Summary
p53 and its related genes, p63 and p73 constitute the p53 gene family. While p53 is the most frequently mutated gene in human tumors, p63 and p73 are rarely mutated or deleted in cancers. Many studies have reported p63/p73 overexpression in human cancers while others showed that a loss of p63/p73 is associated with tumor progression and metastasis. Thus, whether p63 or p73 is a tumor suppressor gene or an oncogene has been a matter of debate. This controversy has been attributed to the existence of multiple splicing isoforms with distinct functions; the full-length TA isoform of p63 has structural and functional similarity to wild-type p53, whereas the ΔNp63 acts primarily in dominant-negative fashion against all family members of p53. Differential activities of TA and ΔN isoforms have been shown in vivo by creating isoform-specific gene knockout mice. All p53, p63, p73 proteins bind to and activate target genes with p53-response elements; p63 also binds to distinct p63-response elements and regulate expression of specific target genes involved in skin, limb, and craniofacial development. Interestingly, several studies have shown that both p63 and p73 are involved in cellular response to cancer therapy and others have indicated that both of these molecules are required for p53-induced apoptosis, suggesting functional interplay among p53 family proteins. Consistent with these findings, aberrant splicing that result in ΔNp63 or ΔNp73 overexpression are frequently found in human cancers, and is associated with poor clinical outcomes of patients in the latter. Thus immunohistochemical staining of tumor specimen with ΔNp73-specific antibody might have diagnostic values in cancer clinics.
Keywords: p63, p73, splicing, alteration, overexpression, knockout, mouse, cancer
Introduction
The p53 tumor suppressor protein integrates endogenous and exogenous signals to modulate cell fate to stress and cellular environments (Toledo 2006; Menendez 2009; Vousden 2009). Upon DNA damage or other cellular stresses, such as oxidative stress, hypoxia, carcinogen exposure, and oncogene overexpression, p53 becomes activated with increased levels. Then, p53 directs a variety of responses, including DNA repair, cell cycle arrest/senescence, apoptosis, and autophagy depending on the input signal and severity of the damage (Green 2006; Rufini 2013). The specific response depends on whether the damage can be repaired or is too serious that death of the cell is required to maintain tissue integrity. The genomic locus for p53 (TP53) is very frequently (~50%) mutated in human cancers, which is associated with therapy resistance and poor prognosis of patients (Xu, 2008; Lai 2012). Since p53 protects humans from damaged and life-threatening cells that may predispose to tumor development, recent research efforts have been made on reconstituting p53 function to effectively treat cancer patients (Frezza & Martins 2012; Senzer 2013).
In the late 1990’s, two other p53 family members, p73 and p63 were discovered (Kaghad 1997; Yang 1998). These three proteins, encoded by the TP53, TP63, and TP73 genes (Trp53, Trp63, and Trp73 in mice, respectively), are transcription factors that bind directly to DNA as tetramers, interact with other transcription factors and the transcription machinery, and together control the expression of genes involved in all aspects of life. It has now become clear that both p63 and p73 are involved in a broad spectrum of biological activities, such as cell proliferation, apoptosis, development, differentiation, senescence, and aging. In particular, p63 has emerged as a critical player in embryonic development, epithelial stem cell maintenance, and differentiation. Both p63 and p73 express as a variety of protein isoforms that originate from two different promoters and extensive gene splicing at the N- and C-termini (Murray-Zmijewski 2006). Moreover, the p63 and p73 genes encode a sterile alpha motif (SAM) domain at the C-terminus that is not found in p53. This domain is responsible for protein-protein interactions and is found in a diverse range of proteins that are involved in developmental regulation. In this chapter, we discuss the structure, splicing isoforms of p63 and p73 in normal and their distinct functions in tumor suppression/proliferation. We also explain their possible interaction with Mdm2 and MdmX. Whether these molecules (p63 and p73) are tumor suppressors or oncoproteins have been a hot topic of debate. Gene knockout studies will tell us the answer; since both of the genes have multiple splicing isoforms, we have put special interest on the phenotypes of splicing isoform - specific gene knockout mouse models. Finally, we summarize the mechanisms and frequencies for alterations of these genes in human cancers and their prognostic significance.
Structure of the p63 and p73 loci
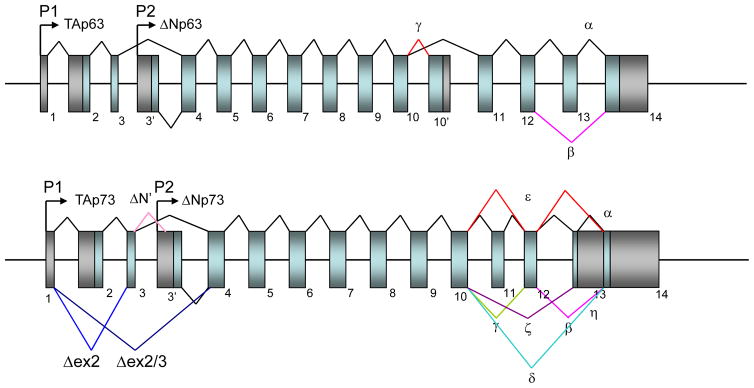
Both p63 and p73 loci (TP63, TP73) generate mRNAs that produce multiple protein products resulting from use of distinct promoters and alternative mRNA splicing (Figure 1) (Kaghad 1997; Yang 1998). Transcription of p63 and p73 occurs from two promoters: one upstream of exon 1 (P1) and the other located within intron 3 (P2). In both proteins, splicing isoforms transcribed from the P1 promoter have an N-terminal transactivation (TA) domain (i.e., TAp63 and TAp73), which is highly homologous to the TA domain of p53, whereas transcripts generated from the P2 promoter lack the N-terminal TA domain (39 amino acids; called ΔNp63 and ΔNp73, respectively; Figure 1) (Kaghad 1997; Yang 1998). The unique structural differences for p63 and p73 are explained below.
Figure 1. Genomic structure of the p63 and p73 loci.
Genomic structures of human p63 and p73 loci. Numbered boxes indicate exons, black shading denotes untranslated sequences, and light blue shading denotes coding regions. Distinct transcription start sites (P1 and P2) are indicated by arrows. N-terminal alternative splicing for p63 and p73 are indicated by blue and light pink lines, and C-terminal splicing events for these proteins are indicated by different colored lines.
p63
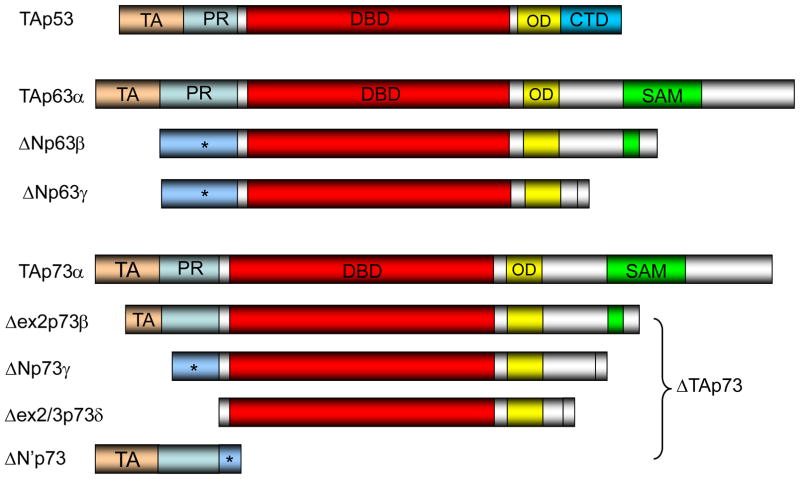
The structure of the genomic locus for p63 is shown in Figure 1, upper panel (Yang 1998). Both mouse and human p63 genes consist of 15 exons spanning around 210kb and 270kb, respectively, on the genome. The human version has been mapped to chromosome 3q27. The structures for the TAp53 protein, representative p63 isoform proteins are shown in Figure 2 (Murray-Zmijewski 2006). Wild-type TAp53 has an N-terminal transactivation domain (TA) for recruitment of core transcriptional factors, a central DNA-binding domain (DBD) for recognition of promoter sequences, an oligomerization domain (OD) for tetramerization, and a short basic stretch of 30 amino acids for regulation of transcriptional activity (Figure 2, top panel). The p63 gene encodes two alternatively spliced isoforms (TA, ΔN) with different ATG at the N-terminus with three alternatively-spliced C-terminal isoforms (α, β, γ), generating 6 different splicing isoforms, i.e., TAp63α, TAp63β, TAp63γ, ΔNp63α, ΔNp63β, ΔNp63γ (Figures). The p63α transcript has all 14 exons while the β transcript lacks exon 13. The γ transcript lacks exons 11–14 by splicing into a unique exon 10′ (Figure 1, top panel). The full-length TA isoform of p63 has structural and functional similarity to wild-type p53, whereas the ΔNp63 acts primarily in dominant-negative fashion against all family members of p53: p53, TAp63 and TAp73. Thus, it is generally assumed that TAp63 is a tumor suppressor gene while ΔNp63 is an oncogene. In addition, the C-terminus of p63 (and also p73) contains a sterile alpha motif (SAM) domain and a transcriptional inhibitory domain (TID) (Figure 2). The SAM domains are small protein–protein interaction modules that are found in a wide variety of proteins, ranging from kinases and transcriptional regulators to cell surface receptors (Schultz 1997; Thanos & Bowie 1999). The TID, an unstructured region C-terminal to the SAM domain, was shown to inhibit the transcriptional activity of p63 by interacting with the TA domain (Serber 2002). These two domains are not found in p53 (Figure 2), suggesting unique functions for p63 and p73.
Figure 2. Structure of the p53 family proteins.
Protein domains of p53 family members. The transactivation (TA) domains shared by p53, TAp63, and TAp73 isoforms are shown in gold. The proline-rich domain (PR: light blue), DNA-binding domain (DBD: red), oligomerization domain (OD: yellow), carboxyl-terminal regulatory domain (CTD: blue), and sterile alpha motif (SAM: green) are shown in colors. The alpha isoforms of p63 and p73 possess a C-terminal SAM domain followed by a transactivational inhibitory domain (TID: silver). TAp63γ/TAp73γ isoforms most closely look like p53. N-terminally truncated ΔN isoforms for p63 possess the unique N-terminal sequence. p73 has four different isoforms at the N terminus (ΔN, ΔN′, Δex2, Δex2/3) dependent on the usage of two different promoters and alternative splicing including exons 2 and 3. ΔN′p73 encodes a small protein having a unique sequence at the C- terminal end, but lacks the DNA-binding domain. * denotes the unique region encoded by exon 3′ (Murray-Zmijewski 2006).
p73
Both mouse and human p73 genes consist of 15 exons spanning around 80kb on the genome. The structure of the genomic locus for p73 is shown in Figure 1, lower panel. The human version has been mapped to chromosome 1p36.33. The p73 gene encodes 4 alternatively spliced isoforms (TA, Δex2, Δex2/3, ΔN) with distinct ATG at the N-terminus and 7 alternatively spliced isoforms at C-terminus (α, β, γ, δ, ε, ζ, and η; Figure 1) (Moll & Slade 2004; Bourdon 2005). In addition, splicing-associated frameshifts yield unique C-terminal sequences for some p63 and p73 isoforms (Courtois 2004; Moll & Slade 2004). This alternative splicing can generate 28 plus one (ΔN′; total 29) different splicing isoforms for p73. Of note, both ΔN and ΔN′ isoforms have unique amino acids at exon 3′ (Figure 1). The difference in the N-terminal region contributes to different protein-protein interactions dependent on the isoform. The p73α transcript has all exons 1–14 while the β transcript lacks exon 13. The γ transcript lacks exon 11, the δ transcript lacks exons 11–13 (Figure 1, lower panel). The ε isoform lacks 11 and 13, ζ lacks exons 11 and 12; η is close to α, but is different at exon 14 (Figure 1). The TAp63γ and TAp73γ isoforms most closely resemble the full-length wild-type TAp53 (Figure 2). In overexpression studies, TAp63γ has been shown to be as potent as p53 in transactivating target gene expression and apoptosis, whereas the most potent transcriptionally active p73 isoform reported is TAp73β (Kaghad 1997; Yang 1998). Since the ΔNp73 acts primarily in dominant-negative fashion against all family members of p53, it is generally accepted that TAp73 is a tumor suppressor gene while ΔNp73 is an oncogene.
Unique C-terminal domains and transcriptional targets for p63 and p73
Both p63α and p73α isoforms also contain a protein–protein interaction domain known as SAM (Figure 2). This is a globular domain composed of four α-helices and a small 310 helix. Although this motif is often found to mediate homodimerization with developmentally regulated proteins, the SAM domain does not contribute to homodimerization in p63 and p73 (Chi 1999). The SAM domains also appear to possess the ability to bind RNA. The post-SAM region known as the transactivational inhibitory domain (TID) has been identified in p63α and p73α isoforms (Serber 2002; Figure 2). This region consisting of ~70 amino acids, which is absent in p53, has been proposed to inhibit transcription of both TAp63α and TAp73α through inter- or intra-molecular association with the TA domain (Serber 2002). Indeed, both of these proteins show decreased potency in transactivation and apoptosis induction as compared to other TA isoforms, and deletion of this region restored transactivating potential for both TAp73α and TAp63α (De Laurenzi 1998, Serber 2002).
Since both p63 and p73 share strong structural, biochemical and biological homologies to p53, they bind specifically to conventional p53 response elements (p53RE: RRRCWWGYYY) and transactivate target genes such as p21Cip1, MDM2, and BAX. In spite of their structural similarities between p53 and p63, the latter functions are greatly different from those of p53. The most striking difference is the apparent involvement of p63 in skin and limb development (Mills 1999; the details of phenotypes will be explained later). Global p63 knockout mice that lack all splicing isoforms exhibit skin and limb defects as well as craniofacial abnormalities, but are not tumor prone. This is in contrast to p53 knockout mice that develop normally, but are prone to develop various cancers from an early age, esp. thymic lymphomas and hemangiosarcomas (Donehower Nature 1992; Jacks 1994). In humans, germ line mutations of p53 cause Li-Fraumeni syndrome, in which affected individuals are very prone to cancer development (Malkin 1990, 1993; Strong 1992). These differences may be due to the differential regulation of target genes by p53 and p63. The p53 and p63 proteins can bind to two or more tandem repeats of RRRCWWGYYY (p53RE) or some other motifs and subsequently activate target gene expression. By using oligonucleotide expression microarray analysis and analyzing the promoters of p63-induced genes, Osada et al. (2005) identified novel p63-specific response elements (p63REs) in the promoter regions of EVPL and SMARCD3. These p63REs exhibit characteristic differences from the canonical p53RE (RRRCWWGYYY) in both the core-binding element (CWWG) as well as the RRR and/or YYY sequences (Osada 2005). Their data indicate that p53 preferentially activates and binds to the RRRCATGYYY sequence, whereas p63 preferentially activates RRRCGTGYYY. Whereas EVPL protein is highly expressed in epithelial cells of the skin and pharynx in the p63+/+ mouse, it is undetectable in these tissues in the p63−/− mouse. Thus p63 can regulate expression of specific target genes such as those involved in skin, limb, and craniofacial development by preferentially activating distinct p63-specific response elements (Osada 2005). Until now, a number of genes have been reported to be targets of p63 and p73, such as REDD1 (regulation of reactive oxygen species), JAG1/JAG2 (Notch signaling), IL4R, ΔNp73, and AQP3 (glycerol and water transporter) (Ellisen 2002; Sasaki 2002; Osada 2005). Among these, ΔNp73 is a splicing variant from the p73 locus, suggesting its autoregulation (Nakagawa 2002). In Notch signaling, Sasaki et al. found that the genes encoding ligands for the Notch receptors (JAG1/2), are up-regulated by p63 and p73 but not by p53 (Sasaki 2002). They identified a p63-binding site in the second intron of the JAG1 gene, which could directly interact with p63 in vivo as demonstrated by chromatin immunoprecipitation. They also found a target of Notch signaling; HES-1 was up-regulated in Jurkat cells with high expression of Notch1 when co-cultured with p63-transfected cells, suggesting that p63 can trigger the Notch signaling pathway in neighboring cells. This suggests a potential molecular mechanism for the involvement of p63 in normal development (Sasaki 2002). Recently it was reported that BRCA1 activates the Notch pathway in breast cells by transcriptional upregulation of Notch ligands and receptors (Buckley 2013). They demonstrated that BRCA1 was localized to an intronic enhancer within the JAG1 gene, an event requiring ΔNp63. This BRCA1/ΔNp63-mediated induction of JAG1 must play important roles in the regulation of breast stem/precursor cells since knockdown of these proteins resulted in increased tumorsphere growth and increased activity of stem cell markers (Buckley 2013). Thus, BRCA1/ΔNp63-mediated transactivation of Notch signaling is a key event in the normal differentiation process in breast tissue.
Regulation of p63 and p73 by Mdm2
The interactions of p53 with Mdm2 and Mdmx, mediated via the TA domain of p53 have been well-documented (Lin 1994; Kussie 1996; Deb 2003; Popowicz 2008). The physiological importance of the regulation of p53 by the Mdm2 and Mdmx ubiquitin ligases as well as the role of its aberrant regulation in tumors has also been reported (Kubbutat 1997; Haupt 1997; Danovi 2004; Linke 2008; Okamoto 2009). Unlike p53, which protects genomic stability, the two homologous proteins of the same family, p63 and p73, regulate developmental processes as described in this chapter.
Since all three p53 family proteins have homologous TA domains, it was speculated that p63 and p73 may be regulated by Mdm in a similar manner as has been reported for p53. The ability of Mdm2 and Mdmx to bind to p73 has been well-documented (Ongkeko 1999) and Zdzalik et al. (2010) provided a detailed kinetic characterization of this interaction. The interaction of Mdm with p63 has also been studied previously, but the results were controversial due to lower affinity for such an interaction (Kojima 2001; Little 2001; Wang 2001; Calabro 2002). In fact, Zdzalik et al. (2010) showed that both Mdm2 and Mdmx form complexes with the p63 TA domain, however the interactions were weaker than those determined for p53 or p73. The interaction of the p63 TA domain is specific and mechanistically similar to that of the p53 TA domain since the p63(Ala) mutant peptide showed no activity in the assays performed. Although the interactions of p73 with Mdm2 and Mdmx have also been studied previously, only the affinity of p73 for Mdm2 has been reported (Schon 2002; Burge 2009). The interaction between p63 and Mdm2 was one order of magnitude weaker than those of Mdm with p53 and p73. Conversely, the affinities of both Mdm2 and Mdmx for p73 were of the same order of magnitude as those for p53, which justifies the conclusion that these proteins truly interact in cells, as has previously been suggested in other studies (Ongkeko 1999, Wang 2001). The weaker interactions of both Mdm2 and Mdmx with p63 explain the inconsistent results reported by different groups on the interactions of those proteins. Clearly, at sufficiently high concentrations, these proteins will form a stable complex, but whether such concentrations are ever encountered under physiological conditions in cells remains a very intriguing question for future studies (Zdzalik 2010). It is also noteworthy that although the affinities of p53 for Mdm2 and Mdmx are similar, both p63 and p73 interact more strongly with Mdmx. Therefore, Mdmx, but not Mdm2, may have a stronger impact on the regulation of intracellular p63 and p73.
Constitutional, all splicing isoforms’ knockout mice for p63, p73
Mills et al. (1999) reported the phenotypes of p63-deficient mice (all splicing isoforms). p63-null mice were born alive but had striking developmental defects. Their limbs were absent or truncated, defects that were caused by a failure of the apical ectodermal ridge differentiation. The skin of p63-null mice did not progress through an early developmental stage lacking stratification with no differentiation markers. Hair follicles, teeth and mammary glands were absent in p63-deficient mice. Thus, in contrast to p53, p63 is essential for several aspects of ectodermal differentiation during embryogenesis. Keyes et al. (2006) studied spontaneous and chemically-induced tumor development using p63+/− mice since p63−/− mice had serious developmental defects, and thus were not suitable for in vivo tumor development studies. They found that p63+/− mice were not tumor prone and mice heterozygous for both p63 and p53 had fewer tumors than p53+/− mice. Furthermore, p63 expression was maintained in carcinomas. These findings demonstrate that p63 plays a markedly different biological role in cancer than p53.
Mice deficient for all p73 splicing isoforms also exhibited profound developmental and immunological defects, including hippocampal dysgenesis, hydrocephalus, chronic infections, and inflammation, as well as abnormalities in pheromone sensory pathways (Yang 2000). It should be noted that mice lacking p73 showed no increased susceptibility to spontaneous tumorigenesis, in contrast to p53-deficient mice (Donehower 1992, Jacks 1994). They speculated that potentially dominant-negative, p73 variants were the predominant expression products of this gene in developing and adult tissues, explaining the mechanistic basis of the hippocampal dysgenesis and the loss of pheromone responses in p73-null mice. In conclusion, p73 plays unique roles in neurogenesis, sensory pathways, and homeostatic control, but its role in tumor suppression is not clear from the global gene deletion model.
Flores et al. (2002) explored the combined role of p63 and p73 in DNA damage-induced apoptosis. The combined absence of p63 and p73 severely impaired the induction of p53-dependent apoptosis in response to DNA damage in E1A-expressing cells and in the developing central nervous system in mice although the p53 locus remained intact. This was explained by the inability of p53 to bind the promoters of apoptosis-associated target genes and to upregulate their transcription in p63−/−; p73−/−; E1A(+) cells and the developing central nervous system (Flores, 2002; Taneja 2011).
Splicing isoform-specific knockout mouse models for p63, p73
p63
The roles of p63 in tumor suppression have been a hot topic of debate. The most intriguing question is whether p63 is a tumor suppressor gene or an oncogene? Many studies have shown p63 overexpression in human cancers (Massion 2003; discussed later in this chapter), while others demonstrate that a loss of p63 is associated with tumor progression and metastasis (Flores 2007). This controversy has been attributed to the existence of multiple splicing isoforms with distinct functions; the full-length TA isoform of p63 has structural and functional similarity to p53 (Figure 2), whereas the ΔNp63 protein acts primarily in dominant-negative fashion against all family members of p53. To study splicing isoform–specific differences of p63 functions in vivo, Su et al. (2009) developed a TAp63 conditional knockout mouse and used it to delete TAp63 in the germline (TAp63−/−; using Zp3-cre or Protamine-cre) or in K14-expressing cells in the basal layer of the epidermis (epidermis-specific TAp63 deletion; using K14cre+). TAp63−/− mice aged prematurely and developed blisters, skin ulcerations, senescence of hair follicle-associated dermal and epidermal cells, and decreased hair morphogenesis, indicating that TAp63 serves to maintain adult skin stem cells by causing cellular senescence and genomic stability, thereby preventing premature tissue aging (Su 2009). The same group followed spontaneous tumor development in TAp63−/−, TAp63+/− and wild-type mice for 2.5 years and found that both TAp63+/− mice and TAp63−/− mice developed carcinomas and sarcomas with significantly shorter lifespan than the wild-type cohort. Consistent with this finding, tumors from TAp63+/− mice retained the wild-type allele of TAp63 suggesting that TAp63 is haplo-insufficient for tumor suppression. Both TAp63+/− and TAp63−/− mice developed highly metastatic tumors, and 10% of these metastases were found in the brain, a rare finding in endogenous mouse tumor models. Although equivalent numbers of carcinomas metastasized in the TAp63−/− and TAp63+/− mice, a greater number of sarcomas metastasized in TAp63+/− mice than in TAp63−/− mice, indicating that heterozygous loss for TAp63 rather than homozygous loss results in a more severe phenotype.
Keyes et al. (2011) observed that ΔNp63α overexpression in mouse embryonic fibroblasts (MEFs) bypassed Ras-mediated senescence and drove tumorigenesis in vivo. They identified chromatin-remodeling protein Leeh as a novel target for ΔN63α, which was an essential mediator of senescence bypass. This bypass of senescence by ΔNp63α promoted stem cell-like proliferation and maintained the survival of keratin 15-positive cells. Thus, ΔNp63α is a novel oncogene that cooperates with Ras to promote tumor development by initiating stem cell proliferation. By contrast, overexpression of TAp63 forms in p53−/− MEFs increased senescence and reduced tumor development in vivo, consistent with a p53-independent effect of TAp63 (Guo 2009).
The TAp63 and ΔNp63 isoforms have special effects in epidermal tissue differentiation (Aberdam 2007). In murine embryonic stem cells, ΔNp63, but not TAp63, is highly expressed in epidermis and is critical for the expression of the cytokeratins K5 and K14, two markers of keratinocyte differentiation, indicating that only ΔNp63 is required for the commitment of ectodermal into epidermal cells (Medawar 2008; Shalom-Feuerstein 2011). In summary, p63 and its splicing variants play specific roles in epidermal commitment, cell proliferation, and senescence bypass; alterations of this intricate balance contribute to tumor development.
p73
Mice with a complete deficiency of p73 exhibited severe neurological and immunological defects due to the absence of all TAp73 and ΔNp73 isoforms as described in the previous section. To study mice deficient for specific p73 protein isoforms, Tak Mak’s group created p73 isoform-specific knockout mice (Tomasini 2008; Wilhlem 2010). Tomasini et al. (2008) created mice in which exons encoding the TAp73 isoforms were specifically deleted at exons 2/3 to establish a TAp73-deficient (TAp73−/−) mice. Mice specifically lacking in TAp73 isoforms showed a phenotype intermediate between the phenotypes of p73−/− and p53−/− mice with respect to the incidence of spontaneous and carcinogen-induced tumors, infertility, and aging, as well as hippocampal dysgenesis. In addition, cells from TAp73−/− mice showed genomic instability associated with enhanced aneuploidy, which could account for the increased incidence of spontaneous tumors in these animals. Hence, TAp73 isoforms exert tumor-suppressive functions indicating an emerging role for Trp73 in the maintenance of genomic stability. Wilhlem et al. (2010) generated mice that were selectively deficient for the ΔNp73 isoform by depleting ΔN form-specific exon 3′ (ΔNp73–/–). These mice were viable and fertile, but showed signs of neurodegeneration. Cells from ΔNp73–/– mice were sensitive to DNA-damaging agents and showed an increase in p53-dependent apoptosis. They found that the ΔNp73 protein localized directly to the site of DNA damage, interacted with the DNA damage sensor protein 53BP1, and inhibited ATM activation and subsequent p53 phosphorylation. This finding may explain why human tumors with high levels of ΔNp73 expression showed resistance to chemotherapy.
In summary, these studies show that TAp63 and TAp73 proteins have specific roles in preventing tumor development in vivo. Conversely the ΔN forms act as oncogenes by preventing senescence and maintaining progenitor cell status. When overexpressed, both TAp63 and TAp73 proteins transactivate subsets of known p53 target genes involved in cell-cycle arrest and apoptosis, such as p21Cip1 and Bbc3 (Zhu 1998; Dohn 2001; Melino 2002, 2011). Of note, both TAp63 and TAp73 also regulate distinct sets of genes that are not transcriptional targets for p53 through unique p63RE as described in the previous section. In contrast, ΔNp63 and ΔNp73 proteins have been shown to function in part as dominant-negative inhibitors of the p53 family, leading to the hypothesis that these isoforms may exhibit proto-oncogenic function. ΔN isoforms inhibit the function of TA forms through 1) direct competition for DNA-binding sites and 2) formation hetero-oligomeric complexes with TAp63/TAp73, and less strongly with p53 (Yang 1998; Davidson 1999; Chi 1999; Grob 2001; Stiewe 2002; Chan 2004). Interestingly, expression of the ΔNp73 is strongly up-regulated by TAp73 and p53, thus creating a feedback loop that tightly regulates the function of TAp73 and more importantly, of p53 (Grob 2001).
Aberrant expression, altered splicing, and mutations of p63 and p73 in human cancer
Alterations of p63 isoforms in human cancers
Both p63 and p73 were initially hypothesized to function as tumor suppressors based on their homology to p53. However, accumulating evidence shows that mutation of either of these genes in human cancer is quite rare (Yoshikawa 1999; Ichimiya 1999), indicating that they are not classical tumor suppressor genes like p53 or RB that meet the Knudson’s two-hit hypothesis (Knudson 1971). Although there have been numerous studies on p63 expression in human cancers, loss of heterozygosity (LOH) of the p63 locus has not been studied extensively in human malignancies (Zaika & El-Rifai 2006), possibly because the genomic locus 3q27–28 is not the site of frequent gene deletion in cancer. Conversely, decreased p63 expression is a common feature of high-grade invasive urothelial carcinomas and associates with reduced β-catenin. Both ΔNp63 and TAp63 are frequently downregulated in bladder cancer and this reduction correlates with a poor prognosis (Park 2000). The majority of prostate cancers show loss of p63, but it is overexpressed in some poorly differentiated tumors and correlates with a poor prognosis (Grismazio 2008). In addition, loss of p63 results in enhanced metastasis in prostate cancer (Tuccu 2012). Koga et al. (2003) studied the expression of p63, β-catenin, and uroplakin III by immunohistochemistry in high-grade invasive bladder carcinomas. Lower p63 expression was significantly associated with higher TNM stage, lymph-node metastasis, reduced β-catenin expression, and thus with poor prognosis. Impaired p63 expression was associated with biological aggressiveness of high-grade invasive urothelial carcinomas. Moreover, loss of p63 expression was a pre-requisite for uroplakin III expression. Their data suggested that p63 plays critical roles in the prevention of tumor progression and biochemical terminal differentiation of urothelial neoplasms (Koga 2003).
Oral lichen planus (OLP) is a relatively common chronic disease of the oral mucosa for which the etiology or pathogenesis is not fully understood. Sniezek et al. (2002, 2004) showed decreased expression of p63 in OLP compared to normal mucosa, a decrease they suggested could explain the hyper-differentiation, or pro-differentiation, seen in OLP. Consistent with these findings, another group reported downregulation of p63 (Ebrahimi 2006) in OLP and GVHD associated with oral inflammation.
The p63 gene maps to chromosome 3q27–28, a region frequently amplified in squamous cell carcinomas (Hibi 2000; DiComco 2002; Massion 2003; Zaika & El-Rifai 2006; Brunelli 2012). Most squamous cell carcinomas retain p63 expression, where it is often overexpressed (Massion 2003; DeYoung 2006; King 2007). Although some controversy exists as to whether p63 is the targeted gene driving amplification of this locus, several groups have reported increased p63 mRNA levels that correlate with an increase in p63 gene copy number in squamous cell carcinomas of the lung, head, and neck (HNSCCs) (Hibi 2000; Tonon 2005). In other cases, overexpression of p63 appears to be independent of genomic DNA amplification of the locus (Redon 2001). In esophageal carcinomas, amplification of the p63 gene was reported in ~20% of squamous cell carcinomas and 10% of adenocarcinomas (Zaika & El-Rifai 2006). Given that the total frequency of tumors in which p63 is upregulated is much higher (>50%), gene amplification is unlikely to be the main mechanism underlying the increased levels of p63. Rather, transcriptional or post-transcriptional changes are involved. Multiple studies have shown that p63 overexpression occurs in up to 80% of primary HNSCCs and also in other squamous cell carcinomas, including those in the lung, nasopharynx, and cervix (Wang 2001; Hu 2002; Weber 2002; Massion 2003). By the use of isoform-specific antibodies, Nylander et al. (2002) mapped expression of the different p63 isoforms within normal oral mucosa and HNSCCs, showing increased expression of p63, mainly the ΔNp63 isoforms, in tumors compared to normal mucosa. They indicated specific roles for the individual isoforms in cell differentiation and neoplasia (Nylander 2002). In invasive breast cancer, the frequency of p63 expression varies, ranging from 0 to 30% (Wang 2002; Reis-Filho 2003; Koker & Kleer 2004). It is now considered that p63 is expressed in at least a subset of breast tumors that are known to have a basal epithelial phenotype (Livasy 2006).
TAp63 vs. ΔNp63 in cancer
In esophageal carcinomas, p63 isoforms are upregulated not only in carcinomas, but also in squamous dysplasias (Zaika & El-Rifai 2006). Although early studies for the detection of p63 did not differentiate among different isoforms, recent studies used isoform-specific RT-PCR coupled with Western blot analysis to quantitatively demonstrate that ΔNp63α is the predominant p63 isoform expressed in squamous cell carcinomas. Using such an approach, it has been reported that tumor-suppressive TAp63 overexpression is rare in HNSCC, and that ΔNp63 mRNA expression was at least 100-fold more abundant than TAp63 mRNA in all cases (Deyoung 2006; Rocco 2006). These findings are consistent with the inability of many investigators to detect TAp63 protein isoforms by Western blot analysis in either primary keratinocytes or HNSCC cells. ΔNp63 is the predominant variant that is found in HNSCCs; however, in Barrett’s esophagus, a disorder in which the stratified epithelium is replaced by a simple columnar epithelium that consists of mucosecretory cells, the p63 gene expression is not highly prominent (Zaika & El-Rifai 2006).
Tumors often have simultaneous transcriptional upregulation of both TAp63 and ΔNp63 isoforms, with ΔNp63 being predominant at protein levels (Massion 2003; DeYoung 2006). This would represent the anti-apoptotic and proliferative effects of ΔNp63 as described in the previous section. Moreover, it was reported that ΔNp63α expression directly correlates with a poor response to cisplatin in HNSCC (Zangen 2005). In pancreatic cancer, Danilov et al. (2011) showed that ΔNp63α enhanced the oncogenic potential of tumor cells through trans-activation of EGFR and 14-3-3σ. Leong et al. (2007) reported that the p63/p73 network mediates chemosensitivity to cisplatin in a subset of primary breast cancers. Thus, p63 is involved in chemosensitivity of multiple types of tumors. In HNSCC, DNA damage by chemotherapy caused a decrease in ΔNp63-mediated transcriptional repression by blocking p63-responsive elements or sequestering TAp63 in less active hetero-tetramers, together with increased expression of p73, thus allowing TAp73-mediated cell death (Rocco 2006). Together, these reports indicate that it is not only the levels of individual p53 family members, but rather the ratio between TA (transcriptionally active, having tumor-suppressor functions) and ΔN (acting as dominant-negative over the TA isoforms, showing oncogenic properties) isoforms that determines the biological outcome.
In lung cancer, amplification of chromosomal region 3q26-3qter is frequently found in tumors. Massion et al. (2003) analyzed p63 gene copy number and expression by immunohistochemistry in tissue microarrays of >200 non-small cell lung cancers (NSCLCs) and correlated them with survival. The p63 genomic locus was amplified in 88% of squamous cell carcinomas, but only in 11% of adenocarcinomas of the lung, indicating clear association of gene amplification with squamous cell lung cancer. The major splicing variant of p63 expressed was ΔNp63α. Furthermore, p63 genomic amplification and protein staining was associated with better survival. They found a significant increase in p63 copy number in pre-invasive lesions graded severe dysplasia or higher. Thus, there is early and frequent genomic amplification of p63 in the development of squamous carcinoma of the lung and patients with NSCLC showing amplification and overexpression of p63 had prolonged survival (Massion 2003). However, two other groups have failed to demonstrate the favorable prognostic value of p63 in lung cancer. Iwata et al. (2005) reported a lack of prognostic significance regarding ΔNp63 immunoreactivity in lung cancer. Uramoto et al. (2006) showed that the expressions of ΔNp63 in lung cancer did not significantly affect survival while patients with a positive ΔNp73 expression had a poorer prognosis in comparison to the negative group. The differential prognostic values of p63 in these, Massion’s and two other studies, can be attributed to the fact that the former study focused on gene copy number of p63 and immunohistochemical staining of p63 (all splicing isoforms) in squamous cell lung cancer while the latter two groups studied the expression of the ΔNp63 protein and survival of non-small cell lung cancer (in Uramoto’s study; squamous cell carcinoma only in Iwata’s study).
ΔNp63α can act as a transcriptional repressor, but the link between the transcriptional functions of p63 and its biological role is still unclear. Barbieri et al. (2006) depleted endogenous p63 by shRNA to investigate the transcriptional programs controlled by p63. Disruption of p63 in squamous cell carcinoma cell lines resulted in down-regulation of transcripts specifically expressed in squamous tissues and a significant alteration of keratinocyte differentiation. They found that depletion of p63 led to up-regulation of markers of non-epithelial tissues (mesenchyme and neural tissue) in squamous cell carcinomas, which were associated with increased capacity for invasion and metastasis in tumors. Furthermore, loss of p63 expression was accompanied by a shift toward mesenchymal morphology and an increase in motility in primary keratinocytes and squamous cell lines (Barbieri 2006). Thus, loss of endogenous p63 results in up-regulation of genes associated with invasion and metastasis, and predisposes to a loss of epithelial markers and acquisition of mesenchymal characteristics. Although the squamous cell carcinoma cell lines they analyzed expressed predominantly ΔNp63, the interpretation of their experimental results is controversial since their shRNA depleted both TAp63 and ΔNp63 at the same time. p63 isoform-specific shRNA should be used to define the roles of each isoform in cell growth, differentiation, invasion and metastasis.
Regulation of gene expression by ΔNp63
Although ΔNp63 lacks the amino-terminal transactivation domain consisting of 39 amino acids that is present in TAp63, ΔNp63 still activates a group of genes that includes, but is not restricted to genes regulated by p53 (Dohn 2001). Helton et al. showed that all NH2-terminally deleted p63 isoforms still retain a potential in transactivation and growth suppression (2006). Interestingly, they showed that ΔNp63β possessed a remarkable ability to suppress cell proliferation and transactivate target genes, which is consistently higher than that seen with ΔNp63α. They showed that an intact DNA-binding domain is required for ΔNp63 function. In addition, they found that the novel transactivation domain for the ΔNp63 variant was composed of the 14 unique ΔN residues along with the adjacent region, including a PXXP motif (Helton 2006). They also showed that a PPXY motif shared by ΔNp63α and ΔNp63β was required for optimal transactivation of target gene promoters (Helton 2006). Very recently, Ceraldo et al. (2013) identified a novel p63 transcriptional target, caspase-1. Caspase-1 is pro-inflammatory caspase, which functions in tumor suppression. They showed that both p63 isoforms (TAp63, ΔNp63) increased caspase-1 expression through physical binding to its promoter. Consistently they also identified a direct correlation between p63 and caspase-1 expression in human cancer data sets. Functional interaction between p63 and caspase-1 represented a predictor of longer survival in human cancers. Together, in addition to dominant-negative effects of ΔNp63 on TA isoforms of p53 family proteins, regulation of gene expression by ΔNp63 variants should be re-evaluated from the viewpoint of tumor suppression.
Alterations of p73 isoforms in human cancers
The p73 gene has been speculated to be classical tumor suppressor genes like p53 when the cDNAs were cloned (Kaghad 1997). In gastrointestinal tumors, LOH for p73 has been reported in 10–40% of the cases (Zaika & El-Rifai 2006) although LOH for p63 has not been reported in cancers. Despite these expectations, subsequent studies have demonstrated that the TP73 locus was not the hot spot of gene deletion in cancers. Rather, studies of multiple tumor types have shown that p73 splicing variants are overexpressed, but not mutated or deleted in human malignancies (Moll & Slade 2004).
To investigate the role of the p73 gene in human carcinogenesis, Han et al. (1999) studied genetic alterations of this gene by analyzing the entire coding exons as well as their surrounding exon-intron boundaries by PCR-CCSP and direct sequencing with primary samples from breast, colorectal, gastric cancers, neuroblastomas, and also with lung and pancreatic cancer cell lines since they are known to have frequent LOH in the 1p region. However, of the 185 cases, somatic missense mutation of glutamine from arginine at codon 269 was found in only one breast cancer. Monoallelic expression of p73 was observed in pancreatic cancer cell lines. Nomoto et al. (1998) analyzed 61 primary lung cancer samples of the p73 locus at 1p36.33 by PCR-SSCP and Southern blotting. Although allelic loss at the 1p36.33 locus was observed in 42% of cases, somatic mutations of the p73 gene were not observed in their samples, suggesting the presence of an as yet to be determined tumor suppressor gene at the locus. In summary, inactivation of the p73 gene is very rare even in cancers involving chromosome 1p (Han 1999).
TAp73 vs. ΔTAp73 in human cancers
Overexpression of p73 mRNA and/or protein relative to neighbor normal tissues has been reported in a variety of tumors, such as neuroblastoma, glioma, ependymoma, breast, lung, colon, stomach, liver, ovarian, bladder, cholangiocellular carcinomas, and myelogenous leukemias (Moll & Slade 2004). Concin et al. (2004) studied the expression profile of all N-terminal isoforms, distinguishing between TAp73 and ΔTAp73 (ΔNp73, ΔN′p73, Δex2p73, and Δex2/3p73) (Figure 2). Ovarian cancers almost universally overexpressed ΔN′p73 compared with normal tissues (95% of cancers). About one-third of tumors also exhibited concomitant up-regulation of TAp73, whereas only a small subgroup of tumors overexpressed ΔNp73 (Concin 2004). Thus, deregulation of the E2F-responsive P1 promoter, rather than the P2 promoter, is mainly responsible for the production of ΔTAp73 in ovarian cancer. A trend was found for better overall survival in patients with low expression of ΔN′p73/ΔNp73, compared with those with high expression. Cancers with wild-type p53 showed significantly higher deregulation of ΔNp73, ΔN′p73, and Δex2/3p73 (transdominant p73) than p53 mutant cancers. Thus, overexpression of transdominant p73 isoforms can function as epigenetic inhibitors of p53 in vivo, thereby alleviating selection pressure for p53 mutations in ovarian cancer (Concin 2004).
Dominguez et al. (2006) analyzed 113 colon and 60 breast cancer patients’ primary samples and reported the association of ΔTAp73 variants and advanced pathologic stage, lymph node metastasis, vascular invasion, presence of polyps, and tumor localization. Overexpression of TP73 variants in tumor tissues indicates that they may be involved in carcinogenesis. The association between upregulation of ΔTAp73 isoforms and poor prognosis suggests that they may be of practical clinical prognostic value. Faridoni-Laurens et al. (2008) analyzed the expression of TAp73 and ΔTAp73 in HNSCC and compared them to the p53 status. They found that all of the p73 isoforms were upregulated in comparison to those in normal adjacent tissue. Although p73 belongs to the gene family of p53, p53 mutations and p73 transcript alterations were not mutually exclusive. All of the HNSCC specimens studied had at least one p53 mutation and/or one ΔTAp73 transcript alteration. Although both the ΔNp73 and the TAp73 transcripts were upregulated in HNSCC, the predominant protein in the cancers expressed was ΔNp73. Furthermore, a trend was found for better overall survival in patients with a low expression of ΔNp73. Thus deregulation of both the p53 and the p73 pathways plays an important role in inducing HNSCC (Faridoni-Laurens 2008).
By using specific polyclonal ΔNp73 antiserum against the exon 3′-specific peptide for p73, ΔNp73 and ΔN′p73 expressions were studied in paraffin-embedded tumor samples from 132 lung cancer samples (Uramoto 2004, 2006). The ΔN/ΔN′p73 protein was detected mainly in the cytoplasm of tumor cells in 77 of 132 patients (58.3%) with lung cancer. Importantly, lung cancer patients with positive ΔN/ΔN′p73 expression had a poorer clinical outcomes than those with negative expression. In addition, multivariate analysis of the clinicopathological characteristics of lung cancer indicated that positive expression of ΔN/ΔN′p73 was a significant independent factor for predicting poor prognosis (P < 0.0001, risk ratio = 3.39). Thus, expression of ΔN/ΔN′p73 will be a useful marker for predicting poor prognosis of patients who undergo resection of lung cancer. Consistent with these findings, overexpression of the N-terminal splice variants (Δex2p73, Δex2-3p73), but not TAp73, was shown to be associated with a poor prognosis in low-grade gliomas (Wager 2006), which should be helpful in decision-making in clinics.
The truncated oncogenic isoform Δex2p73 is expressed in hepatocellular carcinomas (HCC); however, the underlying mechanisms regulating this process are unknown. Castillo et al. (2009) used human normal and diseased liver tissue samples to examine the association between activation of epidermal growth factor receptor (EGFR) by its ligand amphiregulin (AR) and the alternative splicing of p73 pre-mRNA into the tumorigenic isoform Δex2p73, via c-Jun N-terminal-kinase-1-mediated signaling. Δex2p73 was expressed in a subset of premalignant cirrhotic livers and in otherwise healthy livers that harbored a primary tumor, as well as in HCC tissues. Δex2p73 expression was correlated with that of the EGFR ligand AR, previously shown to have a role in hepatocarcinogenesis. Autocrine activation of the EGFR by AR triggered c-Jun N-terminal kinase-1 activity and inhibited the expression of the splicing regulator Slu7, leading to the accumulation of Δex2p73 transcripts in HCC cells. Their study provided a mechanism for the generation of pro-tumorigenic Δex2p73 during liver tumorigenesis via activation of EGFR signaling by AR and c-Jun N-terminal kinase-1 activity, leading to inhibition of the splicing regulator Slu7 (Castillo 2009). This is a unique report that showed the specific role of a particular splicing factor in aberrant p73 splicing.
The molecular mechanisms underlying overexpression of ΔNp63 or ΔNp73 in cancers in comparison to normal tissues need further investigation. Methylation-mediated silencing of the P1 promoter for TAp73 was reported in lymphoblastic leukemias and Burkitt’s lymphomas (Corn 1999; Kawano 1999). These findings indicate that either ΔNp63 or ΔNp73 overexpression or TAp73 promoter silencing is required to inactivate the tumor-suppressive activity of TAp73. Although TAp73 isoforms were paradoxically overexpressed (18–30 folds) in HNSCC tumor cells in comparison to non-transformed keratinocytes, ΔNp63α was also overexpressed in these tumors and was physically associated with TAp73, thereby inhibiting p73-dependent pro-apoptotic activity (Deyoung 2006; Rocco 2006). BRCA1-deficient tumor cells exhibit increased sensitivity to cisplatin, and patients with BRCA1-associated ovarian carcinomas had better outcomes with platinum-based chemotherapy compared with sporadic cases. Ibrahim et al. (2010) reported that BRCA1-deficient ovarian carcinoma cells exhibited hypermethylation within the P1 promoter for p73, which included the binding site for the p73 transcriptional repressor ZEB1, leading to the abrogation of ZEB1-binding and increased expression of transactivating p73 isoforms (TAp73), explaining increased cisplatin sensitivity of BRCA1-deficient ovarian carcinomas. Thus, TAp73 might represent a response predictor and potential therapeutic target for enhancing chemosensitivity in ovarian cancer.
Although promoter methylation is the major mechanism of p73 inactivation in hematopoietic malignancies (Alexandrova & Moll 2012), the situation is different in epithelial tumors - carcinomas. Daskalos et al. (2011) studied the DNA methylation status of both P1 and P2 promoters as a means of epigenetic transcriptional control of their corresponding isoforms in 102 primary NSCLCs and reported that the P2 hypomethylation-associated overexpression of ΔNp73 mRNA is a frequent event, particularly among squamous cell carcinomas. P2 hypomethylation strongly correlated with long interspersed nuclear element-1 element hypomethylation, indicating that ΔNp73 overexpression may be a consequence of global DNA hypomethylation. Guan et al. analyzed p73 in prostate cancer and found that ΔNp73 was significantly increased in 20 of 33 prostate carcinomas (Guan & Chen 2005). However, none of the specimens expressed ΔN′p73. The positive expression of ΔNp73 correlated with the Gleason score in prostate cancer. Interestingly, prostate cancer samples with wild-type p53 had significantly higher expression of ΔNp73 than p53 mutant cancers. These data suggested a potential role for ΔNp73 in prostate cancer progression.
Diaz et al. (2010) conducted a translational study to evaluate whether 1,25(OH)(2) vitamin D(3) downregulates TP73 variants in colon and breast cancers (Diaz 2010). They reported that ectopic survivin expression led to an increase in all of the TAp73, ΔNp73, ΔEx2p73, and ΔEx2-3p73 transcripts. In these cancers, direct correlations were observed between TP73 variants and survivin levels. Interestingly, 1,25(OH)(2) vitamin D(3) negatively regulated survivin and TP73 variants in these tumors. Thus positive regulation of TP73 isoforms by survivin may exist, which raised the possibility that the downregulation of TP73 isoforms may be possible with 1,25(OH)(2)D(3) through survivin.
In summary, although somatic point mutations are rarely found in p73 in human cancers, aberrant splicing that result in ΔTAp73 overexpression are very frequently found. Since these proteins have transdominant activity on all p53 family proteins, it is speculated that this abnormal splicing contributes to human carcinogenesis, esp. in ovarian, breast, lung, and prostate cancers, HNSCCs, and hematological malignancies. Published results indicate that ΔTAp73 overexpression is associated poor clinical outcomes at least in lung cancer and HNSCCs. Of note, it may be possible to correct aberrant expression of p73 isoforms in cancer through the use of 1,25(OH)(2)D(3).
Conclusive remarks
Judging from the very low frequency of mutations for p63 and p73 in human cancers, these are not classical tumor suppressor genes, but the possibility remains that these are haplo-insufficient tumor suppressors, just like p27Kip1, PTEN, or DMP1 (Quon & Berns, 2001, Berger & Pandolffi 2011; Inoue 2001, 2007; Mallakin 2007; Sugiyama 2008; Taneja 2010; Zhu 2013; Fry 2013). Detailed analyses with specific primers are required to determine whether these are true tumor suppressors. Accumulating pieces of evidence suggest that TA- and ΔN- isoforms play distinct roles in cell cycle progression, apoptosis, and tumor development/prevention. Detection of each isoform by Western blotting or immunohistochemistry with specific antibodies or real-time PCR-mediated quantification of each splicing isoform will be needed to determine the prognostic value of each splicing isoform in cancer. Of note, both ΔNp73 and ΔN′p73 have unique amino acid sequences generated from the exon 3′ that is absent in TAp73. This has made it possible to generate ΔN/ΔN′p73-specific antibodies that can be used in diagnostic immunohistochemistry.
Not many studies have been done to elucidate the mechanisms of overexpression of ΔN isoforms of p63 and p73 in human cancers. Identification of critical splicing factors and characterization of signaling pathways that contribute to this process will be critical to correct the errors for splicing for these genes in human cancers. Finally, specific targeting of ΔN- isoforms with antisense DNA, stabilized RNA, shRNA may have therapeutic values in treating human cancer overexpressing these splicing isoforms with oncogenic activity.
Acknowledgments
K. Inoue has been supported by NIH/NCI 2R01CA106314, ACS RSG-07-207-01-MGO, and KG080179.
References
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. Review. [DOI] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–37. doi: 10.1038/nrc2730. Review. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. Review. [DOI] [PubMed] [Google Scholar]
- Green DR, Chipuk JE. p53 and metabolism: Inside the TIGAR. Cell. 2006 Jul 14;126(1):30–2. doi: 10.1016/j.cell.2006.06.032. Review. [DOI] [PubMed] [Google Scholar]
- Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013 Feb 18; doi: 10.1038/onc.2012.640. Review. [DOI] [PubMed]
- Xu Y. Induction of genetic instability by gain-of-function p53 cancer mutants. Oncogene. 2008 Jun 5;27(25):3501–7. doi: 10.1038/sj.onc.1211023. [DOI] [PubMed] [Google Scholar]
- Lai D, Visser-Grieve S, Yang X. Tumour suppressor genes in chemotherapeutic drug response. Biosci Rep. 2012 Aug;32(4):361–74. doi: 10.1042/BSR20110125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Martins CP. From tumor prevention to therapy: empowering p53 to fight back. Drug Resist Updat. 2012 Oct;15(5–6):258–67. doi: 10.1016/j.drup.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Senzer N, Nemunaitis J, Nemunaitis D, Bedell C, Edelman G, Barve M, Nunan R, Pirollo KF, Rait A, Chang EH. Phase I study of a systemically delivered p53 nanoparticle in advanced solid tumors. Mol Ther. 2013 May;21(5):1096–103. doi: 10.1038/mt.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997 Aug 22;90(4):809–19. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dötsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998 Sep;2(3):305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006 Jun;13(6):962–72. doi: 10.1038/sj.cdd.4401914. Review. [DOI] [PubMed] [Google Scholar]
- Schultz J, Ponting CP, Hofmann K, Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997 Jan;6(1):249–53. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos CD, Bowie JU. p53 Family members p63 and p73 are SAM domain-containing proteins. Protein Sci. 1999 Aug;8(8):1708–10. doi: 10.1110/ps.8.8.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber Z, Lai HC, Yang A, Ou HD, Sigal MS, Kelly AE, Darimont BD, Duijf PH, Van Bokhoven H, McKeon F, Dötsch V. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol Cell Biol. 2002 Dec;22(24):8601–11. doi: 10.1128/MCB.22.24.8601-8611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Slade N. p63 and p73: roles in development and tumor formation. From tumor prevention to therapy: empowering p53 to fight back. Mol Cancer Res. 2004 Jul;2(7):371–86. Review. [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005 Sep 15;19(18):2122–37. doi: 10.1101/gad.1339905. Epub 2005 Aug 30. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois S, Caron de Fromentel C, Hainaut P. p53 protein variants: structural and functional similarities with p63 and p73 isoforms. Oncogene. 2004 Jan 22;23(3):631–8. doi: 10.1038/sj.onc.1206929. Review. [DOI] [PubMed] [Google Scholar]
- Chi SW, Ayed A, Arrowsmith CH. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 1999 Aug 16;18(16):4438–45. doi: 10.1093/emboj/18.16.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998 Nov 2;188(9):1763–8. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999 Apr 22;398(6729):708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992 Mar 19;356(6366):215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994 Jan 1;4(1):1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990 Nov 30;250(4985):1233–8. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- Malkin D. p53 and the Li-Fraumeni syndrome. Cancer Genet Cytogenet. 1993 Apr;66(2):83–92. doi: 10.1016/0165-4608(93)90233-c. Review. [DOI] [PubMed] [Google Scholar]
- Strong LC, Williams WR, Tainsky MA. The Li-Fraumeni syndrome: from clinical epidemiology to molecular genetics. Am J Epidemiol. 1992 Jan 15;135(2):190–9. doi: 10.1093/oxfordjournals.aje.a116271. Review. [DOI] [PubMed] [Google Scholar]
- Osada M, Park HL, Nagakawa Y, Yamashita K, Fomenkov A, Kim MS, Wu G, Nomoto S, Trink B, Sidransky D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol. 2005 Jul;25(14):6077–89. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002 Nov;10(5):995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T. The p53 family member genes are involved in the Notch signal pathway. J Biol Chem. 2002 Jan 4;277(1):719–24. doi: 10.1074/jbc.M108080200. Epub 2001 Oct 18. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Takahashi M, Ozaki T, Watanabe Ki K, Todo S, Mizuguchi H, Hayakawa T, Nakagawara A. Autoinhibitory regulation of p73 by Delta Np73 to modulate cell survival and death through a p73-specific target element within the Delta Np73 promoter. Mol Cell Biol. 2002 Apr;22(8):2575–85. doi: 10.1128/MCB.22.8.2575-2585.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley NE, Nic An Tsaoir CB, Blayney JK, Oram LC, Crawford NT, D’Costa ZC, Quinn JE, Kennedy RD, Harkin DP, Mullan PB. BRCA1 is a key regulator of breast differentiation through activation of Notch signaling with implications for anti-endocrine treatment of breast cancers. Nucleic Acids Res. 2013;41:8601–14. doi: 10.1093/nar/gkt626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55 kD protein. Genes Dev. 1994;8:1235–46. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–53. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Deb SP. Cell cycle regulatory functions of the human oncoprotein MDM2. Mol Cancer Res. 2003 Dec;1(14):1009–16. Review. [PubMed] [Google Scholar]
- Popowicz GM, Czarna A, Holak TA. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle. 2008;7:2441–3. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–43. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–8. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710–4. doi: 10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- Ongkeko WM, Wang XQ, Siu WY, Lau AW, Yamashita K, Harris AL, Cox LS, Poon RY. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol. 1999;9(15):829–32. doi: 10.1016/s0960-9822(99)80367-4. Jul 29–Aug 12. [DOI] [PubMed] [Google Scholar]
- Zdzalik M, Pustelny K, Kedracka-Krok S, Huben K, Pecak A, Wladyka B, Jankowski S, Dubin A, Potempa J, Dubin G. Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle. 2010 Nov 15;9(22):4584–91. doi: 10.4161/cc.9.22.13871. [DOI] [PubMed] [Google Scholar]
- Kojima T, Ikawa Y, Katoh I. Analysis of molecular interactions of the p53-family p51(p63) gene products in a yeast two-hybrid system: homotypic and heterotypic interactions and association with p53-regulatory factors. Biochem Biophys Res Commun. 2001;281:1170–5. doi: 10.1006/bbrc.2001.4486. [DOI] [PubMed] [Google Scholar]
- Little NA, Jochemsen AG. Hdmx and Mdm2 can repress transcription activation by p53 but not by p63. Oncogene. 2001;20:4576–80. doi: 10.1038/sj.onc.1204615. [DOI] [PubMed] [Google Scholar]
- Wang X, Arooz T, Siu WY, Chiu CH, Lau A, Yamashita K, et al. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 2001;490:202–8. doi: 10.1016/s0014-5793(01)02124-x. [DOI] [PubMed] [Google Scholar]
- Calabro V, Mansueto G, Parisi T, Vivo M, Calogero RA, La Mantia G. The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J Biol Chem. 2002;277:2674–81. doi: 10.1074/jbc.M107173200. [DOI] [PubMed] [Google Scholar]
- Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- Burge S, Teufel DP, Townsley FM, Freund SM, Bycroft M, Fersht AR. Molecular basis of the interactions between the p73 N terminus and p300: effects on transactivation and modulation by phosphorylation. Proc Natl Acad Sci USA. 2009;106:3142–7. doi: 10.1073/pnas.0900383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzalik M, Pustelny K, Kedracka-Krok S, Huben K, Pecak A, Wladyka B, Jankowski S, Dubin A, Potempa J, Dubin G. Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle. 2010;9:4584–91. doi: 10.4161/cc.9.22.13871. [DOI] [PubMed] [Google Scholar]
- Keyes WM, Vogel H, Koster MI, Guo X, Qi Y, Petherbridge KM, Roop DR, Bradley A, Mills AA. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc Natl Acad Sci USA. 2006 May 30;103(22):8435–40. doi: 10.1073/pnas.0602477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000 Mar 2;404(6773):99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002 Apr 4;416(6880):560–4. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- Taneja P, Zhu S, Maglic D, Fry EA, Kendig RD, Inoue K. Transgenic and knockout mice models to reveal the functions of tumor suppressor genes. Clin Med Insights Oncol. 2011;5:235–57. doi: 10.4137/CMO.S7516. Epub 2011 Jul 28. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR, Pietenpol JA, Carbone DP, Gonzalez AL. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003 Nov 1;63(21):7113–21. [PubMed] [Google Scholar]
- Flores ER. The roles of p63 in cancer. Cell Cycle. 2007 Feb 1;6(3):300–4. doi: 10.4161/cc.6.3.3793. Epub 2007 Feb 3. Review. [DOI] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, Biernaskie JA, Sinha S, Prives C, Pevny LH, Miller FD, Flores ER. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009 Jul 2;5(1):64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes WM, Pecoraro M, Aranda V, Vernersson-Lindahl E, Li W, Vogel H, Guo X, Garcia EL, Michurina TV, Enikolopov G, Muthuswamy SK, Mills AA. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011 Feb 4;8(2):164–76. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H, Mills AA. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009 Dec;11(12):1451–7. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar A, Virolle T, Rostagno P, de la Forest-Divonne S, Gambaro K, Rouleau M, Aberdam D. DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One. 2008;3(10):e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, Melino G, Aberdam D. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011 May;18(5):887–96. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberdam D, Gambaro K, Medawar A, Aberdam E, Rostagno P, de la Forest Divonne S, Rouleau M. C R Biol. 2007 Jun-Jul;330(6–7):479–84. doi: 10.1016/j.crvi.2007.03.007. Epub 2007 Apr 16. Review. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008 Oct 1;22(19):2677–91. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, Kaplan DR, Miller FD, Mak TW. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010 Mar 15;24(6):549–60. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998 Nov 15;58(22):5061–5. [PubMed] [Google Scholar]
- Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001 May 31;20(25):3193–205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002 Aug;2(8):605–15. doi: 10.1038/nrc861. Review. [DOI] [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011 Sep;18(9):1487–99. doi: 10.1038/cdd.2011.81. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison TS, Vagner C, Kaghad M, Ayed A, Caput D, Arrowsmith CH. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J Biol Chem. 1999 Jun 25;274(26):18709–14. doi: 10.1074/jbc.274.26.18709. [DOI] [PubMed] [Google Scholar]
- Grob TJ, Novak U, Maisse C, Barcaroli D, Lüthi AU, Pirnia F, Hügli B, Graber HU, De Laurenzi V, Fey MF, Melino G, Tobler A. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001 Dec;8(12):1213–23. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- Stiewe T, Zimmermann S, Frilling A, Esche H, Pützer BM. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res. 2002 Jul 1;62(13):3598–602. [PubMed] [Google Scholar]
- Chan WM, Siu WY, Lau A, Poon RY. How many mutant p53 molecules are needed to inactivate a tetramer? Mol Cell Biol. 2004 Apr;24(8):3536–51. doi: 10.1128/MCB.24.8.3536-3551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H, Nagashima M, Khan MA, McMenamin MG, Hagiwara K, Harris CC. Mutational analysis of p73 and p53 in human cancer cell lines. Oncogene. 1999 Jun 3;18(22):3415–21. doi: 10.1038/sj.onc.1202677. [DOI] [PubMed] [Google Scholar]
- Ichimiya S, Nimura Y, Kageyama H, Takada N, Sunahara M, Shishikura T, Nakamura Y, Sakiyama S, Seki N, Ohira M, Kaneko Y, McKeon F, Caput D, Nakagawara A. p73 at chromosome 1p36.3 is lost in advanced stage neuroblastoma but its mutation is infrequent. Oncogene. 1999 Jan 28;18(4):1061–6. doi: 10.1038/sj.onc.1202390. [DOI] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971 Apr;68(4):820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaika AI, El-Rifai W. The role of p53 protein family in gastrointestinal malignancies. Cell Death Differ. 2006 Jun;13(6):935–40. doi: 10.1038/sj.cdd.4401897. Review. [DOI] [PubMed] [Google Scholar]
- Park BJ, Lee SJ, Kim JI, Lee CH, Chang SG, Park JH, Chi SG. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer research. 2000;60:3370–74. [PubMed] [Google Scholar]
- Grisanzio C, Signoretti S. p63 in prostate biology and pathology. J Cell Biochem. 2008;103:1354–68. doi: 10.1002/jcb.21555. [DOI] [PubMed] [Google Scholar]
- Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, Muller PA, Dötsch V, Kehrloesser S, Sayan BS, Giaccone G, Lowe SW, Takahashi N, Vandenabeele P, Knight RA, Levine AJ, Melino G. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA. 2012;109:15312–7. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga F, Kawakami S, Fujii Y, Saito K, Ohtsuka Y, Iwai A, Ando N, Takizawa T, Kageyama Y, Kihara K. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin Cancer Res. 2003 Nov 15;9(15):5501–7. [PubMed] [Google Scholar]
- Sniezek JC, Matheny KE, Burkey BB, Netterville JL, Pietenpol JA. Expression of p63 and 14–3–3sigma in normal and hyperdifferentiated mucosa of the upper aerodigestive tract. Otolaryngol Head Neck Surg. 2002 Jun;126(6):598–601. doi: 10.1067/mhn.2002.125302. [DOI] [PubMed] [Google Scholar]
- Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope. 2004 Dec;114(12):2063–72. doi: 10.1097/01.mlg.0000149437.35855.4b. [DOI] [PubMed] [Google Scholar]
- Ebrahimi M, Wahlin YB, Coates PJ, Sjöström B, Nylander K. Decreased expression of p63 in oral lichen planus and graft-vs.-host disease associated with oral inflammation. J Oral Pathol Med. 2006 Jan;35(1):46–50. doi: 10.1111/j.1600-0714.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA. 2000 May 9;97(10):5462–7. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como CJ, Urist MJ, Babayan I, Drobnjak M, Hedvat CV, Teruya-Feldstein J, Pohar K, Hoos A, Cordon-Cardo C. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002 Feb;8(2):494–501. [PubMed] [Google Scholar]
- Brunelli M, Bria E, Nottegar A, Cingarlini S, Simionato F, Caliò A, Eccher A, Parolini C, Iannucci A, Gilioli E, Pedron S, Massari F, Tortora G, Borze I, Knuutila S, Gobbo S, Santo A, Tondulli L, Calabrò F, Martignoni G, Chilosi M. True 3q chromosomal amplification in squamous cell lung carcinoma by FISH and a CGH molecular analysis: impact on targeted drugs. PLoS One. 2012;7(12):e49689. doi: 10.1371/journal.pone.0049689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung MP, Johannessen CM, Leong CO, Faquin W, Rocco JW, Ellisen LW. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006 Oct 1;66(19):9362–8. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- King KE, Weinberg WC. p63: defining roles in morphogenesis, homeostasis, and neoplasia of the epidermis. Mol Carcinog. 2007 Aug;46(8):716–24. doi: 10.1002/mc.20337. [DOI] [PubMed] [Google Scholar]
- Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, Martin ES, Yang Z, Ji H, Chin L, Depinho RA. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA. 2005a Jul 5;102(27):9625–30. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonon G, Brennan C, Protopopov A, Maulik G, Feng B, Zhang Y, Khatry DB, You MJ, Aguirre AJ, Martin ES, Yang Z, Ji H, Chin L, Wong KK, Depinho RA. Common and contrasting genomic profiles among the major human lung cancer subtypes. Cold Spring Harb Symp Quant Biol. 2005b;70:11–24. doi: 10.1101/sqb.2005.70.021. [DOI] [PubMed] [Google Scholar]
- Redon R, Muller D, Caulee K, Wanherdrick K, Abecassis J, du Manoir S. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001 May 15;61(10):4122–9. [PubMed] [Google Scholar]
- Wang TY, Chen BF, Yang YC, Chen H, Wang Y, Cviko A, Quade BJ, Sun D, Yang A, McKeon FD, Crum CP. Histologic and immunophenotypic classification of cervical carcinomas by expression of the p53 homologue p63: a study of 250 cases. Hum Pathol. 2001 May;32(5):479–86. doi: 10.1053/hupa.2001.24324. [DOI] [PubMed] [Google Scholar]
- Hu H, Xia SH, Li AD, Xu X, Cai Y, Han YL, Wei F, Chen BS, Huang XP, Han YS, Zhang JW, Zhang X, Wu M, Wang MR. Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int J Cancer. 2002 Dec 20;102(6):580–3. doi: 10.1002/ijc.10739. [DOI] [PubMed] [Google Scholar]
- Weber A, Bellmann U, Bootz F, Wittekind C, Tannapfel A. Expression of p53 and its homologues in primary and recurrent squamous cell carcinomas of the head and neck. Int J Cancer. 2002 May 1;99(1):22–8. doi: 10.1002/ijc.10296. [DOI] [PubMed] [Google Scholar]
- Nylander K, Vojtesek B, Nenutil R, Lindgren B, Roos G, Zhanxiang W, Sjöström B, Dahlqvist A, Coates PJ. Differential expression of p63 isoforms in normal tissues and neoplastic cells. J Pathol. 2002 Dec;198(4):417–27. doi: 10.1002/path.1231. [DOI] [PubMed] [Google Scholar]
- Wang X, Mori I, Tang W, Nakamura M, Nakamura Y, Sato M, Sakurai T, Kakudo K. p63 expression in normal, hyperplastic and malignant breast tissues. Breast Cancer. 2002;9(3):216–9. doi: 10.1007/BF02967592. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Milanezi F, Amendoeira I, Albergaria A, Schmitt FC. Distribution of p63, a novel myoepithelial marker, in fine-needle aspiration biopsies of the breast: an analysis of 82 samples. Cancer. 2003 Jun 25;99(3):172–9. doi: 10.1002/cncr.11061. [DOI] [PubMed] [Google Scholar]
- Koker MM, Kleer CG. p63 expression in breast cancer: a highly sensitive and specific marker of metaplastic carcinoma. Am J Surg Pathol. 2004 Nov;28(11):1506–12. doi: 10.1097/01.pas.0000138183.97366.fd. [DOI] [PubMed] [Google Scholar]
- Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006 Feb;19(2):264–71. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006 Jan;9(1):45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Zangen R, Ratovitski E, Sidransky D. DeltaNp63alpha levels correlate with clinical tumor response to cisplatin. Cell Cycle. 2005 Oct;4(10):1313–5. doi: 10.4161/cc.4.10.2066. Epub 2005 Oct 1. [DOI] [PubMed] [Google Scholar]
- Danilov AV, Neupane D, Nagaraja AS, Feofanova EV, Humphries LA, DiRenzo J, Korc M. DeltaNp63alpha-mediated induction of epidermal growth factor receptor promotes pancreatic cancer cell growth and chemoresistance. PLoS One. 2011;6(10):e26815. doi: 10.1371/journal.pone.0026815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007 May;117(5):1370–80. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T, Uramoto H, Sugio K, Fujino Y, Oyama T, Nakata S, Ono K, Morita M, Yasumoto K. A lack of prognostic significance regarding DeltaNp63 immunoreactivity in lung cancer. Lung Cancer. 2005 Oct;50(1):67–73. doi: 10.1016/j.lungcan.2005.03.039. [DOI] [PubMed] [Google Scholar]
- Uramoto H, Sugio K, Oyama T, Nakata S, Ono K, Nozoe T, Yasumoto K. Expression of the p53 family in lung cancer. Anticancer Res. 2006 May-Jun;26(3A):1785–90. [PubMed] [Google Scholar]
- Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006 Aug 1;66(15):7589–97. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- Helton ES, Zhu J, Chen X. The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J Biol Chem. 2006 Feb 3;281(5):2533–42. doi: 10.1074/jbc.M507964200. [DOI] [PubMed] [Google Scholar]
- Celardo I, Grespi F, Antonov A, Bernassola F, Garabadgiu AV, Melino G, Amelio I. Caspase-1 is a novel target of p63 in tumor suppression. Cell Death Dis. 2013 May 23;4:e645. doi: 10.1038/cddis.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Semba S, Abe T, Makino N, Furukawa T, Fukushige S, Takahashi H, Sakurada A, Sato M, Shiiba K, Matsuno S, Nimura Y, Nakagawara A, Horii A. Infrequent somatic mutations of the p73 gene in various human cancers. Eur J Surg Oncol. 1999 Apr;25(2):194–8. doi: 10.1053/ejso.1998.0626. [DOI] [PubMed] [Google Scholar]
- Nomoto S, Haruki N, Kondo M, Konishi H, Takahashi T, Takahashi T, Takahashi T. Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res. 1998 Apr 1;58(7):1380–3. [PubMed] [Google Scholar]
- Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, Daxenbichler G, Zeimet A, Zeillinger R, Marth C, Moll UM. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004 Apr 1;64(7):2449–60. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- Domínguez G, García JM, Peña C, Silva J, García V, Martínez L, Maximiano C, Gómez ME, Rivera JA, García-Andrade C, Bonilla F. DeltaTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J Clin Oncol. 2006 Feb 10;24(5):805–15. doi: 10.1200/JCO.2005.02.2350. Epub 2005 Dec 27. [DOI] [PubMed] [Google Scholar]
- Faridoni-Laurens L, Tourpin S, Alsafadi S, Barrois M, Temam S, Janot F, Koscielny S, Bosq J, Bénard J, Ahomadegbe JC. Involvement of N-terminally truncated variants of p73, deltaTAp73, in head and neck squamous cell cancer: a comparison with p53 mutations. Cell Cycle. 2008 Jun 1;7(11):1587–96. doi: 10.4161/cc.7.11.5894. [DOI] [PubMed] [Google Scholar]
- Uramoto H, Sugio K, Oyama T, Nakata S, Ono K, Morita M, Funa K, Yasumoto K. Expression of deltaNp73 predicts poor prognosis in lung cancer. Clin Cancer Res. 2004 Oct 15;10(20):6905–11. doi: 10.1158/1078-0432.CCR-04-0290. [DOI] [PubMed] [Google Scholar]
- Wager M, Guilhot J, Blanc JL, Ferrand S, Milin S, Bataille B, Lapierre F, Denis S, Chantereau T, Larsen CJ, Karayan-Tapon L. Prognostic value of increase in transcript levels of Tp73 DeltaEx2–3 isoforms in low-grade glioma patients. Br J Cancer. 2006 Oct 23;95(8):1062–9. doi: 10.1038/sj.bjc.6603410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo J, Goñi S, Latasa MU, Perugorría MJ, Calvo A, Muntané J, Bioulac-Sage P, Balabaud C, Prieto J, Avila MA, Berasain C. Amphiregulin induces the alternative splicing of p73 into its oncogenic isoform DeltaEx2p73 in human hepatocellular tumors. Gastroenterology. 2009 Nov;137(5):1805–15. e1–4. doi: 10.1053/j.gastro.2009.07.065. [DOI] [PubMed] [Google Scholar]
- Corn PG, Kuerbitz SJ, van Noesel MM, Esteller M, Compitello N, Baylin SB, Herman JG. Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt’s lymphoma is associated with 5′ CpG island methylation. Cancer Res. 1999 Jul 15;59(14):3352–6. [PubMed] [Google Scholar]
- Kawano S, Miller CW, Gombart AF, Bartram CR, Matsuo Y, Asou H, Sakashita A, Said J, Tatsumi E, Koeffler HP. Loss of p73 gene expression in leukemias/lymphomas due to hypermethylation. Blood. 1999 Aug 1;94(3):1113–20. [PubMed] [Google Scholar]
- Ibrahim N, He L, Leong CO, Xing D, Karlan BY, Swisher EM, Rueda BR, Orsulic S, Ellisen LW. BRCA1-associated epigenetic regulation of p73 mediates an effector pathway for chemosensitivity in ovarian carcinoma. Cancer Res. 2010 Sep 15;70(18):7155–65. doi: 10.1158/0008-5472.CAN-10-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Moll UM. Role of p53 family members’ p73 and p63 in human hematological malignancies. Leuk Lymphoma. 2012 Nov;53(11):2116–29. doi: 10.3109/10428194.2012.684348. [DOI] [PubMed] [Google Scholar]
- Daskalos A, Logotheti S, Markopoulou S, Xinarianos G, Gosney JR, Kastania AN, Zoumpourlis V, Field JK, Liloglou T. Global DNA hypomethylation-induced ΔNp73 transcriptional activation in non-small cell lung cancer. Cancer Lett. 2011 Jan 1;300(1):79–86. doi: 10.1016/j.canlet.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Guan M, Chen Y. Aberrant expression of DeltaNp73 in benign and malignant tumours of the prostate: correlation with Gleason score. J Clin Pathol. 2005 Nov;58(11):1175–9. doi: 10.1136/jcp.2005.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz R, González-Sancho JM, Soldevilla B, Silva J, García JM, García V, Peña C, Herrera M, Gómez I, Bonilla F, Domínguez G. Differential regulation of TP73 isoforms by 1α,25-dihydroxyvitamin D3 and survivin in human colon and breast carcinomas. Genes Chromosomes Cancer. 2010 Dec;49(12):1135–42. doi: 10.1002/gcc.20821. [DOI] [PubMed] [Google Scholar]
- Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001 Nov 15;15(22):2917–21. doi: 10.1101/gad.949001. Review. [DOI] [PubMed] [Google Scholar]
- Berger AH, Pandolfi PP. Haplo-insufficiency: a driving force in cancer. J Pathol. 2011 Jan;223(2):137–46. doi: 10.1002/path.2800. Review. [DOI] [PubMed] [Google Scholar]
- Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001 Nov 15;15(22):2934–9. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression. Oncogene. 2007 Jun 28;26(30):4329–35. doi: 10.1038/sj.onc.1210226. Epub 2007 Jan 22. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallakin A, Sugiyama T, Taneja P, Matise LA, Frazier DP, Choudhary M, Hawkins GA, D’Agostino RB, Jr, Willingham MC, Inoue K. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell. 2007 Oct;12(4):381–94. doi: 10.1016/j.ccr.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Frazier DP, Taneja P, Kendig RD, Morgan RL, Matise LA, Lagedrost SJ, Inoue K. Signal transduction involving the dmp1 transcription factor and its alteration in human cancer. Clin Med Oncol. 2008;2:209–19. doi: 10.4137/cmo.s548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja P, Maglic D, Kai F, Sugiyama T, Kendig RD, Frazier DP, Willingham MC, Inoue K. Critical roles of DMP1 in human epidermal growth factor receptor 2/neu-Arf-p53 signaling and breast cancer development. Cancer Res. 2010 Nov 15;70(22):9084–94. doi: 10.1158/0008-5472.CAN-10-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Mott RT, Fry EA, Taneja P, Kulik G, Sui G, Inoue K. Cooperation between Dmp1 loss and cyclin D1 overexpression in breast cancer. Am J Pathol. 2013 Oct;183(4):1339–50. doi: 10.1016/j.ajpath.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry EA, Taneja P, Maglic D, Zhu S, Sui G, Inoue K. Dmp1α inhibits HER2/neu-induced mammary tumorigenesis. PLoS One. 2013;8(10):e77870. doi: 10.1371/journal.pone.0077870. [DOI] [PMC free article] [PubMed] [Google Scholar]