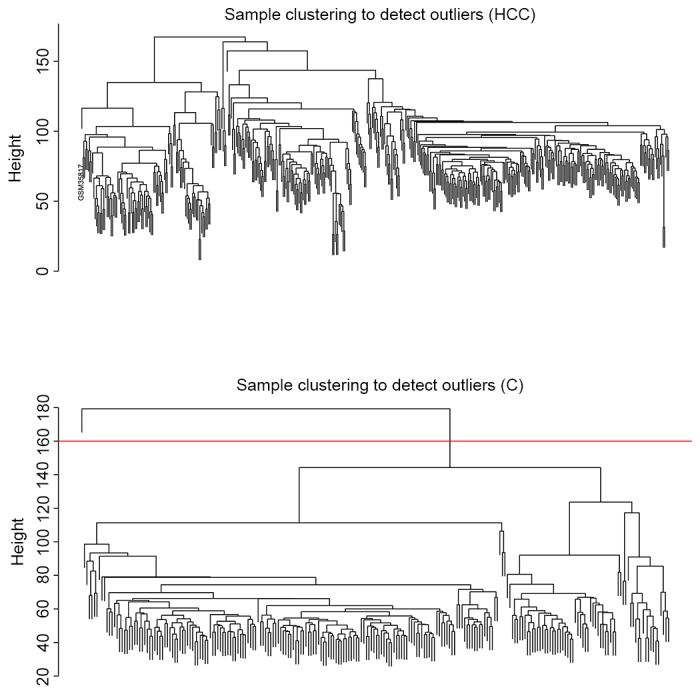
Clustering diagram of samples based on their Euclidean distance to detect outlier samples. The top panel is the clustering diagram for dataset HCC. The bottom panel is the clustering diagram for dataset C. Red line indicates the cut tree cutoff at height 160, so that one outlier sample in dataset C was removed. HCC, hepatocellular carcinoma; C, cirrhosis.
Gene co-expression network and modules
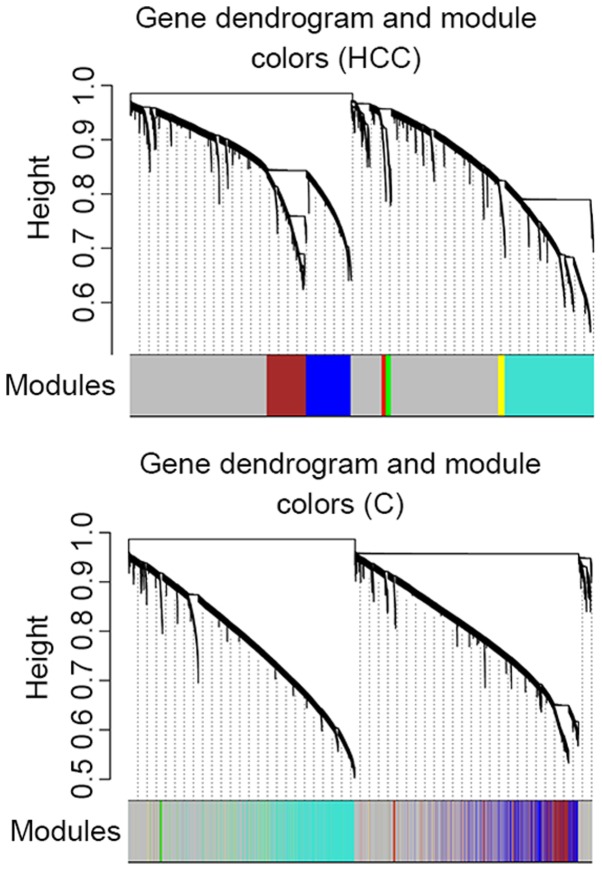
A total of 6 modules were identified in the weighted gene co-expression network of dataset HCC (blue, brown, turquoise, green, red and yellow; Fig. 4; Table II). The cluster analysis result of dataset C revealed that turquoise, red, blue and green modules demonstrated a high preservation (Fig. 4).
Figure 4.
Clustering diagram and modules identified by Weighted Gene Co-expression Network Analysis. The clustering diagram and 6 modules in different colors (blue, brown, turquoise, green, red and yellow) presented in the left panel for dataset HCC. Grey color indicates uncharacterized genes. Right panel presents the clustering result for dataset C. HCC, hepatocellular carcinoma; C, cirrhosis.
Table II.
Modules identified by weighted gene co-expression network analysis-based clustering.
| Module | No. of genes |
|---|---|
| Blue | 3,635 |
| Brown | 2,121 |
| Turquoise | 5,360 |
| Green | 755 |
| Red | 606 |
| Yellow | 1,004 |
The preservation of these modules was examined with modulePreservation from package WGCNA and the results are presented in Table III. Unclassified genes were included in module grey whereas random genes were included in module gold. Theoretically, the z-score of conservative module should be greater than that of modules grey and gold. Modules blue, brown and turquoise demonstrated a greater preservation compared with module yellow, which exhibited the lowest preservation.
Table III.
Preservation of modules between the hepatocellular carcinoma and cirrhosis datasets.
| Module | No. of genes | Z-score |
|---|---|---|
| Blue | 400 | 43.785690 |
| Brown | 400 | 42.285353 |
| Turquoise | 400 | 40.996754 |
| Grey | 400 | 22.025369 |
| Green | 139 | 16.709992 |
| Red | 101 | 12.126042 |
| Gold | 100 | 9.438428 |
| Yellow | 168 | 6.539301 |
Module grey contains all uncharacterized genes; module gold contains random genes.
Clustering of modules
Cluster analysis and principle component analysis were performed for the modules. As presented in Fig. 5, two clusters were revealed, one containing modules red and green and the other containing the remaining 5 modules.
Figure 5.
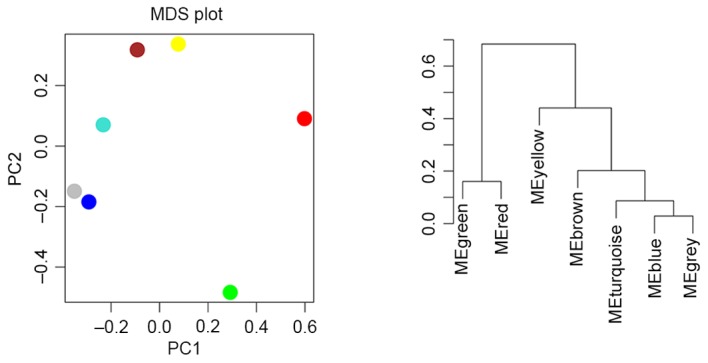
Cluster analysis and PCA of the 6 modules. ME, Module Eigengene; PCA, principle component analysis; MDS, multidimensional scaling.
Functional annotations of modules
Significantly over-represented GO terms and KEGG pathways were assigned for genes with MM >0.6. Functional annotation networks were constructed with the plug-in EnrichmentMap of Cytoscape. A node represented a GO term. If two nodes shared >50% genes, a line connected the two nodes. Cluster analysis was performed for nodes based on connectivity using another plug-in, ClusterOne of Cytoscape. A node cluster should have no less than 10 nodes. The clusters with less than 10 nodes were labeled artificially according to similarity of annotations.
Module blue (Fig. 6) was associated with transcription (regulation of transcription, RNA slicing and nuclear lumen), mitosis (histone protein methylation, chromosome organization, cell cycle and microtubule cytoskeleton), as well as protein ubiquitination and adenosine triphosphate binding. Module brown (Fig. 7) had similar biological functions to module blue and was associated with mitosis. Module turquoise (Fig. 8) was associated with ion channels, cation transportation and cation homeostasis. Module yellow (Fig. 9) demonstrated similarities with module turquoise and was additionally associated with secretion and regulation of cyclase activity.
Figure 6.
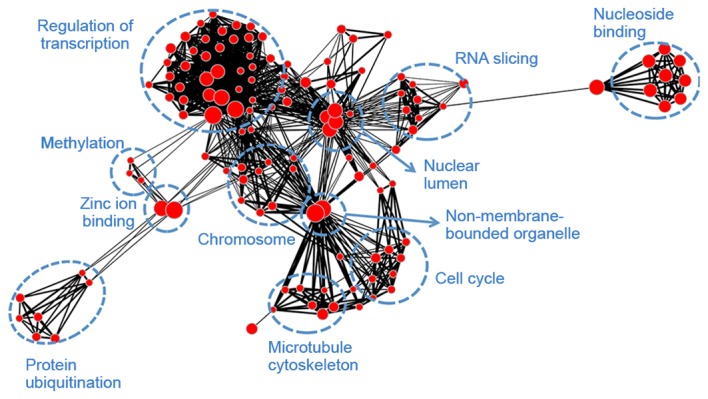
Gene ontology-annotation enrichment map for module blue. Nodes represent annotation terms and lines indicate the linked two annotation terms share >50% common genes.
Figure 7.
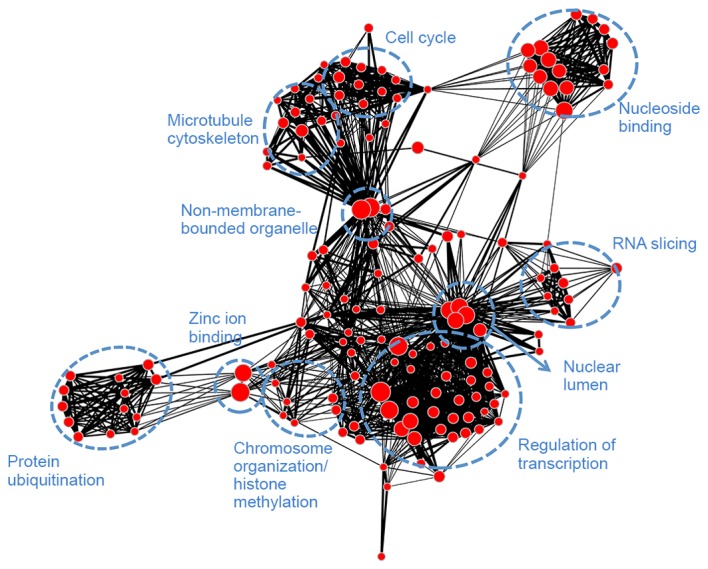
Gene ontology-annotation enrichment map for module brown. Nodes represent annotation terms and lines indicate the linked two annotation terms share >50% common genes.
Figure 8.
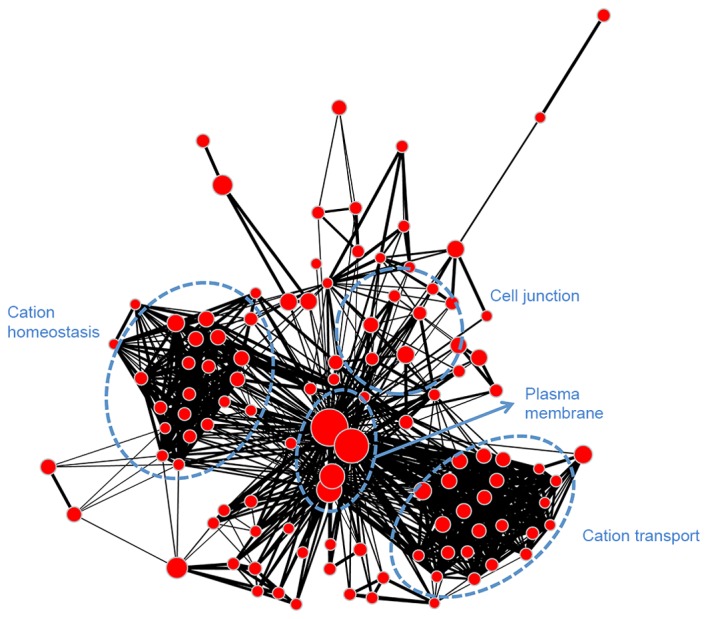
Gene ontology-annotation enrichment map for module turquoise. Nodes represent annotation terms and lines indicate the linked two annotation terms share >50% common genes.
Figure 9.
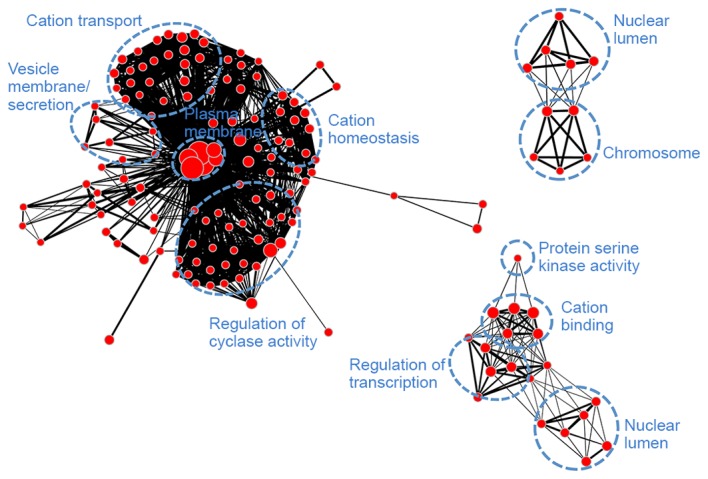
Gene ontology-annotation enrichment map for module yellow (left), green (top right) and red (bottom right). Nodes represent annotation terms and lines indicate the linked two annotation terms share >50% common genes.
Hub genes
The top 10 hub genes of modules were identified for databases HCC and C (Table IV). The top 20 hub genes of modules for database HCC are presented in Table V, in which the overlapped hub genes with database C were marked as bold. The number of overlapping hub genes may reflect the preservation of the module. Module yellow demonstrated low preservation and thus these hub genes may distinguish cirrhosis from HCC. Therefore, functional annotations were given for these genes (P<0.05; Table VI). Cytokine activity and cytokine-cytokine receptor interaction were significantly over-represented in the hub genes. Chemokine (C-C motif) ligand 22 (CCL22), cardiotrophin 1 (CTF1) and interleukin-19 (IL-19) were three cytokines identified that may be important in the progression of HCC.
Table IV.
Hub genes identified in hepatocellular carcinoma and cirrhosis datasets.
| Module | Hub genes |
|---|---|
| Blue | CUL1, EPN1, LSM14B, METTL2B, NACA2, PDAP1, POM121, TIGD6, ZNF440, ZNF480 |
| Brown | AFF4, CHM, EXOSC1, FOXN2, PPP4R2, SLC39A9, SMEK2, THRAP3, WRNIP1, ZNF770 |
| Turquoise | ADRBK1, CXXC1, DTX2P1-UPK3BP1-PMS2P11, EPOR, GDPD2, KRTAP5-8, PAX8, PNPLA2, PRB1, WAS |
| Grey | ATG16L1, EPN1, GPATCH1, MAP2K7, METTL2B, PRDM10, PRMT7, RBBP5, SBNO1, ZNF37A |
| Green | ERN1, FGD2, H2AFJ, HIPK3, KRTAP1-3, LNPEP, PACS1, PACS2, SH3GLB2, ZSWIM1 |
| Red | ARL15, CBX4, ETV3, HSD17B1, KCNK3, LIN7A, MAGIX, TMOD3, WHSC1L1, ZKSCAN8 |
| Yellow | C6orf15, CETN1, FAM155B, GJA8, KCNS1, NTRK3, OR1F1, PLCD1, SLC39A2, TMPRSS15 |
Table V.
Hub genes identified in the hepatocellular carcinoma dataset.
| Module | No. of overlapped genes | Hub genes |
|---|---|---|
| Blue | 9 | ALOX15, AMELX, CYP2F1, EPN1, HNRNPF, LSM14B, METTL2B, NACA2, PDAP1, POM121, POMZP3, PRR14L, PRR7, STYK1, TIGD6, ZBTB6, ZFP69B, ZNF440, ZNF480, ZNF674 |
| Brown | 10 | AFF4, CHM, DDX6, EXOSC1, FAM204A, FBXO22, FOXN2, MAP3K1, MRPL52, NR2C2, PAFAH1B2, PPP4R2, QSER1, RARS2, SLC39A9, SMEK2, THRAP3, USP19, WRNIP1, ZNF770 |
| Turquoise | 10 | ADRBK1, CACNA1G, CHRNB2, CXXC1, DMWD, DTX2P1-UPK3BP1-PMS2P11, EPOR, GDPD2, GLP1R, GRIK5, KCNJ5, KIR2DL1, KRTAP5-8, NCR2, PADI1, PAX8, PNPLA2, PRB1,WAS, WDTC1 |
| Grey | 2 | CEP295, EDC4, EIF3B, GGA3, GPATCH1, GPATCH2, ILKAP, LSM14B, METTL2B, MSL2, OCRL, PDAP1, POM121, POMZP3, PRR14L, UBE3B, ZBTB6, ZNF440, ZNF480, ZNF573 |
| Green | 10 | EFNA2, ELK4, ERN1, ERVH-6, FGD2, H2AFJ, HIPK3, KIF13A, KRTAP1-3, LATS1, LNPEP, PACS1, PACS2, PIK3R2, PTPN14, RBFOX1, SH3GLB2, STRN, WNT7B, ZSWIM1 |
| Red | 10 | ARL15, CBX4, ELK4, ETV3, FGD2, HSD17B1, IL37, KCNK3, KIF13A, LIN7A, MAGIX, MMP28, MTUS2, NCS1, RASA2, RFX7, TAF13, TMOD3, WHSC1L1, ZKSCAN8 |
| Yellow | 0 | ADAM11, BCL11A, CCL22, CLDN17, CTF1, CXorf36, EPB42, FRMD1, FSCN3, GAD2, GFRA4, IL19, KRT2, NTNG1, OR10J1, PP13, PRG3, PSG6, SEZ6L, SMCP |
Table VI.
Functional annotations of hub genes in module yellow.
| Term | P-value (<0.05) | Genes |
|---|---|---|
| GO:0005125~cytokine activity | 0.011 | CCL22, CTF1, IL19 |
| GO:0005576~extracellular region | 0.029 | CCL22, PSG6, CXORF36, CTF1, IL19, NTNG1, GFRA4 |
| GO:0001775~cell activation | 0.030 | PRG3, BCL11A, KRT2 |
| GO:0031225~anchored to membrane | 0.030 | GAD2, NTNG1, GFRA4 |
| GO:0048870~cell motility | 0.034 | CCL22, SMCP, KRT2 |
| GO:0051674~localization of cell | 0.034 | CCL22, SMCP, KRT2 |
| hsa04060:cytokine-cytokine receptor interaction | 0.035 | CCL22, CTF1, IL19 |
GO, gene ontology.
Discussion
A total of five gene expression datasets of HCC and cirrhosis were collected and analyzed in the present study. A weighted gene co-expression network was used for dataset HCC, from which 6 modules were identified. Five out of the 6 modules indicated considerable preservation in dataset C and were revealed to be associated with transcription, mitosis, protein ubiquitination and cation homeostasis.
The other module may be closely involved in the progression of HCC. Functional enrichment analysis demonstrated that module yellow was associated with cation transport, secretion and regulation of cyclase activity. Hub genes of module yellow were obtained and revealed to be involved in cytokine-cytokine receptor interactions and cell motility. It has been suggested that cytokine-mediated signal transduction may contribute to the development of HCC. Various critical genes were identified, including CCL22, CTF1 and IL-19. CCL22 binds to chemokine receptor type 4 and may be important in the trafficking of activated T lymphocytes to inflammatory sites and other aspects of activated T lymphocyte physiology. CCL22 was revealed to enhance tumor migration and associate with venous infiltration in HCC (23). Yang et al (24) demonstrated that transforming growth factor-β-miR-34a-CCL22 signaling-induced regulatory T cell recruitment promotes venous metastases of HCC. Therefore, it may be regarded as a potential therapeutic target for metastatic HCC (25). IL-19 is upregulated in breast cancer and promotes tumor progression (26). Guo et al (27) reported that the copy number of IL-19 is significantly increased in HCC. Oleksyk et al (28) reported that polymorphism of IL-19 may be involved in natural clearance of the hepatitis C virus in the African-American population. Therefore, it has been hypothesized that IL-19 may be important in the progression of HCC. In addition, various hub genes require further research to verify their roles in the development of HCC. B-cell lymphoma/leukemia 11A (BCL11A) is a C2H2 type zinc-finger protein that is essential for normal lymphoid development (29) and is implicated in lymphoid malignancy (30) as well as non-small cell lung cancer (31). Claudin (CLDN) 17 is a member of the claudin family, which is a family of integral membrane proteins and tight junction strand components. Various members of the claudin family have been implicated in HCC. Suppression of CLDN10 may inhibit HCC invasion (32). CLDN10 expression levels are associated with recurrence of primary HCC (33). CLDN6 and 9 function as additional co-receptors for hepatitis C virus (34). BCL11A and CLDN17 may be involved in the progression of HCC.
In conclusion, package WGCNA was used to analyze HCC and cirrhosis gene expression data. A total of 6 gene modules were identified, including one module that was closely associated with HCC. Furthermore, a variety of critical genes were revealed, further investigation of which may help to advance understanding of the pathogenesis of HCC.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831–10839. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- 3.Azechi H, Nishida N, Fukuda Y, Nishimura T, Minata M, Katsuma H, Kuno M, Ito T, Komeda T, Kita R, et al. Disruption of the p16/cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001;60:346–354. doi: 10.1159/000058531. [DOI] [PubMed] [Google Scholar]
- 4.Sudo T, Utsunomiya T, Mimori K, Nagahara H, Ogawa K, Inoue H, Wakiyama S, Fujita H, Shirouzu K, Mori M. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer. 2005;92:1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt CM, McKillop IH, Cahill PA, Sitzmann JV. Increased MAPK expression and activity in primary human hepatocellular carcinoma. Biochem Biophys Res Commun. 1997;236:54–58. doi: 10.1006/bbrc.1997.6840. [DOI] [PubMed] [Google Scholar]
- 6.Huynh H, Nguyen TT, Chow KH, Tan PH, Soo KC, Tran E. Over-expression of the mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK in hepatocellular carcinoma: Its role in tumor progression and apoptosis. BMC Gastroenterol. 2003;8:3. doi: 10.1186/1471-230X-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newell P, Toffanin S, Villanueva A, Chiang DY, Minguez B, Cabellos L, Savic R, Hoshida Y, Lim KH, Melgar-Lesmes P, et al. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J Hepatol. 2009;51:725–733. doi: 10.1016/j.jhep.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang Y, Yang Z, Zhong R. Primary biliary cirrhosis and cancer risk: A systematic review and meta-analysis. Hepatology. 2012;56:1409–1417. doi: 10.1002/hep.25788. [DOI] [PubMed] [Google Scholar]
- 9.Park YN, Chae KJ, Kim YB, Park C, Theise N. Apoptosis and proliferation in hepatocarcinogenesis related to cirrhosis. Cancer. 2001;92:2733–2738. doi: 10.1002/1097-0142(20011201)92:11<2733::AID-CNCR10126>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Laouirem S, Le Faouder J, Alexandrov T, Mestivier D, Léger T, Baudin X, Mebarki M, Paradis V, Camadro JM, Bedossa P. Progression from cirrhosis to cancer is associated with early ubiquitin post-translational modifications: Identification of new biomarkers of cirrhosis at risk of malignancy. J Pathol. 2014;234:452–463. doi: 10.1002/path.4398. [DOI] [PubMed] [Google Scholar]
- 11.Arrieta O, Rodriguez-Diaz JL, Rosas-Camargo V, Morales-Espinosa D, de Leon S Ponce, Kershenobich D, Leon-Rodriguez E. Colchicine delays the development of hepatocellular carcinoma in patients with hepatitis virus-related liver cirrhosis. Cancer. 2006;107:1852–1858. doi: 10.1002/cncr.22198. [DOI] [PubMed] [Google Scholar]
- 12.Yu DC, Liu J, Chen J, Shao JJ, Shen X, Xia HG, Li CJ, Xue B, Ding YT. GGPPS1 predicts the biological character of hepatocellular carcinoma in patients with cirrhosis. Bmc Cancer. 2014;14:248. doi: 10.1186/1471-2407-14-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nam SW, Park JY, Ramasamy A, Shevade S, Islam A, Long PM, Park CK, Park SE, Kim SY, Lee SH, et al. Molecular changes from dysplastic nodule to hepatocellular carcinoma through gene expression profiling. Hepatology. 2005;42:809–818. doi: 10.1002/hep.20878. [DOI] [PubMed] [Google Scholar]
- 14.Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512.e2. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia HL, Ye QH, Qin LX, Budhu A, Forgues M, Chen Y, Liu YK, Sun HC, Wang L, Lu HZ, et al. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:1133–1139. doi: 10.1158/1078-0432.CCR-06-1025. [DOI] [PubMed] [Google Scholar]
- 16.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 17.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. Bmc Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment Map: A network-based method for gene-set enrichment visualization and interpretation. PloS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 23.Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, Ng KT, Forbes SJ, Guan XY, Poon RT, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62:607–616. doi: 10.1016/j.jhep.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 24.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, et al. TGF-β-miR-34a-CCL22 signaling-induced treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budhu A, Wang XW. Transforming the microenvironment: A trick of the metastatic cancer cell. Cancer Cell. 2012;22:279–280. doi: 10.1016/j.ccr.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Hsing C-H, Cheng HC, Hsu YH, Chan CH, Yeh CH, Li CF, Chang MS. Upregulated IL-19 in breast cancer promotes tumor progression and affects clinical outcome. Clin Cancer Res. 2012;18:713–725. doi: 10.1158/1078-0432.CCR-11-1532. [DOI] [PubMed] [Google Scholar]
- 27.Guo X, Yanna, Ma X, An J, Shang Y, Huang Q, Yang H, Chen Z, Xing J. A meta-analysis of array-CGH studies implicates antiviral immunity pathways in the development of hepatocellular carcinoma. Plos One. 2011;6:e28404. doi: 10.1371/journal.pone.0028404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oleksyk TK, Thio CL, Truelove AL, Goedert JJ, Donfield SM, Kirk GD, Thomas DL, O'Brien SJ, Smith MW. Single nucleotide polymorphisms and haplotypes in the IL10 region associated with HCV clearance. Genes Immun. 2005;6:347–357. doi: 10.1038/sj.gene.6364188. [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 30.Satterwhite E, Sonoki T, Willis TG, Harder L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, et al. The BCL11 gene family: Involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.V98.12.3413. [DOI] [PubMed] [Google Scholar]
- 31.Jiang B, Zhang X, Su J, Meng W, Yang XN, Yang JJ, Zhou Q, Chen ZY, Chen ZH, Xie Z, et al. BCL11A overexpression predicts survival and relapse in non-small cell lung cancer and is modulated by microRNA-30a and gene amplification. Mol Cancer. 2013;12:61. doi: 10.1186/1476-4598-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ip YC, Cheung ST, Lee YT, Ho JC, Fan ST. Inhibition of hepatocellular carcinoma invasion by suppression of claudin-10 in HLE cells. Mol Cancer Ther. 2007;6:2858–2867. doi: 10.1158/1535-7163.MCT-07-0453. [DOI] [PubMed] [Google Scholar]
- 33.Cheung ST, Leung KL, Ip YC, Chen X, Fong DY, Ng IO, Fan ST, So S. Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res. 2005;11:551–556. [PubMed] [Google Scholar]
- 34.Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]