Abstract
Background:
Colorectal cancer (CRC) is common and associated with significant mortality. Current screening methods for CRC lack patient compliance. microRNAs (miRNAs), identified in body fluids, are negative regulators of gene expression and are dysregulated in many cancers, including CRC. This paper summarises studies identifying blood-based miRNAs dysregulated in CRC compared with healthy controls in an attempt to evaluate their use as a screening tool for the diagnosis of CRC.
Methods:
A search of electronic databases (PubMed and EMBASE) and grey literature was performed between January 2002 and April 2016. Studies reporting plasma or serum miRNAs in the diagnosis of CRC compared with healthy controls were selected. Patient demographics, type of patient sample (serum or plasma), method of miRNA detection, type of normalisation, and the number of significantly dysregulated miRNAs identified were recorded. Statistical evaluation of dysregulated miRNAs using sensitivity, specificity, and area under the curve (AUC) was performed.
Results:
Thirty-four studies investigating plasma or serum miRNAs in the diagnosis of CRC were included. A total of 31 miRNAs were found to be either upregulated (n=17) or downregulated (n=14) in CRC cases as compared with controls. Fourteen studies identified panels of ⩾2 dysregulated miRNAs. The highest AUC, 0.943, was identified using a panel of 4 miRNAs with 83.3% sensitivity and 93.1% specificity. Meta-analysis of studies identifying a single dysregulated miRNA in CRC cases compared with controls was performed. Overall sensitivity and specificity of 28 individual miRNAs in the diagnosis of CRC were 76% (95% CI 72%–80%) and 76% (95% CI 72%–80%), respectively, indicating good discriminative ability of miRNAs as biomarkers for CRC. These data did not change with sensitivity analyses.
Conclusions:
Blood-based miRNAs distinguish patients with CRC from healthy controls with high sensitivity and specificity comparable to other common and invasive currently used screening methods for CRC. In future, miRNAs may be used as a relatively non-invasive blood-based marker for detection of CRC.
Keywords: miRNA, colorectal cancer, diagnosis, blood-based biomarker, screening
Colorectal cancer (CRC) is a common cancer with substantial mortality. A significant proportion of CRC arises from preexisting colorectal adenomas (CAA; polyps) through the adenoma–carcinoma sequence. In this sequence, a stepwise pattern of genetic changes drive normal colonic epithelium to invasive cancer. Many deaths could be avoided if precancerous adenomas or early-stage cancers could be identified and treated prior to the development of more advanced malignancy (Muller and Sonnenberg, 1995; Zauber et al, 2012). Current screening tools for detection of CRC include colonoscopy, faecal-based tests (occult blood testing and DNA-based stool tests), and plasma-based assays. As these methods each have disadvantages, including morbidity (Lohsiriwat, 2010), low sensitivity and specificity (Hundt et al, 2009), expense (Calderwood and Jacobson, 2013), and poor patient compliance (Logan et al, 2012), there is need for a more accurate, less invasive screening tool to permit early detection and intervention.
Biological markers or biomarkers are defined as ‘a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention' (Naylor, 2003). Biomarkers, such as blood, urine, and cerebrospinal fluid, can be used for screening, diagnosis, and predicting prognosis in human diseases (Mayeux, 2004). Plasma and serum are components of blood and are commonly used in diagnostic assays. Serum is of similar composition to plasma except for lacking clotting factors. Both plasma and serum contain hormones, glucose, electrolytes, antibodies, antigens, and nutrients making them ideal media in diagnostic testing (Maton et al, 1993). The most commonly used blood-based CRC biomarker, carcinoembryonic antigen, which is used for postoperative surveillance and for monitoring response to therapy, lacks sensitivity and specificity for screening or for the detection of recurrent CRC (Fakih and Padmanabhan, 2006).
microRNAs (miRNAs) are small, non-protein-coding RNA molecules that regulate gene expression by complementary binding to the 3′ untranslated region of target mRNA. They cause target degradation, translational repression, or gene silencing and thus affect subsequent protein expression. miRNAs exhibit important regulatory functions related to cell differentiation, development, and growth (Meltzer, 2005; Croce, 2008). They have also been shown to be dysregulated in a number of cancers, including CRC, by influencing oncogenes and tumour-suppressor genes (Zhang et al, 2007; Chiang et al, 2012). miRNAs have been identified in body fluids, such as plasma, saliva, urine, and faeces (Chevillet et al, 2014). They have been shown to be actively released from cells in microvesicles, exosomes, or bound to proteins. miRNAs are inherently stable accounting for the emerging use as potential biomarkers for human disease and as targets for disease intervention (Kanaan et al, 2012; Mishra, 2014).
The purpose of this systematic review and meta-analysis is to assess published studies describing the use of plasma- or serum-based miRNAs as biomarkers for the diagnosis of CRC, to assess the accuracy of these miRNAs as biomarkers, and to identify those miRNAs for which there is the most evidence of potential use as a specific and sensitive blood-based assay for the diagnosis of CRC.
Materials and methods
This study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analysis guidelines (Mahid et al, 2006; Moher et al, 2009). Two reviewers (JC, NG) were independently involved with study selection, data extraction, and quality assessment.
Study selection
An electronic search of PubMed (NCBI, Bethesda, MD, USA) and EMBASE (Elsevier, Amsterdam, The Netherlands) databases was performed for relevant articles published between January 2002 and April 2016 using the following medical subject heading terms: (‘plasma' OR ‘serum') AND (‘microRNA' OR ‘miRNAs' OR ‘miR*') AND (‘colorectal cancer' OR ‘colorectal carcinoma' OR ‘colon cancer' OR ‘colorectal adenoma'). This time period was based on a preliminary search that revealed no serum or plasma miRNA studies in patients with CRC undertaken prior to 2002. A grey literature search of meeting proceedings and abstracts was performed to detect any relevant unpublished work (Mahid et al, 2006). Meeting proceedings from the following organisations were searched for abstracts relevant to plasma miRNA and CRC for the period 2010 and 2016: American Society for Clinical Oncology, the American Society of Colon and Rectal Surgeons, the American Association for Cancer Research, and the Society for Surgical Oncology.
No language restrictions were imposed. Duplicates were removed. Based on the title and abstract, manuscripts of interest were obtained for full-text review. Only full-text manuscripts were included. Additional articles were identified by manually searching the references of original and review articles.
Inclusion and exclusion criteria
Inclusion and exclusion criteria were developed by the investigative team. Human studies investigating plasma- or serum-based miRNAs as a method for diagnosis of CRC using patients with CRC as compared with healthy controls were included. Studies evaluating treatment response were excluded, as were studies evaluating tissue miRNA or miRNA in other body fluids. The primary outcome measure was determination of significantly dysregulated serum or plasma miRNA in patients with CRC as compared with healthy controls.
Data extraction, meta-analysis, and quality assessment
Each manuscript was assessed independently by two investigators (JC and NG). Disagreements among reviewers were resolved by consensus. Data extracted included the following: authors, publication year, country, type of blood-based fluid (serum or plasma), characteristics of the study population (both case and control), study design (qRT-PCR detection method), whether miRNA screening was performed, number of miRNAs assessed, listing of the specific dysregulated miRNAs in patients with CRC as compared with controls, and outcome of statistical analyses including details of miRNA analysis, such as type of reference miRNA utilised. Studies reporting on single miRNA were included in the meta-analysis and were evaluated according to the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) checklist (Whiting et al, 2011). QUADAS-2 is designed to assess the risk of bias and applicability of studies of diagnostic accuracy. It consists of four domains: patient selection, index test, reference standard, and flow and timing. Each is assessed with respect to the risk of bias and the first three domains assessed with respect to applicability.
Statistical analysis
Analysis was based on the accuracy of the identified miRNAs to diagnose the presence of CRC as determined using Receiver Operator Characteristic (ROC) curves as Area Under the Curve (AUC) value and sensitivity and specificity, where available (Carter et al, 2016a). Studies that identified the sensitivity and specificity of individual miRNA in diagnosing CRC were included in the meta-analysis. Owing to the presumed heterogeneity of studies, a random-effects model (DerSimonian–Laird method) was used (Mahid et al, 2006). Forest plots were constructed using STATA version 11 (StataCorp LP, College, Station, TX, USA). The heterogeneity of included studies was assessed using I2 and χ2 statistics and was judged to be significant if I2 was >50% or P<0.05. Sensitivity analyses were performed for (1) studies judged to have low risk and low concern following QUADAS-2 assessment (see above), (2) based upon the type of sample used (i.e., plasma or serum), (3) the method of qRT-PCR detection (TaqMan or SYBR Green), and (4) the type of reference control used for normalisation (internal or external).
Results
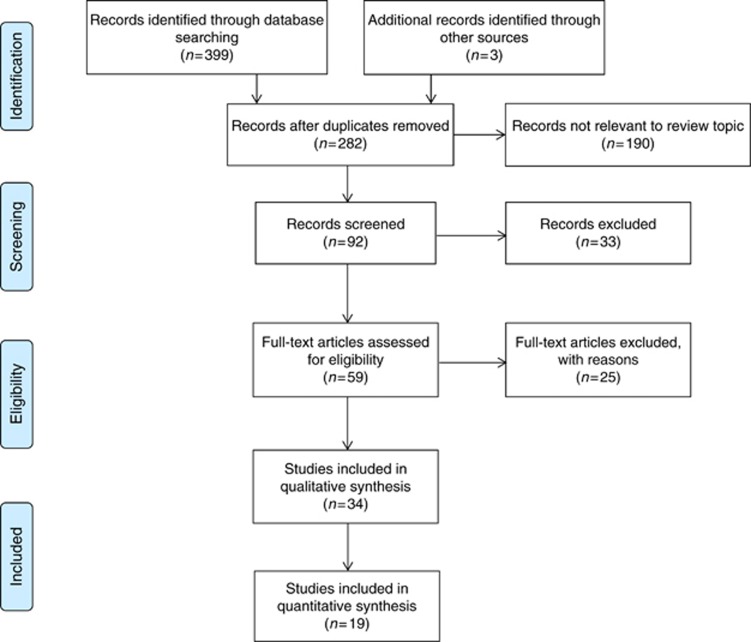
Three hundred and ninety-nine records from the database search and 3 from the grey literature were initially identified (Figure 1); 282 records remained after duplicates were removed and a further 190 were considered not relevant to the review topic. Ninety-two records were screened, and after abstract review, 33 were subsequently excluded owing to a lack of reporting on miRNA, results using cancers other than CRC, non-human subjects, and/or inability to obtain full abstracts or articles. Full-text articles were obtained for the remaining 59 articles that were deemed relevant. A further 25 articles were excluded for the following reasons: review article (n=14), abstract only (n=2), not plasma or serum (n=1), not diagnostic (n=5), and not CRC (n=3). Thirty-four studies investigating plasma or serum miRNAs in the diagnosis of CRC were included for the final systematic review (Ng et al, 2009; Huang et al, 2010; Pu et al, 2010; Ahmed et al, 2012; Kanaan et al, 2012; Wang and Zhang, 2012; Wang et al, 2012b; Brunet Vega et al, 2013; Giraldez et al, 2013; Hofsli et al, 2013; Kanaan et al, 2013; Liu et al, 2013; Luo et al, 2013; Toiyama et al, 2013; Yu et al, 2013; Zhang et al, 2013; Basati et al, 2014; Du et al, 2014; Perilli et al, 2014; Xu et al, 2014; Zanutto et al, 2014; Zheng et al, 2014; Basati et al, 2015; Chen et al, 2015; Fang et al, 2015; Ghanbari et al, 2015; Ho et al, 2015; Ramzy et al, 2015; Wang et al, 2015; Li et al, 2015; Li et al, 2016; Sun et al, 2016; Wang et al, 2016; Yu et al, 2016) and of these 19 were included in the meta-analysis (Ng et al, 2009; Huang et al, 2010; Pu et al, 2010; Kanaan et al, 2012; Wang and Zhang, 2012; Wang et al, 2012b; Liu et al, 2013; Toiyama et al, 2013; Zhang et al, 2013; Basati et al, 2014; Du et al, 2014; Xu et al, 2014; Basati et al, 2015; Chen et al, 2015; Fang et al, 2015; Li et al, 2016; Sun et al, 2016; Wang et al, 2016; Yu et al, 2016). Of these 34 studies, 20 (59%) used plasma (Ng et al, 2009; Huang et al, 2010; Pu et al, 2010; Ahmed et al, 2012; Kanaan et al, 2012; Wang et al, 2012b; Giraldez et al, 2013; Kanaan et al, 2013; Luo et al, 2013; Zhang et al, 2013; Du et al, 2014; Perilli et al, 2014; Xu et al, 2014; Zanutto et al, 2014; Chen et al, 2015; Fang et al, 2015; Ghanbari et al, 2015; Wang et al, 2015; Li et al, 2016; Sun et al, 2016) and 14 (41%) used serum samples (Wang and Zhang, 2012; Brunet Vega et al, 2013; Hofsli et al, 2013; Liu et al, 2013; Toiyama et al, 2013; Yu et al, 2013; Basati et al, 2014; Zheng et al, 2014; Basati et al, 2015; Ho et al, 2015; Ramzy et al, 2015; Li et al, 2015; Wang et al, 2016; Yu et al, 2016). Studies were conducted in North America, South East Asia, or Europe. Study characteristics are shown in Table 1.
Figure 1.
Flow diagram of the literature search process and study inclusion.
Table 1. Characteristics of studies included in systematic review.
|
Study population |
||||||||
|---|---|---|---|---|---|---|---|---|
| First author | Year | Country | CRC (no. of patients) | Control (no. of patients) | Age (mean±s.d./(range)), years | Gender male: female | Specimen | |
| Ng | 2009 | China | 130 | 75 | CRC | 71 (42–91) | 68 : 62 | Plasma |
| Control | 69 (45–85) | 39 : 36 | ||||||
| Huang | 2010 | China | 100 | 59 | CRC | 61±11 | 51 : 49 | Plasma |
| Control | 58±12 | 31 : 28 | ||||||
| Pu | 2010 | China | 103 | 37 | CRC | 58 (39–84) | 66 : 37 | Plasma |
| Control | 32 (17–77) | 19 : 18 | ||||||
| Kanaan | 2012 | USA | 50 | 50 | CRC | 60±11 | 21 : 29 | Plasma |
| Control | 61±9 | 21 : 29 | ||||||
| Wang Q | 2012 | China | 100 | 68 | CRC | 61 (21–84) | 50 : 50 | Plasma |
| Control | 57 (36–85) | 35 : 33 | ||||||
| Wang B | 2012 | China | 32 | 39 | CRC | 63 (45–80) | 17 : 15 | Serum |
| Control | 46 | 30 : 9 | ||||||
| Ahmed | 2012 | USA | 10 | 5 | CRC | — | — | Plasma |
| Control | — | — | ||||||
| Kanaan | 2013 | USA | 45 | 26 | CRC | 64 | 28 : 17 | Plasma |
| Control | 60±11 | 11 : 15 | ||||||
| Zhang G | 2012 | China | 78 | 86 | CRC | 61.4±13.6 | 43 : 35 | Plasma |
| Control | 60.3±11.8 | 53 : 33 | ||||||
| Luo | 2013 | Germany | 130 | 244 | CRC | 67.55 | 70 : 60 | Plasma |
| Control | 61.93 | 105 : 139 | ||||||
| Giraldez | 2013 | Spain | 63 | 73 | CRC | — | — | Plasma |
| Control | 61.35 | 37 : 36 | ||||||
| Brunet Vega | 2013 | Spain | 30 | 26 | CRC | 68.1±11 | 18 : 12 | Serum |
| Control | 64.1±7.4 | 13 : 13 | ||||||
| Liu | 2013 | China | 200 | 80 | CRC | 57.09 (20–89) | 126 : : 74 | Serum |
| Control | 57.71 (28–89) | 42 : 38 | ||||||
| Yu H | 2013 | China | 30 | 30 | CRC | — | — | Serum |
| Control | — | — | ||||||
| Toiyama | 2013 | USA | 186 | 53 | CRC | 67.5±7.5 | 106 : 80 | Serum |
| Control | 64±12.9 | 27 : 26 | ||||||
| Hofsli | 2013 | Norway | 70 | 20 | CRC | 70.5 | 32 : 38 | Serum |
| Control | ⩾50 | 10 : 10 | ||||||
| Perilli | 2014 | Italy | 20 | 10 | CRC | — | — | Plasma |
| Control | — | — | ||||||
| Basati | 2014 | Iran | 40 | 40 | CRC | 55.35±10.13 | 21 : 19 | Serum |
| Control | 55.00±10.35 | 22 : 18 | ||||||
| Zheng | 2014 | China | 307 | 226 | CRC | 57.68 | 180 : 127 | Serum |
| Control | 53.15 | 128 : 98 | ||||||
| Du | 2014 | China | 49 | 49 | CRC | — | — | Plasma |
| Control | — | — | ||||||
| Xu | 2014 | China | 94 | 46 | CRC | 64.9 | 54 : 40 | Plasma |
| Control | 65.8 | 22 : 24 | ||||||
| Zanutto | 2014 | Italy | 65 | 70 | CRC | — | — | Plasma |
| Control | — | — | ||||||
| Chen | 2015 | China | 100 | 79 | CRC | 58.91 | 60 : 40 | Plasma |
| Control | 60.20 | 44 : 35 | ||||||
| Ramzy | 2015 | Egypt | 25 | 10 | CRC | — | — | Serum |
| Control | — | — | ||||||
| Ho | 2015 | USA | 11 | 10 | CRC | 68.0 | 8 : 3 | Serum |
| Control | 54.5 | 4 : 6 | ||||||
| Li J | 2015 | China | 175 | 130 | CRC | 58.7 | 113 : 62 | Serum |
| Control | 54.1 | 70 : 50 | ||||||
| Li L | 2015 | China | 200 | 400 | CRC | 66.3±11.8 | 135 : 65 | Plasma |
| Control | 65.5±10.8 | 283 : : 117 | ||||||
| Fang | 2015 | China | 111 | 130 | CRC | 60 | 59 : 52 | Plasma |
| Control | — | — | ||||||
| Basati | 2015 | Iran | 55 | 55 | CRC | 58.52±10 | 30 : 25 | Serum |
| Control | 57.87±10.15 | 31 : 24 | ||||||
| Ghanbari | 2015 | Iran | 61 | 24 | CRC | 64.13±8.67 | 34 : 27 | Plasma |
| Control | 61.96±8.67 | 14 : 10 | ||||||
| Wang S | 2015 | China | 124 | 117 | CRC | 64 | 54 : 70 | Plasma |
| Control | 43 | 65 : 52 | ||||||
| Wang W | 2016 | China | 268 | 102 | CRC | 58 (49–66) | 151 : 117 | Serum |
| Control | 56 (48–66) | — | ||||||
| Sun | 2016 | USA | 227 | 57 | CRC | 55±7.8 | 119 : 108 | Plasma |
| Control | 54±6.3 | 27 : 30 | ||||||
| Yu J | 2016 | China | 165 | 30 | CRC | 60.7±11.6 | 78 : 87 | Serum |
| Control | 59.4±14.5 | 15 : 15 | ||||||
Abbreviation: CRC, colorectal cancer.
A total of 6010 patients were included; 3454 CRC patients, and 2556 healthy controls. Seven studies also included evaluation of 420 patients with CAA, the precursor lesion to CRC. From the studies that reported patient demographics, 1792 men and 1410 women were included among CRC cases and 1198 men and 942 women were included among healthy controls (Table 1).
All patients included in this review had the diagnosis of CRC confirmed by histopathology. Patients were excluded from individual studies if they had a diagnosis of familial adenomatous polyposis, hereditary non-polyposis CRC or other inflammatory condition or if they received preoperative radiotherapy or chemotherapy. Of the 19 studies included in meta-analysis, 18 studies staged CRC according to either the American Joint Committee on Cancer or the Union for International Cancer Control Tumor-Node-Metastasis staging system. The remaining study staged CRC cases according to the Dukes' classification system.
Identification of dysregulated miRNAs in the plasma or serum
Sixteen of the 34 studies performed miRNA screening studies (Ng et al, 2009; Ahmed et al, 2012; Kanaan et al, 2012; Wang et al, 2012b; Brunet Vega et al, 2013; Giraldez et al, 2013; Hofsli et al, 2013; Kanaan et al, 2013; Luo et al, 2013; Zheng et al, 2014; Xu et al, 2014; Ghanbari et al, 2015; Ho et al, 2015; Wang et al, 2015; Li et al, 2015; Sun et al, 2016) in order to identify dysregulated miRNAs in the plasma or serum of CRC cases compared with healthy controls, whereas the remaining 18 studies identified miRNAs for assessment based on the literature or based on their own prior studies (Huang et al, 2010; Pu et al, 2010; Wang and Zhang, 2012; Liu et al, 2013; Yu et al, 2013; Toiyama et al, 2013; Zhang et al, 2013; Basati et al, 2014; Du et al, 2014; Perilli et al, 2014; Zanutto et al, 2014; Basati et al, 2015; Chen et al, 2015; Fang et al, 2015; Ramzy et al, 2015; Li et al, 2016; Wang et al, 2016; Yu et al, 2016). A total of 617 miRNAs were differentially expressed between CRC and healthy controls in these screening studies, and of these, 69 miRNAs were found to be significantly dysregulated in validation or subsequent studies. A further 36 miRNAs were shown to have either increased or decreased expression in CRC, but no statistical analyses were performed (Ahmed et al, 2012; Hofsli et al, 2013) (Supplementary Table S1).
Significantly dysregulated miRNAs in the plasma or serum of CRC cases
Thirty-one miRNAs were found to be either upregulated (n=17, miR-17-3p, miR-18a, miR-19a-3p, miR-20a, miR-21, miR-29a, miR-92, miR-96, miR-106a, miR-182, miR-200c, miR-206, miR-210, miR-221, miR-223-3p, miR-372, miR-378) (Ng et al, 2009; Huang et al, 2010; Pu et al, 2010; Kanaan et al, 2012; Wang and Zhang, 2012; Brunet Vega et al, 2013; Liu et al, 2013; Toiyama et al, 2013; Zhang et al, 2013; Basati et al, 2014; Du et al, 2014; Perilli et al, 2014; Xu et al, 2014; Zanutto et al, 2014; Zheng et al, 2014; Chen et al, 2015; Yamada et al, 2015; Li et al, 2015; Sun et al, 2016; Wang et al, 2016; Yu et al, 2016) or downregulated (n=14, miR-24, miR-26a-5p, miR-29b, miR-30b, miR-142-3p, miR-145, miR-194, miR-218, miR-320a, miR-375, miR-422a, miR-423-5p, miR-601, miR-760) (Wang et al, 2012b; Yu et al, 2013; Xu et al, 2014; Zheng et al, 2014; Basati et al, 2015; Fang et al, 2015; Ghanbari et al, 2015; Ho et al, 2015; Ramzy et al, 2015; Li et al, 2015; Li et al, 2016) in CRC cases as compared with controls (Table 2). Six upregulated miRNAs (miR-17-3p, miR-18a, miR-21, miR-29a, miR-92, miR-106a) and two downregulated miRNAs (miR-29b, miR-145) were identified by more than one study (Ng et al, 2009; Huang et al, 2010; Kanaan et al, 2012; Wang and Zhang, 2012; Brunet Vega et al, 2013; Liu et al, 2013; Toiyama et al, 2013; Zhang et al, 2013; Basati et al, 2014; Du et al, 2014; Zanutto et al, 2014; Zheng et al, 2014; Basati et al, 2015; Chen et al, 2015; Ramzy et al, 2015; Yamada et al, 2015; Li et al, 2015; Li et al, 2016). miR-21 was the most frequently identified dysregulated miRNA (Kanaan et al, 2012; Wang and Zhang, 2012; Liu et al, 2013; Toiyama et al, 2013; Basati et al, 2014; Du et al, 2014; Zanutto et al, 2014; Yamada et al, 2015).
Table 2. Significantly dysregulated miRNAs in the plasma and serum of patients with CRC as compared with controls.
| miRNA | Expression | Area under the curve (AUC) | Sensitivity | Specificity | Study |
|---|---|---|---|---|---|
|
Single miRNA | |||||
| miR-17-3p | Upregulated | 0.717 | 64% | 70% | Ng et al, 2009 |
| 0.781 | —a | — | Li et al, 2015 | ||
| miR-18a | Upregulated | 0.804 | 73.1% | 79.1% | Zhang et al, 2013 |
| — | — | — | Brunet Vega et al, 2013 | ||
| miR-19a-3p | Upregulated | 0.849 | — | — | Zheng et al, 2014 |
| miR-20a | Upregulated | 0.590 | 46% | 73.42% | Chen et al, 2015 |
| miR-21 | Upregulated | 0.910 | 90% | 90% | Kanaan et al, 2012 |
| 0.850 | 87.5% | 74.4% | Wang and Zhang, 2012 | ||
| 0.802 | 65% | 85% | Liu et al, 2013 | ||
| 0.927 | 82.8% | 90.6% | Toiyama et al, 2013 | ||
| 0.870 | 77% | 78% | Basati et al, 2014 | ||
| 0.877 | 76.2% | 93.2% | Du et al, 2014 | ||
| 0.647 | — | — | Zanutto et al, 2014 | ||
| miR-29a | Upregulated | 0.844 | 69% | 89.1% | Huang et al, 2010 |
| — | — | — | Brunet Vega et al, 2013 | ||
| miR-92 | Upregulated | 0.885 | 89% | 70% | Ng et al, 2009 |
| miR-92a | 0.838 | 84% | 71.2% | Huang et al, 2010 | |
| miR-92a-3p | 0.786 | 65.5% | 82.5% | Liu et al, 2013 | |
| 0.890 | — | — | Zheng et al, 2014 | ||
| miR-96 | Upregulated | 0.740 | 65.4% | 73.3% | Sun et al, 2016 |
| miR-106a | Upregulated | 0.605 | 74% | 44.4% | Chen et al, 2015 |
| 0.808 | — | — | Li et al, 2015 | ||
| miR-182 | Upregulated | — | — | — | Perilli et al, 2014 |
| miR-200c | Upregulated | 0.749 | 64.1% | 73.3% | Zhang et al, 2013 |
| miR-206 | Upregulated | 0.705 | — | — | Xu et al, 2014 |
| miR-210 | Upregulated | 0.821 | 74.6% | 73.5% | Wang et al, 2016 |
| miR-221 | Upregulated | 0.606 | 86% | 41% | Pu et al, 2010 |
| miR-223-3p | Upregulated | 0.871 | — | — | Zheng et al, 2014 |
| miR-372 | Upregulated | 0.854 | 81.9% | 73.3% | Yu et al, 2016 |
| miR-378 | Upregulated | 0.796 | — | — | Zanutto et al, 2014 |
| miR-24 | Downregulated | 0.839 | 78.38% | 83.85% | Fang et al, 2015 |
| miR-26a-5p | Downregulated | 0.670 | — | — | Ghanbari et al, 2015 |
| miR-29b | Downregulated | 0.743 | 61.4% | 72.5% | Li et al, 2015 |
| 0.870 | 77% | 75% | Basati et al, 2015 | ||
| miR-30b | Downregulated | — | — | — | Ho et al, 2015 |
| miR-142-3p | Downregulated | 0.710 | — | — | Ghanbari et al, 2015 |
| miR-145 | Downregulated | — | — | — | Ramzy et al, 2015 |
| 0.874 | — | — | Li et al, 2015 | ||
| miR-194 | Downregulated | 0.850 | 72% | 80% | Basati et al, 2015 |
| miR-218 | Downregulated | — | — | — | Yu et al, 2013 |
| miR-320a | Downregulated | 0.886 | 92.79% | 73.08% | Fang et al, 2015 |
| miR-375 | Downregulated | 0.749 | 76.92% | 64.63% | Xu et al, 2014 |
| miR-422a | Downregulated | 0.843 | — | — | Zheng et al, 2014 |
| miR-423-5p | Downregulated | 0.833 | 91.89% | 70.77% | Fang et al, 2015 |
| miR-601 | Downregulated | 0.747 | 69.2% | 72.4% | Wang et al, 2012b |
| miR-760 | Downregulated | 0.788 | 80% | 72.4% | Wang et al, 2012b |
|
Two miRNA panels | |||||
| miR-19a, -19b | Upregulated | 0.820 | 78.57% | 77.36% | Giraldez et al, 2013 |
| miR-21, -92a | Upregulated | 0.847 | 68% | 91.2% | Liu et al, 2013 |
| miR-29a, -92a | Upregulated | 0.883 | 83% | 84.7% | Huang et al, 2010 |
| miR-200c, -18a | Upregulated | 0.839 | 84.6% | 75.6% | Zhang et al, 2013 |
| miR-431, -139-3p | Upregulated | 0.829 | 91% | 57% | Kanaan et al, 2013 |
| miR-601, -760 | Downregulated | 0.792 | 83.3% | 69.1% | Wang et al, 2012b |
| miR-375, -206 | Upregulated and Downregulatedb | 0.846 | — | — | Xu et al, 2014 |
|
Three miRNA panels | |||||
| miR-19a, -19b, -15b | Upregulated | 0.840 | 78.57% | 79.25% | Giraldez et al, 2013 |
| miR-486, -25, -1180 | Upregulated | — | — | — | Ho et al, 2015 |
| miR-24, -320a, -423-5p | Downregulated | 0.899 | 92.79% | 70.77% | Fang et al, 2015 |
| miR-145, -106a, -17-3p | Upregulated and Downregulated | 0.886 | 78.5% | 82.8% | Li et al, 2015 |
| miR-409-3p, -7, -93 | Upregulated and Downregulated | 0.897 | 82% | 89% | Wang et al, 2015 |
|
Panels with ⩾4 miRNA | |||||
| miR-18a, -19a, -19b, -15b, 29a, -335 | Upregulated | 0.70–0.80 | — | — | Giraldez et al, 2013 |
| miR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145 | Upregulated | 0.745 | — | — | Luo et al, 2013 |
| miR-423-5p, -210, -720, -320a, -378, -92a, -29a, 155 | Upregulated | — | — | — | Hofsli et al, 2013 |
| miR-7, -17-3p, -20a, -21, -92a, -96, -183, -196a, -214 | Upregulated | — | — | — | Ahmed et al, 2012 |
| miR-29a, -92a, -601, -760 | — | 0.943 | 83.3% | 93.1% | Wang et al, 2012b |
| miR-124, -127-5p, -138, -143, -146a, -222 | Downregulated | — | — | — | Ahmed et al, 2012 |
| miR-106a, -143, -103, -199-3p, -151-5p, -107, -191, -423-3p, -221, -382, -409-3p, -652, let 7d | Downregulated | — | — | — | Hofsli et al, 2013 |
Abbreviations: CRC=colorectal cancer; miRNA=microRNA.
Not reported.
Upregulated and downregulated—where the individual miRNAs were either upregulated or downregulated in CRC compared with healthy controls.
Seven studies identified a two miRNA panel (Huang et al, 2010; Wang et al, 2012b; Giraldez et al, 2013; Kanaan et al, 2013; Liu et al, 2013; Zhang et al, 2013; Xu et al, 2014), five studies identified a three miRNA panel (Giraldez et al, 2013; Fang et al, 2015; Ho et al, 2015; Wang et al, 2015; Li et al, 2015), and five studies identified a panel consisting of four or more miRNAs (Ahmed et al, 2012; Wang et al, 2012b; Giraldez et al, 2013; Hofsli et al, 2013; Luo et al, 2013) that was utilised in the diagnosis of CRC (Table 2).
Area under the curve, sensitivity, and specificity
The most common method of reporting diagnostic accuracy of dysregulated miRNAs was using AUC and sensitivity and specificity as determined from ROC curves. Of the identified miRNAs, AUC values ranged from 0.590 to 0.943, sensitivity ranged from 46% to 92.79% and specificity from 41% to 93.2%. The highest AUC, sensitivity, and specificity combination was reported for a panel of four miRNAs, miR-29a, miR-92a, miR-601, and miR-760 (AUC 0.943, sensitivity 83.3%, specificity 93.1%) (Wang et al, 2012b) (Table 2).
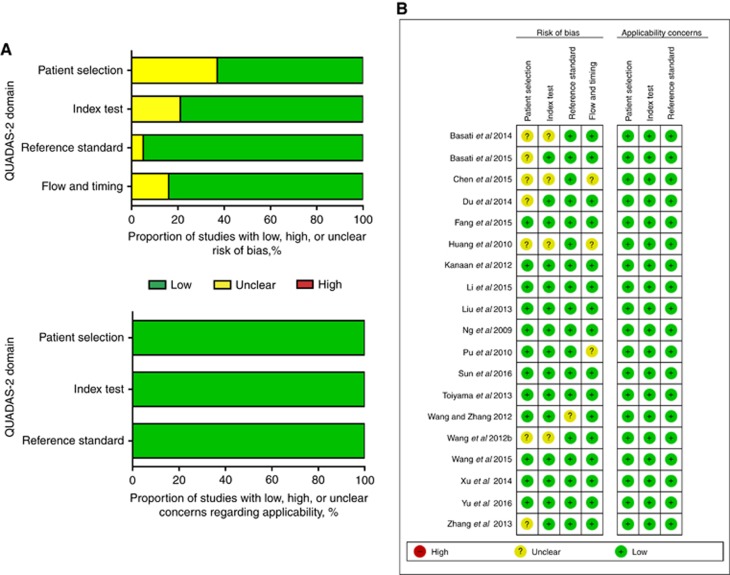
Quality assessment
Quality assessment results of the studies reporting upon single miRNA included in meta-analysis are shown in Figures 2A and B using the QUADAS-2 evaluation tool.
Figure 2.
QUADAS-2 assessment. Investigators' assessment regarding each domain for included studies. Risk of bias and applicability concerns' (A) graph and (B) summary.
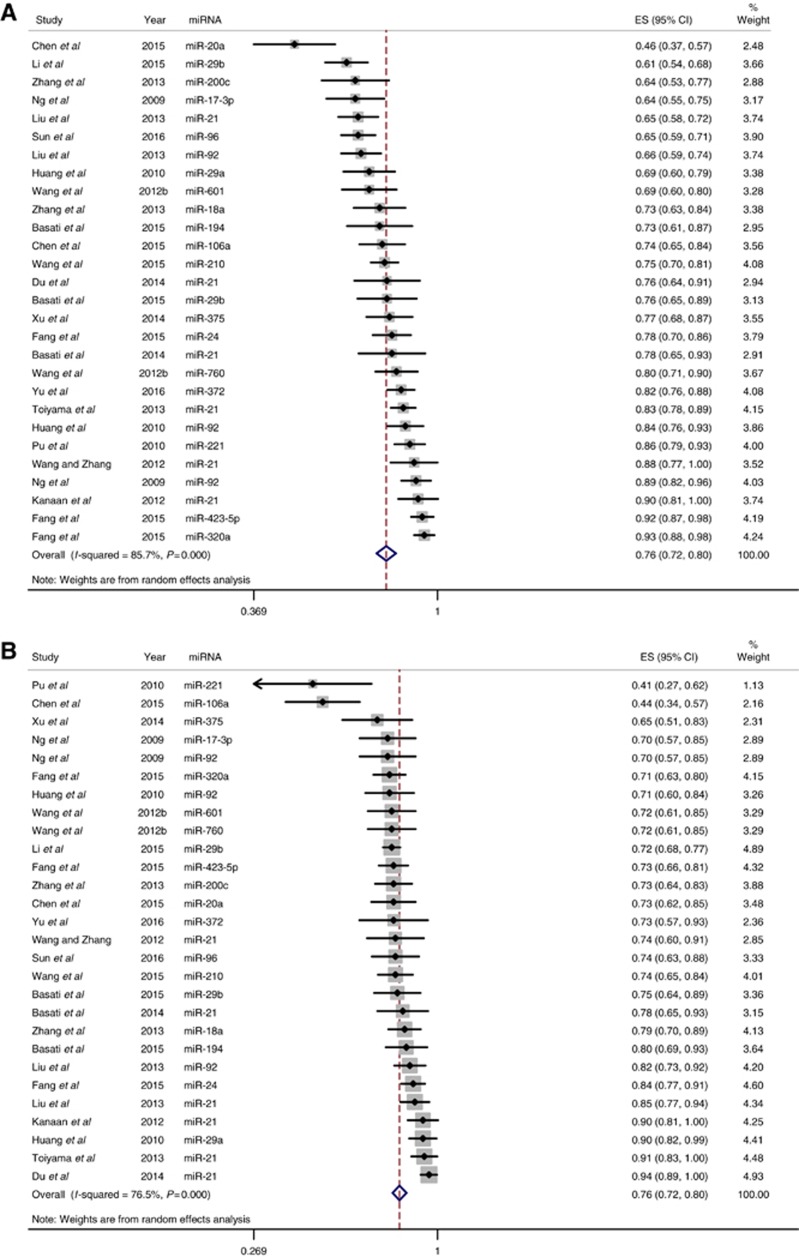
Meta-analysis
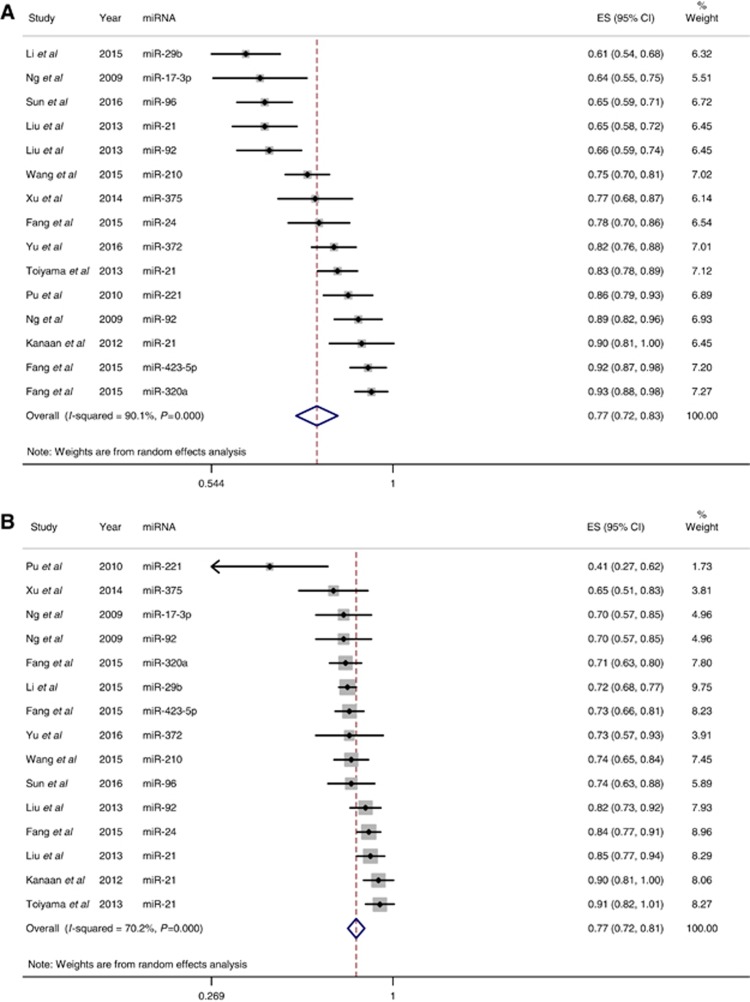
Studies that identified a single dysregulated miRNA in CRC cases compared with controls, along with the corresponding AUC and sensitivity and specificity for the identified miRNA, were included in the meta-analysis. Using these criteria, 28 individual miRNAs were identified in 19 studies (Supplementary Table S2). The sensitivity (±95% confidence intervals (CIs)) and specificity (±95% CIs) for each miRNA are shown in the corresponding forest plots (Figures 3A and B). Overall sensitivity and specificity of the 28 individual miRNAs in the diagnosis of CRC were 76% (95% CI 72–80%) and 76% (95% CI 72–80%), respectively. These results indicate good discriminative ability of the use of miRNAs as biomarkers for the detection of CRC. Sensitivity analyses were performed for studies of good quality (Figures 4A and B), type of patient sample used (plasma vs serum, Supplementary Figures S1a and b and S2a and b), type of miRNA detection method (SYBR green vs TaqMan, Supplementary Figures S3a and b and S4a and b), and type of miRNA reference used for normalisation (internal vs external, Supplementary Figures S5a and b and S6a and b). Compared with the sensitivity and specificity obtained when including all studies, sensitivity analysis using only high-quality studies did not significantly alter sensitivity and specificity (77% vs 76% and 77% vs 76%, respectively). This was also true for the remaining sensitivity analyses, where there was no improvement in sensitivity or specificity.
Figure 3.
Forest plots for studies on individual miRNA assays used in the diagnosis of CRC among the 19 studies included in the meta-analysis. (A) Estimates of sensitivity. (B) Estimates of specificity. A full color version of this figure is available at the British Journal of Cancer journal online.
Figure 4.
Analysis performed for low-risk/low-concern studies with respect to QUADAS-2 quality assessment. (A) Quality sensitivity analysis. Overall sensitivity was 77% (95% CI 72–83%). (B) Quality specificity analysis. Overall specificity was 77% (95% CI 72–81%). A full color version of this figure is available at the British Journal of Cancer journal online.
Discussion
This systematic review of 34 manuscripts that utilise plasma- or serum-based miRNAs in the diagnosis of CRC has identified 617 miRNAs reported to be dysregulated in 3454 cases of CRC as compared with 2556 healthy controls. A total of 31 single miRNAs were identified as significantly dysregulated between CRC cases and controls. Eight of the dysregulated miRNAs were identified and validated by more than one study; six were upregulated: miR-17-3p, miR-18a, miR-21, miR-29a, miR-92, and miR-106a and two were downregulated: miR-29b and miR-145. A further 23 significantly dysregulated miRNAs were identified by only one study. Conceptually, the increased expression of specific miRNAs in the presence of disease is accepted, assuming that the cancer is the source of the dysregulated miRNA export into the blood. However, the source of the miRNA dysregulation is not necessarily known, and miRNAs that are consistently downregulated only in the presence of disease should not necessarily be discounted as reliable biomarkers, provided there is an acceptable negative predictive value for the elevated levels of that miRNA in the absence of disease. This would, for example, be true of a miRNA whose target was a known oncogene.
Significant advances have occurred in the field of tumour-associated miRNAs since their first discovery in plasma (Mitchell et al, 2008; Chevillet et al, 2014) as the number of studies investigating their expression has markedly increased in recent years. Although colonoscopy remains the gold standard screening test for the diagnosis of CRC (Muller and Sonnenberg, 1995; Citarda et al, 2001; Zauber et al, 2012; Nishihara et al, 2013), the evaluation of miRNA within plasma and/or serum of circulating blood has provided a new area for biomarker research.
microRNAs are small, naturally occurring, non-protein-coding RNA that are transcribed in the nucleus as large RNA precursors called primary miRNAs (pri-miRNAs) (Lee et al, 2004). These pri-miRNAs are enzymatically cleaved in the nucleus by the enzyme Drosha into precursor miRNA (pre-miRNA) (Han et al, 2004). The resulting approximately 70-nucleotide pre-miRNA are folded into stem-loop structures. These pre-miRNAs are then exported into the cytoplasm by the GTP-dependent transport protein exportin 5 (Yi et al, 2003). Once in the cytoplasm, they undergo additional processing by the enzyme Dicer to generate the mature double-stranded miRNA approximately 22 nucleotides in length. Dicer also initiates the formation of the RNA-induced silencing complex (RISC) (Hammond, 2005). It is in this final part of miRNA biogenesis that the leading miRNA strand is incorporated into the RISC complex, which is then guided into the complementary 3′ or 5′ untranslated region of the target mRNA. The RISC is responsible for the gene silencing observed owing to miRNA expression and RNA interference (Filipowicz et al, 2005).
miRNAs are stable in extracellular fluid as they are protected from RNases by virtue of being bound to argonaute proteins (Meister, 2013). Discovery of dysregulated miRNA expression has been identified in oesophageal, lung, liver, pancreatic, bladder, ovarian, and gastric cancers (Chiang et al, 2012). miRNAs have been identified in numerous body fluids, such as plasma, saliva, faeces, and urine, where they are actively released from cells within microvesicles, exosomes, or bound to proteins (Kanaan et al, 2012; Chevillet et al, 2014). These factors suggest that miRNAs can be used as biomarkers for cancer and other disease states (Turchinovich et al, 2011).
miRNAs can function as either tumour suppressors or oncogenes. Overexpression of miRNAs in cancer may function as oncogenes and promote oncogenesis by downregulating tumour suppressors or other genes involved in cell differentiation. In contrast, underexpressed miRNAs in cancer may function as tumour suppressors by inhibiting oncogenesis via downregulation of proteins with oncogenic qualities (Zhang et al, 2007; Shenouda and Alahari, 2009). The miR-17-92 cluster, also known as oncomiR-1 was the first miRNA oncogene to be described (Li et al, 2014). Six miRNAs form this miR-17-92 cluster: miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92a-1, and are known to be dysregulated in many cancers (Mogilyansky and Rigoutsos, 2013). miR-17-92 has also been found to be overexpressed in both the tissue and serum of CRC patients (Dews et al, 2006; Ng et al, 2009). In our systematic review, five of the six members of this cluster, with the exception of miR-19b-1, were found to be significantly upregulated in the serum or plasma of CRC patients as compared with controls in eight studies (Ng et al, 2009; Huang et al, 2010; Brunet Vega et al, 2013; Liu et al, 2013; Zhang et al, 2013; Zheng et al, 2014; Chen et al, 2015; Li et al, 2015). The most predictive miRNA was miR-92a, with AUC values ranging from 0.786 to 0.890, from 65.5% to 89% sensitivity, and from 70% to 82.5% specificity. The remaining member of the cluster, miR-19b-1, was also significantly upregulated in CRC patients when combined as a two miRNA panel together with miR-19a, with an AUC of 0.82, 78.57% sensitivity, and 77.36% specificity (Giraldez et al, 2013). These data support the oncogenic role of the miR-17-92 cluster in CRC.
We identified seven different studies that demonstrated significant upregulation of miR-21 in CRC patients as compared with healthy controls. Kanaan et al (2012) identified miR-21 as a biomarker for CRC. Validated CRC tissue miRNAs were evaluated in a plasma test set consisting of 30 CRC patients and 30 healthy controls. The most dysregulated tissue miRNAs were then validated in different cohort of 20 CRC patients with 20 age- and race-matched subjects without CRC. In the plasma test group, miR-21 differentiated CRC patients from controls with AUC 0.910 and 90% specificity and sensitivity (Kanaan et al, 2012). Following this, 6 groups reproduced similar results with AUCs ranging from 0.647–0.927, 65–87.5% sensitivity, and 74.4–93.2% specificity. Although these data were promising, dysregulation of miR-21 has also been described in many other cancers (Chan et al, 2005; Iorio et al, 2005; Volinia et al, 2006; Iorio et al, 2007; Lui et al, 2007; Meng et al, 2007; Tetzlaff et al, 2007; Hu et al, 2011). There is therefore need for a plasma miRNA profile specific for CRC.
miR-29a is another miRNA that has been reported to be upregulated in CRC patients in three studies (Huang et al, 2010; Brunet Vega et al, 2013; Yamada et al, 2015). miR-29a is part of the miR-29 family (miR-29a, miR-29b, miR-29c). Interestingly, another member of the miR-29 family, miR-29b, was observed to be downregulated in CRC compared with control subjects in two studies (Basati et al, 2015; Li et al, 2016). Members of the miR-29 family exhibit differential regulation, implying that their functional relevance may not be identical (Kriegel et al, 2012). miR-29b has been shown to function as a tumour suppressor in various cancers and diseases (Yan et al, 2015), while miR-29a has been shown to act as a tumour suppressor in lung cancer (Fabbri et al, 2007) but as an oncogene in CRC, ovarian, and breast cancer (Gebeshuber et al, 2009; Resnick et al, 2009).
In order to reduce CRC-associated mortality, the ideal biomarker would be able to identify individuals with both early-stage CRC and its precursor lesion, the colorectal advanced adenoma (CAA), from healthy control subjects. CAA have previously been defined as adenomas with a villous component or with high-grade dysplasia >0.75 cm size in diameter (Carter et al, 2016b). In the studies included in this review, seven studies evaluated miRNAs found to be significantly dysregulated in CRC patients and in 420 patients with a diagnosis of CAA. Five of these seven studies reported miRNA panels significantly upregulated in CAA as compared with healthy controls (Huang et al, 2010; Kanaan et al, 2013; Liu et al, 2013; Toiyama et al, 2013; Zheng et al, 2014). One study also identified a panel of 5 miRNAs that distinguished between CRC and CAA with AUC 0.856 (Kanaan et al, 2013). The remaining two studies found no significant differences in miRNA profile between CAA and healthy controls (Giraldez et al, 2013; Luo et al, 2013) (Supplementary Table S3).
Commercially available non-invasive screening tests have recently been developed in an attempt to have the same detection accuracy of CRC compared with the current gold standard screening method of colonoscopy (2016; Imperiale et al, 2014; Marshall et al, 2010; Nichita et al, 2014). These tests, both blood based and faecal based, measure validated markers of CRC and include DNA markers and gene expression profiles. To date, none have a good ability to detect the precursor to CRC, CAA. We have developed a plasma-based miRNA screening assay comprising of combinations of 7 miRNAs, which is able to detect any neoplasia from controls with AUC 0.91, colorectal neoplasia from other cancers with AUC 0.79, and CRC from CAA with AUC 0.98. In addition, our assay is able to differentiate CRC from controls with AUC 0.98, sensitivity 90%, and specificity 100%, CAA from controls with AUC 0.823, sensitivity 65%, and specificity 95%, and CRC and CAA from controls with AUC 0.835, sensitivity 80%, and specificity 85% (Carter et al, 2016b). This miRNA panel, although in its infancy, provides much promise in the development of a non-invasive screening test for detection of CRC.
Inevitably, studies included in the systematic review and meta-analysis will vary with regards to clinical characteristics, study design, and type of study, and these factors contribute sources of heterogeneity. Clinical characteristics of control subjects were often poorly defined. Of the individual studies included in the meta-analysis, five defined healthy controls by colonoscopic confirmed absence of CRC, including absence of adenomas. The remaining 14 studies defined healthy controls as having the absence of malignancy or inflammatory condition or a benign physical examination without other qualification or elaboration. Similarly, 11 studies reported no significant differences in age or sex between CRC cases and healthy controls, 5 reported to age-sex matching between cases and controls, and 3 did not comment on age-sex matching status.
There was significant heterogeneity of the studies included in this review (I2 85.7% for sensitivity, and 76.5% for specificity, P<0.001). Heterogeneity is expected in meta-analyses, one reason being differing sample sizes between studies (Higgins, 2008). Many of the studies included a relatively small number of participants. For the studies included in meta-analysis, the study populations ranged from 32 to 268 for CRC cases and from 30 to 400 for control cases. This may reflect the strict inclusion criteria adopted by investigators but potentially carries the risk of type I error. In future, studies with larger numbers of patients, along with different populations of patients in terms of genetics, geography and race, will increase the validity of our conclusions regarding the reliability of plasma miRNA assays. In addition, reference miRNA controls used varied between studies, with both internal and external miRNAs used for normalisation of results. This may account for the differing miRNAs identified. Similarly, miRNAs assessed were extracted from either plasma or serum samples. Some have reported differing miRNA concentrations depending on whether serum or plasma are used as the source material (Wang et al, 2012a). Unlike previous studies, miRNAs measured in the serum of patient samples yielded better results than for plasma in this review. Although there is little difference in composition between plasma and serum, for the development of a blood-based biomarker, a standard blood component is preferred. A limitation to this review is the validity of the results given the significant heterogeneity of the studies. We believe it is, however, still reasonable to interpret these results as the overwhelming majority of the studies trend towards high sensitivity and specificity of miRNAs as biomarkers for the diagnosis of CRC.
Despite the large numbers of studies investigating plasma or serum miRNAs in patients with CRC or CAA, there still remains variation as to which significantly dysregulated miRNAs are identified. Some miRNAs were found to be significantly dysregulated in multiple studies, yet not all of these studies performed in miRNA screening prior to testing and validation, for example, miR-21, was significantly dysregulated in seven studies; however, only one of these performed initial miRNA screening. In addition, of the 28 miRNAs included in the meta-analysis, only 3 were reported as significantly dysregulated by more than 1 study: miR-21, miR-29b and miR-92.
The majority of studies were performed in either North America or South East Asia. It is possible that genetics, geography, and race may affect miRNA expression differently in these groups of patients, resulting in the observed differences in significantly dysregulated miRNAs. With the recent availability of blood-based screening tests for CRC, it is important to focus on quality and study reporting standards.
Conclusion
This systematic review identifies numerous miRNAs associated with colorectal carcinogenesis. miRNAs can distinguish patients with CRC from healthy controls with high sensitivity and specificity, however; no single stand-alone miRNA has yet been identified as an ideal biomarker for the diagnosis of CRC. In order to develop a diagnostic test for the detection of CRC, we propose that a panel of miRNAs with high sensitivity and specificity should be chosen and tested in a large population of subjects. This would include a combination of the most commonly identified miRNA, those miRNA most predictive of CRC, and the most unique miRNA to CRC, in order to develop a non-invasive test with sensitivity and specificity comparable to current screening methods for CRC. In addition, standardisation of study methodology such as the use of serum samples, TaqMan qRT-PCR detection, and an external miRNA reference control should be implemented in future studies.
With deaths from CRC among the leading cause of cancer deaths worldwide, there is an emphasis to increase screening, and therefore identify early, less invasive stage of disease, to improve survival. For this purpose, miRNAs have the potential in the future to be used as a relatively non-invasive, inexpensive, blood-based marker for detection of CRC.
Acknowledgments
This work was made possible in part by the John W and Barbara Thruston Atwood Price Trust. The funders had no role in study concept, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- (2016) Test: About ColoVantage [Internet]. Questdiagnostics.com [Online]. Available at http://www.questdiagnostics.com/home/physicians/testing-services/by-test-name/colovantage/about (accessed 16 March 2016).
- Ahmed FE, Amed NC, Vos PW, Bonnerup C, Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE, Allison RR (2012) Diagnostic microRNA markers to screen for sporadic human colon cancer in blood. Cancer Genomics Proteomics 9: 179–192. [PubMed] [Google Scholar]
- Basati G, Emami Razavi A, Abdi S, Mirzaei A (2014) Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Med Oncol 31: 205. [DOI] [PubMed] [Google Scholar]
- Basati G, Razavi AE, Pakzad I, Malayeri FA (2015) Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer. Tumour Biol 37: 1781–1788. [DOI] [PubMed] [Google Scholar]
- Brunet Vega A, Pericay C, Moya I, Ferrer A, Dotor E, Pisa A, Casalots A, Serra-Aracil X, Oliva JC, Ruiz A, Saigi E (2013) microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep 30: 320–326. [DOI] [PubMed] [Google Scholar]
- Calderwood AH, Jacobson BC (2013) Colonoscopy quality: metrics and implementation. Gastroenterol Clin North Am 42: 599–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JV, Pan J, Rai SN, Galandiuk S (2016. a) ROC-ing along: evaluation and interpretation of receiver operating characteristic curves. Surgery 159: 1638–1645. [DOI] [PubMed] [Google Scholar]
- Carter JV, Roberts HL, Pan J, Rice JD, Burton JF, Galbraith NJ, Eichenberger MR, Jorden J, Deveaux P, Farmer R, Williford A, Kanaan Z, Rai SN, Galandiuk S (2016. b) A highly predictive model for diagnosis of colorectal neoplasms using plasma microRNA: improving specificity and sensitivity. Ann Surg 264: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033. [DOI] [PubMed] [Google Scholar]
- Chen WY, Zhao XJ, Yu ZF, Hu FL, Liu YP, Cui BB, Dong XS, Zhao YS (2015) The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer. Int J Clin Exp Pathol 8: 7092–7101. [PMC free article] [PubMed] [Google Scholar]
- Chevillet JR, Lee I, Briggs HA, He Y, Wang K (2014) Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules 19: 6080–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y, Song Y, Wang Z, Liu Z, Gao P, Liang J, Zhu J, Xing C, Xu H (2012) microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp Ther Med 3: 560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citarda F, Tomaselli G, Capocaccia R, Barcherini S, Crespi M Italian Multicentre Study Group (2001) Efficacy in standard clinical practice of colonoscopic polypectomy in reducing colorectal cancer incidence. Gut 48: 812–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM (2008) Oncogenes and cancer. N Engl J Med 358: 502–511. [DOI] [PubMed] [Google Scholar]
- Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A (2006) Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 38: 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Liu S, Gu D, Wang Q, Zhu L, Kang M, Shi D, Chu H, Tong N, Chen J, Adams TS, Zhang Z, Wang M (2014) Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis 35: 2723–2730. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 104: 15805–15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakih MG, Padmanabhan A (2006) CEA monitoring in colorectal cancer. What you should know. Oncology (Williston Park) 20: 579–587. [PubMed] [Google Scholar]
- Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, Lin L, You P, Li J, Dai Z, Liang X, Su Y, Hu Q, Wang F, Zhang ZY (2015) Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res 34: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol 15: 331–341. [DOI] [PubMed] [Google Scholar]
- Gebeshuber CA, Zatloukal K, Martinez J (2009) miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 10: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari R, Mosakhani N, Asadi J, Nouraee N, Mowla SJ, Yazdani Y, Mohamadkhani A, Poustchi H, Knuutila S, Malekzadeh R (2015) Downregulation of plasma miR-142-3p and miR-26a-5p in patients with colorectal carcinoma. Iran J Cancer Prev 8: e2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez MD, Lozano JJ, Ramirez G, Hijona E, Bujanda L, Castells A, Gironella M (2013) Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol 11: 681–8 e3. [DOI] [PubMed] [Google Scholar]
- Hammond SM (2005) Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett 579: 5822–5829. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP (2008) Commentary: Heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol 37: 1158–1160. [DOI] [PubMed] [Google Scholar]
- Ho GY, Jung HJ, Schoen RE, Wang T, Lin J, Williams Z, Weissfeld JL, Park JY, Loudig O, Suh Y (2015) Differential expression of circulating microRNAs according to severity of colorectal neoplasia. Transl Res 166: 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsli E, Sjursen W, Prestvik WS, Johansen J, Rye M, Trano G, Wasmuth HH, Hatlevoll I, Thommesen L (2013) Identification of serum microRNA profiles in colon cancer. Br J Cancer 108: 1712–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Correa AM, Hoque A, Guan B, Ye F, Huang J, Swisher SG, Wu TT, Ajani JA, Xu XC (2011) Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer 128: 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127: 118–126. [DOI] [PubMed] [Google Scholar]
- Hundt S, Haug U, Brenner H (2009) Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann Intern Med 150: 162–169. [DOI] [PubMed] [Google Scholar]
- Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM (2014) Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 370: 1287–1297. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM (2007) MicroRNA signatures in human ovarian cancer. Cancer Res 67: 8699–8707. [DOI] [PubMed] [Google Scholar]
- Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S (2012) Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg 256: 544–551. [DOI] [PubMed] [Google Scholar]
- Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S (2013) A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg 258: 400–408. [DOI] [PubMed] [Google Scholar]
- Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M (2012) The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 44: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Wang C, Deng T, Liang H, Wang Y, Huang D, Fan Q, Wang X, Ning T, Liu R, Zhang CY, Zen K, Chen X, Ba Y (2015) Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci Rep 5: 12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Guo Y, Chen Y, Wang J, Zhen L, Guo X, Liu J, Jing C (2016) The diagnostic efficacy and biological effects of microRNA-29b for colon cancer. Technol Cancer Res Treat 15: 772–779. [DOI] [PubMed] [Google Scholar]
- Li M, Guan X, Sun Y, Mi J, Shu X, Liu F, Li C (2014) miR-92a family and their target genes in tumorigenesis and metastasis. Exp Cell Res 323: 1–6. [DOI] [PubMed] [Google Scholar]
- Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, Sun XF (2013) Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol 34: 2175–2181. [DOI] [PubMed] [Google Scholar]
- Logan RF, Patnick J, Nickerson C, Coleman L, Rutter MD, Von Wagner C English Bowel Cancer Screening Evaluation Committee (2012) Outcomes of the Bowel Cancer Screening Programme (BCSP) in England after the first 1 million tests. Gut 61: 1439–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohsiriwat V (2010) Colonoscopic perforation: incidence, risk factors, management and outcome. World J Gastroenterol 16: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, FIRE A (2007) Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 67: 6031–6043. [DOI] [PubMed] [Google Scholar]
- Luo X, Stock C, Burwinkel B, Brenner H (2013) Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One 8: e62880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahid SS, Hornung CA, Minor KS, Turina M, Galandiuk S (2006) Systematic reviews and meta-analysis for the surgeon scientist. Br J Surg 93: 1315–1324. [DOI] [PubMed] [Google Scholar]
- Marshall KW, Mohr S, Khettabi FE, Nossova N, Chao S, Bao W, Ma J, Li XJ, Liew CC (2010) A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer 126: 1177–1186. [DOI] [PubMed] [Google Scholar]
- Maton A, Hopkins J, Mclaughlin C, Johnson S, Warner M, Lahart D, Wright J (1993) Human Biology and Health. Prentice Hall: Englewood Cliffs, New Jersey, USA. [Google Scholar]
- Mayeux R (2004) Biomarkers: potential uses and limitations. NeuroRx 1: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G (2013) Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 14: 447–459. [DOI] [PubMed] [Google Scholar]
- Meltzer PS (2005) Cancer genomics: small RNAs with big impacts. Nature 435: 745–746. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ (2014) Non-coding RNAs as clinical biomarkers for cancer diagnosis and prognosis. Expert Rev Mol Diagn 14: 917–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilyansky E, Rigoutsos I (2013) The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ 20: 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG Group, P. (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol 62: 1006–1012. [DOI] [PubMed] [Google Scholar]
- Muller AD, Sonnenberg A (1995) Prevention of colorectal cancer by flexible endoscopy and polypectomy. A case-control study of 32 702 veterans. Ann Intern Med 123: 904–910. [DOI] [PubMed] [Google Scholar]
- Naylor S (2003) Biomarkers: current perspectives and future prospects. Expert Rev Mol Diagn 3: 525–529. [DOI] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58: 1375–1381. [DOI] [PubMed] [Google Scholar]
- Nichita C, Ciarloni L, Monnier-Benoit S, Hosseinian S, Dorta G, Ruegg C (2014) A novel gene expression signature in peripheral blood mononuclear cells for early detection of colorectal cancer. Aliment Pharmacol Ther 39: 507–517. [DOI] [PubMed] [Google Scholar]
- Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT (2013) Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 369: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli L, Vicentini C, Agostini M, Pizzini S, Pizzi M, D'Angelo E, Bortoluzzi S, Mandruzzato S, Mammano E, Rugge M, Nitti D, Scarpa A, Fassan M, Zanovello P (2014) Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget 5: 6611–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY (2010) Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 25: 1674–1680. [DOI] [PubMed] [Google Scholar]
- Ramzy I, Hasaballah M, Marzaban R, Shaker O, Soliman ZA (2015) Evaluation of microRNAs-29a, 92a and 145 in colorectal carcinoma as candidate diagnostic markers: an Egyptian pilot study. Clin Res Hepatol Gastroenterol 39: 508–515. [DOI] [PubMed] [Google Scholar]
- Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE (2009) The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol 112: 55–59. [DOI] [PubMed] [Google Scholar]
- Shenouda SK, Alahari SK (2009) MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev 28: 369–378. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu Y, Cogdell D, Calin GA, Sun B, Kopetz S, Hamilton SR, Zhang W (2016) Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget 10: 11434–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, Livolsi VA, Baloch ZW (2007) Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol 18: 163–173. [DOI] [PubMed] [Google Scholar]
- Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A (2013) Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 105: 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res 39: 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhang Q (2012) The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol 138: 1659–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Yuan Y, Cho JH, Mcclarty S, Baxter D, Galas DJ (2012. a) Comparing the microRNA spectrum between serum and plasma. PLoS One 7: e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang L, Huang D, Tan C, Sheng W, Du X (2012. b) Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One 7: e44398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xiang J, Li Z, Lu S, Hu J, Gao X, Yu L, Wang L, Wang J, Wu Y, Chen Z, Zhu H (2015) A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer 136: 152–161. [DOI] [PubMed] [Google Scholar]
- Wang W, Qu A, Liu W, Liu Y, Zheng G, Du L, Zhang X, Yang Y, Wang C, Chen X (2016) Circulating miR-210 as a diagnostic and prognostic biomarker for colorectal cancer. Eur J Cancer Care (Engl) e-pub ahead of print 22 February 2016; doi:10.1111/ecc.12448. [DOI] [PubMed]
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM QUADAS-2 Group (2011) QUADAS-2: a revised tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med 155: 529–536. [DOI] [PubMed] [Google Scholar]
- Xu L, Li M, Wang M, Yan D, Feng G, An G (2014) The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer 14: 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, Higurashi T, Yukawa N, Amanuma Y, Kikuchi O, Muto M, Ueno Y, Nakajima A, Chiba T, Boland CR, Goel A (2015) Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res 21: 4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Guo Q, Fu FJ, Wang Z, Yin Z, Wei YB, Yang JR (2015) The role of miR-29b in cancer: regulation, function, and signaling. Onco Targets Ther 8: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J (2013) Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol 6: 2904–2911. [PMC free article] [PubMed] [Google Scholar]
- Yu J, Jin L, Jiang L, Gao L, Zhou J, Hu Y, Li W, Zhi Q, Zhu X (2016) Serum miR-372 is a diagnostic and prognostic biomarker in patients with early colorectal cancer. Anticancer Agents Med Chem 16: 424–431. [DOI] [PubMed] [Google Scholar]
- Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F, Perrone F, Leo E, Pilotti S, Verderio P, Gariboldi M, Pierotti MA (2014) Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer 110: 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, Van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, Stewart ET, Waye JD (2012) Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med 366: 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA (2007) microRNAs as oncogenes and tumor suppressors. Dev Biol 302: 1–12. [DOI] [PubMed] [Google Scholar]
- Zhang GJ, Zhou T, Liu ZL, Tian HP, Xia SS (2013) Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma. Mol Clin Oncol 1: 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, Li J, Wang C (2014) Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer 111: 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.