Abstract
Salvia miltiorrhiza is a well-known material of traditional Chinese medicine. Understanding the regulatory mechanisms of phenolic acid biosynthesis and metabolism are important for S. miltiorrhiza quality improvement. We report here that S. miltiorrhiza contains 19 polyphenol oxidases (PPOs), forming the largest PPO gene family in plant species to our knowledge. Analysis of gene structures and sequence features revealed the conservation and divergence of SmPPOs. SmPPOs were differentially expressed in plant tissues and eight of them were predominantly expressed in phloem and xylem, indicating that some SmPPOs are functionally redundant, whereas the others are associated with different physiological processes. Expression patterns of eighteen SmPPOs were significantly altered under MeJA treatment, and twelve were yeast extract and Ag+-responsive, suggesting the majority of SmPPOs are stress-responsive. Analysis of high-throughput small RNA sequences and degradome data showed that miR1444-mediated regulation of PPOs existing in P. trichocarpa is absent from S. miltiorrhiza. Instead, a subset of SmPPOs was posttranscriptionally regulated by a novel miRNA, termed Smi-miR12112. It indicates the specificity and significance of miRNA-mediated regulation of PPOs. The results shed light on the regulation of SmPPO expression and suggest the complexity of SmPPO-associated phenolic acid biosynthesis and metabolism.
Polyphenol oxidases (PPOs) are copper-binding enzymes widely distributed in plants. They utilize oxygen to catalyze exclusively the dehydrogenation of catechols (i.e., o-diphenols) to the corresponding o-quinones (known as the diphenol oxidase/catecholase/o-diphenolase activity; EC 1.10.3.1) or act as bifunctional enzymes to catalyze the oxidation of monophenols to o-diphenol intermediates (known as the tyrosinase/cresolase/monophenolase activity; EC 1.14.18.1) and the subsequent oxidation of o-diphenols to the corresponding o-quinones1. The highly reactive o-quinones can then polymerize with themselves or react with functional groups of proteins to produce black, brown or red pigments. It is the major cause of fruit browning and discoloration of plants during processing2.
Postharvest browning of fruit and vegetables may cause nutritional loss and has a negative effect on consumer acceptability3. On the other hand, the brown color developed by PPOs is of great importance in the quality of black tea and cacao4. Due to the economic significance, studies on plant PPOs have mainly concentrated on their roles in postharvest discoloration of plants and on preventing PPO-mediated browning reactions. Although it has been over a century of intensive research on PPOs, the physiological function of PPOs in plants is still not well understood5.
Accumulating evidence suggests that PPOs are involved in plant response to biotic and abiotic stresses5. Transgenic tomato with reduced PPO activity showed high drought resistance than wild-type and PPO-over-expressed plants6. PPOs in Populus are Cu- and Zn-responsive and involved in metal ion-associated gene networks7. Over-expression of a potato PPO cDNA in tomato resulted in decreased susceptibility to Pseudomonas syringae pv. Tomato8, while down-regulation of PPOs in tomato and dandelion led to enhanced disease susceptibility6,9. Forest tent caterpillar larvae reared on transgenic hybrid aspen foliage over-expressing PPO genes showed decreased growth rates10. Consistently, negative correlation between PPO activity and larvae consumption and growth was observed when cotton bollworm, beet armyworm and common cutworm were fed on PPO-transgenic tomato plants11,12, and jasmonate-dependent induction of PPO activity in tomato foliage was found to be important for defense against beet armyworm13. Additionally, PPOs are involved in seeds against herbivores, bacteria and fungi and proposed to be vital for seed survival in the soil14. In addition to being stress-responsive, a subset of PPOs is targeted by miR1444 for direct cleavage in P. trichocarpa15. MiR1444-mediated posttranscriptional regulation of PPOs plays significant roles in copper homeostasis and stress responses7,15. However, miR1444 exists only in Salicaceae plants. The regulatory role of miRNAs in PPOs from other plant species is largely unknown.
Although PPOs appear to be vital in plants, the roles of PPOs played may vary among species. It is attributed to the diversity of PPO sequences5. A plant PPO protein typically consists of three domains: an N-terminal targeting signal, a dicopper centre, and a C-terminal region16. The N-terminal targeting signal is usually a chloroplast transit peptide (cTP) responsible for the translocation of PPO proteins to the thylakoid lumen1; while in Antirrhinum majus aureusidin synthase 1 (AmAS1) and Populus trichocarpa PtrPPO13, it is a secretory pathway signal peptide (SP) directing PPO proteins to vacuola1,17. The dicopper centre includes two copper-binding domains, CuA and CuB, each of which is approximately 50 amino acids in length16. Sequence comparison suggests that CuA is more variable than CuB and this variation may affect substrate preferences16. The C-terminal region consists of a 50 amino acid PPO1_DWL domain and a 140–150 amino acid PPO1_KFDV domain16. The functional significance of this region is not known, but it is susceptible to proteolytic cleavage in bean and sugarcane16.
Salvia miltiorrhiza Bunge is a well-known material of various traditional Chinese medicines (TCMs) widely used in the treatment of cardiovascular, hyperlipidemia, cerebrovascular and acute ischemic stroke diseases18. It is also an emerging model system for medicinal plant biology19. Hydrophilic phenolic acids are a group of the main active pharmaceutical ingredients of S. miltiorrhiza. Understanding the regulatory mechanisms of phenolic acid biosynthesis and metabolism are important for S. miltiorrhiza quality improvement20. Although the significance of PPOs has been shown in various plants, no information is available for PPOs in S. miltiorrhiza. Here, we report genome-wide prediction, molecular cloning and expression analysis of 19S. miltiorrhiza SmPPO genes. We show the conservation and divergence of SmPPOs in gene structures, sequence features and expression patterns. We found that the majority of SmPPOs were stress-responsive and a novel miRNA was involved in posttranscriptional regulation of SmPPOs. The results shed light on the regulation of SmPPO expression and suggest the complexity of PPO-related phenolic acid biosynthesis and metabolism in S. miltiorrhiza.
Results
Prediction and molecular cloning of S. miltiorrhiza SmPPO genes
Through genome-wide computational search and subsequent gene model prediction, a total of 12 full-length and 7 partial SmPPOs were obtained. To verify the prediction and get full-length coding sequences (CDSs) of all 19 SmPPOs, molecular cloning approaches, including 5′ and 3′ RACE and PCR amplification of coding regions, were performed. The identified SmPPOs were named SmPPO1–SmPPO19, respectively. The deduced SmPPO proteins have amino acid numbers from 564 to 625, isoelectric points (pI) from 5.23 to 8.60, and molecular weights (Mw) from 49.0 to 66.0 kDa. All of the cloned SmPPO CDSs have been submitted to GenBank under the accession numbers shown in Table 1.
Table 1. Sequence features of SmPPOs.
| Gene name | Accession no. | ORF (bp) | AA len | pI | Mw (Da) |
|---|---|---|---|---|---|
| SmPPO1 | KX458045 | 1773 | 590 | 6.56 | 66010.64 |
| SmPPO2 | KX458046 | 1713 | 570 | 6.06 | 64457.11 |
| SmPPO3 | KX458047 | 1701 | 566 | 6.49 | 63976.95 |
| SmPPO4 | KX458048 | 1695 | 564 | 6.41 | 64061.26 |
| SmPPO5 | KX458049 | 1716 | 571 | 6.55 | 63827.14 |
| SmPPO6 | KX458050 | 1746 | 581 | 6.25 | 65018.72 |
| SmPPO7 | KX458051 | 1734 | 577 | 5.90 | 64231.44 |
| SmPPO8 | KX458052 | 1704 | 567 | 6.42 | 64143.58 |
| SmPPO9 | KX458053 | 1716 | 571 | 8.41 | 64190.41 |
| SmPPO10 | KX458054 | 1728 | 575 | 5.23 | 64820.16 |
| SmPPO11 | KX458055 | 1704 | 567 | 5.66 | 63326.35 |
| SmPPO12 | KX458056 | 1727 | 580 | 5.69 | 65575.18 |
| SmPPO13 | KX458057 | 1878 | 625 | 8.60 | 70294.13 |
| SmPPO14 | KX458058 | 1701 | 566 | 6.39 | 63573.64 |
| SmPPO15 | KX458059 | 1657 | 551 | 5.91 | 61922.95 |
| SmPPO16 | KX458060 | 1767 | 588 | 5.61 | 65957.20 |
| SmPPO17 | KX458061 | 1719 | 572 | 5.77 | 64851.20 |
| SmPPO18 | KX458062 | 1692 | 563 | 5.28 | 63775.88 |
| SmPPO19 | KX458063 | 1710 | 569 | 6.30 | 64738.34 |
ORF, open reading frame; AA len, the number of amino acid residues; pI, theoretical isoelectric point; Mw, molecular weight.
Phylogenetic analysis of SmPPOs and PPOs from other plant species
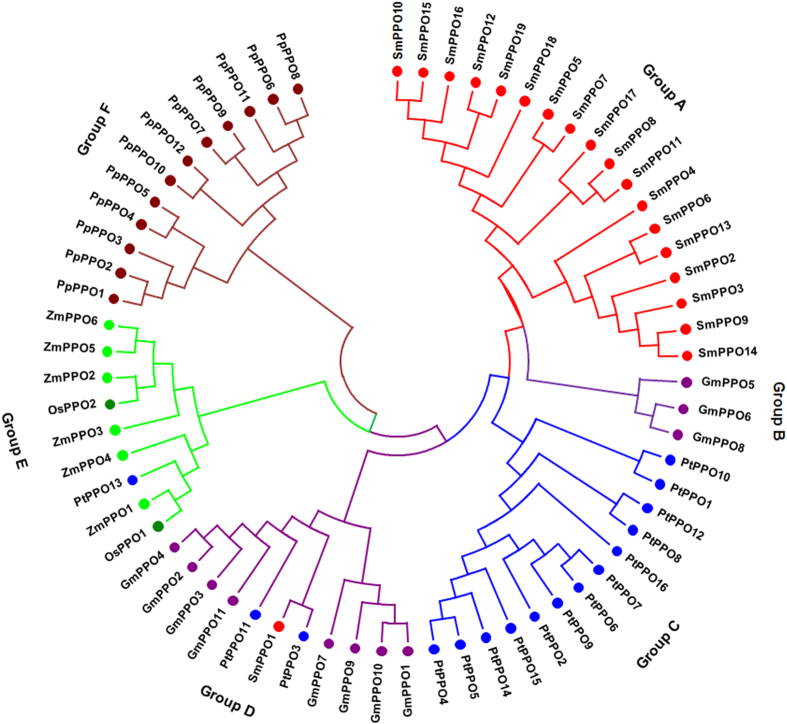
To examine the phylogenetic relationship among 70 PPO proteins from S. miltiorrhiza, P. trichocarpa, Physcomitrella patens, rice (Oryza sativa), maize(Zea mays) and soybean (Glycine max), an unrooted Neighbor-Joining (NJ) tree was constructed (Fig. 1). PPO proteins can be divided into six subgroups (Fig. 1). Phylogenetic analysis reveals species-specific PPO subgroups, which is consistent with previous observation16. In P. patens, S. miltiorrhiza and P. trichocarpa, PPO expansion is largely a consequence of lineage-specific gene duplication and subsequent divergence. 12 PPO sequences from P. patens are clustered into a group. 18 of 19 SmPPOs belong to single group. 14 PtPPOs form a monophyletic group. It indicates that the expansion and diversification of these PPOs was occurred independently in different lineage. OsPPO1, OsPPO2 and PtPPO13 are clustered with ZmPPOs in group E, whereas PtPPO3, PtPPO11 and SmPPO1 are grouped with GmPPOs in group D. It indicates that these PPOs from different plant species share common ancestors.
Figure 1. Phylogenetic relationship of PPO proteins in six plant species.
The phylogenetic tree was built using the neighbor-joining (NJ) method implemented in MEGA7.0 with 1000 bootstrap replicates. PPOs from S. miltiorrhiza (Sm), P. trichocarpa (Pt), Glycine max (Gm), Oryza sativa (Os), Zea mays (Zm) and Phycomitrella patens (Pp) are indicated by bullet points with different colors.
In total, 17 SmPPO genes belonging to six paralogous groups were identified from the 19 cloned SmPPO genes (Table 2). In order to examine the divergence of these paralogs, Ka and Ks were calculated for open reading frames (ORFs) and coding sequences of CuA domain, CuB domain, DWL domain, KFDV domain (Supplementary Fig. S1). On average, the Ka values of ORF, CuA domain, CuB domain, DWL domain, KFDV domain were 0.29, 0.15, 0.23, 0.29, and 0.26, respectively. The Ka values were not significantly different, except for the CuA domain, as revealed by the t-test (P < 0.05). The Ks values from ORF, CuA domain, CuB domain, DWL domain, and KFDV domain were similar (0.75, 0.60, 0.65, 0.70 and 0.64, respectively). The values of Ka and Ks suggest that SmPPOs are highly conserved. The average Ka/Ks ratio of CuA domain (0.25) is much smaller than the average Ka/Ks ratio of other four sources (Ks values of ORF, CuB domain, DWL domain, KFDV domain were 0.38, 0.35, 0.41 and 0.40, respectively). The Ka/Ks ratios indicate strong purifying selection on SmPPOs, and the CuA domain is more conserved than CuB domain.
Table 2. Paralogous groups of SmPPOs in S. miltiorrhiza.
| Paralogous group | Gene name |
|---|---|
| 1 | SmPPO10, SmPPO15, SmPPO16 |
| 2 | SmPPO12, SmPPO18, SmPPO19 |
| 3 | SmPPO5, SmPPO7 |
| 4 | SmPPO8, SmPPO11, SmPPO17 |
| 5 | SmPPO2, SmPPO3, SmPPO9, SmPPO14 |
| 6 | SmPPO6, SmPPO13 |
Conservation and divergence of SmPPO gene structures, conserved domains and motifs
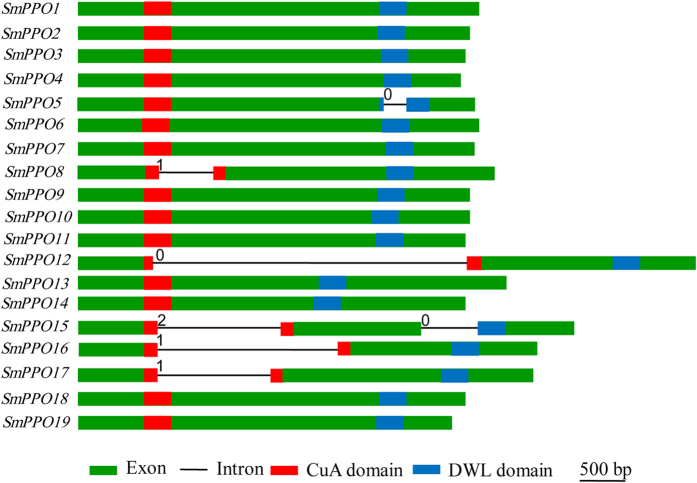
Gene structure analysis showed that the intron number in the coding regions of 19 SmPPO genes varied between 0 and 2 with the majority (63%) to be 0 (Fig. 2). Consistently, most PtPPO genes have no introns in the coding regions16. Only a few eudicot PPOs contain introns, such as the cherimoya PPO gene16 and four Salicaceae PPOs21. It suggests the similarity of PPOs in gene structures. Further examining the position of introns showed that SmPPO8, SmPPO12, SmPPO16 and SmPPO17 contained an intron located in the region encoding CuA domain. SmPPO5 has an intron in the PPO_DWL domain-encoding region. SmPPO15 contains two introns. One is located in the CuA domain-encoding region, whereas the other one is located in the PPO_DWL domain-encoding region (Fig. 2).
Figure 2. Intron-exon structures of SmPPO genes.
Red boxes, blue boxes, green boxes and lines indicate CuA domain, PPO_DWL domain, exons and introns, respectively. The numbers indicate intron phases.
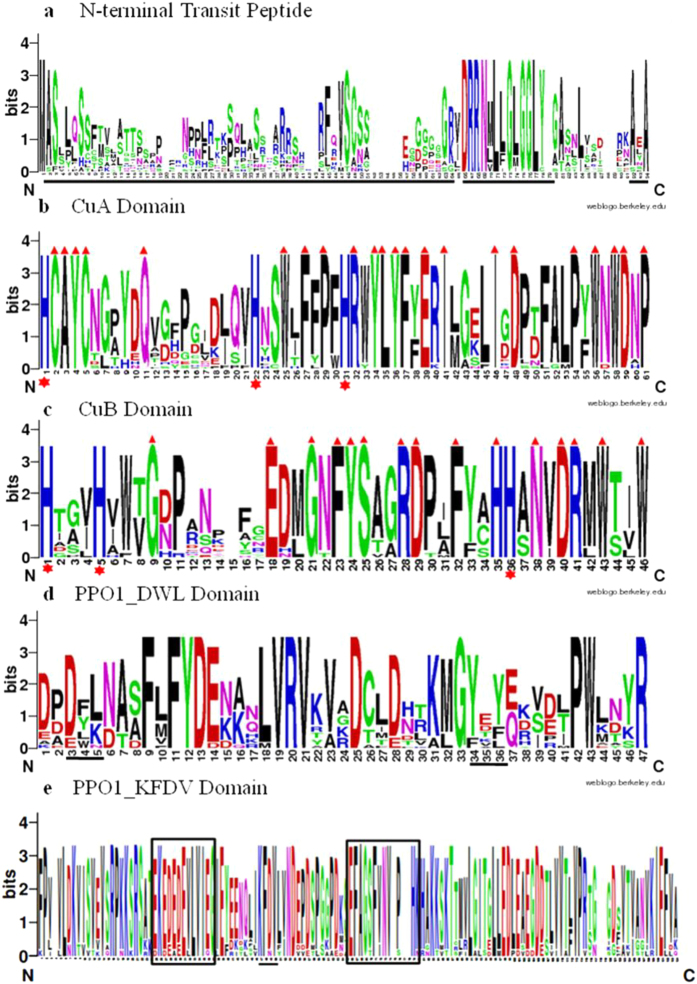
Plant PPO proteins typically consist of three domains: an N-terminal targeting signal, a dicopper centre, and a C-terminal region16. These domains are highly conserved in plants. In order to elucidate sequence features of domains and the degree of conservation of each residue, multiple sequence alignment was performed and sequence logos for these domains of SmPPOs were created using WebLogo. The results showed that the distribution of residues in domains of SmPPOs was quite similar to other plant PPOs16. The N-terminal region of SmPPOs, varying from 80 to 90 amino acids (Fig. 3a), harbors the chloroplast transit peptide (cTP) (Table 3). The cTP (~60 aa) is rich in serine residues. All chloroplast transit peptides of SmPPOs display conserved motifs for a thylakoid transfer domain (TTD) and an alanine cleavage motif (AxA) (Fig. 3a). The dicopper centre includes two copper-binding domains, CuA and CuB (Fig. 3b,c), each of which is approximately 50 amino acids in length16. The highly conserved histidine residues were found in CuA and CuB domains. The CuA domain of SmPPOs contains a highly conserved HCAYC motif (HCAYC) (Fig. 3b), which also exists in other plant PPOs16. The second Cys in this motif is predicted to form a thioether bond with the second conserved histidine of the CuA domain. The sequences between the HxxxC motif and the second conserved histidine are highly variable. In addition to the three histidines, several other amino acids located downstream of the third conserved histidine in the CuA domain are also conserved. It includes arginine, tyrosine, leucine, phenylalanine, glutamic acid, aspartic acid, proline, and tryptophan. In the CuB domain, there are two conserved histidine residues (HxxxH motif) (Fig. 3c). The CuB domain contains several highly conserved residues (Fig. 3c). Sequence comparison suggests that CuB is more variable than CuA in SmPPOs (Fig. 3b,c). The C-terminal portion of SmPPOs shows highly conserved features. It has the PPO1_DWL domain and the PPO1_KFDV domain (Fig. 3d,e)16. PPO1_DWL domain is approximately of 50 amino acids in length (Fig. 3d). It contains a conserved DWL sequence motif and a tyrosine motif (YxY) (Fig. 3d). The PPO1_KFDV domain has a highly conserved sequence motif, KFDV (Fig. 3e). Additionally, the EEEEEVLVI motif enriched in glutamic acid residues and the EFAGSF motif are present in many SmPPOs (Fig. 3e) and other land plant PPOs16. The functional importance of these domains and motifs remains to be elucidated.
Figure 3. Highly conserved domains in SmPPO proteins.
(a) The N-terminal transit peptide. The transit peptide sequences are underlined. (b) The CuA domain. Red triangle indicates absolutely conserved residues. Red stars indicate three conserved histidine residues. (c) The CuB domain. Absolutely conserved residues and conserved histidine residues are indicated by red triangle and stars, respectively. (d) The PPO1_DWL domain. The DWL motif and the tyrosine (YxY) motif are underlined. (e) The PPO1_KFDV domain. The thylakoid transfer domain, the KFDV motif and the alanine (AxA) cleavage motif are underlined. The conserved EEEEEVLVI and EFAGSF motifs are boxed.
Table 3. TargetP prediction of SmPPO localization.
| Name | cTP | mTP | SP | other | Loc | RC |
|---|---|---|---|---|---|---|
| SmPPO1 | 0.966 | 0.022 | 0.028 | 0.084 | C | 1 |
| SmPPO2 | 0.911 | 0.162 | 0.021 | 0.029 | C | 2 |
| SmPPO3 | 0.808 | 0.136 | 0.022 | 0.091 | C | 2 |
| SmPPO4 | 0.727 | 0.200 | 0.080 | 0.046 | C | 3 |
| SmPPO5 | 0.975 | 0.030 | 0.008 | 0.060 | C | 1 |
| SmPPO6 | 0.969 | 0.067 | 0.004 | 0.076 | C | 1 |
| SmPPO7 | 0.977 | 0.018 | 0.009 | 0.077 | C | 1 |
| SmPPO8 | 0.966 | 0.126 | 0.019 | 0.023 | C | 1 |
| SmPPO9 | 0.765 | 0.099 | 0.032 | 0.078 | C | 2 |
| SmPPO10 | 0.977 | 0.078 | 0.014 | 0.028 | C | 1 |
| SmPPO11 | 0.966 | 0.126 | 0.019 | 0.023 | C | 1 |
| SmPPO12 | 0.979 | 0.118 | 0.007 | 0.021 | C | 1 |
| SmPPO13 | 0.929 | 0.132 | 0.035 | 0.009 | C | 2 |
| SmPPO14 | 0.699 | 0.127 | 0.046 | 0.076 | C | 3 |
| SmPPO15 | 0.954 | 0.096 | 0.007 | 0.065 | C | 1 |
| SmPPO16 | 0.979 | 0.053 | 0.022 | 0.024 | C | 1 |
| SmPPO17 | 0.979 | 0.088 | 0.118 | 0.007 | C | 1 |
| SmPPO18 | 0.980 | 0.039 | 0.021 | 0.077 | C | 1 |
| SmPPO1 | 0.966 | 0.022 | 0.028 | 0.084 | C | 1 |
cTP, chloroplast transit peptide; mTP, mitochondrial targeting peptide; SP, signal peptide; Loc, protein localization; C, chloroplast; RC, reliability class from 1 to 5, where 1 indicates the strongest prediction.
Expression profiles of SmPPO genes in S. miltiorrhiza
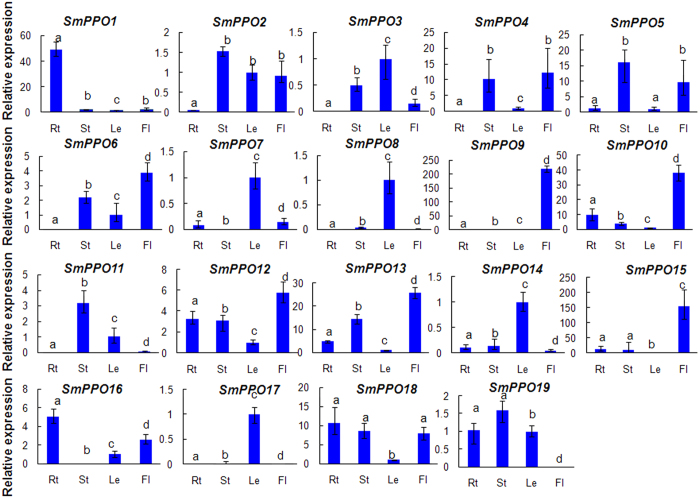
In order to preliminarily elucidate the physiological roles of SmPPOs, we detected the levels of SmPPO transcripts in roots, stems, leaves and flowers of S. miltiorrhiza plants. Differential expression was observed (Fig. 4). SmPPO1 and SmPPO16 were predominantly expressed in roots. SmPPO11 was predominantly expressed in stems. SmPPO3, SmPPO7, SmPPO8, SmPPO14 and SmPPO17 were strongly expressed in leaves. SmPPO6, SmPPO9, SmPPO10, SmPPO12, SmPPO13 and SmPPO15 exhibited the highest expression in flowers. The other 5 SmPPOs were mainly expressed in at least two tissues analyzed. Differentially expressed SmPPOs may be involved in different physiological processes.
Figure 4. Expression of SmPPOs in roots (Rt), stems (St), leaves (Le) and flowers (Fl) of S. miltiorrhiza.
The expression levels were quantified using the quantitative RT-PCR method. Transcript levels in leaves were arbitrarily set to 1 and the levels in other tissues were given relative to this. Error bars represent standard deviations of mean value from three biological and three technical replicates. ANOVA (analysis of variance) was calculated using SPSS. P < 0.05 was considered statistically significant.
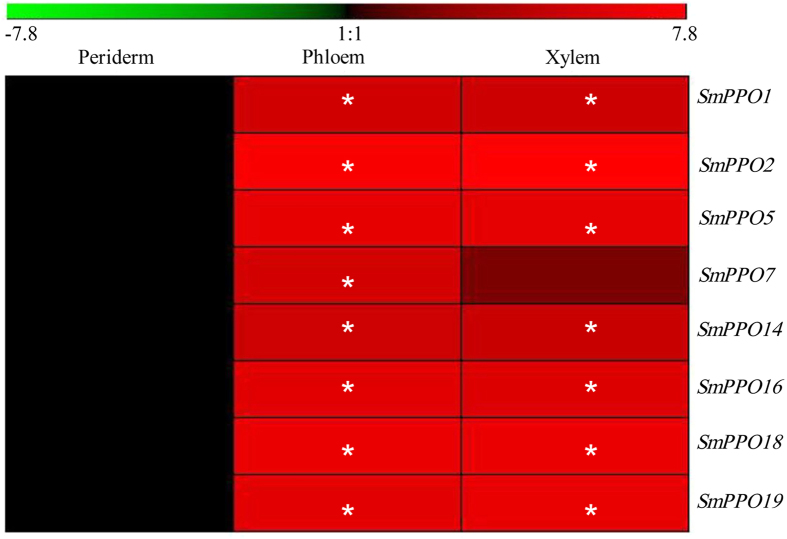
S. miltiorrhiza roots are well-known materials of various TCMs. The pharmacologically active phenolic acid, lithospermic acid B, accumulates mainly in the phloem and xylem of S. miltiorrhiza roots22. In order to elucidate the role of SmPPOs in S. miltiorrhiza roots, transcriptome-wide analysis were carried out. RNA-seq reads from periderm, phloem and xylem of S. miltiorrhiza roots were downloaded from GenBank and then mapped to SmPPOs using SOAP2.022,23. The results showed that 8 SmPPOs were expressed in S. miltiorrhiza roots with RPKM value greater than 224. All of them exhibited higher expression levels in the phloem and xylem than the peridem (Fig. 5), indicating their potential in lithospermic acid B biosynthesis and metabolism.
Figure 5. Expression of SmPPOs in periderm, phloem and xylem of S. miltiorrhiza roots.
RNA-seq reads were mapped to the cloned ORFs of SmPPOs. Genes with RPKM value greater than 2 were analyzed for differential expression using Fisher’s exact test. P < 0.05 was considered as differentially expressed. *Indicates significant differential expression compared with the level in periderm.
Responses of SmPPOs to MeJA treatments
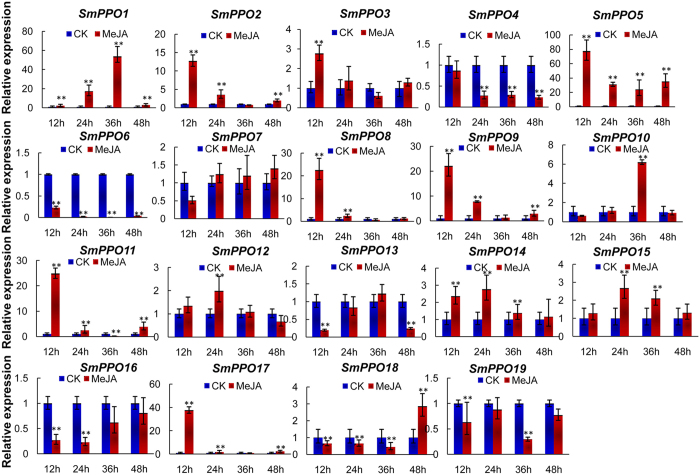
MeJA is an effective elicitor of tanshinone and phenolic acid production in S. miltiorrhiza and participates in plant response to stress24,25. To gain knowledge of SmPPOs in response to MeJA, the expression of SmPPOs in leaves of S. miltiorrhiza plantlets with or without MeJA treatment was performed using the qRT-PCR method. Significant expression level changes were observed for 18 of the 19 SmPPOs at one or more time-points a time-point of MeJA treatment (Fig. 6). A total of 11 SmPPOs, including SmPPO1, SmPPO2, SmPPO3, SmPPO5, SmPPO8, SmPPO9, SmPPO10, SmPPO12, SmPPO14, SmPPO15 and SmPPO17, were significantly up-regulated at least a time-point of MeJA treatment. Among them, SmPPO1 and SmPPO5 were significantly up-regulated at all time-points of MeJA treatment. Five SmPPOs, including SmPPO4, SmPPO6, SmPPO13, SmPPO16, and SmPPO19, were significantly down-regulated at one or more time-points time-point of MeJA treatment. Among them, SmPPO6 was down-regulated at all four time-points. The responses of SmPPO11 and SmPPO18 to MeJA treatment were fluctuated. SmPPO11 was up-regulated at the time-points of 12-, 24- and 48-h-treatment, while down-regulated at the time-point of 36-h-treatment. SmPPO18 was down-regulated at the time-points of 12-, 24- and 36-h-treatment, while up-regulated at the time-point of 48-h-treatment. No significant changes were observed for SmPPO7 after MeJA treatment. The results suggest that the majority of SmPPOs are involved in response to MeJA treatment in S. miltiorrhiza.
Figure 6. Quantitative RT-PCR analysis of SmPPO gene expression in S. miltiorrhiza leaves treated with MeJA.
Fold changes of SmPPOs in leaves of S. miltiorrhiza plantlets treated with MeJA for 12, 24, 36 and 48 h are shown. The level of transcripts in leaves treated with carrier solution (CK) was arbitrarily set to 1 and the levels in leaves treated with MeJA were given relative to this. Mean values and standard deviations were obtained from three biological and three technical replicates. ANOVA (analysis of variance) was calculated using SPSS. P < 0.05(*) and P < 0.01(**) were considered statistically significant and extremely significant, respectively.
Yeast extract and Ag+-responsive SmPPOs
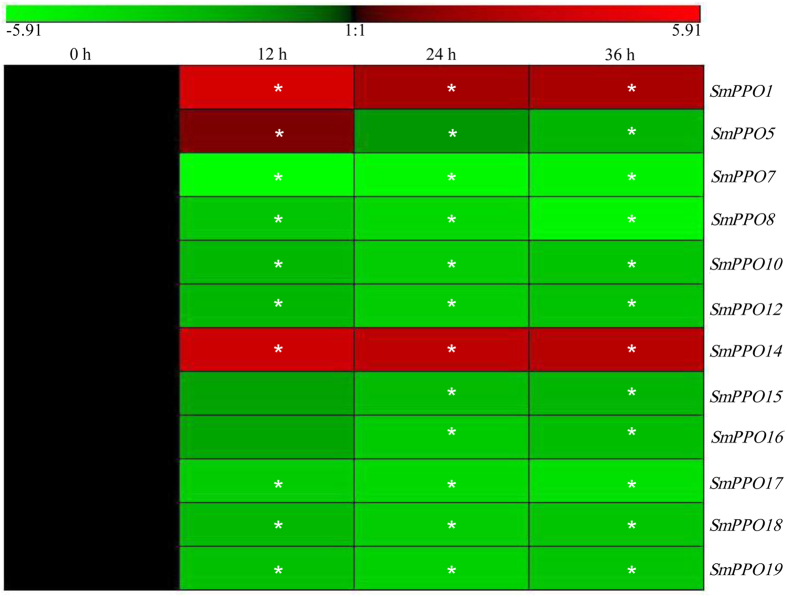
It has been shown that PPOs are involved in plant response to stress5. In order to determine whether SmPPOs play a role in plant defense, we carried out a transcriptome-wide analysis of SmPPO expression in response to the treatment of yeast extract (100 μg/ml) and Ag+ (30 μM), a combination of biotic and abiotic stresses24. RNA-seq data of S. miltiorrhiza hairy roots treated with or without yeast extract (100 μg/ml) and Ag+ (30 μM) were downloaded from GenBank and then mapped to SmPPOs using the SOAP2.0 software23,24. Using a cutoff of RPKM value greater than 224, a total of 12 SmPPOs were found to be expressed in hairy roots. Compared with the level in non-treated control, all of the 12 SmPPOs expressed in hairy roots were differentially expressed at all three time-points of yeast extract and Ag+ treatment (Fig. 7). SmPPO5 was up-regulated at the time-point of 12-h-treatment, whereas down-regulated after 24- and 36-h-treatment. SmPPO1 and SmPPO14 were significantly up-regulated at all time points. The other 9 SmPPOs were significantly down-regulated at all time points (Fig. 7). It suggests that over 60% of SmPPOs are yeast extract and Ag+-responsive.
Figure 7. Responses of SmPPO genes to yeast extract and Ag+ treatment.
RNA-seq reads were mapped to the cloned ORFs of SmPPO genes. Genes with RPKM value greater than 2 were analyzed for differential expression using Fisher’s exact test. P < 0.05 was considered as differentially expressed. *Indicates significant differential expression compared with the level in hairy roots without treatment.
SmPPOs are not regulated by miR1444 in S. miltiorrhiza
In Salicaceae, a subset of PPOs are regulated by a microRNA, termed miR14447,15,21. This microRNA is a lineage-specific young microRNA15. MiR1444-mediated regulation of PtPPOs is significant in copper homeostasis and stress responses in P. trichocarpa7. In order to examine whether SmPPOs are regulated by miR1444, we analyzed published and our own high throughput sequencing data of small RNAs from roots, stems, leaves and flowers of S. miltiorrhiza26,27. No miR1444 sequence was found. We next analyzed the whole genome sequence of S. miltiorrhiza28. Consistently, no MIR1444 precursor sequence was identified. Taken together, we conclude that miR1444 does not exist in S. miltiorrhiza and SmPPOs are not regulated by miR1444.
Identification of Smi-miR12112 targeting 15 SmPPOs for cleavage in S. miltiorrhiza
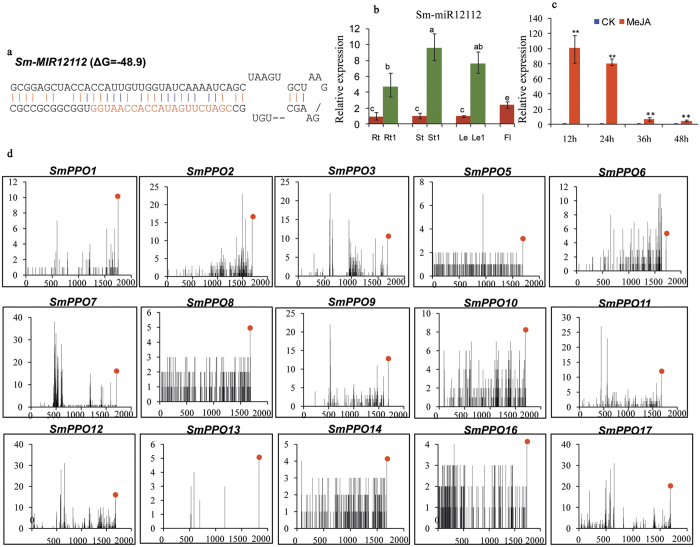
In order to elucidate whether SmPPOs are regulated by other microRNAs, we searched S. miltiorrhiza small RNAs potentially targeting SmPPOs for cleavage using psRNATarget29. With the maximum expectation of 3.0 applied in the target search, a total of 54 small RNAs with sequence reads greater than four were identified. These small RNAs were aligned with the whole genome sequence of S. miltiorrhiza28. Secondary structures were predicted for the retrieved genomic DNA sequences using mfold30 as described previously31. The structures were manually checked and miRNAs were annotated by applying the criteria suggested by Meyers et al.32. As a result, we finally identified a miRNA stem-loop structure designated as Smi-MIR12112 (Fig. 8a). Smi-miR12112 is a novel miRNA that has not been reported previously.
Figure 8. Smi-miR12112 targets SmPPOs for cleavage.
(a) Predicted hairpin structures of Smi-miR12112. Mature miRNA sequences are indicated in red. The secondary structure of Smi-miR12112 was predicted by the mfold program using the default parameters30. (b) Expression of Smi-miR12112 in S. miltiorrhiza. Relative expression of Smi-miR12112 was quantified in total RNA isolated from roots (Rt), stems (St), leaves (Le) and flowers (Fl) of 2-year-old, field-grown S. miltiorrhiza Bunge (line 993) and roots (Rt1), stems (St1) and leaves (Le1) of two-month-old plants cultivated in vitro by quantitative real-time RT-PCR and normalized to the level of 5.8S rRNA in the sample. (c) Expression of Smi-miR12112 in S. miltiorrhiza leaves treated with MeJA. Fold changes of SmPPOs in leaves of S. miltiorrhiza plantlets treated with MeJA for 12, 24, 36 and 48 h are shown. The level of transcripts in leaves treated with carrier solution (CK) was arbitrarily set to 1 and the levels in leaves treated with MeJA were given relative to this. Mean values and standard deviations were obtained from three biological and three technical replicates. ANOVA (analysis of variance) was calculated using SPSS. P < 0.01(**) were considered statistically extremely significant. (d) Validation of Smi-miR12112-directed cleavage using degradome analysis. X-axis shows the nucleotide (nt) position of targets and Y-axis shows the reads obtained by degradome sequencing. Each black spot represents a degradome fragment mapped to the target gene. The red spots indicate that the products are resulted from Smi-miR12112-directed cleavage.
Using the poly(A) adaptor RT-PCR method33, we analyzed the expression of Smi-miR12112 in roots, stems, leaves and flowers of 2-year-old, field-grown S. miltiorrhiza Bunge (line 993) and roots, stems and leaves of two-month-old plants cultivated in vitro. Smi-miR12112 exhibited higher expression level in young roots, young stems and young leaves compared with the level in mature roots, mature stems, mature leaves and flowers (Fig. 8b). High level of expression in young tissues indicates that Smi-miR12112 plays more important roles in young tissues than mature ones. In order to determine whether Smi-miR12112 is transcriptionally regulated by MeJA, we analyzed the expression of Smi-miR12112 in leaves of S. miltiorrhiza plantlets with or without MeJA treatment using the qRT-PCR method. The results showed that the level of Smi-miR12112 was up-regulated at all four time-points (Fig. 8c). Differential response of Smi-miR12112 (Fig. 8c) and SmPPOs (Fig. 6) to MeJA treatment suggest the complexity of the miRNA and SmPPO-associated gene regulatory networks.
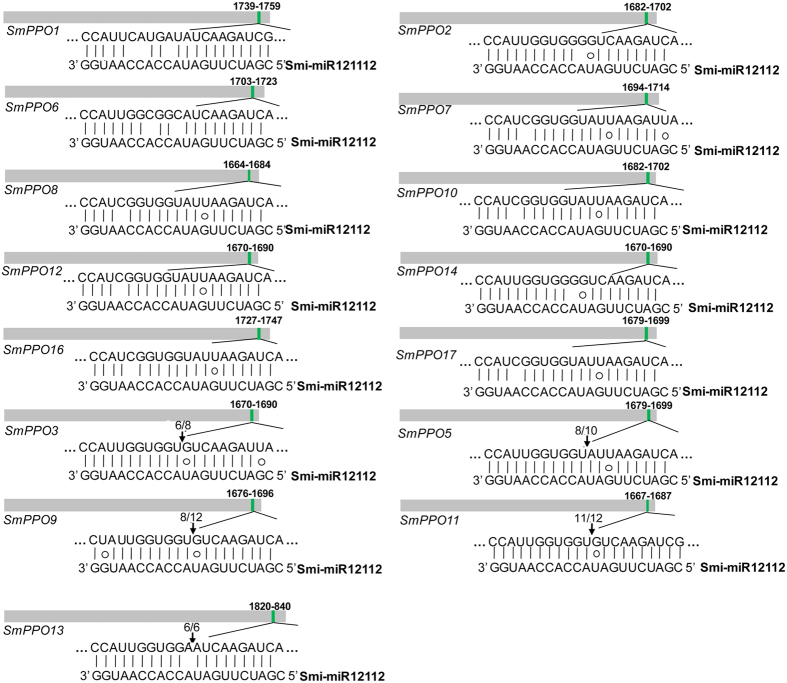
Computational target prediction showed that 15 of the 19 identified SmPPOs contained a sequence near-perfectly complementary to Smi-miR12112 (Fig. 9). The sequence locates in a region encoding KFDV, a conserved domain of PPO proteins. Large proportion of SmPPOs having putative miRNA target sites could be due to deep conservation of sequence in the KFDV region. Plant mature miRNAs usually guide RNA-induced silencing complexes (RISCs) to cleave target mRNAs at the tenth complementary nucleotide from the 5′ end of the miRNA34. Based on this ruler, we analyzed our S. miltiorrhiza degradome sequencing data set for SmPPO cDNA fragments resulted from Smi-miR12112-mediated cleavage. The results confirmed the 15 SmPPOs to be targets of Smi-miR12112 (Fig. 8c). To further validate miRNA-mediated cleavage of predicted targets, we carried out rapid amplification of 5′ complementary DNA ends (5′-RACE). The results showed that five SmPPOs, including SmPPO3, SmPPO5, SmPPO9, SmPPO11 and SmPPO13, were indeed cleaved by Smi-miR12112 in vivo (Fig. 9). It verifies the results from computational prediction.
Figure 9. Locations of Smi-miR12112 complementary sequences in SmPPOs.
Heavy grey lines represent ORFs. Smi-miR12112 complementary sites (green) with the nucleotide positions of SmPPO are indicated. The mRNA sequence of each complementary site from 5′ to 3′ and the mautre Smi-miR12112 sequence from 3′ to 5′ are shown in the expanded regions. Watson-Crick pairing (vertical dashes) and G:U wobble pairing (circles) are indicated. Vertical arrows indicate the 5′ termini of miRNA-guided cleavage products, as identified by 5′-RACE, with the frequency of clones shown.
Discussion
PPOs exist in land plants, fungi and some bacteria, such as P. patens17,35, P. trichocarpa1,7,8 potato36, tomato37, walnut38, eggplant39, sugarcane17, litchi40, Vicia faba41, and grapevine42. PPOs are encoded by a gene family with member numbers varied significantly among species17. The non-vascular moss, P. patens, contains thirteen PPO members36. Soybean has eleven. Sorghum (Sorghum bicolor) has eight. Maize (Zea mays) and purple false brome (Brachypodium distachyon) contain six. Fox millet (Setaria italica) has four. Rice contains two. Cucumber, cassava, castor bean and walnut have one6,38. No PPO was detected in Arabidopsis, Brassica napus and green algae (Chlorella, Stigeoclonium, Microspora, Ulva and Spirogyra)17. The lack of a PPO gene in various plant species points to the ecological or secondary metabolic functions for PPOs16. Consistently, the number of PPO genes in an organism is not directly related to the size of its genome. For instance, the Selaginella moellendorffii genome (~100 Mbp) is one of the smallest plant genomes known; however, the number of PPO genes, 11, in this species is relatively greater16. Analysis of the P. trichocarpa genome assembly v3.0 (http://www.phytozome.net/poplar.php#B) showed the existence of 15 full-length PtPPO genes. In this study, we identified a total of 19 full-length PPO genes in S. miltiorrhiza. It suggests that S. miltiorrhiza contains the largest PPO gene family in plant species with the whole genome sequence available.
It has been shown that land plant PPO genes originate in bacteria via an ancient horizontal gene transfer event43. The transferred PPO gene may expand through gene duplication or lost through chromosome rearrangement or fragment deletion during plant evolution, resulting in significant variation in PPO gene numbers in different plant species. The lack of PPO genes in Arabidopsis suggests that PPO genes may be lost through deletion and mutation during chromosome rearrangement. Consistently, phylogenetic and gene structure analysis indicated that PPO genes are relatively conserved across different species. For instance, most PPO proteins share similar domains and motifs across plants. The intron/exon structures of PPO genes are also largely conserved. The evolution history suggests that PPO genes from different organisms may share conserved molecular functions.
Recent studies implicate that PPOs are involved in the biosynthesis of phenolic acids. PPOs from Portulaca grandiflora and other plants in the Caryophyllales are able to hydroxylate tyrosine to 3, 4-dihydroxyphenylalanine (DOPA) and are considered to be one of the key enzymes for the biosynthesis of water-soluble pigment betalains, which replace the anthocyanins in flowers and fruits of most Caryophyllales plants44,45,46,47,48. A vacuole-localized PPO from snapdragon (Antirrhinum majus), named AmAS1, specifically catalyzes the formation of aurones from chalcones, a class of plant flavonoids responsible for the yellow coloration of flowers17. Larreatricin hydroxylase (LtLH), a typical PPO from creosote bush (Larrea tridentate), specifically hydroxylates (+)-larreatricin to (+)-3′-hydroxylarreatricin and is thought to play a central role in the biosynthesis of the creosote bush 8–8′ linked lignans49. In walnut, PPO is involved in the phenylpropanoid pathway and the tyramine pathway and acts as an indirect regulator of cell death38. Phenolic acids are a group of bioactive compounds in S. miltiorrhiza. The SmPPOs invovled in phenolic acid biosynthesis are currently unknown. Further analysis of SmPPO functions through genetic transformation will definitely shed lights on the mechanism of phenolic acid biosynthesis in S. miltiorrhiza. In addition to be involved in phenolic acid biosynthesis, the diversified expression patterns indicate that SmPPOs may also play significant roles in other physiological processes. Evidence obtained in this study includes that the expression of SmPPOs exhibits apparent tissue specificity (Fig. 4), and the majority of SmPPOs are responsive to MeJA treatment (Fig. 6) and yeast extract and Ag+ treatment (Fig. 7).
miRNAs are a class of small endogenous non-coding RNAs with size about 21 nucleotides. They play vital roles in multiple developmental and physiological processes in various organisms through sequence-specific regulation of target genes at the transcriptional or post-transcriptional level50. MiRNA-mediated posttranscriptional regulation is important for the function of a subset of PPOs in P. trichocarpa and grapevine15,51. In P. trichocarpa, at least 13 PtPPOs are regulated by miR14447. In grapevine, VvPPO is regulated by miR05851. Although miRNAs have been studied extensively in the past several years, only few documents have been reported for miRNAs in S. miltiorrhiza26,27,52,53,54,55. The functions of most S. miltiorrhiza miRNAs are still unknown. Analysis of small RNA data revealed the loss of miR1444 in S. miltiorrhiza. Instead, a novel PPO-targeting miRNA, designated as Smi-miR12112, exists in S. miltiorrhiza. It targets to 15 of the 19 identified SmPPOs in a region encoding the conserved KFDV domain. The target site of Smi-miR12112 is different from those of Pt-miR1444s and Vv-miR058, which locate in the regions encoding the conserved CuB domain and the thylakoid transfer domain, respectively. Since miRNA target sites generally occur outside of family-defining domains56, the origination and evolution of PPO conserved domain-targeting miRNAs may be under strong negative selection. It suggests the significance of miRNA-mediated posttranscriptional regulation of PPOs in plants.
Materials and Methods
Plant materials and treatments
Salvia miltiorrhiza Bunge (line 99–3) with whole genome sequence available was grown in a field nursery at the Institute of Medicinal Plant Development. Flowers, leaves, stems and roots were harvested from 2-year-old plants in June when S. miltiorrhiza is booming, and stored in liquid nitrogen until use. Leaves were treated with MeJA (200 μM) for 12 h, 24 h, 36 h, and 48 h and collected as described in a previous study57. Plantlets treated with carrier solution were used as controls. Three independent biological replicates were carried out for each experiment.
Gene prediction and cDNA cloning
The amino acid sequences of 15 P. trichocarpa PPOs (PtPPOs) were downloaded from the P. trichocarpa genome assembly v3.0 (http://www.phytozome.net/poplar.php#B). To obtain sequencing data for all of the SmPPO genes, tBLASTn searches were performed on the genome database of S. miltiorrhiza line 99-3 (http://www.ndctcm.org/)28. An e-value cut-off of e−10 was applied. Gene model prediction was carried out for the retrieved genomic DNA sequences on the GENSCAN web server (http://genes.mit.edu/GENSCAN.html). The predicted gene models were examined and comparatively analyzed with the other genome database of S. miltiorrhiza (http://www.herbal-genome.cn)58.
To clone the full-length SmPPO cDNAs, rapid amplification of 5′ (5′-RACE) and 3′ (3′-RACE) cDNAs ends was performed using the SMARTTM RACE cDNA amplification kit (TaKaRa Bio, Otsu, Japan). The nesting and the nested PCR amplification was performed on cDNA reverse-transcribed from total RNA using primers listed in Supplementary Tables S1 and S2. Full-length coding sequences were amplified by PCR using a combination of gene-specific forward primers and reverse primers (Supplementary Table S3). PCR products were purified, cloned and sequenced.
Bioinformatic analysis and phylogenetic tree construction
The theoretical isoelectric point (pI) and molecular weight (Mw) were analyzed using the Compute pI/Mw tool on the ExPASy server (http://web.expasy.org/compute_pi/)59. The conserved domain of SmPPO proteins was searched against the Conserved Domain Database (CDD, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The expected e-value threshold of 1.0 and the maximum size of hits to be 500 amino acids were applied60. Sequence logos were created on the WebLogo server (http://weblogo.berkeley.edu/logo.cgi)61.
Protein sequences of PPOs from P. trichocarpa (PtPPOs), Glycine max (GmPPOs), Oryza sativa (OsPPOs), Zea mays (ZmPPOs) and Physcomitrella patens (PpPPOs) were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) and NCBI (http://www.ncbi.nlm.nih.gov/protein/) (Supplementary Table S4). The phylogenetic tree was constructed using MEGA 7.0, with 1000 bootstrap replicates62.
Paralog identification and synteny analysis
Paralog groups were identified using BLASTP with the following criteria applied. It includes E value ≤10–40, cumulative identity percentage (CIP) ≥60%, and cumulative alignment length percentage (CALP) ≥70%. The paralog groups were recognized as homologous SmPPO gene groups among all of the SmPPO genes identified in the S. miltiorrhiza genome. cDNA sequences of each paralogous group and orthologous group were subjected to multiple sequence alignments. The numbers of nonsynonymous substitutions per nonsynonymous site (Ka) and synonymous substitutions per synonymous site (Ks) were calculated using MEGA 7.062.
Quantitative real-time reverse transcription-PCR (qRT-PCR)
The first cDNA strand was reversely transcribed using SuperScript Ш Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). qRT-PCR analysis of SmPPOs in flowers, leaves, stems and roots of 2-year-old plants and in plantlets treated with MeJA was carried out as described previously57. Gene-specific primers were designed using the tool of IDT designing primers (http://www.idtdna.com/scitools/Applications/RealTimePCR/) (Supplementary Table S5). The length of amplicons was between 80 bp and 200 bp. SmUBQ10 was used as an internal control as previously described54,57. The expression of Smi-miR12112 was analyzed using the poly(A) adaptor RT-PCR method as described previously33. Real-time PCR was performed using 5′-CGATCTTGATACCACCAATGG-3′ as the forward primers and 5′-GCGAGCACAGAATTAATACGAC-3′ as the reverse primer. 5.8S rRNA gene was selected as a reference7. Gene expression data from three biological replicates was standardized as described previously63. ANOVA (analysis of variance) was calculated using SPSS (Version 19.0, IBM, USA). P < 0.05 was considered statistically significant, and P < 0.01 was considered extremely significant.
Analysis of SmPPOs expression using RNA-seq data
RNA-seq data from various S. miltiorrhiza tissues was downloaded from GenBank. The tissues include periderm, phloem and xylem of roots (SRR1640458), hairy roots, and hairy roots treated with yeast extract (100 μg/ml) and Ag+ (30 μM) (SRR924662)22,24. RNA-seq reads were mapped to SmPPOs using SOAP2.023 and analyzed as described previously57. SmPPOs with the RPKM value greater than 2 were analyzed for differential expression using Fisher’s exact test. P < 0.05 was considered as differentially expressed.
Identification of S. miltiorrhiza miRNAs with perfect or near-perfect complementarity to SmPPOs
S. miltiorrhiza small RNAs with the potential to target SmPPOs for cleavage were predicted using psRNATarget29. The nineteen SmPPO genes identified were used as target transcript candidates. The maximum expectations of 3.0 and the target accessibility-allowed maximum energy to unpair the target site of 25.0 were applied. The identified small RNAs were aligned with the genome of S. miltiorrhiza27. Secondary structures of S. miltiorrhiza genomic DNA sequences with small RNAs aligned were predicted using mfold30. In each case, the lowest energy structure was analyzed as described previously30.
Degradome and experimental verification of miRNA-directed cleavage of SmPPOs
Degradome analysis of miRNA-targeted SmPPOs was carried out using SOAP2.023. To map miRNA cleavage sites in SmPPO targets, the modified RNA ligase-mediated rapid amplification of 5′ cDNAs method (5′RLM-RACE) was performed using the SMARTTM RACE cDNA amplification kit (TaKaRa Bio, Otsu, Japan). The nesting and the nested primers used in this experiment are listed in Supplementary Table S6. Nesting PCR amplification was performed under the following program of touchdown PCR: 94 °C for 5 min, 5 cycles of amplification 94 °C for 30 s and 72 °C for 3 min, 5 cycles of amplification at 94 °C for 30 s, 70 °C for 30 s and 72 °C for 3 min, 25 cycles of amplification at 94 °C for 30 s, 56 °C for 30 s and 72 °C for 3 min, followed by a final extension at 72 °C for 10 min. Nested PCR amplification was carried out under the following conditions: 94 °C for 5 min, 25 cycles of amplification at 94 °C for 30 s, 56 °C for 30 s and 72 °C for 3 min, followed by a final extension at 72 °C for 10 min. PCR products were purified, cloned and sequenced.
Additional Information
How to cite this article: Li, C. et al. Characterization of the polyphenol oxidase gene family reveals a novel microRNA involved in posttranscriptional regulation of PPOs in Salvia miltiorrhiza. Sci. Rep. 7, 44622; doi: 10.1038/srep44622 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We appreciate Prof. Xian’en Li at the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College for providing S. miltiorrhiza plants. This work was supported by grants from the National Key Research and Development Program of China (Grant No. 2016YFD0600104), the Natural Science Foundation of China (Grant Nos 31370327, 31570667 and 81603225), the Beijing Natural Science Foundation (Grant No. 5152021), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-3-016), and PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (Grant No. 3332016072).
Footnotes
The authors declare no competing financial interests.
Author Contributions C.L. analyzed the data, performed RACE and coding sequence (CDS) cloning, and participated in writing the manuscript. D.L. contributed to RNA extraction, RACE, coding sequence (CDS) cloning, and qRT-PCR. J.L. participated in bioinformatics analysis. F.S. prepared the plantlets treated with methyl jasmonate (MeJA). S.F. designed the experiment, participant in bioinformatics analysis, and wrote the manuscript. All authors have read and approved the version of manuscript.
References
- Tran L. T. & Constabel C. P. The polyphenol oxidase gene family in poplar: phylogeny, differential expression and identification of a novel, vacuolar isoform. Planta 234, 799–813 (2011). [DOI] [PubMed] [Google Scholar]
- Bittner S. When quinones meet amino acids: chemical, physical and biological consequences. Amino Acids 30, 205–224 (2006). [DOI] [PubMed] [Google Scholar]
- Vámos-Vigyázó L. Polyphenol oxidase and peroxidase in fruits and vegetables. Critical Reviews in Food Science & Nutrition 15, 49–127 (1981). [DOI] [PubMed] [Google Scholar]
- Stodt U. W. et al. Investigation of processes in black tea manufacture through model fermentation (oxidation) experiments. Journal of Agricultural & Food Chemistry 62, 7854–7861 (2014). [DOI] [PubMed] [Google Scholar]
- Mayer A. M. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry 67, 2318–2331 (2006). [DOI] [PubMed] [Google Scholar]
- Thipyapong P., Hunt M. D. & Steffens J. C. Antisense downregulation of polyphenol oxidase results in enhanced disease susceptibility. Planta 220, 105–117 (2004). [DOI] [PubMed] [Google Scholar]
- Lu S., Yang C. & Chiang V. L. Conservation and diversity of microRNAassociated copper-regulatory networks in Populus trichocarpa. Journal of Integrative Plant Biology 53, 879–891 (2011). [DOI] [PubMed] [Google Scholar]
- Li L. & Steffens J. C. Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta 215, 239–247 (2002). [DOI] [PubMed] [Google Scholar]
- Richter C., Dirks M. E., Gronover C. S., Pruefer D. & Moerschbacher B. M. Silencing and heterologous expression of ppo-2 indicate a specific function of a single polyphenol oxidase isoform in resistance of dandelion (Taraxacum officinale) against Pseudomonas syringae pv. tomato. Molecular Plant-Microbe Interactions 25, 200–210 (2012). [DOI] [PubMed] [Google Scholar]
- Wang J. & Constabel C. P. Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 220, 87–96 (2004). [DOI] [PubMed] [Google Scholar]
- Mahanil S., Attajarusit J., Stout M. J. & Thipyapong P. Overexpression of tomato polyphenol oxidase increases resistance to common cutworm. Plant Science 174, 456–466 (2008). [Google Scholar]
- Bhonwong A., Stout M. J., Attajarusit J. & Tantasawat P. Defensive role of tomato polyphenol oxidases against cotton bollworm (Helicoverpa armigera) and beet armyworm (Spodoptera exigua). Journal of Chemical Ecology 35, 28–38 (2009). [DOI] [PubMed] [Google Scholar]
- Bosch M., Berger S., Schaller A. & Stintzi A. Jasmonate-dependent induction of polyphenol oxidase activity in tomato foliage is important for defense against Spodoptera exigua but not against Manduca sexta. BMC Plant Biology 14, 257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst E. P., Okubara P. A., Anderson J. V. & Morris C. F. Polyphenol oxidase as a biochemical seed defense mechanism. Frontiers in Plant Science 5, 689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Sun Y. H. & Chiang V. L. Stress-responsive microRNAs in Populus. Plant Journal 55, 131–151 (2008). [DOI] [PubMed] [Google Scholar]
- Tran L. T., Taylor J. S. & Constabel C. P. The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genomics 13, 395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E. et al. Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles. Plant Journal 45, 133–143 (2006). [DOI] [PubMed] [Google Scholar]
- Li Y. G., Song L., Liu M., Hu Z. B. & Wang Z. T. Advancement in analysis of Salvia miltiorrhizae Radix et Rhizoma (Danshen). Journal of Chromatography 1216, 1941–1953 (2009). [DOI] [PubMed] [Google Scholar]
- Song J. Y. et al. Salvia miltiorrhiza as medicinal model plant. Yao Xue Xue Bao 48, 1099–1106 (2013). [PubMed] [Google Scholar]
- Hou X., Shao F., Ma Y. & Lu S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: genome-wide characterization, molecular cloning and expression analysis. Molecular Biology Reports 40, 4301–4310 (2013). [DOI] [PubMed] [Google Scholar]
- Wang M., Li C. & Lu S. Origin and evolution of MIR1444 genes in Salicaceae. Scientific Reports 7, 39740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant Journal 82, 951–961 (2015). [DOI] [PubMed] [Google Scholar]
- Li R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009). [DOI] [PubMed] [Google Scholar]
- Gao W. et al. Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genomics 15, 73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y. et al. Methyl jasmonatedramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiologia Plantarum 137, 1–9 (2009). [DOI] [PubMed] [Google Scholar]
- Xu X. et al. Deep sequencing identifies tissue-specific microRNAs and their target genes involving in the biosynthesis of tanshinones in Salvia miltiorrhiza. PLoS One 9, e111679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F., Qiu D. & Lu S. Comparative analysis of the Dicer-like gene family reveals loss of miR162 target site in SmDCL1 from Salvia miltiorrhiza. Scientific Reports 5, 9891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Molecular Plant 6, 949–952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhuang Z. & Zhao P. X. Computational analysis of miRNA targets in plants: current status and challenges. Briefings in Bioinformatics 12, 115–121 (2011). [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. et al. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17, 2186–2203 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B. C. et al. Criteria for annotation of plant microRNAs. Plant Cell 20, 3186–3190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R. & Chiang V. L. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39, 519–525 (2005). [DOI] [PubMed] [Google Scholar]
- Rhoades M. W. et al. Prediction of plant microRNA targets. Cell 110, 513–520 (2002). [DOI] [PubMed] [Google Scholar]
- Richter H., Lieberei R. & Von Schwartzenberg K. Identification and characterisation of a bryophyte polyphenol oxidase encoding gene from Physcomitrella patens. Plant Biology 7, 283–291 (2005). [DOI] [PubMed] [Google Scholar]
- Thygesen P. W., Dry I. B. & Robinson S. P. Polyphenol oxidase in potato (a multigene family that exhibits differential expression patterns). Plant Physiology 109, 525–531 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven R., Ronning C., Giovannoni J., Martin G. & Tanksley S. Deductions about the number, organization and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araji S. et al. Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiology 164, 1191–1203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty S. M., Chandrashekar A. & Venkatesh Y. P. Eggplant polyphenol oxidase multigene family: Cloning, phylogeny, expression analyses and immunolocalization in response to wounding. Phytochemistry 72, 2275–2287 (2011). [DOI] [PubMed] [Google Scholar]
- Wang J., Liu B., Xiao Q., Li H. & Sun J. Cloning and expression analysis of litchi (Litchi Chinensis Sonn.) polyphenol oxidase gene and relationship with postharvest pericarp browning. PLoS One 9, e93982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary J. W., Lax A. R. & Flurkey W. H. Cloning and characterization of cDNAs coding for Vicia faba polyphenol oxidase. Plant Molecular Biology 20, 245–253 (1992). [DOI] [PubMed] [Google Scholar]
- Dry I. B. & Robinson S. P. Molecular cloning and characterization of grape berry polyphenol oxidase. Plant Molecular Biology 26, 495–502 (1994). [DOI] [PubMed] [Google Scholar]
- Llorente B. et al. Selective pressure against horizontally acquired prokaryotic genes as a driving force of plastid evolution. Scientific Reports 6, 19036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner U., Schliemann W., Böhm H. & Strack D. Tyrosinase involved in betalain biosynthesis of higher plants. Planta 208, 114–124 (1999). [Google Scholar]
- Strack D., Vogt T. & Schliemann W. Recent advances in betalain research. Phytochemistry 62, 247–269 (2003). [DOI] [PubMed] [Google Scholar]
- Gandía-Herrero F., Escribano J. & García-Carmona F. Characterization of the monophenolase activity of tyrosinase on betaxanthins: the tyramine-betaxanthin/dopamine-betaxanthin pair. Planta 222, 307–318 (2005). [DOI] [PubMed] [Google Scholar]
- Gandía-Herrero F., Escribano J. & García-Carmona F. Betaxanthins as substrates for tyrosinase. An approach to the role of tyrosinase in the biosynthetic pathway of betalains. Plant Physiology 138, 421–432 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandía-Herrero F., Jiménez-Atiénzar M., Cabanes J., Escribano J. & García-Carmona F. Fluorescence detection of tyrosinase activity on dopamine-betaxanthin purified from Portulaca oleracea (common purslane) flowers. Journal of Agricultural & Food Chemistry 57, 2523–2528 (2009). [DOI] [PubMed] [Google Scholar]
- Cho M. H. et al. (+)-Larreatricin hydroxylase, an enantio-specific polyphenol oxidase from the creosote bush (Larrea tridentate). Proceedings of the National Academy of Sciences of the United States of America 100, 10641–10646 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C. A. & Martienssen R. A. The developmental role of microRNA in plants. Current opinion in plant biology 8, 38–44 (2005). [DOI] [PubMed] [Google Scholar]
- Ren G. et al. Cloning, expression, and characterization of miR058 and its target PPO, during the development of grapevine berry stone. Gene 548, 166–173 (2014). [DOI] [PubMed] [Google Scholar]
- Shao F. & Lu S. Genome-wide identification, molecular cloning, expression profiling and posttranscriptional regulation analysis of the Argonaute gene family in Salvia miltiorrhiza, an emerging model medicinal plant. BMC Genomics 14, 512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. Journal of Integrative Plant Biology 56, 38–50 (2014). [DOI] [PubMed] [Google Scholar]
- Li C. & Lu S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics 15, 277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Identification and characterization of Salvia miltiorrhizain miRNAs in response to replanting disease. PLoS One 11, e0159905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E. et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nature Genetics 36, 1282–1290 (2004). [DOI] [PubMed] [Google Scholar]
- Li C., Li D., Shao F. & Lu S. Molecular cloning and expression analysis of WRKY, transcription factor genes in Salvia miltiorrhiza. BMC Genomics 16, 200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. et al. Hybrid de novo genome assembly of the Chinese herbal plant danshen (Salvia miltiorrhiza Bunge). GigaScience 4, 62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjellqvist B., Basse B., Olsen E. & Celis J. E. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539 (1994). [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research 39, D225–D229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M. & Brenner S. E. WebLogo: A sequence logo generator. Genome Reseach 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G. & Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E., Leyns L. & Vandesompele J. Standardization of real-time PCR gene expression data from independent biological replicates. Analytical Biochemistry 379, 127–129 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.