Abstract
In recent years, it has become clear that mechanical cues play an integral role in maintaining stem cell functions. Here we discuss how integrating physical approaches and engineering principles in stem cell biology, including culture systems, preclinical models, and functional assessment, may improve clinical application in regenerative medicine.
Introduction
Nearly a century ago, D’Arcy Thompson underscored the importance of mechanics in understanding growth and formation of biological structures, and recent advances in multiple areas of science are now allowing researchers to explore how the mechanics of the microenvironment impact stem cell biology. The relevance of this topic is highlighted by efforts to utilize stem cells therapeutically. Stem cells have been explored extensively in regenerative medicine based on their promise to make or replenish functional tissues in a sustained manner. This promise was first demonstrated by blood regeneration after intravenous injection of hematopoietic stem and progenitor cells (HSPCs), which remain the only type of stem cells used clinically to treat patients with hematopoietic disorders. While this progress has opened the door to prospectively isolate functional stem cells followed by in vivo validation through transplantation, the stem cells generally lose key functions once isolated, limiting directed ex vivo tissue engineering or organogenesis using stem cells. This limitation had led to extensive efforts to identify many specific molecules and cell types from native microenvironments or niches that regulate different aspects of stem cell functions. Advances in biomaterials have broadened the repertoire of how these components can be organized and presented in synthetic scaffolds to control stem cells. These efforts, especially within the last decade, have led to a key paradigm in stem cell biology: stem cells generate forces and subsequently sense physical properties of the matrix through adhesion, which then activates signaling cascades to control stem cell functions. Thus, in designing niches, it is increasingly appreciated that recapitulating their mechanics is a key biologically relevant cue to quantitatively direct stem cell functions. In addition, some of the cellular and molecular components in microenvironments are now being reinterpreted in the context of physical forces and motions that govern stem cell functions.
Here, we summarize the recent progress in understanding mechanosensing of stem cells, discuss its applications to preclinical models of stem cell therapy, and consider how these insights may be used to translate stem cells into clinical applications.
Physical Biology of Stem Cells
To understand how stem cells sense the mechanics of their microenvironments, it is important to look at how stem cells are intrinsically wired for mechanosensing, what physical parameters of the microenvironment stem cells can sense, and how molecular circuits convert cell-extrinsic physical parameters into long-term functions.
The actin cytoskeleton is an active material that plays a key role in generating, transmitting, and responding to mechanical forces. It is anchored to the cell membrane and composed of myosin-II motors, actin filaments, and crosslinker proteins. Micropipette aspiration in combination with fluorescent tagging of structural proteins has proven to be useful in studying how cells are wired for mechanosensing by applying precisely controlled forces to cells and measuring membrane deformation and dynamic localization of proteins. In the stem cell field, this approach was used to show that the myosin-IIB isoform is more sensitive to mechanical stress than the myosin-IIA isoform and that this difference leads to functional consequences during HSPC differentiation: the force-induced segregation of myosin-IIB during asymmetric division promotes self-renewal in one daughter cell, while constitutive myosin-IIA promotes differentiation in the other daughter cell (Shin et al., 2014). Therefore, mechanosensitivity of stem cells and their lineages may initially depend on their intrinsic material properties, which could be programmed during differentiation and adapted by external forces.
Cells have been cultured mostly on plastic dishes, but their native environments exhibit a range of matrix mechanics even within the same organ. For example, bone marrow varies from very soft tissue to stiff bone. Recent advances have introduced precise control of physical parameters to the reconstituted matrices and hydrogels used to culture cells on or in solid environments, revealing their critical roles in directing stem cell differentiation. Research in this area has been particularly focused on the mesenchymal lineages. The role of matrix mechanics on stem cell functions was first investigated when Engler et al. (2006) tuned mechanical properties of hydrogels functionalized with adhesion ligands on 2D elastic materials. Using this approach, it was revealed that matrix stiffness directs mesenchymal stem cell (MSC) differentiation through focal adhesions and myosin-II contractility (Engler et al., 2006). In this study, MSCs were shown to differentiate into branch-forming cells on soft matrices, whereas they differentiate into osteolineages on stiff matrices. While 2D culture is relatively simple and has been useful for the study of cell-surface interactions, cells within tissues usually encounter matrices in 3D space. As early as the 1970s, studies were being done in 3D culture by embedding cells in hydrogels to investigate colony formation of hematopoietic lineages. However, advances in chemistry and soft matter rheology were needed to fabricate 3D hydrogels with precise and independent control of mechanical and chemical properties to allow definitive studies to be performed. By using an ionically crosslinked alginate hydrogel functionalized with the RGD integrin peptide, Huebsch et al. (2010) showed that matrix stiffness directs 3D MSC differentiation in a similar manner to what was observed on 2D environments, but by altering integrin clustering rather than cell morphology. In a covalently crosslinked hyaluronic acid hydrogel, however, it was shown that MSCs are differentiated into adipocytes independently of matrix stiffness, and once matrix is degraded, MSCs spread and undergo osteogenesis (Khetan et al., 2013). Whether this latter finding is related to the changes in hydrogel mechanical properties resulting from degradation is unclear, but combining these studies with a recent finding that the stress relaxation of gels regulates spreading, proliferation, and differentiation of MSCs (Chaudhuri et al., 2015) suggests that matrix-stiffness-driven differentiation in 3D requires a labile environment where cells can generate traction forces and reorganize ligand binding. While new insights are emerging continuously, significant data indicate that matrix viscoelasticity is a potent physical parameter that regulates stem cell differentiation.
Although evidence suggests that force can be transmitted from the matrix to the nucleus through physical connections between cytoskeletal and nucleoskeletal proteins, how matrix stiffness influences long-term gene expression and cell fate is just beginning to be understood at the molecular level. Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) were shown to play a functional role in MSC differentiation by promoting expression of mechanosensitive genes upon matrix stiffening (Dupont et al., 2011). Importantly, it was shown that matrix stiffening causes nuclear stiffening in MSCs by stabilizing the turnover of the nucleoskeletal protein lamin-A through phosphorylation, which then increases nuclear localization of YAP and other mechanosensitive transcription factors to drive osteogenesis and further transcribe lamin-A (Swift et al., 2013). Overall, these studies are beginning to provide mechanistic understanding of how matrix stiffness directs stem cell differentiation.
Mechanobiology in Preclinical Models of Stem Cell Therapy
While physical parameters of materials used in cell culture and therapy have been considered “ancillary” and mostly from the toxicity perspective, the progress in mechanobiology suggests that some of the parameters could act as active cues that influence cell functions. This is particularly important when materials are used for culturing donor cells prior to injection or co-implanted with donor cells. The majority of current stem cell trials have used marrow-derived MSCs, which are isolated by adherence and ex vivo expansion on rigid plastic. However, the culture history can potentially dictate subsequent stem cell functions, based on evidence that MSCs cultured for longer periods on a particular substrate stiffness become committed to this matrix-defined lineage even when later presented with other soluble induction cues (Engler et al., 2006). Similarly, while the major functions of scaffolds have traditionally been thought to increase in vivo residence times of donor stem cells and present functional ligands, the stiffness of scaffolds could also determine therapeutic efficacy (Huebsch et al., 2015). It is thus important to consider the mechanical properties of both pre-transplantation adhesion scaffolds and materials used for cell delivery.
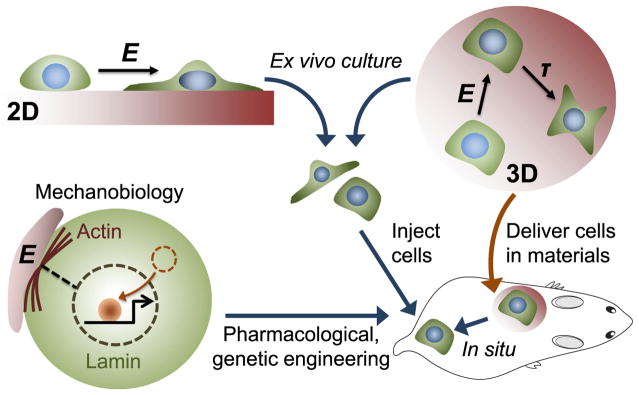
While the fundamental understanding of how stem cells sense matrix mechanics has been achieved mostly by the study of MSCs, the first efforts in translating this understanding to preclinical animal models have been made with HSPCs and muscle stem cells (MuSCs) where in vivo functional assays are more rigorously defined. Culturing HSPCs on highly elastic tropoelastin increases their number as assessed by in vivo limiting dilution transplantation (Holst et al., 2010). When human hematopoietic cells were perturbed to generate low contractile forces by being cultured on soft matrices or with pharmacological inhibition of myosin-II, there was significant enrichment of functional hematopoietic stem cells as validated by a mouse xenograft transplantation model (Shin et al., 2014). Similarly, priming skeletal muscle stem cells on a soft hydrogel substrate that mimics the elasticity of muscle enhances their self-renewal and muscle regeneration capability in vivo (Gilbert et al., 2010). While all of these studies employed ex vivo culture of stem cells on 2D hydrogels followed by in vivo transplantation of cells alone, a recent study encapsulated human MSCs in 3D void-forming hydrogels and implanted them into a rat xenograft cranial defect model to show that matrix stiffness regulates bone regeneration in situ (Huebsch et al., 2015). This finding suggests that mechanical cues can be used along with biochemical cues to program stem cells for therapeutic tissue regeneration (Figure 1).
Figure 1. Translating Stem Cell Mechanobiology to Preclinical Models.
Stem cells can be programmed ex vivo either on 2D or 3D hydrogels with tunable matrix stiffness (E) prior to injection. 3D hydrogels can also be used for the transplantation of stem cells to the body. Matrix degradation (τ) can regulate stem cell functions as well. Molecular pathways in stem cell mechanosensing can potentially be modulated by pharmacological or genetic strategies to impact regenerative capacity.
Toward Clinical Translation of Stem Cell Mechanobiology
With over 300 clinical trials currently exploring the utility of stem cells in regenerative medicine, there is an emerging need to strengthen insights on how stem cells will work in patients. Mechanobiology and biophysical engineering could offer novel insights and quantitative methods, respectively, which may help improve upon the existing pharmacological framework currently designed for small molecules to evaluate safety, potency, and efficacy of stem cell products. To achieve the goal of translating mechanobiology into stem cell therapy, it will be important to consider the following points.
In Vivo Relevance of Mechanobiology
Since most of the findings in this field are based on ex vivo mechanical control of stem cells, it will be important to validate their significance in situ or in vivo for eventual clinical translation. If the fate impact of a particular mechanical cue (Engler et al., 2006) holds true in vivo, it will be important to investigate whether it is possible to prospectively isolate stem cell subpopulations with distinct mechanical memories. These kinds of investigations may help define more homogeneous stem cell sources for therapy; most previous studies have derived human MSCs from primary tissues without cell sorting. In addition, introducing recombinant mechanosensing proteins with reporters into in situ stem cell populations in transgenic animal models will facilitate investigations into how stem cells sense and generate physical forces during development and diseases in vivo.
Mechanobiology and Pharmacodynamics of Stem Cells
To predict how stem cells function in the body, it will be useful to adapt the current theoretical framework behind pharmacodynamics. For cell-based products, at least three parameters are important: (1) interactions between external signals and cell receptors (input); (2) signal transduction pathways; and (3) functional phenotypes (output). It is important to appreciate that biophysical forces could potentially regulate all of these three parameters. First, shear forces and the actin cytoskeleton can regulate conformational changes of receptors, which will likely influence dose-response. How other mechanical cues, such as matrix mechanics, influence dose-response to biochemical cues, and vice versa, remains to be elucidated. Second, systems biology approaches are now being leveraged to identify and model signaling pathways that are selectively activated in response to mechanical parameters, including matrix stiffness (Swift et al., 2013). How these mechanotransduction pathways synergize or antagonize other pathways activated by biochemical cues remains to be elucidated. Third, the functional output of cells is very complex, but one useful parameter to focus on is protein secretion, since the majority of clinical trials with MSCs rely on their ability to produce anti-inflammatory and trophic factors. Defining how culture methods, scaffolds, and mechanical cues influence this process will further inform pharmacodynamics of stem cells.
Mechanical Deformability and Pharmacokinetics of Stem Cells
For a therapy to be successful, stem cells must be delivered to the correct location or locations where the mode of action occurs. The success of cell delivery will likely be determined not only by what route of administration is used, but also by how donor cells traffic under shear and permeate through small constraining pores in tissues and capillaries. As the latter is highly dependent on the ability of cells to mechanically deform (Shin et al., 2014; Swift et al., 2013), it will be informative to characterize the physical deformability of candidate stem cell products under shear and correlate it with the cell biodistribution and engraftment in vivo. Focused profiling of structural proteins could then be performed to identify molecular biophysical markers that may eventually help predict appropriate delivery of stem cells.
Mechanobiology in Other Applications Complementary to Cell Therapy
Induced pluripotent stem cells (iPSCs) are being extensively explored due to their promise in on-demand production of autologous tissue sources. However, lineage programming is considered more difficult with iPSCs than native stem cells due to epigenetic differences. Mechanical parameters can potentially help prime iPSCs prior to in vivo applications. In addition, incorporating mechanical parameters into high-throughput screening will be useful in the discovery of existing or new drugs that can augment stem cell efficacy. Recapitulating mechanical parameters in disease or “organ-on-a-chip” models may be helpful to predict potency of stem cells targeting specific organs or tissues with altered mechanics during disease progression.
In conclusion, progress in mechanobiology to date highlights the power of combining biophysical engineering with cell and molecular biology to reveal fundamental insights in mechanical control of stem cell functions. This progress is providing inspiration to stem cell therapies and likely will impact regulatory sciences as well.
References
- Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee HP, Lippens E, Duda GN, Mooney DJ. Nat Mater. 2015 doi: 10.1038/nmat4489. in press Published online November 30, 2015. http://dx.doi.org/10.1038/nmat4489. [DOI] [PMC free article] [PubMed]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J, Watson S, Lord MS, Eamegdool SS, Bax DV, Nivison-Smith LB, Kondyurin A, Ma L, Oberhauser AF, Weiss AS, Rasko JE. Nat Biotechnol. 2010;28:1123–1128. doi: 10.1038/nbt.1687. [DOI] [PubMed] [Google Scholar]
- Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N, Lippens E, Lee K, Mehta M, Koshy ST, Darnell MC, Desai RM, Madl CM, Xu M, Zhao X, et al. Nat Mater. 2015;14:1269–1277. doi: 10.1038/nmat4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JW, Buxboim A, Spinler KR, Swift J, Christian DA, Hunter CA, Léon C, Gachet C, Dingal PC, Ivanovska IL, et al. Cell Stem Cell. 2014;14:81–93. doi: 10.1016/j.stem.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]