Abstract
mRNA splicing is an important mechanism to regulate mRNA expression. Abnormal regulation of this process may lead to lung cancer. Here, we investigated the associations of 11,966 single-nucleotide polymorphisms (SNPs) in 206 mRNA splicing-related genes with lung cancer risk by using the summary data from six published genome-wide association studies (GWASs) of Transdisciplinary Research in Cancer of the Lung (TRICL) (12,160 cases and 16,838 controls) and another two lung cancer GWASs of Harvard University (984 cases and 970 controls) and deCODE (1,319 cases and 26,380 controls). We found that a total of 12 significant SNPs with false discovery rate (FDR) ≤0.05 were mapped to one novel gene PRPF6 and two previously reported genes (DHX16 and LSM2) that were also confirmed in this study. The six novel SNPs in PRPF6 were in high linkage disequilibrium and associated with PRPF6 mRNA expression in lymphoblastoid cells from 373 Europeans in the 1000 Genomes Project. Taken together, our studies shed new light on the role of mRNA splicing genes in the development of lung cancer.
Lung cancer is a major challenge to human health and caused by multiple environment and genetic factors1. Smoking, radon gas, asbestos and air pollution have been established as the environment risk factors2, and genetic factors have also been clearly illustrated in familial3 and segregation studies4. Single nucleotide polymorphisms (SNPs) are the most common genetic variants in the human genome and have been shown to be associated with risk of human diseases, including cancers5. With a large sample size of study subjects, multiple-stage replication and high-throughput chips, genome-wide association studies (GWASs) have been shown to be a robust way of detecting genetic variants involved in susceptibility to cancer. To date, GWASs in different ethnic groups have successfully identified 30 loci in 12 chromosomal regions (3q28, 5p15.33, 6p21.1, 6p21.33, 6q22.1, 7p15.3, 10q25.2, 13q12.12,13q13.1, 15q25.1, 17q24.2, 22q12.2) that confer susceptibility to lung cancer6,7,8,9,10,11,12,13,14,15,16,17, among which, loci at 5p15.33, 6p21.33 and 15q25.1 were found to be risk factors for lung cancer in European descents.
Despite the great success achieved by GWASs, the identified SNPs by GWASs still only explain a small fraction of heritability of lung cancer, a phenomenon called “missing heritability”18. Therefore, many complementary approaches have been developed to improve the study power of GWASs. For example, one of such approaches is the pathway-based association analysis through additional imputation to increase the number of SNPs to be analyzed, which could detect the risk-associated genes with multiple, independent and small effect sizes. This approach could alleviate issues due to insufficient chip coverage of earlier GWASs, reduce the multiple-testing burden of GWAS, achieve consistent results across studies and provide understanding of genetic findings with some functional relevance19,20,21,22.
Among the biological candidate pathways for lung carcinogenesis, mRNA splicing, a modification of the pre-mRNA transcript in which introns are removed and exons are joined, is of significant importance23. In most of the circumstances, this pathway functions normally for the human genome to generate proteomic diversity sufficient for various biological processes. However, cancer cells may take advantage of this mechanism to produce aberrant proteins with added, deleted, or altered functional domains that contribute to carcinogenesis including lung cancer24. Recent studies had shown that somatic mutations and germline variants of some mRNA splicing-related genes were associated with development of lung cancer. For instance, Shen et al. found several SNPs in mRNA splicing associated genes (SRSF7, PTBP2 and HNRNPQ) were associated with lung cancer risk in a Chinese population25. To date, however, only a limited number of candidate genes and SNPs in the pathway have ever been studied and reported. In the present study, we systematically selected 206 mRNA splicing-related genes and comprehensively investigated associations between 11,966 genetic variants of these genes and lung cancer risk using eight published lung-cancer GWAS datasets from the Transdisciplinary Research in Cancer of the Lung (TRICL) Consortium.
Results
Overall associations between SNPs in mRNA splicing-related genes and lung cancer risk in the TRICL Consortium
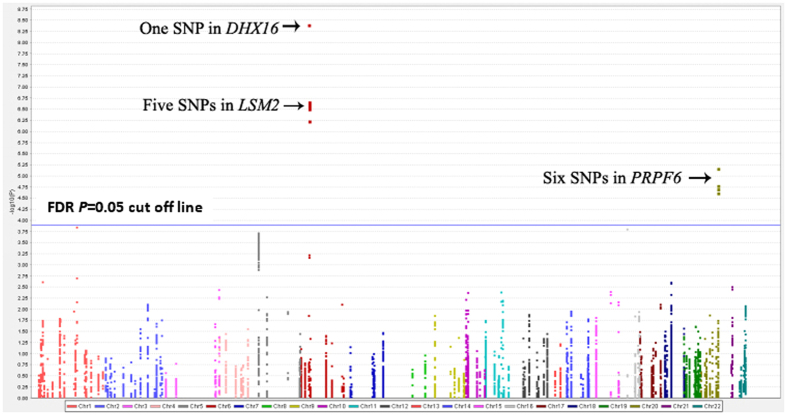
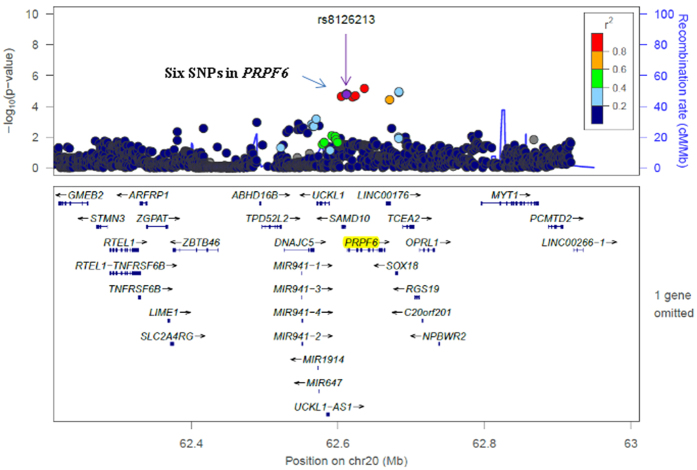
Overall, 11,966 SNPs in 206 mRNA splicing-related genes in six GWAS datasets were selected from the TRICL Consortium (an imputated dataset that included 5,472,374 common SNPs), and their associations with lung cancer risk are shown in the Manhattan plot (Fig. 1). The sample size for each study had been listed in Table 1. After FDR correction (PFDR corrected ≤ 0.05), the 12 SNPs in three genes remained statistically significant (Fig. 1). The most significant SNP for each gene was: rs75100087 in PRPF6 (P = 6.83E-06); rs115420460 in DHX16 (P = 3.93E-09) and rs114312980 in LSM2 (P = 2.19E-07). Their basic information and associations with lung cancer risk are listed in Table 2. SNPs in DHX16 (rs115420460) and LSM2 (rs114312980, rs115489726, rs115801685, rs114637560, rs115834633) were excluded from further analysis due to their locations within the previously identified lung cancer susceptible region Chr6p21.33 and in high LD with previously reported lung cancer GWAS SNPs. Specifically, as shown in Supplement Figure 2, rs115420460 in DHX16 was in high LD (r2 > 0.6) with the previously reported lung cancer GWAS SNP rs43247989. Similarly, Supplement Figure 3 shows that all of the five SNPs in LSM2 were in high LD (r2 > 0.8) with SNP rs3117582 that was identified from a the previously published lung cancer GWAS14. Therefore, we focused on the remaining six newly identified SNPs in PRPF6 (Table 2) for further analysis. Regional plot and recombination rates of PRPF6 (rs8126213 ± 500 kb) in the TRICL consortium are presented in Fig. 2.
Figure 1. Association results of 11,966 SNPs in 206 mRNA splicing-related genes and lung cancer risk in the TRICL Consortium.
SNPs are plotted on the X-axis according to their positions on each chromosome. The association P values with lung cancer risk are shown on the Y-axis (as −log10 P values). The 12 SNPs from three genes (DHX16, LSM2 and PRPF6) were identified after the FDR correction.
Table 1. Characteristic information of the study populations.
| Study | Controls | Lung cancer patients | ||
|---|---|---|---|---|
| All | AD10 | SQ11 | ||
| ICR1 | 5200 | 1952 | 465 | 611 |
| MDACC2 | 1134 | 1150 | 619 | 306 |
| IARC3 | 3791 | 2533 | 517 | 911 |
| NCI4 | 5736 | 5713 | 1841 | 1447 |
| Toronto5 | 499 | 331 | 90 | 50 |
| GLC6 | 478 | 481 | 186 | 97 |
| TRICL7 | 16838 | 12160 | 3718 | 3422 |
| Harvard8 | 970 | 984 | 597 | 216 |
| deCODE9 | 26380 | 1319 | 547 | 259 |
| All combined | 44188 | 14463 | 4862 | 3897 |
1ICR: the Institute of Cancer Research Genome-wide Association Study, UK;
2MDACC: the MD Anderson Cancer Center Genome-wide Association Study, US;
3IARC: the International Agency for Research on Cancer Genome-wide Association Study, France;
4NCI: the National Cancer Institute Genome-wide Association Study, US;
5Toronto: the Lunenfeld-Tanenbaum Research Institute Genome-wide Association Study, Toronto, Canada;
6GLC: German Lung Cancer Study, Germany;
7TRICL: GWASs datasets combined by six GWASs of ICR, MDACC, IARC, NCI, Toronto and GLC;
8Harvard: Harvard Lung Cancer Study, US;
9deCODE: Icelandic Lung Cancer Study, Iceland;
10AD: adenocarcinoma;
11SQ: squamous cell carcinoma.
Table 2. mRNA splicing-related genes SNPs and lung cancer risk in the TRICL1 Consortium with FDR corrected P ≤ 0.05.
| ID | SNP | Chr: Position2 | GENE | Allele3 | EAF4 | Q5 | I2 5 | Effects6 | OR (95% CI)7 | P7 | FDR8 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs115420460 | 6: 30618906 | DHX16 | A/G | 0.11 | 0.67 | 0.00 | ++++++ | 1.18 (1.12–1.25) | 3.93E-09 | <0.0001 |
| 2 | rs114312980 | 6: 31768799 | LSM2 | A/C | 0.11 | 0.23 | 26.77 | ++++++ | 1.20 (1.12–1.29) | 2.19E-07 | 0.0007 |
| 3 | rs115489726 | 6: 31766660 | LSM2 | C/T | 0.11 | 0.24 | 25.69 | ++++++ | 1.20 (1.12–1.29) | 2.54E-07 | 0.0007 |
| 4 | rs115801685 | 6: 31772093 | LSM2 | C/A | 0.11 | 0.23 | 27.36 | ++++++ | 1.20 (1.12–1.29) | 2.61E-07 | 0.0007 |
| 5 | rs114637560 | 6: 31765864 | LSM2 | T/A | 0.15 | 0.27 | 22.42 | +−++++ | 1.14 (1.07–1.21) | 3.04E-07 | 0.0007 |
| 6 | rs115834633 | 6: 31765984 | LSM2 | G/A | 0.11 | 0.20 | 30.79 | ++++++ | 1.20 (1.12–1.30) | 5.75E-07 | 0.0011 |
| 7 | rs116165844 | 20: 62610556 | PRPF6 | G/T | 0.13 | 0.89 | 0.00 | −−−−−− | 0.89 (0.85–0.94) | 1.62E-05 | 0.0197 |
| 8 | rs8126213 | 20: 62611478 | PRPF6 | G/A | 0.13 | 0.89 | 0.00 | −−−−−− | 0.89 (0.85–0.94) | 1.64E-05 | 0.0197 |
| 9 | rs147176547 | 20: 62613001 | PRPF6 | C/G | 0.14 | 0.98 | 0.00 | −−−−−− | 0.90 (0.85–0.94) | 1.65E-05 | 0.0197 |
| 10 | rs112219537 | 20: 62620029 | PRPF6 | G/A | 0.13 | 0.96 | 0.00 | −−−−−− | 0.90 (0.85–0.94) | 2.37E-05 | 0.0237 |
| 11 | rs113450630 | 20: 62623703 | PRPF6 | C/T | 0.14 | 0.98 | 0.00 | −−−−−− | 0.90 (0.85–0.94) | 1.93E-05 | 0.021 |
| 12 | rs75100087 | 20: 62636139 | PRPF6 | C/T | 0.14 | 0.97 | 0.00 | −−−−−− | 0.89 (0.85–0.94) | 6.83E-06 | 0.0117 |
1TRICL: GWASs datasets combined by six GWASs of ICR, MDACC, IARC, NCI, Toronto and GLC.
2Based on NCBI build 37 of the human genome.
3Reference allele/effect allele.
4EAF, effect allele frequency.
5Fixed-effects models were used when no heterogeneity was found between studies (Q > 0.10 and I2 < 25.0); otherwise, random-effects models were used.
6+ means positive association, and − means negative association.
7Meta-analysis of additive results from six lung cancer GWASs.
8FDR, false discovery rate.
Figure 2. Regional plots and recombination rates of PRPF6 (rs8126213 ± 500 kb) in the TRICL Consortium.
PRPF6 rs8126213 was shown in purple and the linkage disequilibrium (LD) values (r2) with the other SNPs are indicated by the heat scale. Other five PRPF6 SNPs are shown in red color, which meant that they were in high LD with rs8126213 (r2 > 0.8).
LD analysis and SNP function annotation
The diagram of six PRPF6 SNPs and their LD plot are shown in Supplement Figure 4b and 4c, respectively. High LD was observed between each pair of the six SNPs (r2 > 0.80), indicating that any of these six SNPs can be a good tag for others. Next, we exploited dbSNP function annotation (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?showRare=on&chooseRs=all&go=Go&locusId=24148) and another two online SNP function prediction tools: SNPfunc (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm) and regulomeDB (http://regulomedb.org/index) to assess their functionality (Supplementary Table 2).
Expression quantitative Trait loci (eQTL) analysis
Expression quantitative trait loci (eQTL) analysis, which directly investigates the correlations between genetic variants and gene expression, has been gradually proved as an effect method to characterize the function of SNPs. In the present study, the eQTL analysis was performed by using the data available from lymphoblastoid cell lines derived from 373 individuals of European descendent (http://www.1000genomes.org/). Finally, all of these six SNPs in PRPF6 were found to be significantly associated with expression levels of PRPF6 in both additive and dominant models (Supplementary Figure 5 and Supplementary Table 2). We also queried the GTEx database (http://www.gtexportal.org/) and found that SNP rs8126213 was significantly correlated with mRNA expression levels of PRPF6 in normal lung tissues (P = 1.50 × 10−5), which was consistent with the results in the lymphoblastoid cell lines. Similar results were found for other five SNPs (Supplementary Figure 6).
Replication of PRPF6 SNP (rs8126213) in another two lung-cancer GWASs
We then selected one of these six PRPF6 SNPs (rs8126213) and investigated its association with lung cancer risk in two additional lung-cancer GWASs of Caucasian origin, Harvard Lung Cancer Study (984 cases and 970 controls) and Icelandic Lung Cancer Study (deCODE from the ILCCO) (1,319 cases and 26,380 controls). However, no significant association was observed in these two studies (P = 0.801 and 0.850, respectively), which may potentially result from a relatively limited sample size of the cases, although their effects were in the same direction as in the TRICL‐ILCCO studies, and the overall effect among all these eight GWASs remained significant (OR = 0.91, 95% CI = 0.87–0.95, P = 6.36E-05 in an additive genetic model) (Table 3). Similar effects were also observed on adenocarcinoma (OR = 0.91, 95% CI = 0.87–0.95, P = 0.002) and squamous cell carcinoma (OR = 0.90, 95% CI = 0.83–0.97, P = 0.005) in the subgroup analysis.
Table 3. Summary association results of PRPF6 rs8126213 (G > A) in all of the eight lung cancer GWASs.
| Study population | Sample size | Overall (N = 14463) | AD (N = 4862) | SQ (N = 3897) | ||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI)* | P* | OR (95% CI)* | P* | OR (95% CI)* | P* | |
| TRICL | 12160 | 16838 | 0.89 (0.85–0.94) | 1.65E-05 | 0.88 (0.81–0.95) | 9.0E-04 | 0.88 (0.82–0.96) | 2.20E-03 |
| ICR | 1952 | 5200 | 0.94 (0.85–1.05) | 0.303 | 0.94 (0.77–1.15) | 0.572 | 0.92 (0.77–1.20) | 0.374 |
| MDACC | 1150 | 1134 | 0.87 (0.73–1.04) | 0.120 | 0.87 (0.71–1.07) | 0.199 | 0.84 (0.64–1.11) | 0.219 |
| IARC | 2533 | 3791 | 0.91 (0.81–1.01) | 0.075 | 0.93 (0.76–1.13) | 0.456 | 0.91 (0.78–1.06) | 0.238 |
| NCI | 5713 | 5736 | 0.87 (0.81–0.95) | 8.36E-04 | 0.86 (0.77–0.97) | 0.012 | 0.85 (0.75–0.96) | 7.92E-03 |
| Toronto | 331 | 499 | 0.86 (0.62–1.20) | 0.379 | 0.77 (0.47–1.27) | 0.314 | 0.97 (0.51–1.83) | 0.923 |
| GLC | 481 | 478 | 0.83 (0.63–1.10) | 0.195 | 0.67 (0.45–0.98) | 0.041 | 1.01 (0.64–1.58) | 0.979 |
| Harvard and deCODE | 2303 | 27350 | 0.98 (0.89–1.09) | 0.772 | 0.97 (0.84–1.12) | 0.683 | 1.02 (0.82–1.26) | 0.875 |
| Harvard | 984 | 970 | 0.97 (0.79–1.20) | 0.801 | 0.94 (0.74–1.18) | 0.576 | 0.97 (0.68–1.40) | 0.886 |
| deCODE | 1319 | 26380 | 0.99 (0.87–1.12) | 0.850 | 0.99 (0.82–1.20) | 0.940 | 1.04 (0.80–1.37) | 0.761 |
| All combined | 14463 | 44188 | 0.91 (0.87–0.95) | 6.36E-05 | 0.91 (0.87–0.95) | 1.70E-03 | 0.90 (0.83–0.97) | 4.80E-03 |
Abbreviations: TRICL: GWASs datasets combined by six GWASs of ICR, MDACC, IARC, NCI, Toronto and GLC; ICR: the Institute of Cancer Research Genome-wide Association Study, UK; MDACC: the MD Anderson Cancer Center Genome-wide Association Study, US; IARC: the International Agency for Research on Cancer Genome-wide Association Study, France; NCI: the National Cancer Institute Genome-wide Association Study, US; Toronto: the Lunenfeld-Tanenbaum Research Institute Genome-wide Association Study, Toronto, Canada; GLC: German Lung Cancer Study, Germany; Harvard: Harvard Lung Cancer Study, US; deCODE: Icelandic Lung Cancer Study, Iceland; AD: adenocarcinoma; SQ: squamous cell carcinoma; *Meta-analysis of results from additive model.
Discussion
mRNA splicing is an important biological mechanism to regulate mRNA expression. Somatic mutations and germline variants of the mRNA splicing-related genes have been found to be associated with various kinds of cancers. For example, SF3B1, U2AF1 and SRSF2 mutations were observed in myeloid and lymphoid lineage tumors26. SF3B1 mutations were commonly found in uveal melanomas27. Mutations affecting spliceosome genes that resulted in defective splicing were attributed to leukemia28. For lung cancer, it had been reported that U2AF1 mutation was found in 3% of lung adenocarcinoma cases29. However, few studies reported a role of genetic variants in the etiology of cancer. For example, only one recent study found that genetic variants in three mRNA splicing-associated genes might modify individual susceptibility to lung cancer in a Chinese population25.
In the present study, we systematically evaluated the associations between 11,966 genetic variants in 206 mRNA splicing-related genes and lung cancer risk using the data from eight published GWAS datasets. To the best of our knowledge, this is the first and largest study focusing on exploring associations between SNPs from mRNA splicing-related genes and risk of lung cancer. Overall, our study identified six SNPs within mRNA splicing-related gene PRPF6 that might play an important role in the development of lung cancer. Furthermore, all of these six SNPs were found to be associated with PRPF6 mRNA expression in both lymphoblastoid cell lines and normal lung tissues of European descendent. Therefore, we proposed that these six SNPs were associated with abnormal expression of PRPF6 and thus modified individual’s susceptibility to lung cancer. However, two major issues of the present study should be addressed. Firstly, the associations of these six SNPs with risk of lung cancer were observed in the six TRICL GWASs but not in Harvard and deCODE GWASs, probably due to a relatively limited sample size of the cases in the replication. Further replication studies are warranted to confirm our results. Secondly, the lung cancer-associated mRNA splicing-related genes identified in the previous Chinese study25 were not replicated in our study, which could be attributed to different genetic background or different environmental exposures as well as other unknown contributing factors between two populations.
PRPF6 is located on chromosome 20q13.33 and is an essential member of the small nuclear ribonucleic proteins (snRNPs), playing an important role in mRNA splicing. For instance, a missense mutation in PRPF6 impairs mRNA splicing and contributes to Autosomal-Dominant Retinitis Pigmentosa30. Its role in the development of cancer had also been reported in previous studies. For example, it was over-expressed in lung adenocarcinomas according to The Cancer Genome Atlas (TCGA) projects31. In addition, PRPF6 was over-expressed in colon cancer32 to drive cancer proliferation by preferential splicing of genes associated with growth regulation33. Abnormal expression of PRPF6 alters the constitutive and alternative splicing of a discrete number of genes, including an oncogenic isoform of the ZAK kinase33 that activates several cancer (including lung cancer)-related signaling pathways, such as those of NF-kB, Wnt/b catenin, AP1, ERK and JNK34,35. It should be mentioned that the protective alleles of these six SNPs were associated with both a decreased lung cancer risk and a decreased mRNA expression of PRPF6, which is consistent with the previous findings of overexpression of the gene in colon and lung cancers. Therefore, based on all these, we hypothesized of abnormal expression of PRPF6 might contribute to development of lung cancer in the following way: the risk allele of PRPF6 was associated with elevated PRPF6 mRNA expression, then the oncogenic isoforms, such as the ZAK kinase, were generated as a result of abnormal splicing and activation in normal lung tissues, which finally resulted in lung carcinogenesis.
In summary, the present large-scale meta-analysis of eight published lung-cancer GWASs consisting of 14,463 lung cancer cases and 44,188 controls revealed a novel lung cancer susceptibility locus in the mRNA splicing-related gene PRPF6 and provided some new insight into genetic architecture and carcinogenic mechanisms of lung cancer. Further validation and functional evaluation of this genetic variant are warranted to verify our findings.
Materials and Methods
Study populations
In the present study, we used the data from eight lung-cancer GWASs with a total of 14,463 lung cancer cases and 44,188 health controls. As shown in Table 1, the analysis included six GWAS datasets from the TRICL consortium (12,160 lung cancer cases and 16,838 controls of European ancestry) and another two datasets of lung cancer GWASs: the Caucasian origin-Harvard Lung Cancer Study (984 cases and 970 controls) and the Icelandic Lung Cancer Study (deCODE from the ILCCO) (1,319 cases and 26,380 controls). As described previously16, the TRICL six lung-cancer GWASs include the MD Anderson Cancer Center (MDACC) GWAS, the Institute of Cancer Research (ICR) GWAS, the National Cancer Institute (NCI) GWAS, the International Agency for Research on Cancer (IARC) GWAS, Lunenfeld-Tanenbaum Research Institute study (Toronto) GWAS and the German Lung Cancer Study (GLC) GWAS.
All of the subjects included in the analysis had provided a written informed consent. All methods were performed in accordance with the relevant guidelines and regulations, and all original studies were approved by the institutional review board from each of the participating institutions.
GWAS genotyping, imputation and quality controls
GWASs included in the current study were performed by the following platforms: Illumina HumanHap 317, 317 + 240 S, 370 Duo, 550, 610 or 1 M arrays. Genotype imputation was conducted based on linkage disequilibrium (LD) information from the 1000 Genomes Project (phase I integrated release 3, March 2012) by IMPUTE2, MaCH or minimac software16. Imputed SNPs with information score <0.40 by IMPUTE2 or r2 < 0.30 by MaCH were excluded from further analysis. In addition, standard quality control on samples by excluding individuals with low call rate (<90%) and extremely high or low heterozygosity (P < 1.0 × 10−4), as well as those estimated to be of non-European ancestry (using the HapMap phase II CEU, JPT/CHB and YRI populations as reference) were conducted among all of the studies.
Genes and SNPs selection
The mRNA splicing-related genes were selected with the combination of two online databases commonly exploited in the gene-set enrichment analysis: Molecular Signatures Database (http://www.broadinstitute.org/gsea/index.jsp) and Genecards (http://www.genecards.org/). Overall, 206 mRNA splicing-related genes were selected (Supplementary Table 1), which included a total of 11,966 genotyped or imputed common SNPs extracted from these genes (include 2-kb upstream and downstream of genes) among the TRICL Consortium with the following inclusion criteria: 1) Genotyping call rate ≥ 90%; 2) Minor allele frequency (MAF) ≥ 5% among Europeans; and 3) Hardy Weinberg Equilibrium exact P value ≥ 10−5. The detailed work-flow can be found in Supplementary Figure 1.
Statistical analysis
The associations between SNPs and lung cancer risk were evaluated using an additive genetic model by R (v2.6), Stata (v10, State College, Texas, US) and PLINK (v1.06) software36. For the studies of ICR, MDACC, IARC, Toronto, NCI and Harvard, top significant principle components were included in the analysis to control for population stratification that might cause inflation of test statistics, while for the deCODE study, genomic control method was used to adjusted for population stratification. No population structure was found in the German Lung Cancer Study (GLC). With the application of the inverse variance method, a meta-analysis under the fixed and random-effects models was performed on the results of a log-additive model for 11,966 SNPs. In order to assess the heterogeneity among studies, the Cochran’s Q statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation due to the heterogeneity were calculated31. A fixed-effects model was applied, if no heterogeneity existed among studies (PQ-test > 0.10 and I2 < 25%); otherwise, a random-effects model was chosen. The Benjamini–Hochberg false discovery rate (FDR) procedure was employed for the correction of multiple testing with a cutoff value ≤ 0.05.
Regional association plots were generated with LocusZoom on the basis of 1000 Genomes European (EUR) reference data (phase I integrated release 3, March 2012)37. Haploview v4.2 was employed to construct the Manhattan and LD plot. All analyses were conducted with SAS (version 9.1.3; SAS Institute, Cary, NC, USA) unless specified otherwise.
In silico SNP function annotation
To prioritize functional SNPs, three in silico tools: dbSNP func annotation (http://www.ncbi.nlm.nih.gov/projects/SNP/), SNPinfo (http://snpinfo.niehs.nih.gov/), and RegulomeDB (http://regulomedb.org/) were employed. Furthermore, the associations between SNPs and PRPF6 mRNA expression were performed using lymphoblastoid cell expression data from 1000 Genomes Project European populations (EUR, 373 individuals) (phase I integrated release 3, March 2012)38 by linear regression model.
Additional Information
How to cite this article: Pan, Y. et al. Associations between genetic variants in mRNA splicing-related genes and risk of lung cancer: a pathway-based analysis from published GWASs. Sci. Rep. 7, 44634; doi: 10.1038/srep44634 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors thank the investigators and participants of all TRICL-ILCCO studies (i.e., ICR, MDACC, IARC, NCI, Toronto, GLC, Harvard and deCODE) for their important contributions. As Duke Cancer Institute members, QW and KO acknowledge support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236). QW was also supported by a start-up fund from Duke Cancer Institute, Duke University Medical Center. Yongchu Pan was sponsored by Collaborative Innovation Center for Cancer Personalized Medicine, Nanjing Medical University. TRICL: This work was supported by the Transdisciplinary Research in Cancer of the Lung (TRICL) Study, U19-CA148127 on behalf of the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network. The Toronto study was supported by Canadian Cancer Society Research Institute (020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award to RH The ICR study was supported by Cancer Research UK (C1298/A8780 andC1298/A8362—Bobby Moore Fund for Cancer Research UK) and NCRN, HEAL and Sanofi-Aventis. Additional funding was obtained from NIH grants (5R01CA055769, 5R01CA127219, 5R01CA133996, and 5R01CA121197). The Liverpool Lung Project (LLP) was supported by The Roy Castle Lung Cancer Foundation, UK. The ICR and LLP studies made use of genotyping data from the Wellcome Trust Case Control Consortium 2 (WTCCC2); a full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Sample collection for the Heidelberg lung cancer study was in part supported by a grant (70–2919) from the Deutsche Krebshilfe. The work was additionally supported by a Helmholtz-DAAD fellowship (A/07/97379 to MNT) and by the NIH (U19CA148127). The KORA Surveys were financed by the GSF, which is funded by the German Federal Ministry of Education, Science, Research and Technology and the State of Bavaria. The Lung Cancer in the Young study (LUCY) was funded in part by the National Genome Research Network (NGFN), the DFG (BI576/2-1; BI 576/2-2), the Helmholtzgemeinschaft (HGF) and the Federal office for Radiation Protection (BfS: STSch4454). Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum Muenchen. Support for the Central Europe, HUNT2/Tromsø and CARET genome-wide studies was provided by Institute National du Cancer, France. Support for the HUNT2/Tromsø genome-wide study was also provided by the European Community (Integrated Project DNA repair, LSHG-CT- 2005-512113), the Norwegian Cancer Association and the Functional Genomics Programme of Research Council of Norway. Support for the Central Europe study, Czech Republic, was also provided by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101). Support for the CARET genome-wide study was also provided by grants from the US National Cancer Institute, NIH (R01 CA111703 and UO1 CA63673), and by funds from the Fred Hutchinson Cancer Research Center. Additional funding for study coordination, genotyping of replication studies and statistical analysis was provided by the US National Cancer Institute (R01 CA092039). The lung cancer GWAS from Estonia was partly supported by a FP7 grant (REGPOT245536), by the Estonian Government (SF0180142s08), by EU RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure’s Roadmap and by University of Tartu (SP1GVARENG). The work reported in this paper was partly undertaken during the tenure of a Postdoctoral Fellowship from the IARC (for MNT). The Environment and Genetics in Lung Cancer Etiology (EAGLE), the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), and the Prostate, Lung, Colon, Ovary Screening Trial (PLCO)studies and the genotyping of ATBC, the Cancer Prevention Study II Nutrition Cohort (CPS-II) and part of PLCO were supported by the Intramural Research Program of NIH, NCI, Division of Cancer Epidemiology and Genetics. ATBC was also supported by US Public Health Service contracts (N01-CN-45165, N01-RC-45035 and N01-RC-37004) from the NCI. PLCO was also supported by individual contracts from the NCI to the University of Colorado Denver (NO1-CN-25514), Georgetown University(NO1-CN-25522), Pacific Health Research Institute (NO1-CN-25515), Henry Ford Health System (NO1-CN-25512), University of Minnesota(NO1-CN-25513), Washington University(NO1-CN-25516), University of Pittsburgh (NO1-CN-25511), University of Utah (NO1-CN-25524), Marshfield Clinic Research Foundation (NO1-CN-25518), University of Alabama at Birmingham (NO1-CN-75022, Westat, Inc. NO1-CN-25476), University of California, Los Angeles (NO1-CN-25404). The Cancer Prevention Study II Nutrition Cohort was supported by the American Cancer Society. The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 andRO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C) and study coordination at the GENEVA Coordination Center (U01 HG004446) for EAGLE and part of PLCO studies. Funding for the MD Anderson Cancer Study was provided by NIH grants (P50 CA70907, R01CA121197, R01CA127219, U19 CA148127, R01 CA55769, and K07CA160753) and CPRIT grant (RP100443). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is funded through a federal contract from the NIH to The Johns Hopkins University (HHSN268200782096C). The Harvard Lung Cancer Study was supported by the NIH (National Cancer Institute) grants CA092824, CA090578, and CA074386. deCODE: The project was funded in part by GENADDICT: LSHMCT-2004-005166), the National Institutes of Health (R01-DA017932).
Footnotes
The authors declare no competing financial interests.
Author Contributions Yongchu Pan, Hongliang Liu and Qingyi Wei conceived and designed the experiments. Yanru Wang, Xiaozheng Kang, Zhensheng Liu and Kouros Owzar helped to analyze the data. Younghun Han, Li Su, Yongyue Wei, Rayjean J. Hung, Yonathan Brhane, John McLaughlin, Paul Brennan, Heike Bickeböller, Albert Rosenberger, Richard S. Houlston, Neil Caporaso, Maria Teresa Landi, Joachim Heinrich, Angela Risch, Xifeng Wu, Yuanqing Ye, David C. Christiani and Christopher I. Amos collected the samples and provided data. All authors participated in the preparation of the manuscript and approval of submittion for publication.
References
- Yang I. A., Holloway J. W. & Fong K. M. Genetic susceptibility to lung cancer and co-morbidities. Journal of thoracic disease 5 Suppl 5, S454–462, doi: 10.3978/j.issn.2072-1439.2013.08.06 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberg A. J., Brock M. V., Ford J. G., Samet J. M. & Spivack S. D. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 143, e1S–29S, doi: 10.1378/chest.12-2345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson S. et al. Familial risk of lung carcinoma in the Icelandic population. Jama 292, 2977–2983, doi: 10.1001/jama.292.24.2977 (2004). [DOI] [PubMed] [Google Scholar]
- Xu H., Spitz M. R., Amos C. I. & Shete S. Complex segregation analysis reveals a multigene model for lung cancer. Human genetics 116, 121–127, doi: 10.1007/s00439-004-1212-9 (2005). [DOI] [PubMed] [Google Scholar]
- Smith C. Genomics: SNPs and human disease. Nature 435, 993, doi: 10.1038/435993a (2005). [DOI] [PubMed] [Google Scholar]
- Amos C. I. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature genetics 40, 616–622, doi: 10.1038/ng.109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J. et al. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nature genetics 44, 895–899, doi: 10.1038/ng.2351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nature genetics 43, 792–796, doi: 10.1038/ng.875 (2011). [DOI] [PubMed] [Google Scholar]
- Hung R. J. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 452, 633–637, doi: 10.1038/nature06885 (2008). [DOI] [PubMed] [Google Scholar]
- Lan Q. et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nature genetics 44, 1330–1335, doi: 10.1038/ng.2456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J. D. et al. Lung cancer susceptibility locus at 5p15.33. Nature genetics 40, 1404–1406, doi: 10.1038/ng.254 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D. et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nature genetics 42, 893–896, doi: 10.1038/ng.667 (2010). [DOI] [PubMed] [Google Scholar]
- Shiraishi K. et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nature genetics 44, 900–903, doi: 10.1038/ng.2353 (2012). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nature genetics 40, 1407–1409, doi: 10.1038/ng.273 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J. et al. Genome-wide association study identifies a novel susceptibility locus at 12q23.1 for lung squamous cell carcinoma in han chinese. PLoS genetics 9, e1003190, doi: 10.1371/journal.pgen.1003190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nature genetics 46, 736–741, doi: 10.1038/ng.3002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. et al. A genome-wide gene-environment interaction analysis for tobacco smoke and lung cancer susceptibility. Carcinogenesis 35, 1528–1535, doi: 10.1093/carcin/bgu076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A. L. & Christiani D. C. Genetic susceptibility to lung cancer–light at the end of the tunnel? Carcinogenesis 34, 487–502, doi: 10.1093/carcin/bgt016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Gene-based meta-analysis of GWAS data identifies independent SNPs in ANXA6 as associated with SLE in Asian populations. Arthritis & rheumatology, doi: 10.1002/art.39275 (2015). [DOI] [PubMed] [Google Scholar]
- Ramanan V. K., Shen L., Moore J. H. & Saykin A. J. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends in genetics: TIG 28, 323–332, doi: 10.1016/j.tig.2012.03.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. & Hakonarson H. Analysing biological pathways in genome-wide association studies. Nature reviews. Genetics 11, 843–854, doi: 10.1038/nrg2884 (2010). [DOI] [PubMed] [Google Scholar]
- Zhong H., Yang X., Kaplan L. M., Molony C. & Schadt E. E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. American journal of human genetics 86, 581–591, doi: 10.1016/j.ajhg.2010.02.020 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. & Manley J. L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer discovery 3, 1228–1237, doi: 10.1158/2159-8290.CD-13-0253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. & Weiss W. A. Alternative splicing in cancer: implications for biology and therapy. Oncogene 34, 1–14, doi: 10.1038/onc.2013.570 (2015). [DOI] [PubMed] [Google Scholar]
- Shen W. et al. Polymorphisms in alternative splicing associated genes are associated with lung cancer risk in a Chinese population. Lung cancer 89, 238–242, doi: 10.1016/j.lungcan.2015.06.010 (2015). [DOI] [PubMed] [Google Scholar]
- Je E. M., Yoo N. J., Kim Y. J., Kim M. S. & Lee S. H. Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. International journal of cancer. Journal international du cancer 133, 260–265, doi: 10.1002/ijc.28011 (2013). [DOI] [PubMed] [Google Scholar]
- Harbour J. W. et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nature genetics 45, 133–135, doi: 10.1038/ng.2523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H. et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 119, 3203–3210, doi: 10.1182/blood-2011-12-399774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120, doi: 10.1016/j.cell.2012.08.029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G. et al. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. American journal of human genetics 88, 643–649, doi: 10.1016/j.ajhg.2011.04.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. Bmj 327, 557–560, doi: 10.1136/bmj.327.7414.557 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L. W. et al. Integrated analysis of genome-wide copy number alterations and gene expression in microsatellite stable, CpG island methylator phenotype-negative colon cancer. Genes, chromosomes & cancer 52, 450–466, doi: 10.1002/gcc.22043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler A. S. et al. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes & development 28, 1068–1084, doi: 10.1101/gad.237206.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R. et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 455, 547–551, doi: 10.1038/nature07179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. J. et al. ZAK inhibits human lung cancer cell growth via ERK and JNK activation in an AP-1-dependent manner. Cancer science 101, 1374–1381, doi: 10.1111/j.1349-7006.2010.01537.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 81, 559–575, doi: 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337, doi: 10.1093/bioinformatics/btq419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65, doi: 10.1038/nature11632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.