Abstract
Microbes associated with deep-sea corals remain poorly studied. The lack of symbiotic algae suggests that associated microbes may play a fundamental role in maintaining a viable coral host via acquisition and recycling of nutrients. Here we employed 16 S rRNA gene sequencing to study bacterial communities of three deep-sea scleractinian corals from the Red Sea, Dendrophyllia sp., Eguchipsammia fistula, and Rhizotrochus typus. We found diverse, species-specific microbiomes, distinct from the surrounding seawater. Microbiomes were comprised of few abundant bacteria, which constituted the majority of sequences (up to 58% depending on the coral species). In addition, we found a high diversity of rare bacteria (taxa at <1% abundance comprised >90% of all bacteria). Interestingly, we identified anaerobic bacteria, potentially providing metabolic functions at low oxygen conditions, as well as bacteria harboring the potential to degrade crude oil components. Considering the presence of oil and gas fields in the Red Sea, these bacteria may unlock this carbon source for the coral host. In conclusion, the prevailing environmental conditions of the deep Red Sea (>20 °C, <2 mg oxygen L−1) may require distinct functional adaptations, and our data suggest that bacterial communities may contribute to coral functioning in this challenging environment.
A growing number of studies support the notion that bacteria associated with multicellular hosts provide important functions related to metabolism, immunity, and environmental adaptation (among others)1. This might be of particular significance in ‘extreme’ environments, such as the deep sea. Nevertheless, only few studies investigated the diversity and function of bacteria associated with deep-sea corals. Similar to shallow corals2, deep-sea coral mucus stimulates microbial activity in the immediate vicinity of the coral host, suggestive of nutrient cycling via a microbial loop and thereby increasing ecosystem productivity3. However, deep-sea coral ecology is fundamentally different from the ecology of shallow-water corals. Deep-sea corals lack light, are exposed to elevated pressures, and usually reside at cold(er) temperatures4. Most importantly, they do not associate with photosynthetic algal symbionts in the genus Symbiodinium, and thereby lack a carbon source that provides the majority of the energetic requirements of shallow-water corals. Consequently, associated microbial communities of deep-sea corals are thought to play an essential role for host metabolism, e.g. by supplementing host feeding via fixing and recycling of nitrogen and carbon5,6,7. In support of this, in shallow-water corals increased bacterial diversity with increasing water depth has been suggested as a mechanism to aid nutrient acquisition8.
Studies of Red Sea corals, along with those in the Persian/Arabian Gulf, are of interest in light of global climate change because corals in this region thrive at summer temperatures (>31 °C) that are hostile for corals from other regions9,10. The tolerance to high temperature extends down to the depths of the Red Sea, where deep-sea corals are found at >20 °C11. The discovery of deep-sea corals at this temperature prompted a reconsideration of environmental limitations of deep-sea corals, previously considered to be limited to temperatures of 4–14 °C4,12. Besides these unusual high temperatures, a study from the central Red Sea by Roder et al.11 reported higher salinity (40.5 PSU) and lower dissolved oxygen (1–2 mg L−1) levels, compared to other deep-sea coral environments.
To date, only few studies investigated microbial communities associated with deep-sea reef communities. At present, studies have focused on octocorals7,13,14,15 and two cold-water scleractinian corals, Lophelia pertusa and Madrepora oculata5,16,17,18,19,20,21,22,23,24,25. These studies support the presence of species-specific microbiomes7,14,23, but at the same time identified highly variable bacterial community structures depending on phenotype, spatial and temporal scales, and even on individual polyps within a colony5,18,19,21,23.
To gain insight into bacterial community structure associated with deep-sea scleractinian corals from the Red Sea, we conducted a 16S rRNA gene survey on specimens from E. fistula, Dendrophyllia sp., and R. typus. We were interested to elucidate how bacterial association differs between coral hosts, and also, whether we find support for a bacterial contribution to functional adaptation to the distinct environment of the Red Sea.
Results
Microbial community composition of deep-sea corals and surrounding seawater
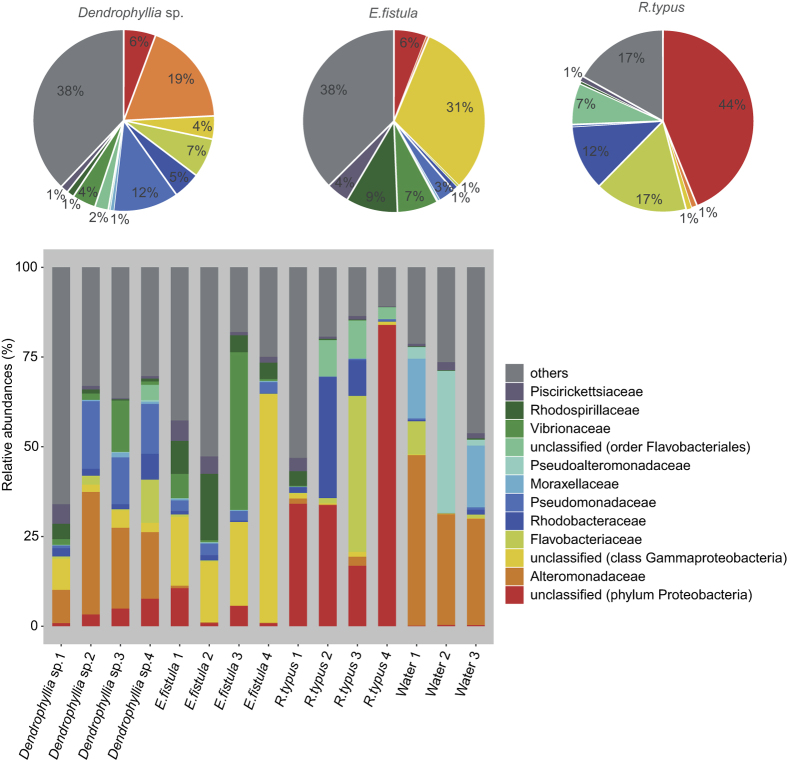
A total of 852,313 sequences were processed and, after filtering, yielded 554,088 sequences across 15 samples (i.e., 4 samples of Dendrophyllia sp., 4 samples of E. fistula, 4 samples of R. typus, and 1 water sample for each of the 3 collection sites). To evaluate bacterial community composition across samples, we classified all sequences to the family level (Fig. 1). The bacterial community composition of coral samples was markedly different from seawater samples that appeared similar across sampling sites (Fig. 1). Coral community composition differed in a species-specific manner with apparent within-species variation (Fig. 1).
Figure 1. Bacterial community composition of deep-sea corals and seawater from the Red Sea on the bacterial family level (Greengenes database, bootstrap ≥60).
Pie charts denote average bacterial abundance across replicates for the three coral species. The taxonomy bar plot denotes relative bacterial abundance across species replicates and seawater. Each color represents one of the 12 most abundant families (overall sequence count) in all samples. All other taxa are grouped under category ‘others’.
Differences and similarities within and between coral species and seawater
In order to identify differences in the bacterial community composition between different deep-sea corals, sequences were subsampled and clustered into OTUs (Operational Taxonomic Units) (Table 1). Species richness, i.e. the number of different bacterial species present in each coral, was estimated using the Chao1 index26. Bacterial species richness estimates ranged from 176 to 1050 taxa for Dendrophyllia sp., from 451 to 899 for E. fistula, from 442 to 675 for R. typus, and from 482 to 599 for seawater. This indicates that species richness in corals was similar to that of the surrounding seawater. Simpson Evenness27 ranged from 0.069 to 0.126 for Dendrophyllia sp., from 0.010 to 0.079 for E. fistula, from 0.020 to 0.031 for R. typus, and from 0.019 to 0.045 for sea water. Species diversity (i.e. Inverse Simpson Index27) ranged between 11.9 and 85.6 for Dendrophyllia sp., 3.2 and 46.7 for E. fistula, 2.7 and 11.9 for R. typus, and 4.6 and 16.0 for sea water samples. The wide range of bacterial richness and diversity across coral species and in particular for Dendrophyllia sp. and E. fistula argues for a considerable genotype dependence on community composition19.
Table 1. Summary statistics of 16S rRNA-based bacterial community composition of deep-sea coral and seawater samples.
| Sample Name | No. of Sequences | No. of OTUs* | Chao 1* | Simpson Evenness* | Inverse Simpson Index* |
|---|---|---|---|---|---|
| Dendrophyllia sp. 1 | 20,984 | 682 | 1050 | 0.126 | 85.6 |
| Dendrophyllia sp. 2 | 4,607 | 173 | 176 | 0.069 | 11.9 |
| Dendrophyllia sp. 3 | 28,345 | 230 | 295 | 0.070 | 16.1 |
| Dendrophyllia sp. 4 | 68,562 | 229 | 262 | 0.083 | 19.1 |
| AVG | 30,625 | 329 | 446 | 0.087 | 33.2 |
| SD | 27,168 | 237 | 406 | 0.027 | 35.1 |
| E.fistula 1 | 58,909 | 657 | 899 | 0.067 | 44.3 |
| E.fistula 2 | 21,485 | 589 | 839 | 0.079 | 46.7 |
| E.fistula 3 | 10,830 | 318 | 451 | 0.019 | 6.1 |
| E.fistula 4 | 33,871 | 337 | 468 | 0.010 | 3.2 |
| AVG | 31,274 | 475 | 664 | 0.044 | 25.1 |
| SD | 20,690 | 173 | 238 | 0.035 | 23.6 |
| R. typus 1 | 12,664 | 452 | 675 | 0.026 | 11.9 |
| R. typus 2 | 39,931 | 202 | 442 | 0.031 | 6.3 |
| R. typus 3 | 63,947 | 241 | 586 | 0.020 | 4.7 |
| R. typus 4 | 56,370 | 191 | 451 | 0.014 | 2.7 |
| AVG | 43,228 | 272 | 538 | 0.023 | 6.4 |
| SD | 22,708 | 122 | 112 | 0.007 | 3.9 |
| Seawater1 | 45,931 | 265 | 506 | 0.024 | 6.4 |
| Seawater2 | 32,959 | 244 | 482 | 0.019 | 4.6 |
| Seawater3 | 54,693 | 358 | 599 | 0.045 | 16.0 |
| AVG | 44,528 | 289 | 529 | 0.029 | 9.0 |
| SD | 10,935 | 61 | 62 | 0.014 | 6.1 |
*Based on subsampled sequences (n = 4,600).
AVG = average; SD = standard deviation.
In all coral species, comparably few OTUs were abundant and made up the majority of all sequences, whereas the majority of OTUs were rare (Supplementary Dataset S1). For instance, for Dendrophyllia sp. we identified 1,050 distinct OTUs, with only 13 OTUs each comprising >1% and 1,037 OTUs comprising <1% of all sequences. The most abundant bacterial taxa, OTU0004 (Pseudomonas veronii) represented 12% of all sequences. In E. fistula, we identified a total of 1,246 OTUs, with 17 OTUs each constituting >1% and 1,229 OTUs constituting <1% of all sequence counts. The most abundant bacterial taxa, OTU0001 (unknown Gammaproteobacterium) comprised 21% of all sequences. Across all R. typus samples, we found 842 distinct OTUs, with 13 OTUs constituting >1% and 829 OTUs with <1% of sequence counts. For this species, OTU00003 (unknown Proteobacterium) comprised 17% of all sequences and constituted the most abundant bacterium. Seawater samples contained a total of 557 distinct OTUs, of which 14 contributed >1% and 543 <1% of all sequences. OTU0002 (Alteromonas sp.) comprised 24% of all sequences.
To further explore differences in bacterial community composition across samples, we plotted a principal coordinate analysis (PCoA) based on a Bray-Curtis dissimilarity matrix (Fig. 2). This showed that sea water samples clustered tightly together and were clearly separated from all coral samples, and coral samples were generally clustering according to species. This was substantiated by AMOVA analysis, which showed that coral were significantly different from seawater (PAMOVA =0.002). To further test whether differences between corals exist, we excluded seawater and performed a PERMANOVA across corals. We identified significant differences between coral species (PPERMANOVA = 0.0019, Pseudo-F = 3.3554, 4,707 unique permutations) and subsequent pair-wise tests showed all coral species differed significantly from each other (Table S1).
Figure 2. Clustering of deep-sea coral and seawater samples based on bacterial abundances.
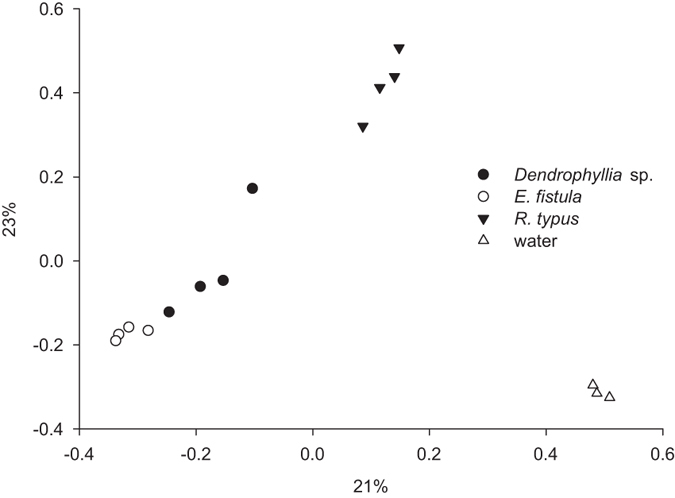
Ordination is based on a Bray-Curtis dissimilarity matrix and Pearson correlation in a principal coordinate analysis (PCoA) plot (R2 = 0.63). Percentages represent the amount variance explained by each dimension.
Core microbiomes
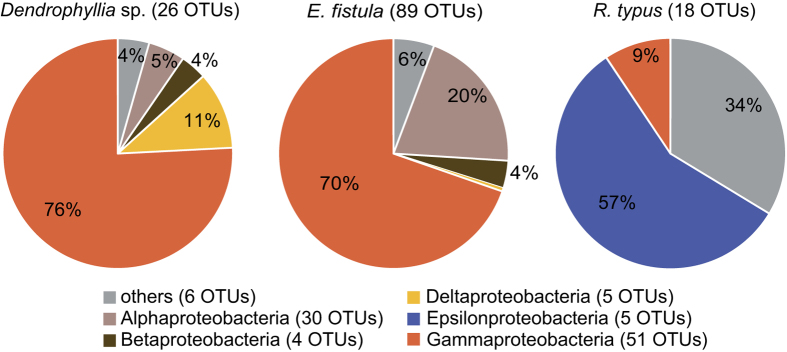
Based on the statistically significant differences between coral species, we next assessed differences in absence/presence of individual bacterial taxa across coral species. To do this, we determined coral species-specific core microbiomes. We considered all those OTUs that were ubiquitously present in each species, indicating putative functional importance. Only few bacteria were core microbiome members, in contrast to the large suite of OTUs associated with each species. We identified 26 bacterial taxa in the core microbiome of Dendrophyllia sp., 89 in that of E. fistula, and 18 in R. typus. Of note, all core microbiomes contained rare and abundant bacteria, comprising average abundances between 2 and 977 sequence counts (Supplementary Dataset S1, Supplementary Dataset S2). Core microbiomes largely encompassed bacterial taxa that were unique to a given coral species and only few members were shared between species. For instance, the core microbiome of Dendrophyllia sp. harbored 9 exclusive OTUs, E. fistula featured 20 unique bacterial taxa, and 3 bacteria were exclusively found in the microbiome of R. typus (Supplementary Dataset S2). In comparison to the core microbiomes of Dendrophylia sp. and E. fistula that were dominated by Gammaproteobacteria, R. typus was dominated by Epsilonproteobacteria (Supplementary Dataset S1, Supplementary Dataset S2, Fig. 3).
Figure 3. Core microbiomes of three deep-sea coral species from the Red Sea at the bacterial taxonomic class level.
Core microbiome members are comprised of bacterial taxa present in 100% of all samples for the species under investigation. Numbers of distinct OTUs in color legend denote bacterial diversity on the class level across coral microbiomes.
Discussion
To date, only few studies describe microbial communities associated with deep-sea scleractinian corals, all of which have been conducted on L. pertusa and M. oculata in Norway, the northern Gulf of Mexico, the Mediterranean, and off the coast of Ireland5,16,18,19,20,21,23,24. Hence, there is a paucity of studies from other species and other environments. The available studies show that coral microbiomes are distinct from the surrounding environment5,16,19,20,21, differ between different species, sites, and sampling times, and even between individual polys of the same colony5,7,18,19,21,23. Here, we assessed microbial community composition of the deep-sea scleractinian corals Dendrophyllia sp., E. fistula, and R. typus from the Red Sea to extend the catalog of available coral species and provide insight into putative indications of functional adaption to this unique and warm environment.
In order to characterize differences in the microbiomes from the three coral species Dendrophyllia sp., E. fistula, and R. typus, we assessed alpha and beta diversity indices and identified differentially abundant bacterial taxa. Microbiomes differed significantly between species as reflected by the presence/absence of bacterial taxa as well as by pronounced differences in the abundance of shared OTUs. Interestingly, only two bacterial taxa were consistently present across all replicates and coral species, i.e. Pseudomonas veronii (OTU0004) and Photobacterium angustum (OTU007), suggesting (1) coral-specific microbiomes and (2) functional importance of these taxa across coral host species. Pseudomonas veronii was previously found in shallow-water corals from the Red Sea and was linked to high salinity environmental conditions28. Interestingly, several species from the same bacterial genus were identified in L. pertusa collected in Norwegian waters24. Photobacterium angustum (previously named Vibrio angustum/fischeri) is described as an endosymbiont in squid, and bacterial species from this genus are shown to inhibit virulence gene expression29,30. Besides these similarities, the core microbiomes of all three coral species differed markedly. Importantly, core microbiome members are suggested to represent ‘true’ bacterial symbionts given their consistent association with coral host taxa8. Only a small proportion of bacterial associates for any coral species were core microbiome members, e.g. 2.5% of all OTUs in Dendrophyllia sp., 7.1% in E. fistula, and 2.0% in R. typus. Nevertheless, these members constituted a substantial portion of all sequences: 51% of all sequences in Dendrophyllia sp. were assigned to microbiome members, 73% in E. fistula, and 29% in R. typus. Unfortunately, a comparison to other deep-sea coral studies remains difficult due to differences in methodology, i.e. cloning-independent (this study) vs. cloning-dependent approaches5,16,18. To our knowledge, there are only two other studies using next-generation sequencing to describe spatial and temporal variable microbiomes of M. oculata and L. pertusa23,25. These studies showed that M. oculata and L. pertusa were dominated by Gammaproteobacteria, Alphaproteobacteria, and Bacteroidetes, and also, that Archaea (Thaumarchaeota marine group I (MGI)) were associated with mucus from L. pertusa. Similarly, all species in our study contained Gammaproteobacteria and Alphaproteobacteria, but Bacteroidetes were absent. In the current study, we did not assess archaeal association due to the choice of 16S rRNA gene primers (see Methods). Microbes associated with corals in this study also seemed remarkably consistent for a given coral species in comparison to a previous study that found pronounced differences between colonies23. This may be due to a lack of temporal replication, but also, might represent the result from more stable environmental conditions in the deep Red Sea. Conversely, sampling sites in the northwestern Mediterranean Sea are exposed to seasonal changes in nutrient availability and physico-chemical parameters (e.g. temperature)23.
Coral habitats from the Red Sea present a unique combination of environmental settings in which coral occur, in particular in regard to depth distribution and dissolved oxygen (DO)9,11,31. Compared to other deep-sea habitats, corals from the Red Sea are exposed to substantially warmer temperatures (>20 °C), considerably lower DO levels (<2 mg L−1), and highly oligotrophic waters10,11. In particular, high water temperatures in combination with low nutrient and oxygen levels are challenging for corals, prompting the suggestion of specific adaptations to the environment of the Red Sea11,31,32. Interestingly, however, deep-sea coral species found in the Red Sea (e.g., E. fistula and R. typus) are not endemic and exist in other deep-sea environments, such as the IndoPacific and Oceania33. It is unclear at present how these corals adapt to the prevailing environmental conditions of the Red Sea. For E. fistula, substantial physiological plasticity has recently been demonstrated34, and it would be intriguing to see whether this plasticity could be linked to flexibility in microbial community structure.
Along this line of thought, microbiomes in shallow water corals are suggested to play an important role for coral health35,36,37,38. This is presumably even more so for azooxanthellate corals5,6,22. In particular, it has been suggested that deep-sea coral associated bacteria may enhance nutrient cycling, serve as nutrition, enable cellulose catabolism, sulphur cycling, and anoxic fermentation5. Most recently, Middelburg et al.6 showed that microbes provide symbiotic nitrogen fixation, chemoautotrophy, and a high capability for nutrient recycling in L. pertusa, but these pathways are not yet linked to specific microbial groups.
Anoxic conditions in micro-niches have been discussed in deep-sea corals5 and the low oxygen levels present in the deep Red Sea may promote association with anaerobic bacteria. We found anaerobic bacteria from the genera Propionibacterium and Planctomyces associated with corals in this study, although their abundances were comparably low (Supplementary Dataset S1). Conversely, we identified bacteria of the families Rhodobacteraceae and Rhodospirillaceae in high abundance. Both families include members connected to anaerobic conditions5. Lastly, bacteria from the genus Vibrio (present at comparably low abundances in our samples) are capable of nitrogen fixation, in particular under low oxygen or anoxic conditions39,40.
Nitrogen fixation and carbon acquisition are prerequisites for corals to survive in low-nutrient environments, such as the Red Sea. In this context, we identified Rhodobacteraceae that are capable of metabolizing nitrogen and carbon from chitin in prey crustaceans. In addition, this bacterial family includes denitrifiers supporting efficient nitrogen cycling in corals5. The abundance of Rhodobacteraceae varied strongly within and between species (152 ± 128 for Dendrophyllia sp., 38 ± 31 for E. fistula, and 512 ± 684 in R. typus), which might align with availability of heterotrophic feeding sources that potentially subsidize for bacterial-assisted metabolic complementation.
Members of the family Rhodobacteraceae have further been identified as key players in oil degradation in the marine environment41. Interestingly, in our samples, besides Rhodobacteraceae, we identified additional bacteria capable of degrading crude oil compounds. For instance, Pseudomonas veronii was present in all samples and commonly at high abundance. P. veronii is capable of degrading a variety of organic compounds including alkylphenols42 and alkyl methyl ketones43, and to absorb heavy metals44, which are also found in crude oil45,46,47. We further identified Methylibium petroleiphilum48, Oleibacter sp.49, Acinetobacter venetianus50, Marinobacter bryozoorum41, Thalassospira xiamenensis51, and Aestuariicella hydrocarbonica (OTU0009, annotation to NCBI ‘nr’, 99% sequence similarity)52, all of which have been described to possess the potential to degrade crude oil (Supplementary Dataset S1). Considering all oil-degraders that could be assigned to the species level, about 30% of all sequences for Dendrophyllia sp., 5% for E. fistula, and only 0.2% for R. typus were on average comprised by these bacteria. Hence, the presence of oil-degraders does not seem to be a general characteristic of deep-sea corals from the Red Sea, although oil degraders have been suggested to play a functional role in (shallow-water) coral holobionts53. Crude oil and its derivatives may therefore constitute a carbon source for deep-sea corals (in particular Dendrophyllia sp.) in the deep Red Sea54, but these assumptions would need to be substantiated by stable isotope data showing incorporation of the hydrocarbon signal into the coral tissue. The presence of anaerobic bacteria, however, seems to be a general hallmark that warrants further study into their contribution to coral functioning in the unique environment of the deep Red Sea.
Conclusion
In this study we characterized microbial communities associated with three deep-sea corals from the Red Sea. All corals exhibited species-specific bacterial associations and specific bacterial taxa provide support to the notion that bacteria contribute to environmental adaption of their coral hosts. These bacteria include obligate or facultative anaerobic bacteria that may aid the coral to survive in the low oxygen and warm environment of the deep Red Sea. We further identified bacteria associated with degrading crude oil compounds, potentially suggesting the utilization of crude oil and its derivatives as a carbon source for deep-sea corals from the Red Sea. To further understand functional implications of bacterial association, microbial characterization of congenerics from these coral species outside the Red Sea is desirable.
Methods
Study site and sample collection
Corals in this study were collected by remotely operated vehicle (ROV) during a central and northern Red Sea expedition (KRSE2013L6) from 11–22 May 2013 on the R/V Aegaeo (operated by the Hellenic Center for Marine Research, Greece). Three coral species, Dendrophyllia sp., Eguchipsammia fistula, and Rhizotrochus typus were collected each with four biological replicates for a total of 12 coral samples. Upon collection, E. fistula and R. typus samples were placed into a custom-made two-compartment container and preserved in RNAlater at depth. Dendrophyllia sp. samples were collected in a plastic basket and brought back to the surface within 90 min of sampling. For each site, one seawater sample was collected from the vicinity of the collection site via Niskin bottles during additional dives. Dendrophyllia sp. was collected between 625 and 630 m depth (N22˚46.167′, E38˚03.102′), E. fistula was collected between 314 and 320 m depth (N22˚17.837′, E38˚53.811′), and R. typus was collected between 970 and 993 m depth (N27˚44.215′, E35˚08.000′). Samples from each coral species were collected on separate ROV dives to prevent cross contamination. Directly after retrieval, coral samples were rinsed with filtered seawater, crushed on liquid nitrogen and stored in cryotubes at −80 °C until DNA extraction (see below). One liter of each seawater sample was filtered over a 0.22 μm Durapore® filter (Millipore, Billerica, USA), and filters were stored at −80 °C until DNA extraction.
DNA extraction and 16S rRNA gene amplicon sequencing
DNA from crushed coral was isolated using the AllPrep DNA/RNA Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. For DNA isolation from seawater samples, 1200 μl of buffer RLT (Qiagen) were used on a quarter of a filter and incubated for 20 minutes on a tabletop rotator (Sb2 rotator, Stuart, Staffordshire, UK). DNA isolation commenced following the manufacturer’s instructions. Sequencing libraries were generated following the Illumina 16S metagenomic sequencing library preparation protocol. 16S rRNA locus was amplified using the primers 784F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGGATTAGATACCCTGGTA-3′) and 1061R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCRRCACGAGCTGACGAC-3′) (Illumina overhang adaptor sequences underlined)55. These primers were found to amplify bacterial 16S rRNA sequences well across a range of coral species56, but are limited in their ability to amplify archaeal 16S rRNA sequences. Hence, archaeal 16S rRNA sequences were removed prior to microbial community composition analysis (see below). Of note, archaea associate with scleractinian corals, but seem to be far less abundant than bacteria57,58. PCRs were run in triplicate at a volume of 20 μl using the Qiagen multiplex PCR Master Mix with ~50–100 ng DNA for coral and ~1–2 ng DNA for seawater samples, respectively. The final primer concentration was adjusted to 0.25 μM. The PCR conditions were set as follows: 95 °C for 15 minutes, followed by 25 cycles of 95 °C for 30 seconds, 55 °C for 90 seconds and 72 °C for 30 seconds, with a final extension time of 10 minutes at 72 °C. Triplicate PCRs for each sample were pooled and amplification was visualized on the Bioanalyzer (Agilent Technologies, Santa Clara, USA). Samples underwent an index PCR amplification following the Illumina 16S metagenomic sequencing library preparation protocol, combined and run on a 2% ultra-pure agarose gel (Ultrapure Agarose, Life Technologies), and cut out on a non-UV transilluminator (Dark Reader, Clare Chemical Research, Dolores, USA) to remove excess primer. DNA was purified following the Zymoclean DNA large fragment recovery kit (Zymo Research, Irvine, USA) with two elutions at 10 μl. The 16S rRNA gene amplicon libraries were sequenced on one lane on the Illumina MiSeq platform with 25% phiX control at the KAUST sequencing facility. The libraries were sequenced using 2 × 300 bp overlapping paired-end reads.
Bacterial community analysis
Bacterial community analysis was conducted using mothur (v.1.36.1)59. Briefly, sequence reads were trimmed and read pairs were merged into contigs. Contigs longer than 306 bp and those with ambiguously called bases were discarded. Sequences that occurred only once were discarded. The sequences were then pre-clustered, allowing for up to a 2 base pair difference between sequences. Chimeras were removed using UCHIME, as implemented in mothur. Sequences were classified with the Greengenes database60 using a 60% bootstrap cutoff, and mitochondria, chloroplasts, archaea, eukaryote, and unknown sequences were removed. All samples were subsampled to 4,600 sequences, corresponding to the lowest number of sequences available for any sample. Bacterial community compositions were visualized using pie charts and a stack column plot based on annotated sequences. Subsampled sequences were clustered into operational taxonomic units (OTUs) with a similarity cutoff of 97%. Alpha diversity indices (Chao1, Simpson Evenness, and Inverse Simpson Index) were calculated using mothur. Beta diversity differences were visualized in a principal coordinate analysis (PCoA) based on a Bray-Curtis dissimilarity matrix and Pearson correlation. Differences between coral and seawater samples were assessed using Analysis of Molecular Variance (AMOVA) in mothur. A one-factorial permutational MANOVA (PERMANOVA) analysis was conducted using PRIMER v6 software (PRIMER–E Ltd, Ivybridge, UK)61 on square root transformed OTU abundance counts. The test design was based on Bray-Curtis similarities, partial sum of squares type III, 9,999 permutations of residuals under a reduced model using Monte-Carlo simulations, and followed by pair-wise tests. Core microbiomes were determined following a conservative approach, considering only OTUs that occurred in every sample of a given coral species.
Declarations
Ethics approval and consent to participate
Experimental research detailed in this study complies with institutional guidelines following KAUST Institutional Biosafety and BioEthics Committee (IBEC).
Additional Information
Accession Codes: Raw sequence data determined in this study have been deposited on NCBI under BioProject Accession No. PRJNA354830 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA354830). Core microbiome OTU reference sequences are available under GenBank Accession numbers KY515469 - KY515585.
How to cite this article: Röthig, T. et al. Microbial community composition of deep-sea corals from the Red Sea provides insight into functional adaption to a unique environment. Sci. Rep. 7, 44714; doi: 10.1038/srep44714 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the crew of the R/V Aegaeo, especially the ROV and submersible team, and all participating scientists (P. Schmitt-Kopplin, N. Hertkorn, C. Roder). We would like to thank CMOR for assistance and support in field operations. This work was supported from baseline funds to CRV and under the Center Competitive Funding (CCF) Program FCC/1/1973-18-01 by the King Abdullah University of Science and Technology (KAUST).
Footnotes
The authors declare no competing financial interests.
Author Contributions C.R.V. designed and conceived the experiments. L.K.Y. and C.R.V. generated data. T.R., L.K.Y. and C.R.V. analyzed and interpreted data. T.R., S.G.K., A.R. and C.R.V. contributed reagents/materials/analysis tools. T.R., L.K.Y., C.R.V. wrote the manuscript. All authors read and approved the final manuscript.
References
- McFall-Ngai M. et al. Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences 110, 3229–3236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C. et al. Degradation and mineralization of coral mucus in reef environments. Marine Ecology Progress Series 267, 159–171 (2004). [Google Scholar]
- Wild C. et al. Organic matter release by cold water corals and its implication for fauna–microbe interaction. Marine Ecology Progress Series 372, 67–75 (2008). [Google Scholar]
- Freiwald A., Fosså J. H., Grehan A., Koslow T. & Roberts J. M. Cold-water coral reefs. UNEP-WCMC, Cambridge, UK 84 (2004). [Google Scholar]
- Neulinger S. C., Järnegren J., Ludvigsen M., Lochte K. & Dullo W.-C. Phenotype-Specific Bacterial Communities in the Cold-Water Coral Lophelia pertusa (Scleractinia) and Their Implications for the Coral’s Nutrition, Health, and Distribution. Applied and Environmental Microbiology 74, 7272–7285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelburg J. J. et al. Discovery of symbiotic nitrogen fixation and chemoautotrophy in cold-water corals. Scientific Reports 5, 17962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler S. N. et al. Coral-Associated Bacterial Diversity is Conserved Across Two Deep-Sea Anthothela Species. Frontiers in Microbiology 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Agreda A., Leggat W., Bongaerts P. & Ainsworth T. D. The Microbial Signature Provides Insight into the Mechanistic Basis of Coral Success across Reef Habitats. mBio 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleypas J. A., McManus J. W. & Meñez L. A. Environmental limits to coral reef development: where do we draw the line? American Zoologist 39, 146–159 (1999). [Google Scholar]
- Raitsos D. E., Pradhan Y., Brewin R. J. W., Stenchikov G. & Hoteit I. Remote Sensing the Phytoplankton Seasonal Succession of the Red Sea. PLoS ONE 8, e64909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roder C. et al. First biological measurements of deep-sea corals from the Red Sea. Scientific Reports 3, 2802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M. S., Orejas C. & Ferrier-Pagès C. Species-specific physiological response by the cold-water corals Lophelia pertusa and Madrepora oculata to variations within their natural temperature range. Deep Sea Research Part II: Topical Studies in Oceanography 99, 36–41 (2014). [Google Scholar]
- Gray M. A., Stone R. P., McLaughlin M. R. & Kellogg C. A. Microbial consortia of gorgonian corals from the Aleutian islands. FEMS Microbiology Ecology 76, 109–120 (2011). [DOI] [PubMed] [Google Scholar]
- Kellogg C. A., Ross S. W. & Brooke S. D. Bacterial community diversity of the deep-sea octocoral Paramuricea placomus. PeerJ 4, e2529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn K., Wu D., Eisen J. A. & Ward N. Characterization of Bacterial Communities Associated with Deep-Sea Corals on Gulf of Alaska Seamounts. Applied and Environmental Microbiology 72, 1680–1683 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov M. M. et al. Phylogenetic survey of metabolically active microbial communities associated with the deep-sea coral Lophelia pertusa from the Apulian plateau, Central Mediterranean Sea. Deep Sea Research Part I: Oceanographic Research Papers 53, 62–75 (2006). [Google Scholar]
- Neulinger S. C. et al. Tissue-Associated “Candidatus Mycoplasma corallicola” and Filamentous Bacteria on the Cold-Water Coral Lophelia pertusa (Scleractinia). Applied and Environmental Microbiology 75, 1437–1444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg C. A., Lisle J. T. & Galkiewicz J. P. Culture-Independent Characterization of Bacterial Communities Associated with the Cold-Water Coral Lophelia pertusa in the Northeastern Gulf of Mexico. Applied and Environmental Microbiology 75, 2294–2303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson L., Agis M., Maier C. & Weinbauer M. G. Community composition of bacteria associated with cold-water coral Madrepora oculata: within and between colony variability. Marine Ecology Progress Series 397, 89–102 (2009). [Google Scholar]
- Schöttner S. et al. Inter- and intra-habitat bacterial diversity associated with cold-water corals. ISME J 3, 756–759 (2009). [DOI] [PubMed] [Google Scholar]
- Schöttner S., Wild C., Hoffmann F., Boetius A. & Ramette A. Spatial Scales of Bacterial Diversity in Cold-Water Coral Reef Ecosystems. PLoS ONE 7, e32093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkiewicz J. P., Pratte Z. A., Gray M. A. & Kellogg C. A. Characterization of culturable bacteria isolated from the cold-water coral Lophelia pertusa. FEMS Microbiology Ecology 77, 333–346 (2011). [DOI] [PubMed] [Google Scholar]
- Meistertzheim A. L. et al. Patterns of bacteria-host associations suggest different ecological strategies between two reef building cold-water coral species. Deep Sea Research Part I: Oceanographic Research Papers 114, 12–22 (2016). [Google Scholar]
- Emblem Å. et al. Mitogenome polymorphism in a single branch sample revealed by SOLiD deep sequencing of the Lophelia pertusa coral genome. Gene 506, 344–349 (2012). [DOI] [PubMed] [Google Scholar]
- van Bleijswijk J. D. L. et al. Microbial assemblages on a cold-water coral mound at the SE Rockall Bank (NE Atlantic): interactions with hydrography and topography. Biogeosciences 12, 4483–4496 (2015). [Google Scholar]
- Chao A. Nonparametric estimation of the number of classes in a population. Scandinavian Journal of statistics, 265–270 (1984). [Google Scholar]
- Simpson E. H. Measurement of Diversity. Nature 163, 688 (1949). [Google Scholar]
- Röthig T., Ochsenkühn M. A., Roik A., van der Merwe R. & Voolstra C. R. Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Molecular Ecology 25, 1308–1323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. C. et al. Genomic taxonomy of vibrios. BMC Evolutionary Biology 9, 1–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson M. et al. Inhibition of Virulence Gene Expression in Staphylococcus aureus by Novel Depsipeptides from a Marine Photobacterium. Marine Drugs 9, 2537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qurban M. A. et al. In-situ observation of deep water corals in the northern Red Sea waters of Saudi Arabia. Deep Sea Research Part I: Oceanographic Research Papers 89, 35–43 (2014). [Google Scholar]
- Dodds L. A., Roberts J. M., Taylor A. C. & Marubini F. Metabolic tolerance of the cold-water coral Lophelia pertusa (Scleractinia) to temperature and dissolved oxygen change. Journal of Experimental Marine Biology and Ecology 349, 205–214 (2007). [Google Scholar]
- Land J. V. d. UNESCO-IOC Register of marine organisms (URMO) 2008). [Google Scholar]
- Roik A., Röthig T., Roder C., Müller P. J. & Voolstra C. R. Captive rearing of the deep-sea coral Eguchipsammia fistula from the Red Sea demonstrates remarkable physiological plasticity. PeerJ 3, e734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R. & Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Micro 5, 355–362 (2007). [DOI] [PubMed] [Google Scholar]
- Ziegler M., Seneca F.O., Yum L.K., Palumbi S. R. & Voolstra C. R. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nature Communications 8, 14213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D. G., Morrow K. M. & Webster N. S. Coral Holobionts. Annual Review of Microbiology 70 (2016). [DOI] [PubMed] [Google Scholar]
- Voolstra C. R. et al. The ReFuGe 2020 consortium - Using ‘omics’ approaches to explore the adaptability and resilience of coral holobionts to environmental change. Frontiers in Marine Science 2 (2015). [Google Scholar]
- Lesser M. P., Mazel C. H., Gorbunov M. Y. & Falkowski P. G. Discovery of symbiotic nitrogen-fixing Cyanobacteria in corals. Science 305, 997–1000 (2004). [DOI] [PubMed] [Google Scholar]
- Urdaci M. C., Stal L. J. & Marchand M. Occurrence of nitrogen fixation among Vibrio spp. Archives of Microbiology 150, 224–229 (1988). [Google Scholar]
- Kostka J. E. et al. Hydrocarbon-Degrading Bacteria and the Bacterial Community Response in Gulf of Mexico Beach Sands Impacted by the Deepwater Horizon Oil Spill. Applied and Environmental Microbiology 77, 7962–7974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajithkumar B., Ajithkumar V. P. & Iriye R. Degradation of 4-amylphenol and 4-hexylphenol by a new activated sludge isolate of Pseudomonas veronii and proposal for a new subspecies status. Research in Microbiology 154, 17–23 (2003). [DOI] [PubMed] [Google Scholar]
- Onaca C., Kieninger M., Engesser K.-H. & Altenbuchner J. Degradation of Alkyl Methyl Ketones by Pseudomonas veronii MEK700. J Bacteriol 189, 3759–3767 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vullo D. L., Ceretti H. M., Daniel M. A., Ramírez S. A. M. & Zalts A. Cadmium, zinc and copper biosorption mediated by Pseudomonas veronii 2E. Bioresource Technology 99, 5574–5581 (2008). [DOI] [PubMed] [Google Scholar]
- Bennett B., Bowler B. F. J. & Larter S. R. Determination of C0−C3 Alkylphenols in Crude Oils and Waters. Analytical Chemistry 68, 3697–3702 (1996). [Google Scholar]
- Gondal M. A., Hussain T., Yamani Z. H. & Baig M. A. Detection of heavy metals in Arabian crude oil residue using laser induced breakdown spectroscopy. Talanta 69, 1072–1078 (2006). [DOI] [PubMed] [Google Scholar]
- Snyder L. R. Nitrogen and oxygen compound types in petroleum. Total analysis of a 400-700.deg. distillate from a California crude oil. Analytical Chemistry 41, 314–323 (1969). [Google Scholar]
- Nakatsu C. H. et al. Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. International Journal of Systematic and Evolutionary Microbiology 56, 983–989 (2006). [DOI] [PubMed] [Google Scholar]
- McGenity T. J., Folwell B. D., McKew B. A. & Sanni G. O. Marine crude-oil biodegradation: a central role for interspecies interactions. Aquatic Biosystems 8, 1–19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach H., Berdichevsky Y. & Gutnick D. An Exocellular Protein from the Oil-Degrading Microbe Acinetobacter venetianus RAG-1 Enhances the Emulsifying Activity of the Polymeric Bioemulsifier Emulsan. Applied and Environmental Microbiology 69, 2608–2615 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu Y., Li L., Ma Y. & Shao Z. Thalassospira xiamenensis sp. nov. and Thalassospira profundimaris sp. nov. International Journal of Systematic and Evolutionary Microbiology 57, 316–320 (2007). [DOI] [PubMed] [Google Scholar]
- Lo N., Kim K. H., Baek K., Jia B. & Jeon C. O. Aestuariicella hydrocarbonica gen. nov., sp. nov., an aliphatic hydrocarbon-degrading bacterium isolated from a sea tidal flat. International Journal of Systematic and Evolutionary Microbiology 65, 1935–1940 (2015). [DOI] [PubMed] [Google Scholar]
- Al-Dahash L. M. & Mahmoud H. M. Harboring oil-degrading bacteria: A potential mechanism of adaptation and survival in corals inhabiting oil-contaminated reefs. Marine Pollution Bulletin 72, 364–374 (2013). [DOI] [PubMed] [Google Scholar]
- Beydoun Z. The hydrocarbon prospects of the Red Sea–Gulf of Aden: a review. Journal of Petroleum Geology 12, 125–144, (1989). [Google Scholar]
- Andersson A. F. et al. Comparative Analysis of Human Gut Microbiota by Barcoded Pyrosequencing. PLoS ONE 3, e2836 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T. et al. The Microbiome of the Red Sea Coral Stylophora pistillata Is Dominated by Tissue-Associated Endozoicomonas Bacteria. Applied and Environmental Microbiology 79, 4759–4762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave M. J. et al. Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J 11, 186–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave M. J., Michell C. T., Apprill A. & Voolstra C. R. Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Scientific Reports 7, 40579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microb 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K. & Gorley R. PRIMER V6: user manual/tutorial. Primer-E Ltd, Ivybridge, UK(2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.