Abstract
Background:
A compromised base excision repair (BER) promotes carcinogenesis by accumulating oxidative DNA-damaged products as observed in MUTYH-associated polyposis, a hereditary colorectal cancer syndrome marked by adenomas and cancers with an accumulation of 8-oxoguanine. Remarkably, DNA global demethylation has been shown to be mediated by BER, suggesting a relevant interplay with early colorectal tumourigenesis. To check this hypothesis, we investigated a cohort of 49 adenomas and 10 carcinomas, derived from 17 MUTYH-associated polyposis patients; as adenoma controls, we used a set of 36 familial adenomatous polyposis and 24 sporadic polyps.
Methods:
Samples were analysed for their mutational and epigenetic status, measured as global LINE-1 (long interspersed nuclear element) and gene-specific LINE-1 MET methylation by mass spectrometry and pyrosequencing.
Results:
MUTYH-associated polyposis adenomas were strikingly more hypomethylated than familial adenomatous and sporadic polyps for both DNA demethylation markers (P=0.032 and P=0.007 for LINE-1; P=0.004 and P<0.0001 for LINE-1 MET, respectively) with levels comparable to those of the carcinomas derived from the same patients. They also had mutations due mainly to KRAS/NRAS p.G12C, which was absent in the controls (P<0.0001 for both sets).
Conclusions:
Our results show that DNA demethylation, together with specific KRAS/NRAS mutations, drives the early steps of oxidative damage colorectal tumourigenesis.
Keywords: colorectal adenomas, DNA hypomethylation, LINE-1/LINE-1 MET methylation, oxidative DNA damage, BER, MUTYH-associated polyposis
Reactive oxygen species (ROS) can have a pivotal role in colorectal tumourigenesis by promoting the accumulation of DNA oxidised bases, mainly 8-oxoguanine (8-oxodG), preferentially repaired by a postreplicative DNA glycosylase belonging to the DNA base excision repair (BER), MUTYH. This enzyme, expressed in the nucleus and mitochondria, specifically counteracts oxidative stress-induced DNA damage by removing adenine misincorporated opposite to 8-oxodG during DNA replication (Ohtsubo et al, 2000).
The involvement of the 8-oxodG:A mispairs in colorectal tumourigenesis is demonstrated by the high risk of developing colorectal cancer (CRC) in patients affected by MUTYH-associated polyposis (MAP), a recessive inherited polyposis linked with biallelic germline mutations of MUTYH (Al-Tassan et al, 2002). The MAP phenotype resembles that of APC-linked attenuated familial adenomatous polyposis (AFAP) with the appearance of a limited number of adenomas (generally 30–100) in the fourth to fifth decade of life. However, unlike AFAP, ∼60% of MAP patients show colorectal cancer at presentation (Nieuwenhuis et al, 2012). MUTYH-associated polyposis carcinogenesis displays peculiar molecular features that characterise disease progression. Chromosomal instability (CIN) is detectable during the early stages in MAP tumours (Cardoso et al, 2006) and the somatic molecular fingerprint of this syndrome is an excess of KRAS c.34G>T transversions, due to the failure of the impaired MUTYH to repair the mismatches induced by the 8-oxoG variant base (Venesio et al, 2013).
Globally acquired DNA hypomethylation is recognised to be an early causal event in colorectal carcinogenesis (King et al, 2014). In addition to the activation of oncogenes, this epigenetic alteration preferentially affects the DNA's repetitive sequences, such as long interspersed nuclear elements (LINE-1 or L1), which represent 17% of the human genome, and leads to the onset of the CIN phenotype (Baba et al, 2010). Over a longer time frame, the hypomethylation-dependent inappropriate expression of heterochromatic sequences and aneuploidy can contribute to the progression of CRCs (Kitkumthorn and Mutirangura, 2011).
DNA demethylation processes were reported to interact with the BER system by different mechanisms including oxidative demethylation, substitution of methylated cytosine and deamination of 5-methyl cytosine (Niehrs, 2009).
Some studies describe a correlation between LINE-1 hypomethylation and colorectal carcinogenesis, reporting an association with clinical, pathological and molecular features (Baba et al, 2010; Sunami et al, 2011). More recently, LINE-1 hypomethylation was recognised as an important feature of early-onset CRCs and a prognostic biomarker for CRCs with a distinct molecular subtype (Ogino et al, 2008; Antelo et al, 2012; Sahnane et al, 2015). Among the specific genes potentially altered by LINE-1 hypomethylation, the MET oncogene contains a LINE-1 sequence in its second intron whose antisense promoter can drive the transcription of a chimeric isoform of the MET gene, LINE-1 MET or L1-MET (Weber et al, 2010; Wolff et al, 2010; Hur et al, 2014; Zhu et al, 2014).
To define the early genetic and epigenetic features in colorectal premalignant lesions characterised by oxidative DNA damage and to assess their potential involvement in driving colorectal carcinogenesis, we investigated a cohort of 49 colorectal adenomas and 10 carcinomas derived from 17 MAP subjects and a control set of 36 familial adenomatous polyposis (FAP/AFAP) and 24 sporadic polyps for their mutational and DNA methylation status.
Materials and methods
Patients and specimens
The study cohort consisted of 17 MAP patients (M1–M17; 9 females, 8 males; median age of 47 years, range 39–79 years) carrying different biallelic MUTYH germline mutations, 17 FAP/AFAP subjects (F1–F17; 10 females and 7 males; median age of 37 years, range 21–61 years) with different APC germline alterations and 21 cases (S1–S21; 10 females and 11 males; median age of 67 years, range 57–84 years) with sporadic colorectal adenomas (Supplementary Tables S1–S4). After genetic counselling, MAP and FAP/AFAP patients underwent MUTYH and APC germline mutation analysis. Germline mutations are reported in Supplementary Tables S1 and S2-–S4. Sixteen out of 34 MUTYH mutations were the most common pathogenic p.Y179C and p.G396D; 10 were truncating and 8 were missense whose pathogenicity was checked in the Leiden Open Variation Database (http://www.lovd.nl/MUTYH). MUTYH mutations have been proven to be biallelic by family segregation. For the sporadic cases, subjects were chosen among those individuals who had undergone the endoscopic screening without a previous report of adenoma detection. In addition, familiarity for polyposis or CRC was excluded according to their clinical history by interview. The present study was carried out according to the research rules of our institutional medical ethical committees on human experimentation, and appropriate written informed consent was collected from all individuals included in the analysis.
All patients were of Italian origin and their corresponding tissue samples were collected from the files of the Department of Pathology of the Ospedale di Circolo-University of Insubria, Varese, and from the archives of the Pathology Unit of Candiolo Cancer Institute (FPO-IRCCS).
Overall, we analysed 49 adenomas and 10 carcinomas from MAP patients, 36 adenomas from FAP/AFAP subjects and 24 polyps from sporadic individuals. In most of the MAP and FAP cases, multiple adenomas from the same patient were investigated. For MAP patients, we could analyse adenomas and carcinomas derived from the same five subjects (cases M1, M6, M16 with adenomas and one carcinoma each, and cases M4 and M9 with adenomas and two carcinomas each), only adenomas in nine cases (M2, M3, M5, M7, M8, M10, M12, M14 and M15) and only carcinoma in three patients (M11, M13 and M17) (Supplementary Tables S1–S4). The histopathological revision was performed by an expert pathologist (C.R.) according to the WHO classification of colorectal tumours (Hamilton et al, 2010). High-grade dysplasia was observed in 7% of MAP adenomas, 6% of FAP/AFAP adenomas and 17% of sporadic adenomas, respectively. The remaining cases showed low-grade dysplasia. A tubular architecture was observed in 71 adenomas (30 MAP, 27 FAP/AFAP and 14 sporadic polyps), while 38 samples were tubulovillous (19 MAP, 9 FAP/AFAP and 10 sporadic). Molecular analyses were performed on formalin-fixed, paraffin-embedded (FFPE) tissue sections using three representative 8-μm-thick sections of tissue samples. The pathologist selected adenoma and carcinoma areas with more than 50% of dysplastic or tumour cells. DNA was extracted after manual dissection using a QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol and quantified using the Qubit 2.0 Fluorimeter (Life Technologies/Thermo Fisher Scientific, Wilmington, DE, USA) following the protocol of High Sensitivity DNA Kit (Life Technologies, Eugene, OR, USA).
Mutational analyses
All the 109 adenomas and the 10 MAP adenocarcinomas were checked for mutations in KRAS, BRAF, NRAS and PIK3CA genes using the mass spectrometry matrix-assisted laser desorption ionisation time of flight method with the MassARRAY System (Agena Bioscience, Hamburg, Germany) with Myriapod Colon Status (Diatech Pharmacogenetics, Jesi, Italy).
L1 and L1-MET methylation studies
The methylation status of global and local LINE-1 sequences was evaluated by bisulfite-PCR and pyrosequencing in 15 samples of histologically normal colonic mucosa derived from healthy individuals obtained from the files of the Department of Pathology of Ospedale di Circolo-University of Insubria (Supplementary Tables S5), and in all MAP, FAP/AFAP, sporadic adenomas and MAP carcinomas. DNA bisulfite conversion was performed using the Epitect Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Global LINE-1 methylation status was assessed through the quantification of the mean methylation percentage of four consecutive CpG sites in the LINE-1 promoter region (GenBank accession number: X58075), as reported previously (Yang et al, 2004; Stefanoli et al, 2014). Intragenic levels of LINE-1 methylation were analysed using the L1-MET assay: the forward PCR primer (5′-GAGATGAATTTAGTATTTTAGATGGAAATG-3) was located inside the LINE-1 promoter, and the reverse primer (5′-biotin-ACAACTCCCATCTACAACTCCCA-3′) was designed within the MET gene intron between exons 2 and 3. The sequencing primer (5′-TTTAGATGGAAATGTAGAAATTAT-3′) amplified a product that includes three CpG sites whose mean methylation percentage was quantified (GenBank accession number: NG_0089961). Each sample was loaded two times for pyrosequencing and fully methylated and unmethylated DNA (Millipore, Billerica, MA, USA) were used as positive and negative controls in each experiment. Run-run variation was 2.1% for each assay and high-resolution melt analysis was performed to validate the pyrosequencing results.
Statistical analyses
Data analysis was carried out with the GraphPad Prism V5.0 software (GraphPad Software, La Jolla, CA, USA). Association analyses were performed using the Fisher's exact test and the independent-sample t-test, whereas Pearson's correlation coefficient (r) was used to measure correlation.
Results
Gene mutation analyses
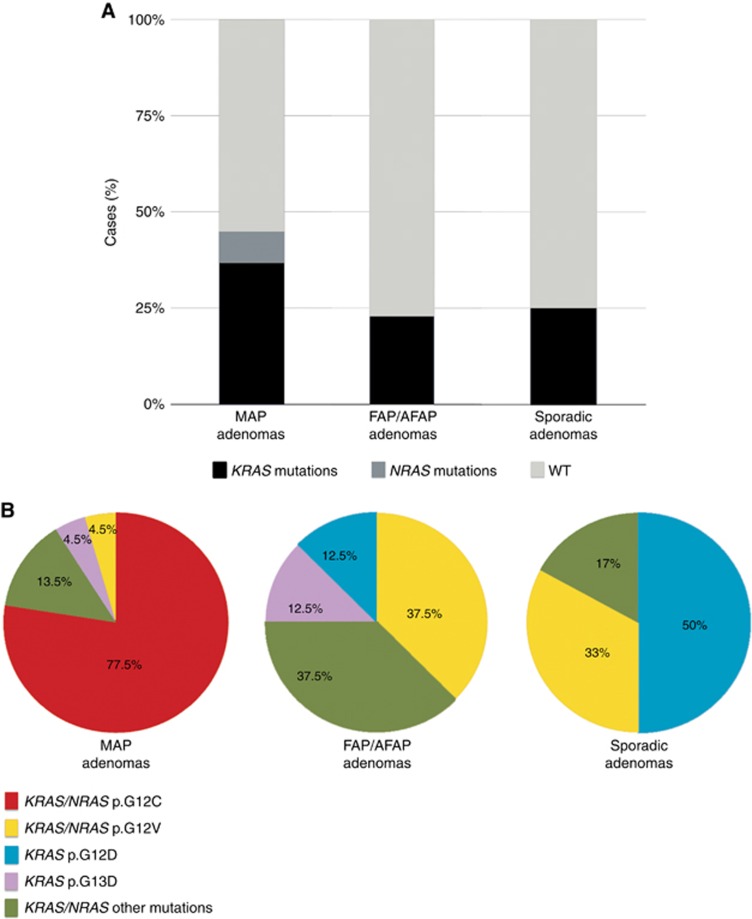
The cohort was preliminary characterised by the mutational status of the most frequently altered colorectal cancer genes, namely KRAS, NRAS, BRAF and PI3KCA. As expected for the KRAS gene (Venesio et al, 2013), alterations were more common in MAP adenomas (37%) compared with that in FAP/AFAP (23%) or sporadic (25%) polyps, whereas NRAS mutations were observed in only 8% of MAP adenomas and no mutations were found in BRAF and PI3KCA genes. Taking into consideration all the alterations, the frequency of MAP-mutated adenomas increased up to 45% (P=0.042 and P=0.1, with respect to FAP/AFAP and sporadic polyps) (Figure 1A and Supplementary Tables S1–S3).
Figure 1.
KRAS/NRAS mutations in the adenoma cohort.(A) Percentage of mutations in MAP FAP/AFAP and sporadic polyps; columns represent the mutation frequencies and KRAS/NRAS mutations or wild-type (WT) status are indicated in grayscale as reported as per the legend below. (B) Spectrum of KRAS/NRAS mutations in different sets of adenomas; the adenoma groups (MAP, FAP/AFAP and sporadic) are reported in pie charts and colours show the different types of KRAS/NRAS alterations as per the legend below. A full color version of this figure is available at the British Journal of Cancer journal online.
The spectrum of KRAS/NRAS mutations was different in the three sets of premalignant lesions: 77.5% of the MAP-mutated adenomas exhibited the c.34G>T transversion in KRAS or NRAS. In particular, this substitution was observed in 83% of KRAS mutations and 50% of NRAS mutations in MAP adenomas. By contrast, this mispair was totally absent in FAP/AFAP and sporadic groups (P<0.0001), which were both enriched for KRAS p.G12V and p.G12D mutations (Figure 1B and Supplementary Tables S1–S3).
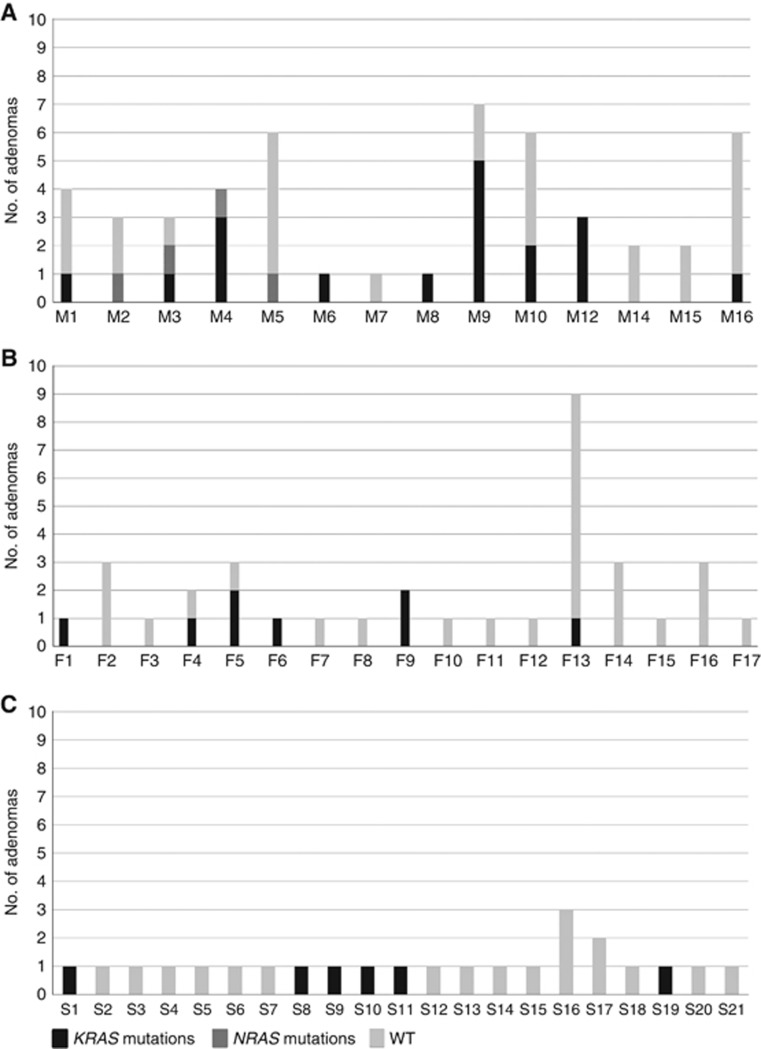
As for the mutational distribution, 79% of the MAP patients with adenomas showed at least one mutated polyp compared with 35% of the FAP/AFAP (P=0.029) and 29% of the sporadic cases (P=0.006) (Figure 2). In particular, 9 out of 14 MAP patients exhibited one or more KRAS/NRAS p.G12C-mutated lesions (Supplementary Table S1).
Figure 2.
(A) Mutational status of adenomas in MAP, (B) FAP/AFAP and (C) sporadic patients. Columns represent the number of analysed adenomas (y axis) for each patient (x axis), whereas types of mutations and wild-type condition are reported in grayscale as per the legend below. WT, wild type.
KRAS/NRAS alterations were found in both tubular and tubulovillous MAP adenomas. However, tubular polyps of this group were more frequently mutated than FAP/AFAP or sporadic adenomas with the same morphology (P=0.006 and P=0.035, respectively; Supplementary Figure S1).
To assess the role of KRAS/NRAS mutations in MAP progression, 10 CRCs from 8 MAP patients were also profiled for the status of the same genes analysed in adenomas (Supplementary Table S4). Mutations were detected in 90% of the examined cases and were mostly due to KRAS p.G12C substitution (89% of the mutated samples; Supplementary Figure S2). None of the carcinomas exhibited NRAS mutations, while five MAP patients (M1, M4, M6, M9 and M16) showed KRAS p.G12C in both adenomas and carcinomas. In addition, three CRCs (M1, M6 and M17) were mutated for KRAS p.G12C and PI3KCA (p.Q546K or p.R38H) (Supplementary Table S4).
DNA hypomethylation profiles
To assess a potential relationship between a compromised BER mechanism and a change in the DNA methylation of MAP lesions, we carried out LINE-1 and L1-MET assays on all the adenomas and MAP carcinomas using normal colonic mucosa samples as controls.
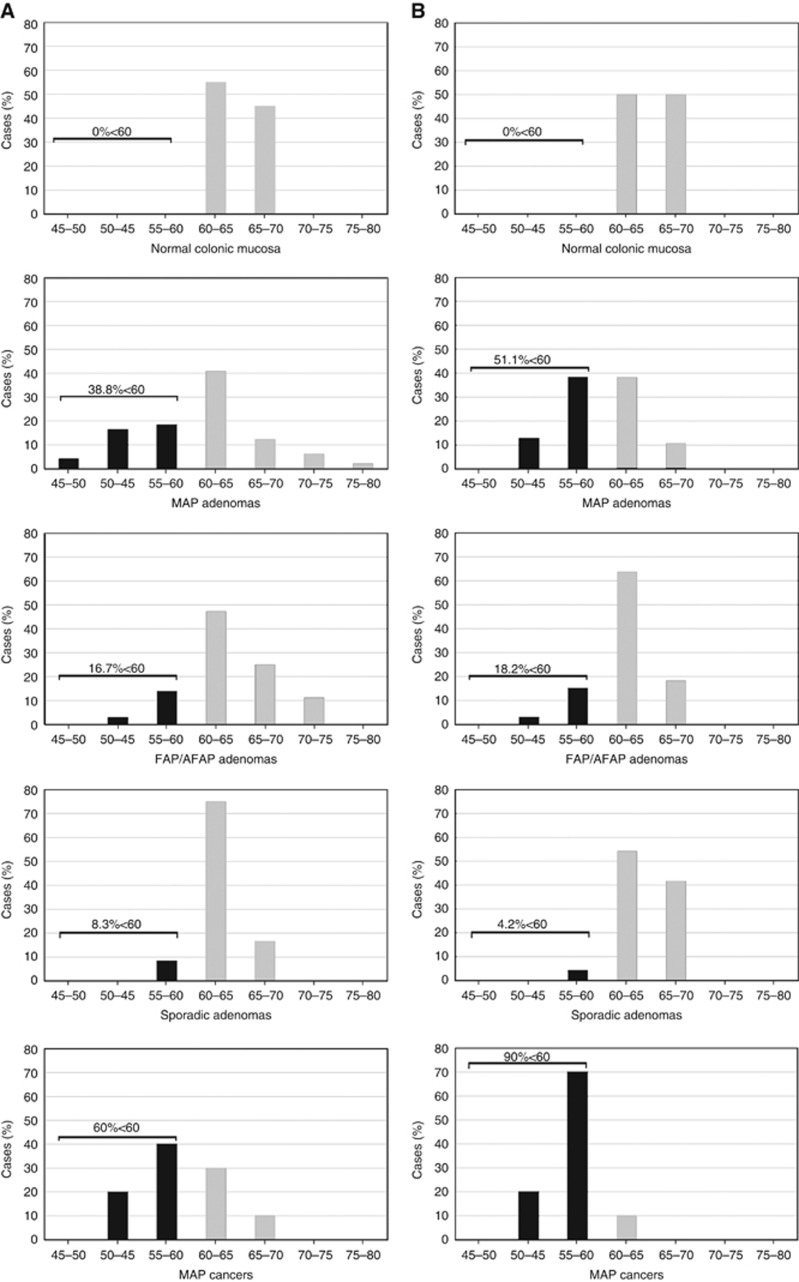
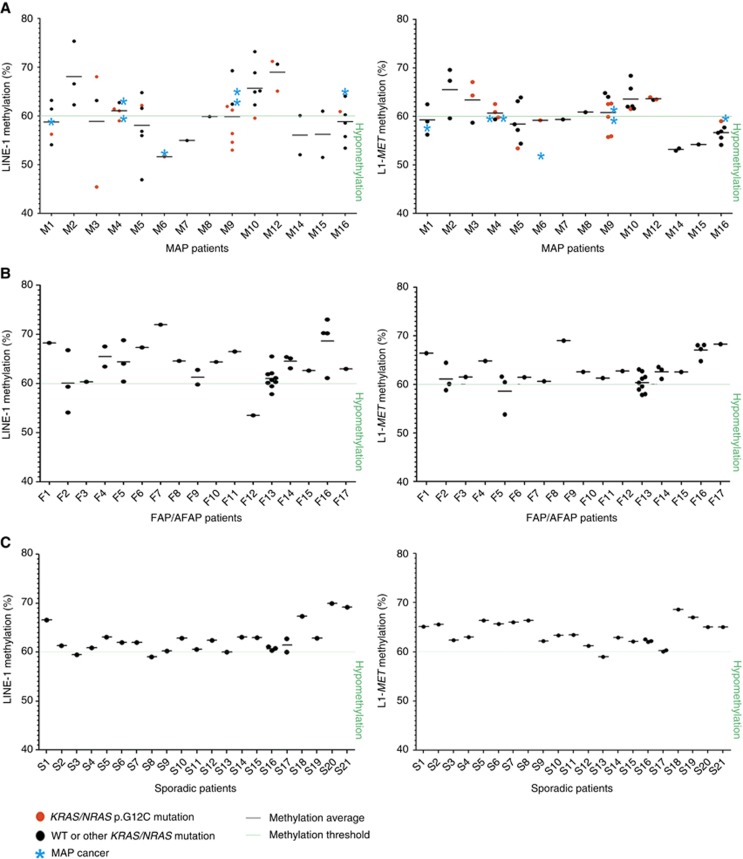
In line with previous reports (Baba et al, 2010; Sahnane et al, 2015), the percentage of LINE-1 methylation in normal mucosa ranged from 60% to 70% with both assays (Figure 3). By using the lowest methylation value of normal mucosa (60%) as the methylation threshold, MAP adenomas exhibited a significantly higher frequency of hypomethylated samples (38.8% and 51.1% of cases with LINE-1 and L1-MET analyses, respectively) compared with FAP/AFAP adenomas (16.7% and 18.2% of cases; P=0.032 and P=0.004, respectively) and to sporadic polyps (8.3% and 4.2% of samples; P=0.007 and P<0.0001, respectively) (Figure 3). A correlation between LINE-1 and L1-MET methylation levels was also assessed (r=0.6; Supplementary Figure S3). Considering the distribution of LINE-1 and L1-MET percentages in the adenomas of each patient, methylation variability was higher in MAP cases than in FAP/AFAP and sporadic patients (Figure 4). Notably, we found that at least one hypomethylated adenoma was detected in most (13 of 14) of the MAP patients with respect to FAP/AFAP (5 of 17) and sporadic (3 of 21) subjects. This finding was observed using the LINE-1 (86% of MAP vs 23.5% of FAP/AFAP and 9.5% of sporadic patients; P=0.001 and P<0.0001, respectively) and/or the L1-MET assay (79% of MAP vs 19% of FAP/AFAP and 5% of sporadic patients; P=0.003 and P<0.0001) (Figure 4). Moreover, in the cases with CRCs (M1, M4, M6, M9 and M16), the mean methylation level of adenomas was mostly comparable to that of each patient's tumour (P=0.4 for LINE-1 and P=0.5 for L1-MET; Figure 4).
Figure 3.
(A) LINE-1 and (B) L1-MET methylation percentages in normal colorectal mucosa, different adenoma groups (MAP, FAP/AFAP and sporadic) and MAP cancers; the methylation threshold was set at 60% as the lowest methylation value of normal mucosa for both LINE-1 and L1-MET; black columns represent hypomethylated samples (< 60%), while grey columns identify samples with a methylation level >60%.
Figure 4.
(A) Dot plots represent LINE-1 and L1-MET methylation percentages in adenomas of single MAP, (B) FAP/AFAP and (C) sporadic patients; black dots identify adenomas and red dots indicate KRAS/NRAS p.G12C-mutated polyps; the green line symbolises the methylation threshold (60%) for LINE-1 and L1-MET assays; bars show the mean values of LINE-1 or L1-MET methylation percentages among adenomas of the same patient and light blue asterisks show the methylation level of the corresponding carcinomas; symbols and colours are reported as per the legend below. WT, wild type. A full color version of this figure is available at the British Journal of Cancer journal online.
With regards to the histology, MAP adenomas with tubular architecture were more frequently hypomethylated than FAP/AFAP (P=0.1 for LINE-1 and P=0.02 for L1-MET analysis) or sporadic polyps with the same morphology (P=0.039 for LINE-1 and P=0.015 for L1-MET assays). No statistical association was found between the hypomehylation level and the site or the dysplasia grade of MAP adenomas.
Although the demethylation was not directly associated with the presence of KRAS/NRAS p.G12C mutations, we found the co-occurrence of mutations and hypomethylation in the same adenoma in several MAP patients (M1, M3, M4, M6, M9 and M10 for LINE-1; M4, M5, M6, M9 and M16 for L1-MET), and at least one mutated and/or one hypomethylated polyp was found in all cases (Figure 4).
Discussion
Oxidative DNA damage is considered a way to promote tumourigenesis and we chose to study colorectal adenomas derived from patients affected by a defective BER system as an appropriate model to investigate the molecular mechanisms underlying this type of carcinogenesis.
In this study, comparing adenomas from a novel cohort of MAP, FAP/AFAP and sporadic patients, we definitely confirmed that the type and frequency of MAPK gene mutations can substantially vary in different models of colorectal carcinogenesis; moreover, in line with previous reports (Jones et al., 2002; van Puijenbroek et al, 2008; Venesio et al, 2013; Aimé et al, 2015), we support the KRAS p.G12C mutation as the somatic hallmark of oxidative DNA damage in MAP disease.
In our cohort, the KRAS p.G12C mutation was by far the most frequent alteration in MAP polyps, and never detectable in comparable lesions from the other subsets. KRAS p.G12C was frequently identified in both adenomas and carcinomas derived from the same MAP patients, suggesting that the adenoma cell clones carrying this specific mutation are positively selected for the adenoma to carcinoma transition. As a consequence, the detection of these alterations in MAP adenomas could be used as a molecular marker for discriminating those patients at higher risk of developing colorectal cancer (Venesio et al, 2013; Rashid et al, 2015). Moreover, the recent development of KRAS p.G12C-specific inhibitors potentially opens new promising prospects for the treatment of patients affected by CRC with this somatic alteration (Ostrem et al, 2013; Patricelli et al, 2016).
Our findings reflect the well-known repair specificity of the MUTYH enzyme and the increased susceptibility to oxidative damage at the first base of the GGT sequence in KRAS codon 12. The peculiarity of the p.G12C mutation in this genomic region is further supported by the same transversion in the homologous BLAST-aligned nucleotide sequence of NRAS gene being detected in two of our MAP wild-type adenomas. So far, no other reports have documented the presence of NRAS mutations in MAP lesions. Although these alterations can be found in 10% of sporadic CRCs, they were shown to regulate homeostasis of colonic cells differently to KRAS alterations (Haigis et al, 2008) and were only occasionally detected in sporadic colorectal adenomas (Vagaja et al, 2015).
As reported by Rashid et al, 2015, we detected a two-fold higher level of somatic mutations in MAP adenomas with respect to FAP/AFAP or sporadic polyps, suggesting that a deficiency of MUTYH could lead to a mutator phenotype. This evidence is supported by the co-occurrence of both KRAS p.G12C and PI3KCA mutations in more than one-third of the examined MAP carcinomas and is consistent with a higher colorectal cancer risk in MAP, even in the presence of a few adenomas (Nieuwenhuis et al, 2012).
Regarding phenotype, KRAS/NRAS mutations were more frequent in MAP adenomas with tubular architecture than in the controls with the same morphology. This can be another characteristic of MAP carcinogenesis since in a survey concerning colorectal sporadic adenomas, KRAS mutations were mainly evidenced in tubulovillous and villous adenomas (Yadamsuren et al, 2012).
Interestingly, MAP adenomas of our cohort were more often hypomethylated than the other groups, for both global genomic and intragenic sequences, as measured by LINE-1 and L1-MET. Although MAP adenomas were heterogeneous for methylation levels of LINE-1 and L1-MET, their mean percentages were generally comparable to the level of the CRCs derived from the same patients, suggesting that DNA demethylation is acquired early and retained in the adenoma–carcinoma transition. Our results are in agreement with studies reporting that DNA global hypomethylation is an early event in colorectal and gastric carcinogenesis (Suter et al, 2004; Kim et al, 2016). For the first time, we demonstrated that significant lower LINE-1 methylation levels may be observed in specific models of colorectal adenomas and that a novel link between a BER defect and DNA hypomethylation may be established in CRC tumourigenesis.
The mechanisms of how oxidative stress decreases DNA methylation have been proposed (Franco et al, 2008). Oxidised DNA lesions that are induced by ROS, such as 8-OHdG in CpG dinucleotides, have been shown to strongly inhibit methylation of adjacent cytosine residues (Weitzman et al, 1994). Moreover, in an oxidative stress condition, with decreased availability of S-adenosylmethionine, depletion of the methyl pool in folate-deficient models has been shown to cause DNA hypomethylation (Miller et al, 1994). Additionally, an unfixed 8-OHdG may introduce a G>T transversion resulting in the loss of CpG dinucleotides (Kuchino et al, 1987). Recently, Kloypan et al (2015) demonstrated that in colorectal cancer LINE-1 hypomethylation is associated with the oxidative stress-mediated activation of the ten-eleven translocation hydroxylase enzymes that cooperate with BER in controlling the formation and replacement of 5-hydroxymethylcytosine (5 hmC). Finally, ROS are considered to be responsible for aberrant DNA methylation in different cancer models (Campos et al, 2007; Lim et al, 2008; Quan et al, 2011; Ziech et al, 2011; Kloypan et al, 2015) and during chronic inflammation and inflammation-associated carcinogenesis, such as ulcerative colitis and Crohn's disease (Iborra et al, 2011; Hartnett and Egan, 2012). All these data suggest that increased oxidative DNA damage and/or lack of DNA repair are strongly correlated not only with genetic but also with aberrant DNA methylation in cancer.
Our study demonstrates that LINE-1/L1-MET hypomethylation is not associated with the presence of KRAS p.G12C or any other mutation. Similarly to KRAS/NRAS mutations, also DNA hypomethylation was more frequent in MAP adenomas with tubular architecture than in FAP/AFAP or sporadic polyps with the same morphology. This observation reinforces the idea that both epigenetic and genetic alterations occur very early in MAP tumourigenesis and is consistent with previous data reported by Cardoso et al (2006), demonstrating that low-grade dysplasia adenomas show higher incidence of aneuploidy in MAP compared with FAP polyps. LINE-1 hypomethylation is suggested to be a key event in cancer development because it results in retrotransposition throughout the genome leading to CIN (Yamada et al, 2005; Rodriguez et al, 2006; Ewing et al, 2015). Moreover, an activated LINE-1 promoter can initiate sense or antisense transcription through either proto-oncogenes or tumour suppressor genes having the potential to induce the illegitimate transcription of the LINE-1 chimeric product (Weber et al, 2010; Wolff et al, 2010; Hur et al, 2014; Zhu et al, 2014). Hypomethylation of the LINE-1 inserted in the second intron of the MET gene has been shown to drive expression of a truncated isoform of the gene with a negative prognostic value in CRC (Hur et al, 2014). In this work, we provide the first evidence for the high frequency of L1-MET hypomethylation in MAP adenomas.
However, to establish DNA hypomethylation as a potential specific molecular marker of MAP carcinogenesis, a higher number of hereditary and sporadic adenomas should be analysed for their epigenetic status. This could deal with the drawback of comparing adenomas from patients who genetically developed polyps at different ages. In this regard, although global DNA hypomethylation has been reported associated with age-related diseases, colorectal cancer-specific changing in LINE-1 methylation was shown not to be affected by aging (Bollati et al, 2009; Baba et al, 2010). In addition, future studies using genome-wide methylation profiling as well as in vitro MAP cell modelling might be useful to demonstrate that repair-based mechanisms are involved in physiological models of DNA demethylation.
In conclusion, our data indicate that, in addition to well-assessed specific mutations, the early steps of oxidative DNA damage-induced colorectal carcinogenesis are characterised by decreased DNA global methylation and specific L1-MET hypomethylation. Finally, our results emphasise the idea that genetic and epigenetic mechanisms strengthen each other in driving colorectal tumourigenesis. Because of the clinical recommendations for recently established aspirin-based chemoprevention strategies (Burn et al, 2011), these results appear to be interesting in improving the identification of individuals who are most likely to benefit from a prophylactic aspirin regimen.
Acknowledgments
This study was supported by the Epigenomics Flagship Project – EPIGEN (to D Furlan) (Project No. 08934412).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Aimé A, Coulet F, Lefevre JH, Colas C, Cervera P, Flejou JF, Lascols O, Soubrier F, Parc Y (2015) Somatic c.34G>T KRAS mutation: a new prescreening test for MUTYH-associated polyposis? Cancer Genet 208: 390–395. [DOI] [PubMed] [Google Scholar]
- Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP (2002) Inherited variants of MYH associated with somatic G:C—>T:A mutations in colorectal tumors. Nat Genet 30: 227–232. [DOI] [PubMed] [Google Scholar]
- Antelo M, Balaguer F, Shia J, Shen Y, Hur K, Moreira L, Cuatrecasas M, Bujanda L, Giraldez MD, Takahashi M, Cabanne A, Barugel ME, Arnold M, Roca EL, Andreu M, Castellvi-Bel S, Llor X, Jover R, Castells A, Boland CR, Goel A (2012) A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS One 7: e45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, Schernhammer ES, Hunter DJ, Giovannucci EL, Fuchs CS, Ogino S (2010) Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumours. Mol Cancer 9: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A (2009) Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 130: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn J, Gerdes AM, Macrae F, Mecklin JP, Moeslein G, Olschwang S, Eccles D, Evans DG, Maher ER, Bertario L, Bisgaard ML, Dunlop MG, Ho JW, Hodgson SV, Lindblom A, Lubinski J, Morrison PJ, Murday V, Ramesar R, Side L, Scott RJ, Thomas HJ, Vasen HF, Barker G, Crawford G, Elliott F, Movahedi M, Pylvanainen K, Wijnen JT, Fodde R, Lynch HT, Mathers JC, Bishop DT CAPP2 Investigators (2011) Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet 378: 2081–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AC, Molognoni F, Melo FH, Galdieri LC, Carneiro CR, D'Almeida V, Correa M, Jasiulionis MG (2007) Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia 9: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso J, Molenaar L, de Menezes RX, van Leerdam M, Rosenberg C, Möslein G, Sampson J, Morreau H, Boer JM, Fodde R (2006) Chromosomal instability in MYH- and APC-mutant adenomatous polyps. Cancer Res 66: 2514–2519. [DOI] [PubMed] [Google Scholar]
- Ewing AD, Gacita A, Wood LD, Ma F, Xing D, Kim MS, Manda SS, Abril G, Pereira G, Makohon-Moore A, Looijenga LH, Gillis AJ, Hruban RH, Anders RA, Romans KE, Pandey A, Iacobuzio-Donahue CA, Vogelstein B, Kinzler KW, Kazazian HH Jr, Solyom S (2015) Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res 25: 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R, Schoneveld O, Georgakilas AG, Panayiotidis MI (2008) Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 266: 6–11. [DOI] [PubMed] [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, Settleman J, Giovannini M, Jacks T (2008) Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and TUMOUR progression in the colon. Nat Genet 40: 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SR BF, Boffetta P, Ilyas M, Morreau H, Nakamura S I, Quirke P, Riboli E, Sobin LH (2010) WHO Classification of Tumours of the Digestive System, Chapter 8 Tumours of the Colon and Rectum. IARC Press: Lyon, France. [Google Scholar]
- Hartnett L, Egan LJ (2012) Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis 33: 723–731. [DOI] [PubMed] [Google Scholar]
- Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A (2014) Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 63: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra M, Moret I, Rausell F, Bastida G, Aguas M, Cerrillo E, Nos P, Beltrán B (2011) Role of oxidative stress and antioxidant enzymes in Crohn's disease. Biochem Soc Trans 39: 1102–1106. [DOI] [PubMed] [Google Scholar]
- Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP (2002) Biallelic germline mutations in MYH predispose to multiple colorectaladenoma and somatic G:C—>T:A mutations. Hum Mol Genet 11: 2961–2967. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Chung WC, Kim DB, Kim Y, Lee JM, Jung JH, Lee YK (2016) Long interspersed nuclear element (LINE)-1 methylation level as a molecular marker of early gastric cancer. Dig Liver Dis 48: 1093–1097. [DOI] [PubMed] [Google Scholar]
- King WD, Ashbury JE, Taylor SA, Yat Tse M, Pang SC, Louw JA, Vanner SJ (2014) A cross-sectional study of global DNA methylation and risk of colorectal adenoma. BMC Cancer 14: 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitkumthorn N, Mutirangura A (2011) Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenet 2: 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloypan C, Srisa-art M, Mutirangura A, Boonla C (2015) LINE-1 hypomethylation induced by reactive oxygen species is mediated via depletion of S-adenosylmethionine. Cell Biochem Funct 33: 375–385. [DOI] [PubMed] [Google Scholar]
- Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S (1987) Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 327: 77–79. [DOI] [PubMed] [Google Scholar]
- Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, Cho JW, Park YM, Jung G (2008) Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology 135: 2128–2140, , 2140e1–e8. [DOI] [PubMed] [Google Scholar]
- Miller JW, Nadeau MR, Smith J, Smith D, Selhub J (1994) Folate-deficiencyinduced homocysteinaemia in rats: disruption of S-adenosylmethionine's coordinate regulation of homocysteine metabolism. Biochem J 298: 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C (2009) Active DNA demethylation and DNA repair. Differentiation 77: 1–11. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis MH, Vogt S, Jones N, Nielsen M, Hes FJ, Sampson JR, Aretz S, Vasen HF (2012) Evidence for accelerated colorectal adenoma–carcinoma progression in MUTYH-associated polyposis? Gut 61: 734–738. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS (2008) LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer 122: 2767–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y (2000) Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res 28: 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KV (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503: 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, Chen JH, Firdaus SJ, Babbar A, Ren P, Liu Y (2016) Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov 6: 316–329. [DOI] [PubMed] [Google Scholar]
- Quan X, Lim SO, Jung G (2011) Reactive oxygen species downregulate catalase expression via methylation of a CpG island in the Oct-1 promoter. FEBS Lett 585: 3436–3441. [DOI] [PubMed] [Google Scholar]
- Rashid M, Fischer A, Wilson CH, Tiffen J, Rust AG, Stevens P, Idziaszczyk S, Julie Maynard J, Williams GT, Mustonen V, Sampson JR, Adams DJ (2015) Adenoma development in familial adenomatous polyposis and MUTYH-associated polyposis: somatic landscape and driver genes. J Pathol 238: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capellà G, Ribas M, Peinado MA (2006) Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res 66: 8462–9468. [DOI] [PubMed] [Google Scholar]
- Sahnane N, Magnoli F, Bernasconi B, Tibiletti MG, Romualdi C, Pedroni M, Ponz de Leon M, Magnani G, Reggiani-Bonetti L, Bertario L, Signoroni S, Capella C, Sessa F, Furlan D (2015) Aberrant DNA methylation profiles of inherited and sporadic colorectal cancer. Clin Epigenet 7: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanoli M, La Rosa S, Sahnane N, Romualdi C, Pastorino R, Marando A, Capella C, Sessa F, Furlan D (2014) Prognostic relevance of aberrant DNA methylation in g1 and g2 pancreatic neuroendocrine tumours. Neuroendocrinology 100: 26–34. [DOI] [PubMed] [Google Scholar]
- Sunami E, de Maat M, Vu A, Turner RR, Hoonet DSB (2011) LINE-1 hypomethylation during primary colon cancer progression. PLoS One 6: e18884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL (2004) Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis 19: 95–101. [DOI] [PubMed] [Google Scholar]
- Vagaja NN, Parry J, McCallum D, Thomas MA, Bentel JM (2015) Are all RAS mutations the same? Coexisting KRAS and NRAS mutations in a caecal adenocarcinoma and contiguous tubulovillous adenoma. J Clin Pathol 68: 657–660. [DOI] [PubMed] [Google Scholar]
- van Puijenbroek M, Nielsen M, Tops CM, Halfwerk H, Vasen HF, Weiss MM, van Wezel T, Hes FJ, Morreau H (2008) Identification of patients with (atypical) MUTYH-associated polyposis by KRAS2 c.34G>T prescreening followed by MUTYH hotspot analysis in formalin-fixed paraffin-embedded tissue. Clin Cancer Res 14: 139–142. [DOI] [PubMed] [Google Scholar]
- Venesio T, Balsamo A, Errichiello E, Ranzani GN, Risio M (2013) Oxidative DNA damage drives carcinogenesis in MUTYH-associated-polyposis by specific mutations of mitochondrial and MAPK genes. Mod Pathol 26: 1371–1381. [DOI] [PubMed] [Google Scholar]
- Weber B, Kimhi S, Howard G, Eden A, Lyko F (2010) Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29: 5775–5784. [DOI] [PubMed] [Google Scholar]
- Weitzman SA, Turk PW, Milkowski DH, Kozlowski K (1994) Free radical adducts induce alterations in DNA cytosine methylation. Proc Natl Acad Sci USA 91: 1261–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EM., Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G (2010) Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet 6: e1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadamsuren EA, Nagy S, Pajor L, Lacza A, Bogner B (2012) Characteristics of advanced- and non advanced sporadic polypoid colorectal adenomas: correlation to KRAS mutations. Pathol Oncol Res 18: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R (2005) Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA 102: 13580–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JPJ (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Utsunomiya T, Ikemoto T, Yamada S, Morine Y, Imura S, Arakawa Y, Takasu C, Ishikawa D, Imoto I, Shimada M (2014) Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann Surg Oncol 21: S729–S735. [DOI] [PubMed] [Google Scholar]
- Ziech D, Franco R, Pappa A, Panayiotidis MI (2011) Reactive oxygen species (ROS)-induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res 711: 167–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.