Abstract
Copper isotopic composition is altered in cancerous compared to healthy tissues. However, the rationale for this difference is yet unknown. As a model of Cu isotopic fractionation, we monitored Cu uptake in Saccharomyces cerevisiae, whose Cu import is similar to human. Wild type cells are enriched in 63Cu relative to 65Cu. Likewise, 63Cu isotope enrichment in cells without high-affinity Cu transporters is of slightly lower magnitude. In cells with compromised Cu reductase activity, however, no isotope fractionation is observed and when Cu is provided solely in reduced form for this strain, copper is enriched in 63Cu like in the case of the wild type. Our results demonstrate that Cu isotope fractionation is generated by membrane importers and that its amplitude is modulated by Cu reduction. Based on ab initio calculations, we propose that the fractionation may be due to Cu binding with sulfur-rich amino acids: methionine and cysteine. In hepatocellular carcinoma (HCC), lower expression of the STEAP3 copper reductase and heavy Cu isotope enrichment have been reported for the tumor mass, relative to the surrounding tissue. Our study suggests that copper isotope fractionation observed in HCC could be due to lower reductase activity in the tumor.
As other transition metals, copper (Cu) is an essential micronutrient for living organisms, but can become toxic if in excess. At low concentrations, Cu is essential for the activity of proteins such as cytochrome c oxidase, which is involved in mitochondrial respiration. At high concentrations, Cu generates reactive oxygen species that are highly toxic1,2. Therefore, living organisms have evolved regulatory mechanisms to control the cellular speciation and content of this metal and avoid its toxicity3. Transmembrane proteins allow controlled cellular import and export, and Cu is transported between different organs tightly bound to organic molecules. In contrast, many cancer types exhibit a dysregulation of cellular Cu homeostasis4,5. Several eukaryotes seem to react similarly to some cancers: Cu concentration in the blood-serum of healthy mice or humans is lower than that of tumor-bearing mice or human patients affected by breast, lung, or colon cancers5, respectively. A way of better understanding dysregulation of transmembrane Cu transport is to use Cu isotopes. Indeed, in a preliminary study, Fosset et al.6 used copper enriched in 65Cu for Cu fluxes tracing in cells. Instead natural variability of 65Cu/63Cu along biochemical pathways, a method similar to that has been used to trace fluxes of various elements through various geological reservoirs7, has been used to trace fluxes in living cells or among organs. It has been found that the 65Cu/63Cu ratio (reported as δ65Cu, i.e. the relative enrichment of 65Cu relative to a reference material: δ65Cu = 1000 × [(65Cu/63Cu)sample/(65Cu/63Cu)standard − 1]) in blood serum decreases with cancer progression8. The red blood cells δ65Cu values were found to be lower for patients with hepatocellular carcinoma (HCC) relative to healthy subjects, and accompanied by an increase of δ65Cu in the tumor mass compared to peritumor tissue9. To date, the explanations of these isotopic variations observed between healthy and HCC patients remain unclear10. Because these variations involve affinity interactions or changes of valence, several steps of Cu transport and translocation between organs, such as Cu reduction, Cu transport through the membrane or Cu fixation on organic ligands, have the potential to generate fractionation. Interestingly, it has been demonstrated that during the development of some cancers, there is a dysregulation of the expression of Cu importers11 and/or of reductases12,13 in cancerous cells. Here we explore the possibility that the dysregulation of Cu homeostasis is responsible for the modification of the cellular Cu isotopic composition.
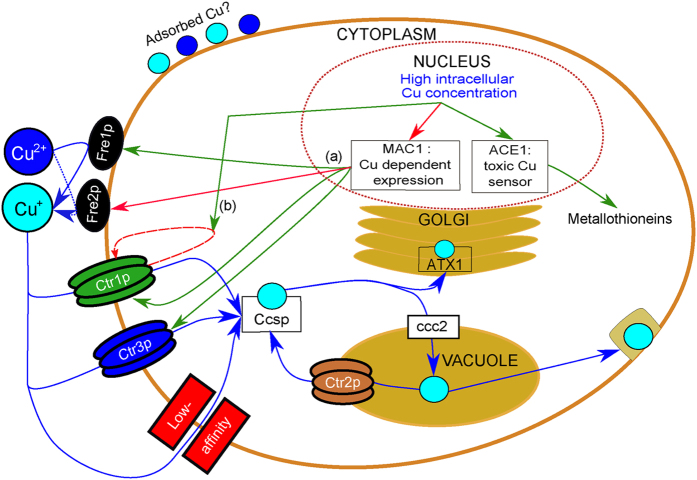
To test this hypothesis, we used the yeast model-organism Saccharomyces cerevisiae, a long-recognized genetic model for Cu metabolism in eukaryotes, which transmembrane Cu transport mechanisms (Fig. 1) are very similar to those of human cells (review by Nevitt et al.3), and which is prone to genetic manipulation. In S. cerevisiae, Cu is first reduced by reductases such as the Fre1 and Fre2 proteins (Fre1p and Fre2p)14,15. Reduced Cu enters the cell through two high-affinity importers Ctr1p16 and Ctr3p17 as well as through low-affinity transporters such as Fet proteins18. The expression of the FRE1, FRE2, CTR1 and CTR3 genes is controlled by the Mac1p transcription factor19. Under low Cu, Mac1p induces the expression of both FRE1 and the high-affinity Cu transporters. Under high Cu conditions, the expression of the MAC1 gene and the activity of Mac1p is inhibited, leading to the lower expression of CTR1, CTR3, and FRE114,20,21,22. CTR1 is further regulated at the protein level through endocytosis and vacuolar degradation23,24,25. Here, we have monitored Cu uptake and isotopic composition of S. cerevisiae mutants, with deletions of high-affinity transporters or impaired Cu reductase activity, in order to identify and quantify the isotopic fractionation associated which each step of the transport process. Our study demonstrates that copper uptake leads to isotopic fractionation between cells and growth-medium, and that this fractionation is generated by Cu importers and modulated by Cu reductases.
Figure 1. Copper pathways in Saccharomyces cerevisiae.
Copper fluxes are represented by blue arrows. Regulatory loops are indicated in red (activation) and green (repression). Monovalent and divalent Cu are represented by dark blue and light blue circles, respectively. The first step of Cu import is the reduction of Cu(II) to Cu(I) by the Fre reductases, followed by the import through either high-affinity transporters represented in green (Ctr1p) and blue (Ctr3p) or low-affinity transporters such as Fet proteins (red). Copper is subsequently transported in the different compartments of the cell cytoplasm by different protein chaperones such as Ccsp, ATX1 or ccc2. Copper transport is regulated by MAC1 which senses high intracellular Cu concentrations and down regulates the expression of the high-affinity transporters CTR1 and CTR3 and of the reductase FRE1, while it activates the expression of the FRE2 reductase3,14,15,16,17,18,21,22,23,24,25,52.
Materials and Methods
Saccharomyces cerevisiae genotypes and culture conditions
Four strains of S. cerevisiae were used in this study: a wild type (DTY165), a Δctr1Δctr3 mutant (MPY17), a Δctr1Δmac1 mutant (SKY34) and a SKY34 strain where FRE1 was expressed under the constitutive promoter of ADH (alcohol dehydrogenase). See detailed genotypes in Table 1. DTY165, MPY17, and SKY34 strains were kindly provided by the Thiele Laboratory (Duke University Medical Center, USA). S. cerevisiae cells were routinely maintained in Yeast-extract Peptone Dextrose (YPD) medium, which contains low concentrations of Cu (0.25 μmol/l). For our experiments, we used the Yeast Nitrogen Base (YNB) growth medium which contains 0.15 μmol/l of Cu.
Table 1. Genotype of the four studied S. cerevisiae strains.
| Strain | Cu importers |
Cu transcription factor | Cu reductases | activity | Genotype | Reference | |
|---|---|---|---|---|---|---|---|
| Ctr1 | Ctr3 | MAC1 | FRE 1 promoter | ||||
 |
(+) | (+) | (+) | wt | +++ | MATα ura3-52 his6 leu2-3,-112 his3-∆200 trp1-901 lys2-801 suc2-∆ | 51 |
 |
(−) | (−) | (+) | wt | +++ | MATα ctr1::ura3::Kanr ctr3::TRP1 his3 lys2-801 CUP1r | 47 |
 |
(−) | (+) | (−) | wt | + | MATα gal1 trp1-1 his3 ade8 CUP1r ctr1::TRP1 mac1 | 47 |
 |
(−) | (+) | (−) | ADH | ++ | MATα gal1 trp1-1 his3 ade8 CUP1r ctr1::TRP1 mac1 pFRE1∆::Kanr-pADH | This study |
(−) knockout, (+) expressed gene.
The relative Cu reductase activity is given by the number of ‘+’ in the 6th column (“activity”). Colors used in this table for each strain are the ones used in the figures.
Genetic manipulations
Expression of FRE1 was re-established in strain SKY34, by substituting the endogenous FRE1 promoter (Mac1p dependent) with the ADH promoter. The PCR product obtained using the plasmid pYM-N6 26 and the following primers (forward: CTAATTTCTCGCATATTCACGCCGACGGAAGAACGAGCCGGATCAATATGCGTACGCTGCAGGTCGAC; reverse: ACCGTAGCAAAAAAAGATATAAATAAGCAGAATAATACACGGGTTCTAACCATCGATGAATTCTCTGTCG) was transformed into SKY34 cells followed by selection on YPD-agar plates containing 200 mg/l of G418 (Gibco). Successful promoter substitution was confirmed by PCR screen using the forward primer AACCCAAACATTTTCGCCGA (upstream the recombination region) and the reverse primer KanB1 TGTACGGGCGACAGTCACAT (annealing within the resistance cassette).
Cell quantification
Cells were quantified by three different approaches. First, cell growth was monitored by measuring the culture optical density (OD) at 600 nm. Second, cells were counted directly using a Thoma cell. Third, in order to normalize the activity of transporters, total cell protein content was measured using a procedure adapted from Zhang et al.27 and Kushnirov et al.28.
Uptake and adsorption experiments
Uptake experiments were started from pre-cultures grown in YPD at 30 °C with 160 rpm shaking and harvested in late exponential phase. Cells were inoculated at a final concentration of 108 cell per ml in fresh 500 ml of YNB containing 2% D-Glucose. The inoculated cultures were doped with 80 μmol/l of Cu (δ65Cu = −0.23 ± 0.06‰, 2σ, N = 33) at the start of the uptake experiment (t = 0) and incubated at 25 °C with shaking. At each time point, 1 ml of the culture was sampled for cell quantification and 50 ml for Cu elemental and isotopic analyses. Cells were pelleted by centrifugation at 10000 rpm and 2 °C. Cells were washed once with YNB and centrifuged. The cell pellet was then processed for metal analysis. For each time point, 1 ml of the culture supernatant was sampled to measure the concentration and isotopic composition of the culture medium. To test the impact of the Cu oxidation state on its import, similar uptake experiments were performed in presence of 1 mM of ascorbic acid, a strong reducing agent.
Cu adsorption on S. cerevisiae cells was monitored as described previously29 using the experimental conditions described above for uptake experiments, except that cells were incubated on ice to inhibit biological activity, e.g. copper uptake by cells.
Preliminary experiments conducted during the development of our experimental settings have shown that the Cu concentration in S. cerevisiae cells did not vary significantly between ca. 120–180 min and 240 min and that the number of cells stayed stable between 0–240 min. Some cellular division could occur after ca. 260 min hence possibly modifying the Cu concentration in S. cerevisiae cells. Thus, to avoid the effect of cell division on Cu concentration, and a potential impact on Cu isotopic composition, the uptake experiments were limited to 0–240 min. The observation of a plateau for Cu concentration reached after 120–180 min, depending on the strain, is in good agreement with previous studies18. Two or three independent experiments, hereafter referred to as “replicate experiments”, were performed, with the exception of experiments involving SKY FRE (see Table S1 for details).
Quantification of cellular Cu reduction activity
Cu reduction was measured using a reductase assay modified from Georgatsou et al.14,30. Briefly, S. cerevisiae cells grown as described above were harvested in the late exponential phase. Cells were inoculated at a concentration of 109 cells/ml of YNB supplemented with 5% D-Glucose, 2 mM bathocuproine disulfonic acid (BCS) - a Cu chelator that becomes red in the presence of Cu(I) -, and 1 mM CuSO4, and incubated with shaking at 25 °C for 40 min. Since BCS-Cu complexes have a stoichiometry of 2:131, our assay contains twice as much BCS as Cu. Reduced Cu was quantified by measuring the optical density at 482 nm14.
Chemical separation and purification procedures
Cell samples were dissolved following a procedure adapted from the study of Moynier et al. on plants32. After weighing, samples were placed in 90 ml PTFE jars and dissolved by sequential addition of concentrated HNO3, then a mixture of concentrated HNO3:H2O2 30% (50:50, v-v). After evaporation, samples were placed in H2O2 30% under UV light for two hours at room temperature. The photolysis induced by UV light destroys organic matter33, which improves the accuracy of the subsequent metal concentration and isotope analyses.
Cellular Cu was separated and purified by anion-exchange chromatography following the procedure described in Maréchal et al.34. Briefly, the sample was loaded on 1.6 ml of macroporous anion-exchange resin (AG-MP1, 100–200 mesh, Bio-Rad) in 1 ml of 7 N HCl - H2O2 0.001%. The matrix was then eluted in 10 ml 7 N HCl - H2O2 0.001%, finally Cu was eluted in 20 ml 7 N HCl - H2O2 0.001%. This process was repeated once to ensure a complete purification of Cu from the matrix. The whole procedure blank was 8 ng, which is well below the amount of Cu in S. cerevisiae samples (>300 ng).
For the growth medium, the separation procedure had to be adapted because of the high organic content of the medium. Briefly, the first purification step was performed with a resin (1.6 ml of AG-MP1, 100–200 mesh, Bio-Rad) conditioned with 20 ml of a mixture of 80% EtOH (96%): 20% (7 N HCl- H2O2 0.001%) (v:v), then the sample was loaded in 1 ml of 7 N HCl-H2O2 0.001%. The matrix was eluted in 50 ml of 80% EtOH (96%): 20% (7 N HCl- H2O2 0.001%) (v:v) followed by the elution of Cu in 30 ml of 7 N HCl-H2O2 0.001%. A second purification step was performed as described above for S. cerevisiae samples.
Elemental and isotopic Cu measurements
Cu concentrations were measured by Quadrupole Inductively-Coupled Plasma Mass-Spectrometry (Q-ICP-MS, Agilent 7500cx) and Cu stable isotope compositions by Multi-Collector Inductively-Coupled Plasma Mass-Spectrometry (MC-ICP-MS, Nu500 HR, Nu Instrument) in wet plasma mode. For this purpose, 50 ml aliquots were taken from the culture medium at ca. 108 cells/ml for each time point (see details in the “Uptake and adsorption experiments” subsection). The instrumental mass fractionation was corrected for using Zn-doping and standard-sample bracketing following the procedure developed by Maréchal et al.34. Samples were diluted to match the concentration of the standard mixture (typically Zn 0.3 ppm - Cu 0.3 ppm). The mass-dependence of isotope fractionation has been systematically checked. The δ65Cu is reported relative to the isotopic solution reference material NIST SRM 976. Measurements were repeated several times over the course of independent MC-ICP-MS sessions (see Supplementary Table S1 for details), hereafter referred to as “replicate measurements”. The long-term external reproducibility was assessed by measuring (1) aliquots of the same S. cerevisiae sample (Supplementary Table S2) in four runs over the course of three months: δ65Cu = −1.47 ± 0.04‰ (2σ, N = 8), and (2) the Cu solution used for S. cerevisiae cultures doping, which was analyzed for each batch of cultures: δ65Cu = −0.23 ± 0.06‰ (2σ, N = 33). Thus, we define the external error for our measurements at 0.06‰ (2σ).
Results
Effect of Cu uptake on Cu isotopes fractionation by S. cerevisiae cells
During uptake experiments, the Cu concentration in the wild type strain (DTY165) and the high-affinity importer knockout strain (MPY17) increased quickly and reached a plateau after approximately 120 minutes (Fig. 2). It is worth noting that a Cu concentration plateau reached after ca. 120 min is in agreement with previous studies conducted on Cu uptake by S. cerevisiae18. It can be considered that this time signals the compensation of Cu import by Cu export. We found that the plateau is remarkably high, reaching 16.6 pmol per 106 cells, in MPY17 - the strain lacking high-affinity importers – relative to DTY165 (9.7 pmol per 106 cells) (Fig. 2).
Figure 2. Net variations of the Cu concentration and isotopic composition in S. cerevisiae for the wild-type (green) and the Δctr1Δctr3 double-knockout strain (red).
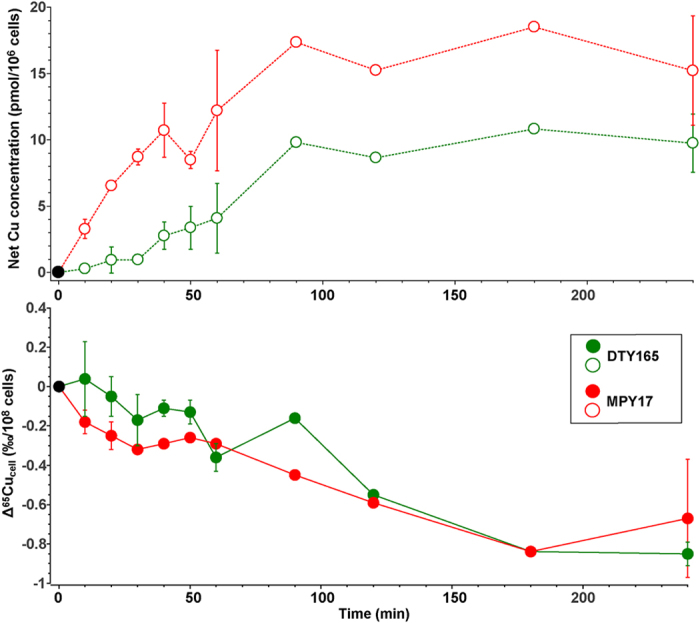
Upper panel: variations of the Cu concentration over 240 min. during uptake experiments. Values are reported as the difference Cu(t) − Cu(t = 0) normalized to 108 cells (see text for details). Lower panel: variations of the Cu isotopic composition (reported as Δ65Cucell = δ65Cu(t) − δ65Cu(t = 0), see text for details) normalized to 108 cells. The error bars are the standard deviation between independent experiments (see Table S1 for details).
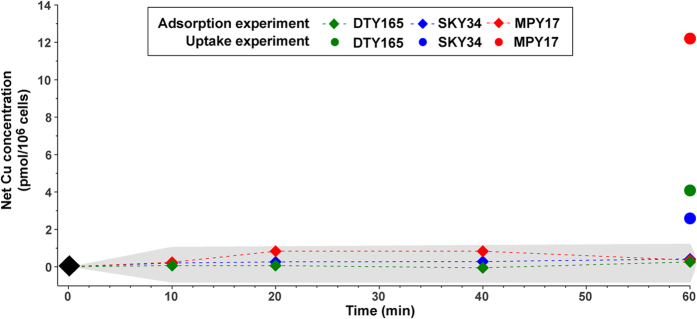
The contribution of Cu adsorption on Cu content in S. cerevisiae cells was evaluated by uptake experiments on ice, which limits active import. Our results show that adsorption on S. cerevisiae cells is very limited and does not vary with time (Fig. 3) and therefore does not contribute significantly to the cellular Cu content.
Figure 3. Cu adsorption on S. cerevisiae cells (diamonds) over 60 min.
Cell contents (circles) at 60 min in uptake experiments are indicated for comparison. The grey range represents the analytical uncertainty on the measurement.
The experimental conditions were designed so that Cu uptake by cells was negligible relative to the metal stock in the growth medium. Concentration in the medium throughout the experiments remained indeed stable, within analytical error ( ± 5%), around the initial concentration (Supplementary Fig. S1). We also confirmed that Cu uptake by cells did not impact the isotopic composition of the medium (Supplementary Fig. S1). The Cu stock in the growth medium can therefore be considered infinite relative to S. cerevisiae uptake. Its isotopic composition (δ65Cumedium) at any given time is equal to that of the Cu doping solution (δ65Cu = −0.23‰ ± 0.06‰, 2σ, N = 33). The cellular isotopic composition relative to the growth medium at time t can be defined as Δ65Cu (t) = δ65Cucell (t) − δ65Cumedium. To compare the evolution of cellular isotopic compositions over time between the various strains, we subtracted the initial Δ65Cu (0) value for each strain to the Δ65Cu (t) at each sampling time t. Thus, we define Δ65Cucell (t) = Δ65Cu (t) − Δ65Cu (0) = δ65Cucell (t) − δ65Cucell (0). Finally, each value was normalized to 108 cells and is thus expressed as ‰/108 cells. An example of the full calculation is given in Table S1.
The Δ65Cucell decreases with time and reaches a plateau at −0.85 ± 0.06‰/108cells for DTY165 and −0.67 ± 0.30‰/108cells for MPY17 (Fig. 2). Therefore, when the plateau is reached, the presence or absence of copper high-affinity transporters does not seem to have a major influence on Cu fractionation in S. cerevisiae.
Impact of Cu oxidation state on S. cerevisiae65Cu/63Cu
In solution, at neutral pH, Cu is present as Cu(II), whereas transport through the membrane involves Cu(I)15. Before entering cells, Cu is reduced by two S. cerevisiae proteins: Fre1p and Fre2p14,15. The impact of the copper oxidation state on fractionation was addressed through three different approaches. We first tested the evolution of Cu uptake and isotopic composition of a MAC1 knockout S. cerevisiae strain (SKY34). Such mutation leads to a lower Cu reduction activity compared to the wild type. Indeed, Mac1p allows FRE1 transcription by binding to its promoter region, thus in a MAC1 knockout the expression of FRE1 is largely suppressed14. It was verified that the overall Cu reduction ability of SKY34 is greatly impaired, being three times lower (1.25 ± 0.003 μmol.l−1.min−1) than in the wild type strain DTY165 (3.62 ± 0.12 μmol.l−1.min−1) (Fig. 4). Our analyses show that the Cu content measured in SKY34 is not significantly different from that measured in the wild type DTY165 (Fig. 5). Although the MAC1 knockout and the lower Cu reduction ability of the SKY34 strain show no effect on Cu uptake, it affects the isotopic composition relative to the wild type or MPY17 cells with deleted high-affinity importers. Indeed, the Δ65CuSKY34 values oscillated around −0.01 ± 0.08‰/108cells (2σ, N = 10) between t = 0 and 240 min (Fig. 5). One possible explanation for this observation is the low availability of Cu(I) for transport.
Figure 4. Copper reductase activity in DTY165, SKY34, and SKY FRE.
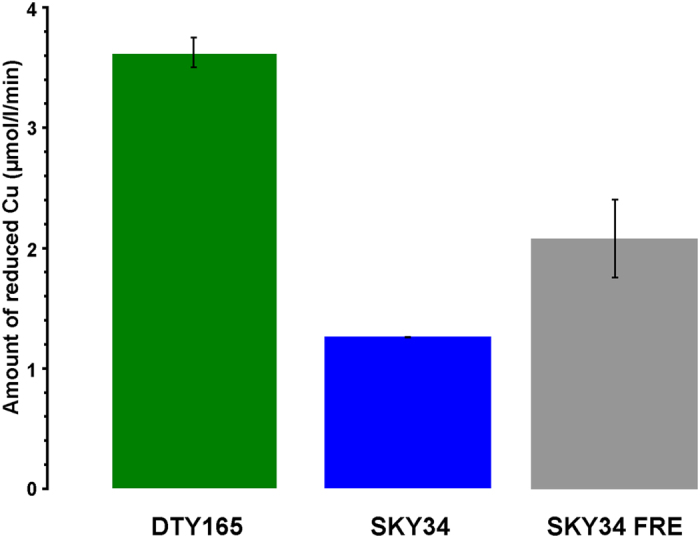
Amount of reduced Cu for the wild type strain (DTY165), the Δctr1Δmac1 knockout strain (SKY34), and the SKY34 strain where FRE1 is under the control of ADH promoter (SKY FRE). All experiments were duplicated. Error bars are the 2σ on the measurements. For SKY 34, error bar is lower than 0.05 μmol/L/min.
Figure 5. Net variations of the Cu concentration and isotopic composition in S. cerevisiae for the wild-type (DTY165, green), the Δctr1Δmac1 double-knockout strain (SKY34, blue), and the Δctr1Δmac1 strain where FRE1 is under the promoter of ADH (SKY FRE, black).
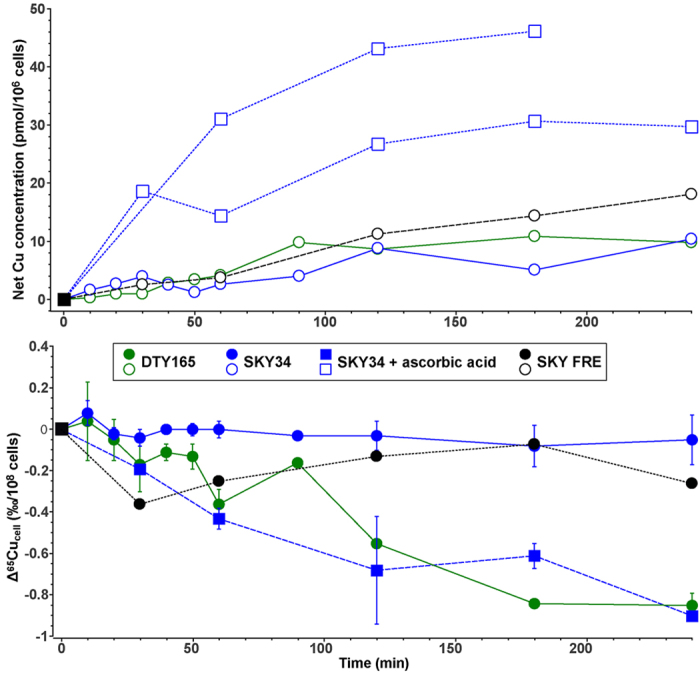
Upper panel: variations of the Cu concentration (see Fig. 2 caption for details) during uptake experiments in absence (circles) or presence of ascorbic acid (squares). Lower panel: Variations of the Cu isotopic composition (see Fig. 2 caption for details) during uptake experiments in absence (circles) or presence of ascorbic acid (squares). The error bars are the standard deviation between independent experiments (see Table S1 for details).
To test whether Cu reduction is the limiting step for Cu uptake and monitor the effect on Cu isotopic fractionation, we cultured SKY34 cells in a growth medium containing exclusively Cu(I). In effect, the addition of ascorbic acid to the growth-medium reduces copper in solution to its monovalent form (see the Experimental Procedures section for details). Copper uptake by SKY34 cells under these conditions was increased by a factor of six and the Δ65Cucell of SKY34 cells reached plateau values (−0.90 ± 0.02‰/108 cells) comparable to those of the DTY165 and MPY17 strains (Fig. 5).
To confirm this link between Cu reduction and higher Cu uptake, we used the SKY FRE strain in which FRE1 expression is rescued. The reduction rate for this strain was 2.07 ± 0.32 μmol.l−1.min−1, which is twice that of SKY34 (Fig. 4). The consequence of this higher reduction rate is that twice as much Cu was taken up by SKY FRE cells (Fig. 5). This indicated that the availability of Cu in its transportable form Cu(I) for the cell is limiting for the uptake of copper by SKY34 cells under normal culture conditions. Under normal culture conditions, the isotopic composition of the SKY FRE strain varies between that of the SKY34 and the wild type DTY165 strains (Fig. 5). We conclude that the greater the ability for the cell to reduce Cu, the stronger the enrichment in light Cu.
Finally, the Δ65Cucell of DTY165 reached its minimal value (−0.85‰/108cells) with small oscillations with a maximum oscillation rate of 7.43 10−3‰/108 cells/min. The maximum oscillation rate for SKY34 is similar: 7.60 10−3‰/108 cells/min. In the strain without high-affinity importers (MPY17), the maximum oscillation rate is slower (3.09 10−3‰/108 cells/min).
Discussion
Copper fractionation by Cu importers during transport into the cell is modulated by the cell reductive ability
In this study, we used S. cerevisiae as a model system to explore the origin of copper fractionation in cells, notably because its Cu transport mechanisms are similar to those of human cells (Fig. 1)3. S. cerevisiae is a versatile genetic model allowing the impact of genetic mutations on cell functions to be tested. The present study demonstrates a preferential 63Cu uptake by yeast, consistent with observations on bacteria and plants29,35. To trace the preference for one of the two Cu isotopes, we used the Δ65Cucell notation (see Results for details). If Δ65Cucell is negative, there is a preferential uptake of 63Cu, i.e. an enrichment in the light Cu isotope. Under steady-state conditions, there is an enrichment in light Cu that after 240 min reaches Δ65Cucell = −0.85 ± 0.06‰/108 cells. Three mechanisms can fractionate copper isotopes in cells: adsorption on cell surface, reduction of divalent to monovalent ions, and transport through the cell membrane (Fig. 1). Adsorption of Cu on organic or mineral surfaces is known to induce isotopic fractionation depending on both the type of surface and the environmental conditions29,36,37,38. The present results clearly show that adsorption on the surface of the cells is very limited (Fig. 3), which rules out adsorption as a mechanism generating Cu fractionation in S. cerevisiae.
In our experiments, the impact of cellular export, notably exocytosis (Fig. 1), can be safely neglected since in S. cerevisiae Cu homeostasis is mainly regulated at the level of the import3. Indeed, after ca. 120–180 min (depending on the strain) and up to 240 min, Cu concentrations stay relatively stable for all the studied S. cerevisiae strains. Cu isotopic compositions do also stay relatively stable after ca. 120–180 min (Figs 2 and 5). Therefore, we assume that, after 180 min, inward and outward Cu fluxes across the S. cerevisiae cell membrane are very small compared to what occurred during the Cu uptake phase, i.e. before 120–180 min, depending on the strain. As a consequence, the Cu isotopic fractionation due to Cu uptake by S. cerevisiae is likely to be very small compared to what is happening during the uptake phase. Thus, we assume that the changes in Cu isotopic fractionation due to Cu uptake can be neglected once the plateau is reached. In addition, strains compared in this study have similar genetic backgrounds for the export. Thus, any potential effect of export on the intracellular Cu isotopic composition should have been similar for all strains. Similarly, after 180 min, the Cu isotopic fractionation due to Cu uptake by S. cerevisiae was likely to be very small compared to what was happening in the phase during which the intracellular concentration increased, i.e. before 120–180 min. This is indeed what we observed (Figs 2 and 5).
It is known from both experimental studies and ab initio calculations that changes in the oxidation state of Cu can lead to fractionation39,40,41,42. To estimate the role of the reductase on Cu isotopic fractionation in S. cerevisiae using biological approaches, we used a Cu reductase deficient mutant (SKY34). In this strain, Cu uptake was similar to the wild type but there was virtually no Cu isotopic fractionation (Δ65CuSKY34 = −0.02 ± 0.07‰/108cells). To distinguish the role of reductase from the role of importers, the copper-reductase deficient SKY34 strain was fed solely with monovalent copper using ascorbic acid to ensure full reduction. In this case, Cu isotope fractionation is similar to that of the wild type strain (Δ65CuSKY34, ascorbic acid = −0.90 ± 0.02‰/108cells, Fig. 5). In the experiments with ascorbic acid, there is no Cu(I) derived from reductase activity. Thus, it is reasonable to assume that Cu fractionation in S. cerevisiae is due to the passage of Cu through transmembrane importers.
To explain the absence of fractionation for SKY34 cultivated under normal growth conditions, we hypothesize that the availability of transportable Cu(I) for the cell is limiting because the reductase activity is twice lower compared to the wild type. As a consequence, all Cu(I) produced by the reductase activity is transported inside the cell by the importers, which does not allow for fractionation of copper isotopes at that step. This hypothesis is corroborated by the fact that when copper is directly provided in excess as monovalent ions to this strain, i.e. in the ascorbic acid uptake experiments, isotopic fractionation does occur and reaches that of the wild type (Fig. 5). When the ability of SKY34 to reduce Cu is enhanced, i.e. in the SKY FRE strain (Fig. 4), the Cu(I) pool increases which permits the fractionation by importers. We indeed observe a decrease of the Δ65Cucell for the SKY FRE strain which falls between the values of the wild type and SKY 34 (Fig. 5). Thus, the ability of cells to reduce Cu controls the quantity of available Cu(I) which in turn modulates the amplitude of the isotopic fractionation. This is the first experimental demonstration of a direct link between the activity of transporters and isotopic fractionation.
Copper isotopic compositions fluctuate with time
Copper transport in S. cerevisiae is tightly regulated. It has been demonstrated that high intracellular Cu concentrations lead to a rapid decrease of the CTR3 gene expression43 and to a rapid decrease of the membrane CTR1 content, which in turn leads to the reduction of copper entry24,25. The extremely rapid and efficient degradation of the transporter proteins leads to a drastic depletion at the membrane, which is compensated by fine-tuning of the expression of the transporter genes. These regulations generate specific and very reproducible oscillations in transcription levels in the first ca. 90 min43. During our monitoring of S. cerevisiae, we observed oscillations of the isotopic composition of copper in strains expressing high-affinity transporters, i.e. the wild-type DTY165 and SKY34 strains (Fig. 5). A possible explanation for these oscillations may be that they occur in response to the oscillations in the transcriptional regulation of the high-affinity transporters and may thus result from the fine-tuning of Cu transport. Demonstration of a direct or indirect connection between copper uptake regulation and the oscillations of the isotopic phenotype of yeast would require additional work using a different experimental setting, and is beyond the scope of this study.
Affinity interactions with the reductases and transporters generate Cu fractionation
Cu(II) present in the growth medium, either free or more likely bound to various ligands5, is reduced by the Fre1p and Fre2p proteins14,15. However, there is no current understanding of how Cu(I) is maintained in its reduced form prior to its uptake by the high-affinity copper importers CTR1 and CTR344. There could be a direct interaction between the reductases and the transporters or intermediate ligands that bind Cu(I). Extracellular Cu(I) is bound to the amino acids present on the N-terminal domain of the high-affinity Cu importers: methionine (Met), histidine (His), and cysteine (Cys)45. CTR1 notably has methionine-rich sites (Mets motifs) that are thought to bind Cu(I) prior to its transport across the membrane46. CTR3 lacks these Met-motifs on its extracellular side but has an abundance of cysteine residues47 that could bind extracellular Cu(I). The precise transmembrane transport mechanism is yet not fully understood. It has been proposed that exchanges between several binding sites allow Cu(I) to move through the membrane to the amino acid residues located at the C terminus of the Ctr proteins where it is presented to cytosolic Cu chaperones46,47,48. The intracellular C domain of CTR1 and CTR3 contains Cys/His motifs that can bind Cu(I)49. Ab initio calculations give a theoretical prediction of the isotopic fractionation occurring between metal ligands. Several Cu(II) ligands have already been studied39,41. Similarly, we can determine an isotopic fractionation between the amino acid complexes that bind extracellular Cu(I) and the cytosolic Cys/His motifs of the importers. We calculated the values of the isotopologue partition functions ratio lnβ for the various Cu(I)-amino acid complexes (Supplementary Table S3) to calculate the theoretical fractionation induced by the transfer of Cu(I) from Met (CTR1) and Cys (CTR3) to His or Cys ligands. Histidine has two coordination sites that we have named His(1) and His(2), with His(1) having the largest lnβ for Cu(I). At the temperature of our experiments (298 K), the calculations predict a 63Cu enrichment due to the transfer of Cu(I) through the transporters for exchanges between Met and His(1) for CTR1 (δ65Cu = −0.78‰), and Cys and His(1) for CTR3 (δ65Cu = −0.73‰). While ab initio calculations represent a simplified approach to the complexity of the biological mechanisms and the match between the experimental results and ab initio calculations is not perfect, the amplitude of the fractionation predicted by ab initio calculation are, to the first order, in agreement with the experimental results presented in this study. Thus, the cellular enrichment in 63Cu in the wild type strain DTY165, in the SKY34 mutant with ascorbic acid, and in the SKY FRE mutant could be explained by the transport of the extracellular Cu(I) bound to the sulfur-rich amino acids methionine and cysteine to the cytosolic histidine-containing motifs of the high-affinity CTR1 and CTR3 transporters. Interestingly, experiments performed with MPY17 which lacks the high-affinity copper transporters show that Cu is fractionated to an extent only slightly lower than that of the wild type (Fig. 2). Although it is difficult to know which low-affinity transporters are responsible for Cu transport in strain MPY17, our observations and ab initio calculation results suggest that several steps of chelation of copper might also be involved in the transmembrane transfer of Cu by these low-affinity transporters.
Using Cu isotopic composition as a prognostic tool for cancer
In this study, we show that a decrease in the overall capacity of the cell to reduce Cu via an impaired transcription of the reductases leads to an increase of the SKY34 cells δ65Cu of ca. 0.9‰ compared to that of the wild type without modification of the intracellular Cu concentration (Fig. 5). This situation is close to what is observed in hepatocellular carcinoma (HCC) patients, for which δ65Cu is higher in the tumor relative to the peritumor, despite no significant differences in Cu concentration9. Such an enrichment in the tumor is mirrored by lower δ65Cu in the serum of HCC patients compared to healthy donors. In human cells, Cu is reduced by FRE homologs, the STEAP proteins50. Intriguingly, in HCC patients, immunohistochemical evidence shows that the level of STEAP3 protein is lower in the tumor mass compared to the surrounding tissue13. Moreover, STEAP3 expression has been shown to be a good prognostic marker of the transition from cirrhosis to HCC12,13. We hypothesize that the mechanism leading to Cu isotope fractionation observed in HCC is similar to the mechanism inferred for S. cerevisiae: a decrease in the ability to reduce Cu via a lower expression of STEAP3 protein leads to higher cellular δ65Cu values compared to the surrounding healthy tissues. It has also been shown that the increase of the δ65Cu values in the tumor is correlated with a decrease of the δ65Cu values in red blood cells9. Thus, monitoring the δ65Cu in red blood cells might constitute a non-invasive prognostic test to quickly detect the transition from liver cirrhosis to carcinoma.
Conclusions
In the present study, we have demonstrated that the main mechanism responsible for the enrichment in light Cu isotope in the model organism S. cerevisiae is the import of Cu via high- (Ctr1p and Ctr3p) and low-affinity transporters. We have also shown that the ability of the cell to reduce Cu modulates the amplitude of the fractionation. Copper adsorption on the membrane does not play a significant role. Based on ab initio calculation results, we propose that Cu fractionation may be due to the binding of Cu(I) with the sulfur-rich amino acids of the metallic center of the transporter, cysteine and methionine and its subsequent transport to the cytosol. Because of the high conservation of Cu transport in eukaryotic cells, our observations can be extended to Cu homeostasis in human cells. During the transition from cirrhosis to hepatocellular carcinoma, the expression of copper reductases is decreased and an enrichment in heavy Cu isotopes in the tumors is observed. Our study suggests that these two observations may be functionally connected and thus δ65Cu could be a good tool to trace changes in Cu fluxes in liver cancer patients.
Additional Information
How to cite this article: Cadiou, J.-L. et al. Copper transporters are responsible for copper isotopic fractionation in eukaryotic cells. Sci. Rep. 7, 44533; doi: 10.1038/srep44533 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
J.L.C. is thankful to the ED PHAST for his PhD salary support. J.L.C., S.P., and P.O. are thankful to the theme “Biosignatures” of the Laboratoire de Géologie de Lyon for financial support for the experiments and to the Instrument National ICP-MS (INSU-CNRS and ENS de Lyon) for support to the instruments. M. Simon, P. Télouk, E. Albalat, and F. Arnaud-Godet are thanked for their help in the clean lab and with the ICP-MS. We are thankful to two anonymous reviewers for the thoughtful and constructive review that improved the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.P. and P.O. conceived the research. J.L.C., S.P., P.O., and V.P.B. designed the experiments. A.S. designed the primers and supervised the yeast genetics. J.L.C. and S.P. designed the geochemical settings. J.L.C., S.P. and P.O. analyzed the data. J.L.C., S.P., P.O., V.P.B., and F.A. interpreted the data. T.F. calculated the partition functions ratio for ab initio calculations. J.L.C., S.P., P.O., and V.P.B. wrote the paper. S.P., J.L.C., and P.O. revised and all authors approved the manuscript.
References
- Lutsenko S. Human copper homeostasis: a network of interconnected pathways. Curr. Op. Chem. Biol. 14, 211–217, doi: 10.1016/j.cbpa.2010.01.003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M. C. Copper and genomic stability in mammals. Mutat. Res. 475, 141–152, doi: 10.1016/S0027-5107(01)00076-8 (2001). [DOI] [PubMed] [Google Scholar]
- Nevitt T., Öhrvik H. & Thiele D. J. Charting the travels of copper in eukaryotes from yeast to mammals. BBA-Mol. Cell Res. 1823, 1580–1593, doi: 10.1016/j.bbamcr.2012.02.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyer D., Masaldan S., La Fontaine S. & Cater M. A. Targeting copper in cancer therapy: ‘Copper That Cancer’. Metallomics 7, 1459–1476, doi: 10.1039/c5mt00149h (2015). [DOI] [PubMed] [Google Scholar]
- Majumder S. et al. The role of copper in drug-resistant murine and human tumors. Biometals 22, 377–384, doi: 10.1007/s10534-008-9174-3 (2009). [DOI] [PubMed] [Google Scholar]
- Fosset C., McGaw B. A., Reid M. D. & McArdle H. J. A non-radioactive method for measuring Cu uptake in HepG2 cells. J. Inorg. Biochem. 99, 1018–1022, doi: 10.1016/j.jinorgbio.2005.01.005 (2005). [DOI] [PubMed] [Google Scholar]
- Albarède F. et al. Precise and accurate isotopic measurements using multiple-collector ICPMS. Geochim. Cosmochim. Acta 68, 2725–2744, doi: 10.1016/j.gca.2003.11.024 (2004). [DOI] [Google Scholar]
- Telouk P. et al. Copper isotope effect in serum of cancer patients. A pilot study. Metallomics 7, 299–308, doi: 10.1039/c4mt00269e (2015). [DOI] [PubMed] [Google Scholar]
- Balter V. et al. Natural variations of copper and sulfur stable isotopes in blood of hepatocellular carcinoma patients. Proc. Nat. Acad. Sci. USA 112, 982–985, doi: 10.1073/pnas.1415151112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarede F., Albalat E. & Telouk P. Instrumental isotope fractionation in multiple-collector icp-ms. J. Anal. At. Spectrom. 30, 1736–1742, doi: 10.1039/c5ja00188a (2015). [DOI] [Google Scholar]
- Holzer A. K. et al. Expression of the human copper influx transporter 1 in normal and malignant human tissues. J. Histochem. Cytochem. 54, 1041–1049, doi: 10.1369/jhc.6A6970.2006 (2006). [DOI] [PubMed] [Google Scholar]
- Caillot F. et al. Down-regulated expression of the TSAP6 protein in liver is associated with a transition from cirrhosis to hepatocellular carcinoma. Histopathology 54, 319–327, doi: 10.1111/j.1365-2559.2009.03224.x (2009). [DOI] [PubMed] [Google Scholar]
- Coulouarn C. et al. Global gene repression in hepatocellular carcinoma and fetal liver, and suppression of dudulin-2 mRNA as a possible marker for the cirrhosis-to-tumor transition. J. Hepatol. 42, 860–869, doi: 10.1016/j.jhep.2005.01.027 (2005). [DOI] [PubMed] [Google Scholar]
- Georgatsou E., Mavrogiannis L. A., Fragiadakis G. S. & Alexandraki D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J. Biol. Chem. 272, 13786–13792, doi: 10.1074/jbc.272.21.13786 (1997). [DOI] [PubMed] [Google Scholar]
- Hassett R. & Kosman D. Evidence for Cu(II) reduction as a component of Cu uptake in Saccharomyces cerevisiae. J. Biol. Chem. 270, 128–134, doi: 10.1074/jbc.270.1.128 (1995). [DOI] [PubMed] [Google Scholar]
- Dancis A., Haile D., Yuan D. S. & Klausnert R. D. The Saccharomyces cerevisiae copper transport protein (Ctr1p). J. Biol. Chem. 269, 25660–25667 (1994). [PubMed] [Google Scholar]
- Knight S. A. B., Labbé S., Kwon L. F., Kosman D. J. & Thiele D. J. A widespread transposable element masks expression of a yeast copper transport gene. Gene. Dev. 10, 1917–1929, doi: 10.1101/gad.10.15.1917 (1996). [DOI] [PubMed] [Google Scholar]
- Hasset R., Dix D. R., Eide D. J. & Kosman D. J. The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 351, 477–484, doi: 10.1042/0264-6021:3510477 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C., Kelleher M., Iyer V. R., Brown P. O. & Winge D. R. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 275, 32310–32316, doi: 10.1074/jbc.M005946200 (2000). [DOI] [PubMed] [Google Scholar]
- Heredia J., Crooks M. & Zhu Z. Phosphorylation and Cu+ Coordination-dependent DNA binding of the transcription factor Mac1p in the regulation of copper transport. J. Biol. Chem. 276, 8793–8797, doi: 10.1074/jbc.M008179200 (2001). [DOI] [PubMed] [Google Scholar]
- Labbé S., Zhu Z. & Thiele D. J. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J. Biol. Chem. 272, 15951–15958, doi: 10.1074/jbc.272.25.15951 (1997). [DOI] [PubMed] [Google Scholar]
- Jungmann J. et al. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 12, 5051–5056 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Sinani D., Kim H. & Lee J. Copper transport activity of yeast Ctr1 is down-regulated via its C terminus in response to excess copper. J. Biol. Chem. 284, 4112–4122, doi: 10.1074/jbc.M807909200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sitaram A. & Burd C. G. Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic 8, 1375–1384, doi: 10.1111/j.1600-0854.2007.00616.x (2007). [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Rabinovich E., Dancis A., Bonifacino J. S. & Klausner R. D. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctrlp in the apparent absence of endocytosis. EMBO J. 15, 3515–3523 (1996). [PMC free article] [PubMed] [Google Scholar]
- Janke C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962, doi: 10.1002/yea.1142 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang T. et al. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28, 795–798, doi: 10.1002/yea.1905 (2011). [DOI] [PubMed] [Google Scholar]
- Kushnirov V. V. Rapid and reliable protein extraction from yeast. Yeast 16, 857–860, doi: (2000 ). [DOI] [PubMed] [Google Scholar]
- Navarrete J. U., Borrok D. M., Viveros M. & Ellzey J. T. Copper isotope fractionation during surface adsorption and intracellular incorporation by bacteria. Geochim. Cosmochim. Acta 75, 784–799, doi: 10.1016/j.gca.2010.11.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgatsou E. & Alexandraki D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 3065–3073 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos C., Guzmán R., López-Fernández E. & Casado Á. Evaluation of the copper(II) reduction assay using bathocuproinedisulfonic acid disodium salt for the total antioxidant capacity assessment: The CUPRAC-BCS assay. Anal. Biochem. 392, 37–44, doi: 10.1016/j.ab.2009.05.024 (2009). [DOI] [PubMed] [Google Scholar]
- Moynier F. et al. Isotopic fractionation and transport mechanisms of Zn in plants. Chem. Geol. 267, 125–130 (2009). [Google Scholar]
- Buldini P. L., Cavalli S. & Sharma J. L. Determination of transition metals in wine by IC, DPASV-DPCSV, and ZGFAAS Coupled with UV Photolysis. J. Agri. Food Chem. 47, 1993–1998 (1999). [DOI] [PubMed] [Google Scholar]
- Maréchal C. N., Telouk P. & Albarède F. Precise analysis of copper and zinc isotopic compositions by plasma-source mass spectrometry. Chem. Geol. 156, 251–273 (1999). [Google Scholar]
- Weinstein C. et al. Isotopic fractionation of Cu in plants. Chem. Geol. 286, 266–271 (2011). [Google Scholar]
- Li D., Liu S.-A. & Li S. Copper isotope fractionation during adsorption onto kaolinite: Experimental approach and applications. Chem. Geol. 396, 74–82, doi: 10.1016/j.chemgeo.2014.12.020 (2015). [DOI] [Google Scholar]
- Balistrieri L. S., Borrok D. M., Wanty R. B. & Ridley W. I. Fractionation of Cu and Zn isotopes during adsorption onto amorphous Fe(III) oxyhydroxide: Experimental mixing of acid rock drainage and ambient river water. Geochim. Cosmochim. Acta 72, 311–328, doi: http://dx.doi.org/10.1016/j.gca.2007.11.013 (2008). [Google Scholar]
- Pokrovsky O. S., Viers J., Emnova E. E., Kompantseva E. I. & Freydier R. Copper isotope fractionation during its interaction with soil and aquatic microorganisms and metal oxy(hydr)oxides: Possible structural control. Geochim. Cosmochim. Acta 72, 1742–1757, doi: http://dx.doi.org/10.1016/j.gca.2008.01.018 (2008). [Google Scholar]
- Fujii T., Moynier F., Blichert-Toft J. & Albarède F. Density functional theory estimation of isotope fractionation of Fe, Ni, Cu, and Zn among species relevant to geochemical and biological environments. Geochim. Cosmochim. Acta 140, 553–576, doi: 10.1016/j.gca.2014.05.051 (2014). [DOI] [Google Scholar]
- Balter V. et al. Contrasting Cu, Fe, and Zn isotopic patterns in organs and body fluids of mice and sheep, with emphasis on cellular fractionation. Metallomics 5, 1470–1482, doi: 10.1039/c3mt00151b (2013). [DOI] [PubMed] [Google Scholar]
- Fujii T., Moynier F., Abe M., Nemoto K. & Albarède F. Copper isotope fractionation between aqueous compounds relevant to low temperature geochemistry and biology. Geochim. Cosmochim. Acta 110, 29–44, doi: 10.1016/j.gca.2013.02.007 (2013). [DOI] [Google Scholar]
- Jouvin D. et al. Stable Isotopes of Cu and Zn in Higher Plants: Evidence for Cu reduction at the root surface and two conceptual models for isotopic fractionation processes. Environ. Sci. Technol. 46, 2652–2660 (2012). [DOI] [PubMed] [Google Scholar]
- Peña M. M., Koch K. A. & Thiele D. J. Dynamic regulation of copper uptake and detoxification genes in Saccharomyces cerevisiae. Molecular and cellular biology 18, 2514–2523 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-E., Nevitt T. & Thiele D. J. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4, 176–185, doi: 10.1038/nchembio.72 (2008). [DOI] [PubMed] [Google Scholar]
- Rubino J. T., Chenkin M. P., Keller M., Riggs-Gelasco P. & Franz K. J. A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics 3, 61–73, doi: 10.1039/c0mt00044b (2011). [DOI] [PubMed] [Google Scholar]
- Rubino J. T., Riggs-Gelasco P. & Franz K. J. Methionine motifs of copper transport proteins provide general and flexible thioether-only binding sites for Cu(I) and Ag(I). J. Biol. Inorg. Chem. 15, 1033–1049, doi: 10.1007/s00775-010-0663-9 (2010). [DOI] [PubMed] [Google Scholar]
- Peña M. M., Puig S. & Thiele D. J. Characterization of the Saccharomyces cerevisiae high affinity copper transporter Ctr3. J. Biol. Chem. 275, 33244–33251, doi: 10.1074/jbc.M005392200 (2000). [DOI] [PubMed] [Google Scholar]
- Öhrvik H. & Thiele D. J. How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Ann. NY Acad. Sci. 1314, 32–41, doi: 10.1111/nyas.12371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S. & Thiele D. J. Molecular mechanisms of copper uptake and distribution. Curr. Op. Chem. Biol. 6, 171–180, doi: http://dx.doi.org/10.1016/S1367-5931(02)00298-3 (2002). [DOI] [PubMed] [Google Scholar]
- Ohgami R. S., Campagna D. R., McDonald A. & Fleming M. D. The Steap proteins are metalloreductases. Blood 108, 1388–1394, doi: 10.1182/blood-2006-02-003681 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-S., Szczypka M., Lu Y.-P., Thiele D. J. & Rea P. A. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione. J. Biol. Chem. 271, 6509–6517, doi: 10.1074/jbc.271.11.6509 (1996). [DOI] [PubMed] [Google Scholar]
- Zhou P. & Thiele D. J. Copper and gene regulation in yeast. Biofactors 4, 105–115 (1993). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.