Abstract
Macromolecular drug candidates and nanoparticles are typically tested in 2D cancer cell culture models, which are often directly followed by in vivo animal studies. The majority of these drug candidates, however, fail in vivo. In contrast to classical small-molecule drugs, multiple barriers exist for these larger molecules that two-dimensional approaches do not recapitulate. In order to provide better mechanistic insights into the parameters controlling success and failure and due to changing ethical perspectives on animal studies, there is a growing need for in vitro models with higher physiological relevance. This need is reflected by an increased interest in 3D tumor models, which during the past decade have evolved from relatively simple tumor cell aggregates to more complex models that incorporate additional tumor characteristics as well as patient-derived material. This review will address tissue culture models that implement critical features of the physiological tumor context such as 3D structure, extracellular matrix, interstitial flow, vascular extravasation, and the use of patient material. We will focus on specific examples, relating to peptide-and protein-conjugated drugs and other nanoparticles, and discuss the added value and limitations of the respective approaches.
Introduction
In current cancer research, drug candidates are still primarily tested in 2D monolayer cultured cell models. If initial experiments are successful, these in vitro studies are often directly followed by animal experiments. However, many compounds that show promising results in vitro perform only poorly in animal models or ultimately fail in clinical trials.1 This lack of success is based on the fact that the complexity of a tumor and its environment is in sharp contrast to the simplicity of immortalized cell lines growing in a monolayer. Also, with the ambition to reduce the number of animal experiments, there is increasing demand for in vitro tumor models that mimic in vivo characteristics more accurately.2,3 Tumor cells that grow in a monolayer culture on a glass or plastic bottom are stretched out, which causes cytoskeletal rearrangements, and have limited cell–cell contact. In contrast, culturing tumor cells in a 3D setting resembles the in vivo architecture and tumor microenvironment more closely due to the more natural morphology of cells, the presence of extracellular matrix, pH and oxygen gradients, and in some cases the presence of interstitial flow.4−6 This complexity leads to increased heterogeneity due to nutrient gradients and to growth characteristics that differ from the two-dimensional situation. In particular, for peptide conjugates and larger biologicals, drug penetration is a limiting factor since cells are less accessible due to their tight 3D arrangement, which cannot be captured by 2D tumor cultures.
In the simplest case, a 3D tumor model consists of aggregates of tumor cells, or spheroids, that mimic small, nonvascularized tumor deposits as they are encountered in several cancer types, such as epithelial ovarian cancer.7 Spheroids increase the relevance of in vitro results compared to 2D cell cultures, since they show a drug resistance pattern more comparable to that of solid tumors.8−10 More complex models were developed alongside the development of organ-on-a-chip microfluidic approaches. These efforts resulted in designs that mimic the in vivo tumor microenvironment better and that reinstate barriers that are encountered by macromolecular drugs.
In practice, the complexity of model systems may become a limiting factor. Therefore, the choice for a tumor model should be based on the balance between predictive power for a specific research question and simplicity and robustness of the model.
Here, we will review tumor models and their applications in the testing of peptide and protein-conjugated drugs and other macromolecular compounds and nanoparticles. First, we will describe the components and characteristics of the tumor microenvironment to illustrate the critical barriers that these molecules face on their way to their final targets, such as the vascular endothelium, the dense extracellular matrix, and the binding site barrier. Then we will discuss possibilities to incorporate these features into 3D tumor models. Instead of providing an exhaustive review of 3D tumor models, we will focus on representative examples relating to macromolecular drug and nanoparticle testing. Even though not all examples directly deal with peptide or protein conjugates, as not all models have yet been tested with such conjugates, the conclusions are also relevant for this class of drug candidates.
Tumor Microenvironment
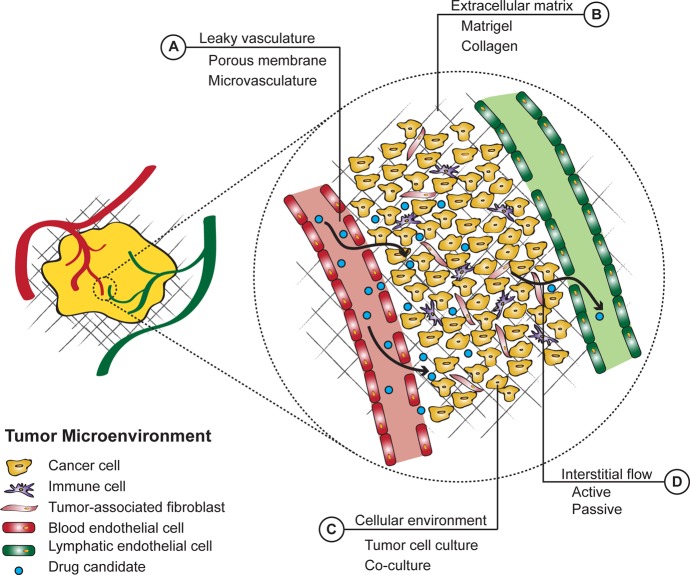
Tumor cells reside in a dynamic environment comprising various cell types such as immune cells and cancer-associated fibroblasts, extracellular matrix (ECM), interstitial flow, and signaling molecules. This tumor microenvironment (TME) plays an important role in tumor progression and metastasis formation, and the physical properties of the TME are greatly influencing drug delivery (Figure 1).
Figure 1.
Schematic representation of the different barriers that a macromolecular drug encounters and options to mimic these in a 3D tumor model. (A) Leaky vasculature is due to a fenestrated endothelium as a consequence of disorganized neovascularization. Porous membranes and self-assembling or aided microvasculature within a microfluidic device are typically used to mimic this barrier. (B) Extracellular matrix is used in static and fluidics based culture methods. (C) Cellular environment is made up of tumor cells and other cells, including cancer-associated fibroblasts and immune cells. Co-cultures of tumor and other cells have been developed for several 3D culture systems. (D) High interstitial fluid flow and high pressure are an effect of leaky vasculature, dense ECM, high cell density, and disorganized lymphatic drainage system. This can be mimicked in microfluidic systems where fluid is either actively pumped (e.g., syringe pump-driven) or passively forced (e.g., gravity flow) through the cellular and ECM layer. (A–C) are by nature present in a tumor explant model and (D) flow characteristics can be mimicked using microfluidics. ECM, extracellular matrix.
The ECM is a compact matrix of proteins, glycosaminoglycans, and signaling molecules that supports its surrounding tissue.11 Generally, the ECM in tumors is denser than in normal tissue, although there are large differences in ECM between different tumor types. Lymphomas and certain neuroblastomas have minor ECM and are therefore considered stroma-poor.12,13 In contrast, other tumors, especially pancreatic cancers, have a very compact matrix, which is a result of the excessive production of matrix molecules. As often seen in different types of cancer, the resulting compact matrix inhibits penetration of large molecules (Figure 1B).14−16
Apart from the physical boundary that the ECM imposes, a high density of target molecules on malignant cells, such as receptors targeted by peptide conjugates, can also inhibit penetration of macromolecular drugs through a mechanism called the binding site barrier (BSB). Originally, the importance of the BSB was recognized because antibodies accumulated mostly in the perivascular region, without penetrating deeper into the tumor. The antibodies were captured by the first antigen they encountered and were therefore unable to enter deeper into the tissue.17,18 Since antibodies typically show a very high affinity toward their target protein, penetration goes along with target saturation, meaning that BSBs are more prominent with low drug concentrations.19 The BSB also exists for non-antibody drug delivery vehicles such as nanoparticles.20 Of note is that nonspecific binding can play an important role in the BSB as well, depending on the targeting moiety.21
Before macromolecular compounds encounter the ECM, they first have to extravasate from blood vessels into the interstitial space. In tumors, the vascular endothelium is often poorly organized, leading to an increased permeability for macromolecules larger than 40 kDa.22,23 The degree of leakiness varies greatly between different tumor types which is a consequence of different numbers and sizes of gaps between endothelial cells (Figure 1A).24 For small molecules, this variability matters little. For larger particles however, leakiness of the endothelium is a critical parameter.22
When endothelial leakiness is combined with inefficient lymphatic drainage, larger particles accumulate in the tumor, whereas small particles diffuse out of the TME (Figure 1D).25 This phenomenon is called the enhanced permeability and retention (EPR) effect and is seen in most tumors. However, the extent of the EPR varies between different tumor types and within individual tumors. As an example, a recent study in dogs found the effect to be much more pronounced in carcinomas than in sarcomas.26 A prime reason for the difference between sarcomas and carcinomas is the higher growth rate of the latter, which leads to more dysfunctional and leaky tumor vessels. Similar differences have been observed between fast- and slow-growing tumors in rat prostate carcinoma models.27
In addition, the extravasation of fluids, together with high cell and ECM density, fibroblast-mediated contraction of the ECM, and dysfunctional lymphatic vessels, leads to an increased interstitial fluid pressure (IFP). High IFP has a negative effect on drug penetration, particularly of larger compounds such as nanoparticles. This effect increases toward the center of tumors, where the IFP is often much greater than at the periphery.6,28 A high IFP thus counteracts the EPR. The presence of a high IFP has been confirmed for many tumors, including breast cancer, colorectal cancer, head and neck cancer, and melanoma.29−33
Multicellular Tumor Spheroids
Multicellular tumor spheroids (MCTSs) constitute the most fundamental 3D culture model that is also amenable to high throughput.5 Generally, MCTSs are clumps of cells with a diameter of 100 to 1000 μm. The 3D arrangement within an MCTS leads to increased cell–cell contacts compared to cells cultured in a monolayer. These differences in cell–cell contacts in turn underlie many of the gene and protein expression changes when compared to their monolayer counterparts.34−37 Also, since the cells adhere to each other rather than to a glass or plastic bottom, they tend to grow slower and show more resistance to chemotherapeutics that are aimed at rapidly proliferating cancer cells.8,9 In spite of their simplicity, MCTSs mimic several relevant in vivo features such as oxygen gradients, matrix deposition, and hindered diffusion.
MCTSs can be generated in several ways and common methods include the hanging drop method, liquid overlay, or spinner flasks.4 Although technically different, all methods have in common that they deprive cells of contacts to a growth-promoting surface so that contacts are established only between cells. Mostly, immortalized cell lines are used to generate MCTSs, but it is also possible to use primary cells such as tumor cells derived from malignant ascites (fluid accumulation in the peritoneal cavity) in epithelial ovarian cancer.38 The addition of ECM materials like collagen, Matrigel, or methylcellulose can promote MCTS formation but is often not necessary from a technical point of view.4,39 Whether to include ECM compounds in a model depends on the tumor type that is being mimicked, since, as mentioned above, in vivo tumors show large variations in stroma content.
There have been several studies in which MCTSs were employed to elucidate the molecular mechanisms of penetration of nanoscale drug delivery systems. For example, small gold nanoparticles of 2 and 6 nm were able to penetrate throughout the MCTS, whereas 15 nm gold nanoparticles accumulated on the rim and penetrated poorly into the spheroid core.40 Concerning the ECM, it was demonstrated that a thick collagen network in MCTSs resulted in low penetration depth for polystyrene nanoparticles and that penetration of particles <100 nm could be significantly enhanced by addition of collagenase. This effect was greater for particles of 40 nm than for particles of 20 nm since the latter were already able to penetrate the spheroids relatively unhindered.15 Particles larger than 100 nm only showed minor penetration into the spheroid (<5% particles in the core), with only minor improvement upon collagenase treatment. These findings indicate that particle size and ECM density are important factors that determine penetration.
In order to further enhance penetration for nanoparticles of a size up to 100 nm, some groups have explored possibilities of functionalizing micelles with different targeting moieties such as antibodies, transferrin, or single-chain variable fragments.41−44 In all these cases, a higher penetration was found for targeted constructs. Nevertheless, delivery via targeting moieties carries the risk of retention by a BSB. This is illustrated by a study from Miao et al., who developed a static spheroid model consisting of a co-culture of UMUC3 bladder carcinoma cells and 3T3 fibroblasts. In this model the researchers used the BSB concept to explain differences in uptake characteristics of lipid-coated calcium phosphate nanoparticles that were either functionalized with the anisamine ligand, which targets the sigma receptor, or remained unfunctionalized. It was found that targeted particles were captured to a larger extent by sigma-receptor expressing tumor-associated fibroblasts as compared to nontargeted particles, which penetrated further into tumor cell spheroids.20 Small particle size (∼18 nm) and high receptor binding affinity contributed to a more prominent BSB. These results were reproduced in an animal model, demonstrating the value of the spheroid model.20 Another example where the spheroid model was validated with in vivo data was for an internalizing-RGD (iRGD) peptide–drug conjugate where iRGD conjugates showed more tumor penetration than RGD conjugates in both models.45 The tumor-homing peptide iRGD targets tumors by binding to integrin, after which it is cleaved by tumor proteases, exposing the positively charged CendR motif. This motif then binds to neuropilin-1, which is also present on many tumors, leading to an increased vascular permeability and the promotion of tumor penetration. Several other studies that employed iRGD conjugates and investigated them both in vitro in a spheroid model and in vivo have provided further validation of the spheroid model. In each case, it has been found that iRGD conjugation enhanced tumor penetration both in vitro and in vivo.45−47
Microfluidics
The examples above demonstrate that also in static culture conditions key characteristics of tumors can be reproduced. Nevertheless, in the absence of fluid flow, the exchange of nutrients, waste products, and other compounds, as well as the uptake of drugs, solely relies on diffusion. In reality, however, the uptake of macromolecular drug candidates, in contrast to small-molecule drugs, depends more on convection than on diffusion.28 The degree of convection in tumors is governed by hydrostatic and osmotic pressure gradients over the endothelial barrier of the tumor vasculature. In healthy tissue, there is a small net outward pressure, but in tumors there can be both an net outward or inward pressure. In general, osmotic pressure is higher inside tumors.28 The degree to which this pressure is counterbalanced by increased hydrostatic pressure in the interstitium, however, varies greatly between tumors, though in most cases it results in a higher level of inward convection, and thus a higher net flow into the tumor.48 The variability can be explained by differences in the tumor or tissue-dependent microenvironmental factors, most of which were discussed before, including the intravascular pressure, the density of cells and matrix components, the tendency of the tissue to expand (i.e., the compliance), and the functionality of the lymph channels.
Through soft lithography and microfluidic technology it is possible to create more advanced tumor models for the study of distribution and activity of peptide and protein conjugates as well as of nanoparticles. These designs can be collectively referred to as tumor-on-a-chip models, a subset of organ-on-a-chip models.49 Overall, with the systems that were developed so far, three major innovative elements were introduced in comparison to static tumor models: compartmentalization, controlled gradients, and perfusion.50 Here, the focus will lie on compartmentalization and perfusion because these are the most relevant for drug delivery studies in 3D models.
In the simplest embodiment, MCTSs are exposed to flow, by continuously perfusing hanging drops, entrapment of cells in microwells, or placing preformed MCTSs in a microfluidic device.51−53 More advanced models also incorporate the ECM as a physical transport barrier in the model, for instance, by seeding cells in a hydrogel.54,55 The most complex models co-culture tumor cells with different cell types, for instance, endothelial cells in order to mimic the endothelial barrier that drug delivery systems need to overcome.56−58
In an early microfluidics study from Ng et al., in which interstitial flow was mimicked using a microfluidic device, the authors found that the penetration of 500 nm nanoparticles in Matrigel with fibroblasts was virtually absent without flow, and increased to ∼60 μm in the presence of flow.54 This considerable difference illustrates the crucial role of including interstitial flow to mimic those situations where this plays a role in vivo. In this study, IFP was considered to be within the physiological range.6,54 However, as the authors also pointed out, the density of Matrigel was probably lower than that of the dense ECM surrounding a tumor, which could lead to an overestimation of particle penetration.54 This example shows that critical degrees of freedom of such systems are the ability to reproduce interstitial flow, IFP, and ECM density for a certain tumor type. In a more recent on-chip dynamic model, Kwak et al. showed that the accumulation of nanoparticles was not influenced so much by the density of the collagen matrix but rather depended on the density of the seeded cells within the matrix (Figure 2A).56
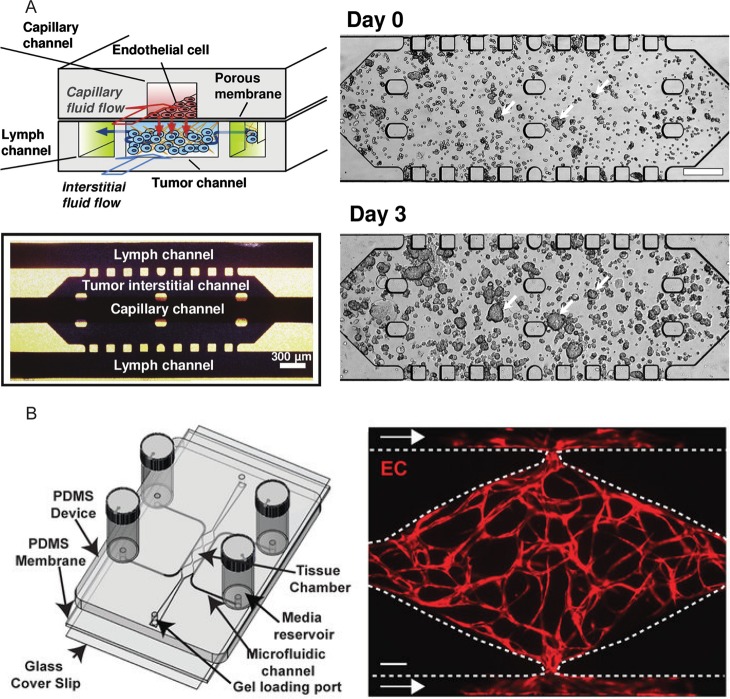
Figure 2.
Examples of 3D culture techniques. (A) Top left: Design of a tumor-microenvironment-on-chip. The two-layered 3D microfluidic platform consisted of a capillary vessel compartment (red) that was positioned on top of a tumor cell-containing compartment (blue), which was in turn separated from two lymph channels (green). All channels were patterned in polydimethylsiloxane (PDMS), which is the most commonly used material for microfluidic chips. In the capillary vessel compartment, a monolayer of endothelial cells was grown on a Matrigel-coated polycarbonate membrane. In this top compartment the nanoparticles were introduced. In the bottom compartment, containing the tumor channel, tumor cells were embedded in a collagen matrix. Bottom left: Top view of the bottom compartment. The position of the capillary channel is indicated, but not visible as it is directly above the tumor interstitial channel. Right: Tumor cell growth after loading (day 0) and after 3 days culture on the microfluidic system. A microscope image of the tumor interstitial channel is presented. Inlets and outlets were connected to fluid columns, thereby introducing height differences which resulted in a fluid flow. The scale bar indicates 300 μm. Reprinted from Kwak et al., Copyright (2014), with permission from Elsevier.56 (B) Left: A schematic overview of a vascularized micro-organ platform. The system consisted of 100-μm-high tissue chambers and microfluidic channels patterned into an 8 mm PDMS layer. This layer was bonded to a 1 mm PDMS layer that was subsequently bonded to a glass coverslip. The central tissue chambers were connected to microfluidic side channels via capillary burst valves that retained the mixture of cells and ECM inside the chambers. Loading of the endothelial cell–ECM suspension was achieved through the indicated gel loading ports. Height differences in the media reservoirs attached to the inlets and outlets enabled a gravity-driven fluid flow through the microfluidic channel toward the tissue chamber. Right: A microscope image of a tissue chamber of the central part of the chip showing a fully developed vascular network after 7 days. Endothelial cells are shown in red and were visualized by confocal microscopy. The supporting stromal cells were not labeled. An outward growth of the endothelial cells into the microfluidic channels could be observed. The scale bar indicates 100 μm. Reprinted from Sobrino et al., Copyright (2016), with permission from The Royal Society of Chemistry.58 ECM, extracellular matrix; PDMS, polydimethylsiloxane.
As indicated before, delivery of macromolecular drugs is also dependent on their ability to extravasate from capillaries surrounding the tumor cells. Therefore, the endothelial barrier has also been implemented into tumor models. Some groups mimicked a leaky endothelial barrier by placing a porous membrane between the fluid and the cellular compartment.56,59 Efficient extravasation only occurred for particles much smaller than the nominal pore size of the membrane.56 A possible explanation is that the nominal pore size represented a maximum, and many pores were smaller. Interestingly, this heterogeneity is also found in patients where leaky vessels in some tumor regions coexist with areas that contain much more well-developed and less leaky endothelial linings.60
Instead of using a fenestrated membrane as a rather simple representation of the endothelial barrier, several other methods have been developed to directly implement microvasculature. These methods vary from self-organizing blood vessels using stem cells and/or organ-specific cells, to microfluidic approaches where endothelial cells are grown on a predefined membrane or ECM-like structure, to hybrid models.57,58,61 These systems vary greatly in complexity and in the way that tumor characteristics are mimicked. A self-organizing, perfusable microvasculature was for example impermeable for FITC-dextran molecules with a molecular weight of 70 kDa, and was therefore less leaky than many tumor vessels in vivo, in which molecules of that size can extravasate.57 The microvasculature functioned very well when incorporated into the tumor model (Figure 2B),58 but since it was not leaky, it would be less predictive when the EPR effect is to be studied.
Ex Vivo Tumor Culture
The models discussed so far employed in vitro adapted cell lines. These approaches have the advantage of standardization, but this advantage comes at the expense of a natural tumor architecture. In this respect, tumor explant cultures are very different from the previously mentioned 3D culture methods. Explant culture does not follow a bottom-up approach in which all components are added separately, but rather uses a top-down approach in which the primary tumor, with all its heterogeneous features, is utilized as a model. The general principle behind this culture method is that primary tumor tissue is excised during surgery and subsequently cultured under laboratory conditions. Critical in these culture systems is, however, the size of the samples. Since the vasculature of the excised tumor does not retain its functionality, the supply of nutrients and oxygen solely relies on diffusion, and lack of these will cause necrosis in the center of larger tumor deposits.
To overcome this problem, two general strategies have been developed. The first strategy is to slice the tumor into ∼300 μm slices. Such slices are thin enough to allow sufficient nutrient supply, but also thick enough to preserve histological features (Figure 3A).62,63 The second method is to microdissect the tumor into cylindrical, spheroid-like, deposits of a few hundred micrometers in diameter (Figure 3B).64 These deposits can then be incorporated into a flow-containing model, with or without an endothelial barrier-mimicking component, employing the techniques discussed above. To our knowledge, however, mimicking the endothelial barrier has not yet been performed in models containing explant cultures.
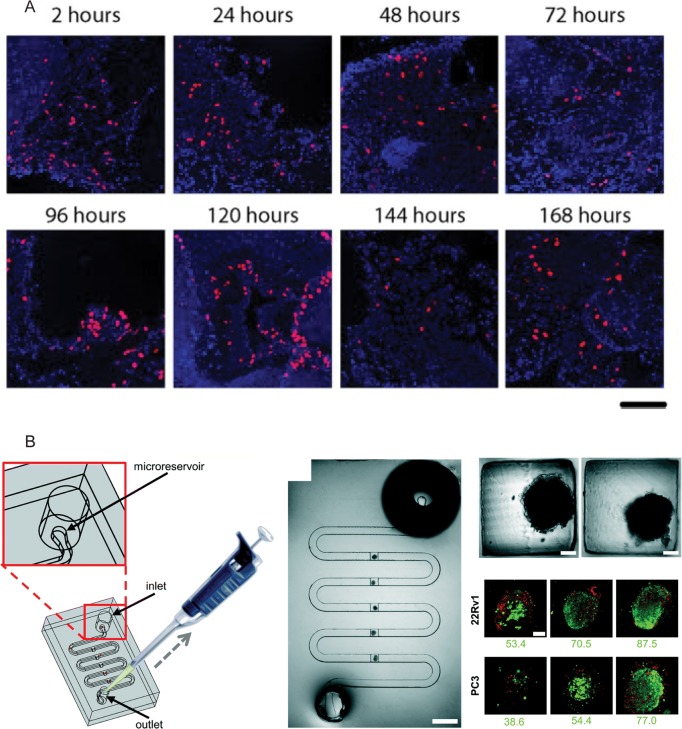
Figure 3.
Examples of tumor explant cultures (A) 7-day culture of 300 μm tumor slices obtained from a single tumor. The tumor slices were cultured under constant orbital movement. Blue, nuclear staining, DAPI; Red, DNA synthesis marker, EdU. The scale bar indicates 100 μm. Reprinted from Naipal et al. (2015), under the Creative Commons Attribution 4.0 International License.62 (B) Microfluidic platform for the study of microdissected tissues. Left: Schematic showing the structure of the microfluidic platform and the loading approach using a micropipette tip. Middle: Picture of the microfluidic device containing microdissected tumor tissues that were captured in the square traps. The scale bar indicates 2 mm. Top right: Zoomed image of microdissected tumor tissues captured in the square traps. The scale bars indicate 100 μm. Bottom right: Microdissected tumor tissues of 22Rv1 or PC3 xenografts were analyzed by confocal microscopy (maximum projection images) and by flow cytometry. The viability of nontreated microdissected tissues for each sample is given below the respective image. The dye for viability was Cell Tracker Green (green) and for dead cells propidium iodide (red). Reprinted from Astolfi et al. (2016), under the Creative Commons Attribution 4.0 International License.64 EdU, 5-ethynyl-2′-deoxyuridine.
The major advantage of explants is that cells from a primary source do not undergo any in vitro adaptation. Another advantage is that the histology and TME of the specimen are left intact which gives the model barrier features that are virtually unchanged from the in vivo situation. Such an accurate representation of extracellular matrix characteristics is currently unfeasible using artificial materials. This key advantage ironically also causes the greatest downside, as there is considerable heterogeneity between different tumor samples and even within a single tumor. The larger variation requires that larger numbers of samples need to be employed in order to reach statistically and/or biologically significant results.
In a study by Dong et al., delivery of chitosan-coated PLGA nanoparticles incorporating antisense oligonucleotides against telomerase was tested on tumor slices generated from tumors of non-small cell lung cancer patients and compared against a monolayer culture of primary cells that originated from the same tumor.65 Indeed, there was heterogeneity within and between samples, and telomerase activity could vary greatly between tumor samples and different cell types within a tumor slice. Telomerase inhibition was, however, effective and relatively comparable to monolayer immortalized cell line culture. Also, the nanoparticles could penetrate throughout the whole tumor slice.
Due to the previously mentioned difficulties and heterogeneity, one may expect that these complex models will complement the standardized methods described above at a later stage of drug development. First, they can provide valuable information on the potential heterogeneity of clinical responses. Second, a thorough assessment of the functional histological context in which a drug candidate shows activity is possible.
Discussion and Future Perspectives
Research published so far demonstrates that in contrast to 2D models, 3D models are able to capture critical features of tumors and their environment, which are necessary to predict the in vivo behavior of macromolecular drugs and targeted conjugates. Very clearly, there is no one-size-fits-all solution. Tumors are highly heterogeneous, leading to enormous differences in the relevance of the EPR effect and of the parameters governing penetration. For all macromolecules, penetration is critical, and while targeted conjugates may enhance cellular internalization rates, the BSB effect can conversely limit effectiveness.
Advances with respect to the incorporation of different barriers in 3D tumor models for the assessment of drug delivery are constantly being made, a development that occurs in parallel with, and may profit from, advances made in model development for other barriers such as skin, lung, and intestine.66 In practice, care has to be taken when choosing a model. For instance, while it is a great achievement that self-organizing microvasculature models can be generated, they do not recapitulate the EPR effect and this should be taken into account when designing a model.
So far, the most popular 3D in vitro model for investigating peptide- and protein-conjugated drugs has without doubt been the static spheroid model, as can be seen in Table 1, where a representative overview is given of the different types of protein- and peptide conjugates that have been studied with this model. Most studies used a simple spheroid model that consisted of a single type of cancer cell, whereas some studies used more advanced spheroid models like complex cocultures that attempted to mimic the endothelial barrier.67−69 A key outcome measure has been the penetration depth. However, the presence of static conditions means that outcomes may have been biased toward systems that diffuse well, rather than those that rely on convection as the main transport mode. Notably, more advanced on-chip 3D models have thus far mainly been performed with “model” nanoparticles such as dextran or polystyrene beads, suggesting a strong focus on the development of these systems even in recent years, rather than on their practical application. Nevertheless, given the arguments that we have laid out in this review, we believe that future studies would benefit from aiming to introduce more barriers and more physiologically relevant conditions.
Table 1. Representative Examples of Peptide- and Protein-Conjugated Drugs That Have Been Tested in Static Spheroid Models.
| conjugate class | (poly)peptide | coupled moiety/particle | refs |
|---|---|---|---|
| peptide-conjugated nanoparticles | CGKRK, CTR, GICP, IL13p, mastoparan, MMP2-sensitive peptide, penetratin, Pep-1, R8, RGD variants, SAPSp, tat, T7, TGN, TH-Lip, tLyp-1, tumstatin | chitosan, iron oxide particle, lipid-based particle, paclitaxel nanocrystallites, PAMAM dendrimer, liposome, polymeric particle (e.g., PEG–PCL, PEG–PLA, PEG–PTMC, PLGA–chitosan), quantum dot | (45−47,67−97) |
| other peptide conjugatesa | RGD | oligonucleotide | (98,99) |
| protein-conjugated nanoparticles | antibody, collagenase, scFv, TRAIL, transferrin | albumin particle, lipid-based particle, liposome, micelle, polystyrene particle | (15,41−44,82,87,100−102) |
| other protein conjugatesa | albumin, antibody, immunotoxin | small-molecule drug, fluorophore, oligonucleotide, radiolabel | (16,98,99,103−106) |
The class of “other conjugates” includes all conjugates that are not nanoparticles.
To make an adequate choice for the design of a 3D model, it is of utmost importance to have detailed information about the barriers that are present in the tumor type that is being modeled. However, the validation that the in vitro model correctly represents the physiological situation is a challenge. Features of patient tumors are not necessarily reflected by animal models, which often employ tumors that grow very rapidly.60 Animal models that deal with spontaneous tumors, rather than xenografts, are more valuable, but typically also more laborious, and, for obvious reasons, few opportunities exist for well-controlled studies in patient tumors in situ.
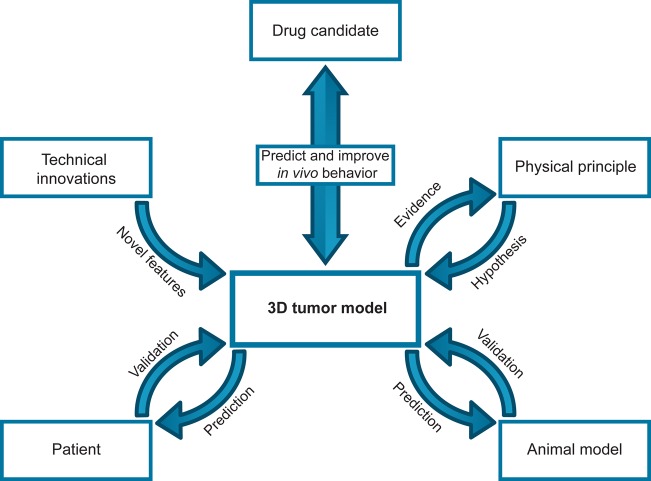
In conclusion, key requirements for the application of 3D tumor models as a predictive tool for macromolecular drug development are the understanding of the tumor physiology and the ability to mimic this in a relevant way. This notion is represented by a workflow in which technical innovation enables the implementation of insights from animal and clinical studies, as well as key physical principles. The cross-validation of the results from 3D tumor models in patient and animal studies then allows for model refinement and improves the ability to provide predictive answers on the efficacy of drug candidates in an in vivo setting (Figure 4).
Figure 4.
Workflow for the development of an optimal 3D model that is highly predictive for the behavior of a potential drug candidate in a clinical setting. The results obtained from studies in a 3D model are used to make a prediction on the outcome of in vivo studies or guide the design of improved drug candidates, and are in turn validated with data from animal and clinical studies. Knowledge of physical principles can be used to further improve the model, and the evidence obtained will in turn lead to more knowledge of physical principles that are difficult to study in animal models and patients. Technical innovations allow the incorporation of novel features, as well as provide avenues for novel or automated analysis methods that can enhance the robustness or increase the throughput of the model.
Acknowledgments
Dr. Verdurmen is supported by the research programme “Talent Scheme” through the project number 863.15.024, which is financed by The Netherlands Organisation for Scientific Research (NWO).
Glossary
Abbreviations
- BSB
binding site barrier
- ECM
extracellular matrix
- EdU
5-ethynyl-2′-deoxyuridine
- EPR
enhanced permeability and retention
- IFP
interstitial fluid pressure
- MCTS
multicellular tumor spheroid
- PAMAM
polyamidoamine
- PDMS
polydimethylsiloxane
- PCL
polycaprolactone
- PEG
polyethylene glycol
- PLA
polylactic acid
- PLGA
poly(lactic-co-glycolic acid)
- PTMC
poly(trimethylene carbonate)
- scFv
single-chain variable fragment
- TME
tumor microenvironment
- TRAIL
TNF-related apoptosis-inducing ligand
The authors declare no competing financial interest.
References
- Hay M.; Thomas D. W.; Craighead J. L.; Economides C.; Rosenthal J. (2014) Clinical development success rates for investigational drugs. Nat. Biotechnol. 32, 40–51. 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- Pampaloni F.; Reynaud E. G.; Stelzer E. H. K. (2007) The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8, 839–845. 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Workman P.; Aboagye E. O.; Balkwill F.; Balmain A.; Bruder G.; Chaplin D. J.; Double J. A.; Everitt J.; Farningham D. A.; Glennie M. J.; et al. (2010) Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer 102, 1555–1577. 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S.; Devi G. R. (2016) Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 163, 94–108. 10.1016/j.pharmthera.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J.; Seidel C.; Ebner R.; Kunz-Schughart L. A. (2009) Spheroid-based drug screen: considerations and practical approach. Nat. Protoc. 4, 309–324. 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- Han B.; Qu C.; Park K.; Konieczny S. F.; Korc M. (2016) Recapitulation of complex transport and action of drugs at the tumor microenvironment using tumor-microenvironment-on-chip. Cancer Lett. 380, 319–329. 10.1016/j.canlet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel E. (2010) Ovarian cancer development and metastasis. Am. J. Pathol. 177, 1053–1064. 10.2353/ajpath.2010.100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner D.; Stok K. S.; Lutolf M. P.; Hutmacher D. W.; Clements J. A.; Rizzi S. C. (2010) Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31, 8494–8506. 10.1016/j.biomaterials.2010.07.064. [DOI] [PubMed] [Google Scholar]
- Lee J. M.; Mhawech-Fauceglia P.; Lee N.; Parsanian L. C.; Lin Y. G.; Gayther S. A.; Lawrenson K. (2013) A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab. Invest. 93, 528–542. 10.1038/labinvest.2013.41. [DOI] [PubMed] [Google Scholar]
- Sutherland R. M. (1988) Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 240, 177–184. 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- Fong E. L.; Harrington D. A.; Farach-Carson M. C.; Yu H. (2016) Heralding a new paradigm in 3D tumor modeling. Biomaterials 108, 197–213. 10.1016/j.biomaterials.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H.; Ambros I. M.; Dehner L. P.; Hata J.; Joshi V. V.; Roald B. (1999) Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer 86, 349–363. . [DOI] [PubMed] [Google Scholar]
- Yasunaga M.; Manabe S.; Tarin D.; Matsumura Y. (2013) Tailored immunoconjugate therapy depending on a quantity of tumor stroma. Cancer Sci. 104, 231–237. 10.1111/cas.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekken C.; Hjelstuen M. H.; Bruland O. S.; de Lange Davies C. (2000) Hyaluronidase-induced periodic modulation of the interstitial fluid pressure increases selective antibody uptake in human osteosarcoma xenografts. Anticancer Res. 20, 3513–3519. [PubMed] [Google Scholar]
- Goodman T. T.; Olive P. L.; Pun S. H. (2007) Increased nanoparticle penetration in collagenase-treated multicellular spheroids. Int. J. Nanomedicine 2, 265–274. [PMC free article] [PubMed] [Google Scholar]
- Sutherland R.; Buchegger F.; Schreyer M.; Vacca A.; Mach J. P. (1987) Penetration and binding of radiolabeled anti-carcinoembryonic antigen monoclonal antibodies and their antigen binding fragments in human colon multicellular tumor spheroids. Cancer Res. 47, 1627–1633. [PubMed] [Google Scholar]
- Juweid M.; Neumann R.; Paik C.; Perez-Bacete M. J.; Sato J.; van Osdol W.; Weinstein J. N. (1992) Micropharmacology of monoclonal antibodies in solid tumors: direct experimental evidence for a binding site barrier. Cancer Res. 52, 5144–5153. [PubMed] [Google Scholar]
- Weinstein J. N.; van Osdol W. (1992) Early Intervention in Cancer Using Monoclonal Antibodies and Other Biological Ligands: Micropharmacology and the “Binding Site Barrier. Cancer Res. 52, 2747s. [PubMed] [Google Scholar]
- Cheng Z.; Al Zaki A.; Hui J. Z.; Muzykantov V. R.; Tsourkas A. (2012) Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science 338, 903–910. 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L.; Newby J. M.; Lin C. M.; Zhang L.; Xu F.; Kim W. Y.; Forest M. G.; Lai S. K.; Milowsky M. I.; Wobker S. E.; et al. (2016) The Binding Site Barrier Elicited by Tumor-Associated Fibroblasts Interferes Disposition of Nanoparticles in Stroma-Vessel Type Tumors. ACS Nano 10, 9243. 10.1021/acsnano.6b02776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Osdol W.; Fujimori K.; Weinstein J. N. (1991) An analysis of monoclonal antibody distribution in microscopic tumor nodules: consequences of a ″binding site barrier″. Cancer Res. 51, 4776–4784. [PubMed] [Google Scholar]
- Dreher M. R.; Liu W.; Michelich C. R.; Dewhirst M. W.; Yuan F.; Chilkoti A. (2006) Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. J. Natl. Cancer Inst. 98, 335–344. 10.1093/jnci/djj070. [DOI] [PubMed] [Google Scholar]
- Fang J.; Nakamura H.; Maeda H. (2011) The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Delivery Rev. 63, 136–151. 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Hashizume H.; Baluk P.; Morikawa S.; McLean J. W.; Thurston G.; Roberge S.; Jain R. K.; McDonald D. M. (2000) Openings between Defective Endothelial Cells Explain Tumor Vessel Leakiness. Am. J. Pathol. 156, 1363–1380. 10.1016/S0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchilin V. (2011) Tumor delivery of macromolecular drugs based on the EPR effect. Adv. Drug Delivery Rev. 63, 131–135. 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Hansen A. E.; Petersen A. L.; Henriksen J. R.; Boerresen B.; Rasmussen P.; Elema D. R.; af Rosenschold P. M.; Kristensen A. T.; Kjaer A.; Andresen T. L. (2015) Positron Emission Tomography Based Elucidation of the Enhanced Permeability and Retention Effect in Dogs with Cancer Using Copper-64 Liposomes. ACS Nano 9, 6985–6995. 10.1021/acsnano.5b01324. [DOI] [PubMed] [Google Scholar]
- Lammers T.; Peschke P.; Kuhnlein R.; Subr V.; Ulbrich K.; Debus J.; Huber P.; Hennink W.; Storm G. (2007) Effect of radiotherapy and hyperthermia on the tumor accumulation of HPMA copolymer-based drug delivery systems. J. Controlled Release 117, 333–341. 10.1016/j.jconrel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Heldin C. H.; Rubin K.; Pietras K.; Ostman A. (2004) High interstitial fluid pressure - an obstacle in cancer therapy. Nat. Rev. Cancer 4, 806–813. 10.1038/nrc1456. [DOI] [PubMed] [Google Scholar]
- Less J. R.; Posner M. C.; Boucher Y.; Borochovitz D.; Wolmark N.; Jain R. K. (1992) Interstitial hypertension in human breast and colorectal tumors. Cancer Res. 52, 6371–6374. [PubMed] [Google Scholar]
- Nathanson S. D.; Nelson L. (1994) Interstitial fluid pressure in breast cancer, benign breast conditions, and breast parenchyma. Ann. Surg. Oncol. 1, 333–338. 10.1007/BF03187139. [DOI] [PubMed] [Google Scholar]
- Curti B. D.; Urba W. J.; Alvord W. G.; Janik J. E.; Smith J. W. 2nd; Madara K.; Longo D. L. (1993) Interstitial pressure of subcutaneous nodules in melanoma and lymphoma patients: changes during treatment. Cancer Res. 53, 2204–2207. [PubMed] [Google Scholar]
- Boucher Y.; Kirkwood J. M.; Opacic D.; Desantis M.; Jain R. K. (1991) Interstitial hypertension in superficial metastatic melanomas in humans. Cancer Res. 51, 6691–6694. [PubMed] [Google Scholar]
- Gutmann R.; Leunig M.; Feyh J.; Goetz A. E.; Messmer K.; Kastenbauer E.; Jain R. K. (1992) Interstitial hypertension in head and neck tumors in patients: correlation with tumor size. Cancer Res. 52, 1993–1995. [PubMed] [Google Scholar]
- Oloumi A.; Lam W.; Banath J. P.; Olive P. L. (2002) Identification of genes differentially expressed in V79 cells grown as multicell spheroids. Int. J. Radiat. Biol. 78, 483–492. 10.1080/09553000210122299. [DOI] [PubMed] [Google Scholar]
- Shiras A.; Bhosale A.; Patekar A.; Shepal V.; Shastry P. (2002) Differential expression of CD44(S) and variant isoforms v3, v10 in three-dimensional cultures of mouse melanoma cell lines. Clin. Exp. Metastasis 19, 445–455. 10.1023/A:1016305611858. [DOI] [PubMed] [Google Scholar]
- Wang F.; Weaver V. M.; Petersen O. W.; Larabell C. A.; Dedhar S.; Briand P.; Lupu R.; Bissell M. J. (1998) Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc. Natl. Acad. Sci. U. S. A. 95, 14821–14826. 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietarska M.; Maugard C. M.; Filali-Mouhim A.; Alam-Fahmy M.; Tonin P. N.; Provencher D. M.; Mes-Masson A. M. (2007) Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC). Mol. Carcinog. 46, 872–885. 10.1002/mc.20315. [DOI] [PubMed] [Google Scholar]
- Lengyel E.; Burdette J. E.; Kenny H. A.; Matei D.; Pilrose J.; Haluska P.; Nephew K. P.; Hales D. B.; Stack M. S. (2014) Epithelial ovarian cancer experimental models. Oncogene 33, 3619–3633. 10.1038/onc.2013.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. Z.; Chang H. Y. (2008) Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 3, 1172–1184. 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- Huang K.; Ma H.; Liu J.; Huo S.; Kumar A.; Wei T.; Zhang X.; Jin S.; Gan Y.; Wang P. C.; et al. (2012) Size-dependent localization and penetration of ultrasmall gold nanoparticles in cancer cells, multicellular spheroids, and tumors in vivo. ACS Nano 6, 4483–4493. 10.1021/nn301282m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarisozen C.; Dhokai S.; Tsikudo E. G.; Luther E.; Rachman I. M.; Torchilin V. P. (2016) Nanomedicine based curcumin and doxorubicin combination treatment of glioblastoma with scFv-targeted micelles: In vitro evaluation on 2D and 3D tumor models. Eur. J. Pharm. Biopharm. 108, 54–67. 10.1016/j.ejpb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Pattni B. S.; Nagelli S. G.; Aryasomayajula B.; Deshpande P. P.; Kulkarni A.; Hartner W. C.; Thakur G.; Degterev A.; Torchilin V. P. (2016) Targeting of Micelles and Liposomes Loaded with the Pro-Apoptotic Drug, NCL-240, into NCI/ADR-RES Cells in a 3D Spheroid Model. Pharm. Res. 33, 2540–2551. 10.1007/s11095-016-1978-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perche F.; Patel N. R.; Torchilin V. P. (2012) Accumulation and toxicity of antibody-targeted doxorubicin-loaded PEG-PE micelles in ovarian cancer cell spheroid model. J. Controlled Release 164, 95–102. 10.1016/j.jconrel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarisozen C.; Abouzeid A. H.; Torchilin V. P. (2014) The effect of co-delivery of paclitaxel and curcumin by transferrin-targeted PEG-PE-based mixed micelles on resistant ovarian cancer in 3-D spheroids and in vivo tumors. Eur. J. Pharm. Biopharm. 88, 539–550. 10.1016/j.ejpb.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.; Zhang X.; Liu Y.; Liu C.; Jiang B.; Jiang Y. (2014) Tumor penetrability and anti-angiogenesis using iRGD-mediated delivery of doxorubicin-polymer conjugates. Biomaterials 35, 8735–8747. 10.1016/j.biomaterials.2014.06.042. [DOI] [PubMed] [Google Scholar]
- Gu G.; Gao X.; Hu Q.; Kang T.; Liu Z.; Jiang M.; Miao D.; Song Q.; Yao L.; Tu Y.; et al. (2013) The influence of the penetrating peptide iRGD on the effect of paclitaxel-loaded MT1-AF7p-conjugated nanoparticles on glioma cells. Biomaterials 34, 5138–5148. 10.1016/j.biomaterials.2013.03.036. [DOI] [PubMed] [Google Scholar]
- Wang X.; Zhen X.; Wang J.; Zhang J.; Wu W.; Jiang X. (2013) Doxorubicin delivery to 3D multicellular spheroids and tumors based on boronic acid-rich chitosan nanoparticles. Biomaterials 34, 4667–4679. 10.1016/j.biomaterials.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Jain R. K. (1987) Transport of molecules in the tumor interstitium: a review. Cancer Res. 47, 3039–3051. [PubMed] [Google Scholar]
- Bhatia S. N.; Ingber D. E. (2014) Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- van Duinen V.; Trietsch S. J.; Joore J.; Vulto P.; Hankemeier T. (2015) Microfluidic 3D cell culture: from tools to tissue models. Curr. Opin. Biotechnol. 35, 118–126. 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Agastin S.; Giang U. B.; Geng Y.; Delouise L. A.; King M. R. (2011) Continuously perfused microbubble array for 3D tumor spheroid model. Biomicrofluidics 5, 24110. 10.1063/1.3596530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackett E. L.; Swofford C. A.; Forbes N. S. (2016) Microfluidic Device to Quantify the Behavior of Therapeutic Bacteria in Three-Dimensional Tumor Tissue. Methods Mol. Biol. 1409, 35–48. 10.1007/978-1-4939-3515-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.; Bang J. H.; Kim Y. E.; Lee S. H.; Kang J. Y. (2012) On-chip anticancer drug test of regular tumor spheroids formed in microwells by a distributive microchannel network. Lab Chip 12, 4135–4142. 10.1039/c2lc40570a. [DOI] [PubMed] [Google Scholar]
- Ng C. P.; Pun S. H. (2008) A perfusable 3D cell-matrix tissue culture chamber for in situ evaluation of nanoparticle vehicle penetration and transport. Biotechnol. Bioeng. 99, 1490–1501. 10.1002/bit.21698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wang J. (2014) Mixed hydrogel bead-based tumor spheroid formation and anticancer drug testing. Analyst 139, 2449–2458. 10.1039/C4AN00015C. [DOI] [PubMed] [Google Scholar]
- Kwak B.; Ozcelikkale A.; Shin C. S.; Park K.; Han B. (2014) Simulation of complex transport of nanoparticles around a tumor using tumor-microenvironment-on-chip. J. Controlled Release 194, 157–167. 10.1016/j.jconrel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya M. L.; Hsu Y. H.; Lee A. P.; Hughes C. C.; George S. C. (2013) In vitro perfused human capillary networks. Tissue Eng., Part C 19, 730–737. 10.1089/ten.tec.2012.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrino A.; Phan D. T.; Datta R.; Wang X.; Hachey S. J.; Romero-Lopez M.; Gratton E.; Lee A. P.; George S. C.; Hughes C. C. (2016) 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 6, 31589. 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K.; Klosterhoff B. S.; Han B. (2016) Characterization of Cell-Type-Specific Drug Transport and Resistance of Breast Cancers Using Tumor-Microenvironment-on-Chip. Mol. Pharmaceutics 13, 2214–2223. 10.1021/acs.molpharmaceut.6b00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers T.; Kiessling F.; Hennink W. E.; Storm G. (2012) Drug targeting to tumors: principles, pitfalls and (pre-) clinical progress. J. Controlled Release 161, 175–187. 10.1016/j.jconrel.2011.09.063. [DOI] [PubMed] [Google Scholar]
- Bogorad M. I.; DeStefano J.; Karlsson J.; Wong A. D.; Gerecht S.; Searson P. C. (2015) Review: in vitro microvessel models. Lab Chip 15, 4242–4255. 10.1039/C5LC00832H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naipal K. A.; Verkaik N. S.; Sanchez H.; van Deurzen C. H.; den Bakker M. A.; Hoeijmakers J. H.; Kanaar R.; Vreeswijk M. P.; Jager A.; van Gent D. C. (2016) Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 16, 78. 10.1186/s12885-016-2119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaira V.; Fedele G.; Pyne S.; Fasoli E.; Zadra G.; Bailey D.; Snyder E.; Faversani A.; Coggi G.; Flavin R.; et al. (2010) Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl. Acad. Sci. U. S. A. 107, 8352–8356. 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astolfi M.; Peant B.; Lateef M. A.; Rousset N.; Kendall-Dupont J.; Carmona E.; Monet F.; Saad F.; Provencher D.; Mes-Masson A. M.; et al. (2016) Micro-dissected tumor tissues on chip: an ex vivo method for drug testing and personalized therapy. Lab Chip 16, 312–325. 10.1039/C5LC01108F. [DOI] [PubMed] [Google Scholar]
- Dong M.; Philippi C.; Loretz B.; Nafee N.; Schaefer U. F.; Friedel G.; Ammon-Treiber S.; Griese E. U.; Lehr C. M.; Klotz U.; et al. (2011) Tissue slice model of human lung cancer to investigate telomerase inhibition by nanoparticle delivery of antisense 2′-O-methyl-RNA. Int. J. Pharm. 419, 33–42. 10.1016/j.ijpharm.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Schweinlin M.; Rossi A.; Lodes N.; Lotz C.; Hackenberg S.; Steinke M.; Walles H.; Groeber F. (2016) Human barrier models for the in vitro assessment of drug delivery. Drug Delivery Transl. Res. 1–11. 10.1007/s13346-016-0316-9. [DOI] [PubMed] [Google Scholar]
- Gao H.; Yang Z.; Zhang S.; Pang Z.; Liu Q.; Jiang X. (2014) Study and evaluation of mechanisms of dual targeting drug delivery system with tumor microenvironment assays compared with normal assays. Acta Biomater. 10, 858–867. 10.1016/j.actbio.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Ho D. N.; Kohler N.; Sigdel A.; Kalluri R.; Morgan J. R.; Xu C.; Sun S. (2012) Penetration of endothelial cell coated multicellular tumor spheroids by iron oxide nanoparticles. Theranostics 2, 66–75. 10.7150/thno.3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L.; Guo X. Y.; Yang T.; Yu M. Z.; Chen D. W.; Wang J. C. (2016) Brain tumor-targeted therapy by systemic delivery of siRNA with Transferrin receptor-mediated core-shell nanoparticles. Int. J. Pharm. 510, 394–405. 10.1016/j.ijpharm.2016.06.127. [DOI] [PubMed] [Google Scholar]
- Hu Q.; Gao X.; Gu G.; Kang T.; Tu Y.; Liu Z.; Song Q.; Yao L.; Pang Z.; Jiang X.; et al. (2013) Glioma therapy using tumor homing and penetrating peptide-functionalized PEG-PLA nanoparticles loaded with paclitaxel. Biomaterials 34, 5640–5650. 10.1016/j.biomaterials.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Ni D.; Ding H.; Liu S.; Yue H.; Bao Y.; Wang Z.; Su Z.; Wei W.; Ma G. (2015) Superior intratumoral penetration of paclitaxel nanodots strengthens tumor restriction and metastasis prevention. Small 11, 2518–2526. 10.1002/smll.201403632. [DOI] [PubMed] [Google Scholar]
- Jiang X.; Xin H.; Gu J.; Xu X.; Xia W.; Chen S.; Xie Y.; Chen L.; Chen Y.; Sha X.; et al. (2013) Solid tumor penetration by integrin-mediated pegylated poly(trimethylene carbonate) nanoparticles loaded with paclitaxel. Biomaterials 34, 1739–1746. 10.1016/j.biomaterials.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Salzano G.; Costa D. F.; Sarisozen C.; Luther E.; Mattheolabakis G.; Dhargalkar P. P.; Torchilin V. P. (2016) Mixed Nanosized Polymeric Micelles as Promoter of Doxorubicin and miRNA-34a Co-Delivery Triggered by Dual Stimuli in Tumor Tissue. Small 12, 4837–4848. 10.1002/smll.201600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H.; Zhang Q.; Yang Y.; Jiang X.; He Q. (2015) Tumor homing cell penetrating peptide decorated nanoparticles used for enhancing tumor targeting delivery and therapy. Int. J. Pharm. 478, 240–250. 10.1016/j.ijpharm.2014.11.029. [DOI] [PubMed] [Google Scholar]
- Kang T.; Jiang M.; Jiang D.; Feng X.; Yao J.; Song Q.; Chen H.; Gao X.; Chen J. (2015) Enhancing Glioblastoma-Specific Penetration by Functionalization of Nanoparticles with an Iron-Mimic Peptide Targeting Transferrin/Transferrin Receptor Complex. Mol. Pharmaceutics 12, 2947–2961. 10.1021/acs.molpharmaceut.5b00222. [DOI] [PubMed] [Google Scholar]
- Gao H.; Yang Z.; Zhang S.; Cao S.; Pang Z.; Yang X.; Jiang X. (2013) Glioma-homing peptide with a cell-penetrating effect for targeting delivery with enhanced glioma localization, penetration and suppression of glioma growth. J. Controlled Release 172, 921–928. 10.1016/j.jconrel.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Cantisani M.; Guarnieri D.; Biondi M.; Belli V.; Profeta M.; Raiola L.; Netti P. A. (2015) Biocompatible nanoparticles sensing the matrix metallo-proteinase 2 for the on-demand release of anticancer drugs in 3D tumor spheroids. Colloids Surf., B 135, 707–716. 10.1016/j.colsurfb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Chen X.; Ying M.; Gao J.; Zhan C.; Lu W. (2016) Glioma-Targeted Drug Delivery Enabled by a Multifunctional Peptide. Bioconjugate Chem. 10.1021/acs.bioconjchem.6b00617. [DOI] [PubMed] [Google Scholar]
- Gao H.; Qian J.; Cao S.; Yang Z.; Pang Z.; Pan S.; Fan L.; Xi Z.; Jiang X.; Zhang Q. (2012) Precise glioma targeting of and penetration by aptamer and peptide dual-functioned nanoparticles. Biomaterials 33, 5115–5123. 10.1016/j.biomaterials.2012.03.058. [DOI] [PubMed] [Google Scholar]
- Liu C.; Yao S.; Li X.; Wang F.; Jiang Y. (2016) iRGD-mediated core-shell nanoparticles loading carmustine and O6-benzylguanine for glioma therapy. J. Drug Target. 1–12. 10.1080/1061186X.2016.1238091. [DOI] [PubMed] [Google Scholar]
- Mei L.; Fu L.; Shi K.; Zhang Q.; Liu Y.; Tang J.; Gao H.; Zhang Z.; He Q. (2014) Increased tumor targeted delivery using a multistage liposome system functionalized with RGD, TAT and cleavable PEG. Int. J. Pharm. 468, 26–38. 10.1016/j.ijpharm.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Qin L.; Wang C. Z.; Fan H. J.; Zhang C. J.; Zhang H. W.; Lv M. H.; Cui S. D. (2014) A dual-targeting liposome conjugated with transferrin and arginine-glycine-aspartic acid peptide for glioma-targeting therapy. Oncol. Lett. 8, 2000–2006. 10.3892/ol.2014.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Ran R.; Chen J.; Kuang Q.; Tang J.; Mei L.; Zhang Q.; Gao H.; Zhang Z.; He Q. (2014) Paclitaxel loaded liposomes decorated with a multifunctional tandem peptide for glioma targeting. Biomaterials 35, 4835–4847. 10.1016/j.biomaterials.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Tang J.; Fu L.; Ran R.; Liu Y.; Yuan M.; He Q. (2013) A pH-responsive alpha-helical cell penetrating peptide-mediated liposomal delivery system. Biomaterials 34, 7980–7993. 10.1016/j.biomaterials.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Kulkarni P.; Haldar M. K.; Katti P.; Dawes C.; You S.; Choi Y.; Mallik S. (2016) Hypoxia Responsive, Tumor Penetrating Lipid Nanoparticles for Delivery of Chemotherapeutics to Pancreatic Cancer Cell Spheroids. Bioconjugate Chem. 27, 1830–1838. 10.1021/acs.bioconjchem.6b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju R.-J.; Li X.-T.; Shi J.-F.; Li X.-Y.; Sun M.-G.; Zeng F.; Zhou J.; Liu L.; Zhang C.-X.; Zhao W.-Y.; et al. (2014) Liposomes, modified with PTDHIV-1 peptide, containing epirubicin and celecoxib, to target vasculogenic mimicry channels in invasive breast cancer. Biomaterials 35, 7610–7621. 10.1016/j.biomaterials.2014.05.040. [DOI] [PubMed] [Google Scholar]
- Sharma G.; Modgil A.; Zhong T.; Sun C.; Singh J. (2014) Influence of short-chain cell-penetrating peptides on transport of doxorubicin encapsulating receptor-targeted liposomes across brain endothelial barrier. Pharm. Res. 31, 1194–1209. 10.1007/s11095-013-1242-x. [DOI] [PubMed] [Google Scholar]
- Zong T.; Mei L.; Gao H.; Cai W.; Zhu P.; Shi K.; Chen J.; Wang Y.; Gao F.; He Q. (2014) Synergistic dual-ligand doxorubicin liposomes improve targeting and therapeutic efficacy of brain glioma in animals. Mol. Pharmaceutics 11, 2346–2357. 10.1021/mp500057n. [DOI] [PubMed] [Google Scholar]
- Biswas S.; Deshpande P. P.; Perche F.; Dodwadkar N. S.; Sane S. D.; Torchilin V. P. (2013) Octa-arginine-modified pegylated liposomal doxorubicin: an effective treatment strategy for non-small cell lung cancer. Cancer Lett. 335, 191–200. 10.1016/j.canlet.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.; Zhang L.; Fu H.; Kuang Q.; Gao H.; Zhang Z.; He Q. (2014) A detachable coating of cholesterol-anchored PEG improves tumor targeting of cell-penetrating peptide-modified liposomes. Acta Pharm. Sin. B 4, 67–73. 10.1016/j.apsb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L.; Jiang Y.; Liu X.; Wang B.; Lv W.; Zhao Y.; Shi H.; Hu Q.; Xin H.; Xu Q.; et al. (2016) Enhanced Antiglioblastoma Efficacy of Neovasculature and Glioma Cells Dual Targeted Nanoparticles. Mol. Pharmaceutics 13, 3506–3517. 10.1021/acs.molpharmaceut.6b00523. [DOI] [PubMed] [Google Scholar]
- Suzuki S.; Itakura S.; Matsui R.; Nakayama K.; Nishi T.; Nishimoto A.; Hama S.; Kogure K. (2017) Tumor microenvironment-sensitive liposomes penetrate tumor tissue via attenuated interaction of the extracellular matrix and tumor cells, and accompanying actin depolymerization. Biomacromolecules 10.1021/acs.biomac.6b01688. [DOI] [PubMed] [Google Scholar]
- Gao W.; Xiang B.; Meng T. T.; Liu F.; Qi X. R. (2013) Chemotherapeutic drug delivery to cancer cells using a combination of folate targeting and tumor microenvironment-sensitive polypeptides. Biomaterials 34, 4137–4149. 10.1016/j.biomaterials.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Waite C. L.; Roth C. M. (2011) Binding and transport of PAMAM-RGD in a tumor spheroid model: The effect of RGD targeting ligand density. Biotechnol. Bioeng. 108, 2999–3008. 10.1002/bit.23255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Zhang X.; Wang M.; Li X.; Mu H.; Wang A.; Liu W.; Li Y.; Wu Z.; Sun K. (2016) Synthesis of a bi-functional dendrimer-based nanovehicle co-modified with RGDyC and TAT peptides for neovascular targeting and penetration. Int. J. Pharm. 501, 112–123. 10.1016/j.ijpharm.2016.01.068. [DOI] [PubMed] [Google Scholar]
- Liang D.-S.; Su H.-T.; Liu Y.-J.; Wang A.-T.; Qi X.-R. (2015) Tumor-specific penetrating peptides-functionalized hyaluronic acid-d-α-tocopheryl succinate based nanoparticles for multi-task delivery to invasive cancers. Biomaterials 71, 11–23. 10.1016/j.biomaterials.2015.08.035. [DOI] [PubMed] [Google Scholar]
- Przysiecka L.; Michalska M.; Nowaczyk G.; Peplinska B.; Jesionowski T.; Schneider R.; Jurga S. (2016) iRGD peptide as effective transporter of CuInZnxS2+x quantum dots into human cancer cells. Colloids Surf., B 146, 9–18. 10.1016/j.colsurfb.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Carver K.; Ming X.; Juliano R. L. (2014) Multicellular tumor spheroids as a model for assessing delivery of oligonucleotides in three dimensions. Mol. Ther.--Nucleic Acids 3, e153. 10.1038/mtna.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming X.; Carver K.; Wu L. (2013) Albumin-based nanoconjugates for targeted delivery of therapeutic oligonucleotides. Biomaterials 34, 7939–7949. 10.1016/j.biomaterials.2013.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao; le Q.; Byeon H. J.; Lee C.; Lee S.; Lee E. S.; Choi Y. W.; Choi H. G.; Park E. S.; Lee K. C.; Youn Y. S. (2016) Doxorubicin-Bound Albumin Nanoparticles Containing a TRAIL Protein for Targeted Treatment of Colon Cancer. Pharm. Res. 33, 615–626. 10.1007/s11095-015-1814-z. [DOI] [PubMed] [Google Scholar]
- Parhi P.; Sahoo S. K. (2015) Trastuzumab guided nanotheranostics: A lipid based multifunctional nanoformulation for targeted drug delivery and imaging in breast cancer therapy. J. Colloid Interface Sci. 451, 198–211. 10.1016/j.jcis.2015.03.049. [DOI] [PubMed] [Google Scholar]
- Ying X.; Wen H.; Lu W. L.; Du J.; Guo J.; Tian W.; Men Y.; Zhang Y.; Li R. J.; Yang T. Y.; et al. (2010) Dual-targeting daunorubicin liposomes improve the therapeutic efficacy of brain glioma in animals. J. Controlled Release 141, 183–192. 10.1016/j.jconrel.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Sapra P.; Damelin M.; Dijoseph J.; Marquette K.; Geles K. G.; Golas J.; Dougher M.; Narayanan B.; Giannakou A.; Khandke K.; et al. (2013) Long-term tumor regression induced by an antibody-drug conjugate that targets 5T4, an oncofetal antigen expressed on tumor-initiating cells. Mol. Cancer Ther. 12, 38–47. 10.1158/1535-7163.MCT-12-0603. [DOI] [PubMed] [Google Scholar]
- Shor B.; Kahler J.; Dougher M.; Xu J.; Mack M.; Rosfjord E.; Wang F.; Melamud E.; Sapra P. (2016) Enhanced Antitumor Activity of an Anti-5T4 Antibody-Drug Conjugate in Combination with PI3K/mTOR inhibitors or Taxanes. Clin. Cancer Res. 22, 383–394. 10.1158/1078-0432.CCR-15-1166. [DOI] [PubMed] [Google Scholar]
- Ingargiola M.; Runge R.; Heldt J. M.; Freudenberg R.; Steinbach J.; Cordes N.; Baumann M.; Kotzerke J.; Brockhoff G.; Kunz-Schughart L. A. (2014) Potential of a Cetuximab-based radioimmunotherapy combined with external irradiation manifests in a 3-D cell assay. Int. J. Cancer 135, 968–980. 10.1002/ijc.28735. [DOI] [PubMed] [Google Scholar]
- Xiang X.; Phung Y.; Feng M.; Nagashima K.; Zhang J.; Broaddus V. C.; Hassan R.; Fitzgerald D.; Ho M. (2011) The development and characterization of a human mesothelioma in vitro 3D model to investigate immunotoxin therapy. PLoS One 6, e14640. 10.1371/journal.pone.0014640. [DOI] [PMC free article] [PubMed] [Google Scholar]