Abstract
Notch is a key signalling pathway playing multiple and varied functions during development. Notch regulates the selection of cells with a neurogenic fate and maintains a pool of yet uncommitted precursors through lateral inhibition, both in insects and in vertebrates. Here, we explore the functions of Notch in the annelid Platynereis dumerilii (Lophotrochozoa). Conserved components of the pathway are identified and a scenario for their evolution in metazoans is proposed. Unexpectedly, neither Notch nor its ligands are expressed in the neurogenic epithelia of the larva at the time when massive neurogenesis begins. Using chemical inhibitors and neural markers, we demonstrate that Notch plays no major role in the general neurogenesis of larvae. Instead, we find Notch components expressed in nascent chaetal sacs, the organs that produce the annelid bristles. Impairing Notch signalling induces defects in chaetal sac formation, abnormalities in chaetae producing cells and a change of identity of chaeta growth accessory cells. This is the first bilaterian species in which the early neurogenesis processes appear to occur without a major involvement of the Notch pathway. Instead, Notch is co-opted to pattern annelid-specific organs, likely through a lateral inhibition process. These features reinforce the view that Notch signalling has been recruited multiple times in evolution due to its remarkable ‘toolkit’ nature.
Keywords: Notch, Platynereis, neurogenesis, chaetogenesis, lateral inhibition
1. Background
Since developmental biology met molecular and cellular biology, considerable efforts have been deployed in understanding signalling pathways function, modularity, architecture and later evolution. Among them, the Notch pathway has been especially investigated in these frameworks. As a direct juxtacrine signalling system, it provides a mechanism for short-range, localized signalling between directly apposing cells in a process called ‘trans-activation’ [1–5]. Upon binding of a ligand (Delta or Jagged) displayed by a neighbouring cell, the Notch transmembrane receptor is cleaved by the γ-secretase complex (presenilin-nicastrin-APH1-PEN2) resulting in the release of the Notch intracellular domain (NICD) into the cytoplasm (additional cleavage of Delta by the γ-secretase complex has been also described in vertebrates [6]). After its translocation to the nucleus of the receiving cell, NICD interacts with the CSL (CBF1, suppressor of hairless (Su(H)), Lag-1)/Ncor/SMRT/histone deacetylase transcriptional complex and activates the transcription of target genes. The same ligands can also interact with the receptor Notch within the same cell (in cis). The Notch pathway functions with a number of ‘core component’ proteins, whose evolutionary emergence has been studied in detail [7]: it was probably already functional as early as in the last common ancestor of all metazoans, but has its roots anchored deep in the eukaryote tree as some non-metazoan unikonts display members of these components.
Expression and functional studies of Notch pathway components during development have been performed in few metazoan species, mostly in deuterostomes and ecdysozoans. In these two lineages, the remarkably pleiotropic Notch pathway affects cell fate choices associated with lineage segregation, cell differentiation, cell proliferation or apoptosis in many tissues and developmental stages [8–11]. The extreme versatility and pleiotropy of Notch signalling seems at first glance to make the task of reconstituting an evolutionary history of its involvement in metazoan development rather daunting. In many ways, the Notch pathway could be described as a developmental ‘Swiss army knife’, a generic tool that has been co-opted multiple times in evolution, in various animal lineages, for unrelated or sometimes convergent functions in development. However, comparisons of the roles of Notch between insect and vertebrate have brought to light interesting similarities that have led to speculations about an ancestral involvement of Notch signalling in neurogenesis [12].
Notch is crucial in the early patterning of neurogenesis both in the fruit fly and in vertebrates. The study of Drosophila neurogenesis in particular has led to uncovering a mechanism called ‘lateral inhibition’. In the fly embryo, the central nervous system originates from delaminating neural progenitors [13,14], called neuroblasts. These divide asymmetrically to give birth to further dividing neuron precursors (the ganglion mother cells). Neuroblasts are active during both embryogenesis and larval development, playing the role of neural stem cells. Delaminating neuroblasts are initially specified in clusters of ectodermal epithelial cells, called ‘proneural clusters’. These cells are initially bipotent, giving either a neuroblast or, for the majority of them, epidermal cells. The cell that will eventually become a neuroblast starts to express higher levels of the ligand transmembrane protein Delta, which binds to the Notch transmembrane protein present at the surface of neighbouring cells. In these neighbouring cells, the activation of the Notch pathway results in the downregulation of both Delta and neurogenic gene expressions, thus inhibiting the neural fate. In vertebrate embryos, early neurogenesis processes take place mostly in the neural tube after it has completed its closure. In the thick pseudostratified epithelium of the neural tube, a gradient of Notch1 expression, maximal apically, keeps neurogenic specification on the apical/ventricular side [15], while differentiating neuron bodies migrate to the basal side. On the apical side, where both symmetrical and asymmetrical cell divisions take place, lateral inhibition by Notch/Delta regulates the pace of neuronal specification, maintaining a pool of neural progenitors [16,17].
This mechanism of lateral inhibition by Notch/Delta has been evidenced in a number of contexts, such as hair cell formation in the inner ear [18]. Interestingly, the inner ear development is also mediated by another mechanism of the Notch pathway called lateral induction, through the alternate ligand Jagged1. Such mechanisms, finely modulated by cis-interactions, exemplify the notion of the Notch pathway as a general and versatile developmental tool that could possibly have been co-opted for playing similar functions. However, there is now considerable evidence that both neurogenesis [19] and nervous system centralization [20] in protostomes and vertebrates share a common origin. In this context, Notch is often seen as a pathway ancestrally involved in neurogenesis patterning in bilaterians. This hypothesis receives support also from cnidarians that have a dispersed nervous system organization [21]. In the sea anemone Nematostella, the Notch pathway appears to play a role in regulating the abundance of differentiating nerve cells, even if the exact roles of specific pathway components remain disputed [22–24]. This pushes the involvement of Notch in nervous system formation back to ancestors of eumetazoans (cnidarians + bilaterians), at the very evolutionary origin of the nervous system.
While plenty of studies focused on the various aspects of Notch pathway mechanisms and regulations among deuterostomes and ecdysozoans, the third large group of bilaterians, i.e. the lophotrochozoans, has been barely studied. The Notch pathway architecture and functions have been investigated so far in two annelid species only. In the leech Helobdella robusta, the Notch pathway has been proposed to be involved in posterior elongation and segment formation [25,26]. In Capitella teleta, the expression patterns of the Notch pathway components (discussed later in the article) in brain, foregut, chaetal sacs and posterior growth zone suggest a possible involvement of this pathway in brain development, chaetogenesis and segmentation processes [27].
In this study, we analysed the functions of the Notch pathway in the marine annelid Platynereis dumerilii, a leading lophotrochozoan model species for evolution and developmental biology. Platynereis dumerilii is considered to be a ‘short branch’ organism that has kept a number of ancestral-looking characters at the genomic [28], anatomical and developmental levels [20,29,30], making it a highly attractive model system for understanding the origin and diversification of these traits. P. dumerilii displays an indirect form of development (for a review, see [31]): embryogenesis first gives rise to a minute spherical trochophore larva with a small number of differentiated larval structures, including a precise arrangement of a few neuronal and sensory cells, such as the apical organ or the posterior pioneer neurons [20,32,33]. Later than this larval nervous system, a wave of massive ‘adult’ neurogenesis takes place in the mid-trochophore stage, at two different locations: the episphere at the animal pole, which will give rise to the anterior brain of the worm, and the ventral neural ectoderm, which will give rise to the ganglia of the ventral nerve cord (VNC) [20]. Ventral neurogenesis takes place in thickening stratified or pseudo-stratified epithelia where mitoses happen on the apical side and neuron differentiations occur on the basal side, similar to what has been described for the vertebrate neuroepithelium [20,34]. In addition, Platynereis neurogenesis involves a number of the same transcription factors that have already been identified in insect and vertebrate models such as the bHLH NeuroD, neurogenin and achaete-scute-related genes [33].
The aim of this study is to explore the roles of the Notch pathway in Platynereis and to shed light on the ancestral role(s) of Notch in lophotrochozoans and bilaterians (we do not describe here a potential role in posterior elongation and segment formation that require further study in the juvenile worm). Taking advantage of small molecule inhibitors, we show here that the Notch signalling pathway does not seem to play a major generic role in the early processes of larval or ‘adult’ neurogenesis in the annelid, even though roles in the formation of a few specific head and trunk neurons cannot be excluded. Instead, at the time when neurogenesis happens, we find a role of Notch in the correct patterning of the cells of chaetal sacs, likely through a lateral inhibition mechanism. Chaetal sacs are the larval and adult organs that are responsible for the formation of chaetae, i.e. locomotory and mechanosensory bristles of the worm. Our results thus challenge the view that the Notch pathway is necessarily involved in nervous system patterning in the same way at a eumetazoan scale. Our data rather support the view of co-options in regulating cell fate specification in multiple contexts.
2. Results
2.1. Notch pathway core components identification: a lophotrochozoan focus
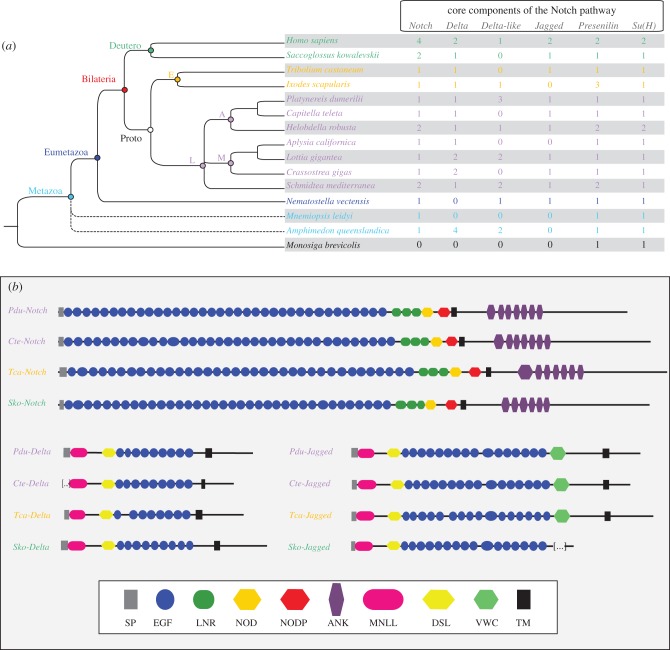
Exhaustive searches on the genome and several transcriptomes of P. dumerilii led us to identify core components of the Notch pathway i.e. the receptor Notch (Pdu-Notch, already identified [28]), the two ‘classical’ ligands Pdu-Delta and Pdu-Jagged, Pdu-Presenilin (component of the γ-secretase complex) and the transcription factor suppressor of hairless (Pdu-SuH; figure 1a; electronic supplementary material, figure S1A). In addition, we found three Delta-like genes (i.e. lacking the minimal domain arrangement defined below). We also identified the intracellular regulator Nrarp (Pdu-Nrarp), a gene encoding a small ankyrin (ANK)-repeat protein that is part of a negative feedback loop that attenuates Notch activity in vertebrates [35], the antagonist Numb (Pdu-Numb) and the post-translational modifier Fringe (Pdu-Fringe) (electronic supplementary material, figure S1A). Platynereis Delta, Jagged and Notch possess conserved domain arrangements that are very similar to other bilaterian species and likely to be ancestral in the bilaterian lineage (figure 1b). Interestingly, we identified two splice variants for Pdu-Delta (Pdu-Deltatv1 and Pdu-Deltatv2; electronic supplementary material, figure S1B). In Pdu-Deltatv2, retention of the last intron led to a shorter sequence lacking the final short ATEV peptide. Despite extensive and specific searches, no other typical Notch, Jagged or Delta proteins have been evidenced.
Figure 1.
Core components of the Notch pathway in metazoans. (a) The presence/absence and number of gene copies at a metazoan scale for the Notch pathway main components. Fifteen species representatives of the metazoans, with a lophotrochozoan focus, are included. Dashed lines show the unresolved phylogenetic positions for sponges and ctenophores. (b) Domain arrangement of Notch, Delta and Jagged proteins in four bilaterian species are schematized. See figure inset for the domain legends. Pdu-Notch presents 36 EGF repeats, three LNR, one NOD domain, one NODP domain and seven ANK repeats. For Pdu-Delta and Pdu-Jagged, we detected the MNLL region, a Delta/Serrate/Lag (DSL) domain (which mediates binding to Notch receptors in bilaterians) and a series of EGF repeats (nine for Pdu-Delta and 16 for Pdu-Jagged). In addition to these domains, Pdu-Jagged also contains a Von Willebrand factor C domain (VWC) characteristic of Serrate/Jagged proteins. The choanoflagellate is in black, the sponge and ctenophore are in light blue, the cnidarian is in blue, lophotrochozoans are in purple, ecdysozoans are in orange and deuterostomians are in green. Deutero, deuterostomes; Proto, protostomes; E, ecdysozoans; L, lophotrochozoans; A, annelids; M, molluscs.
The origin and evolution of the components and auxiliary factors of the Notch pathway have previously been investigated at a large scale [7]. We performed here a detailed search of Notch pathway core components in metazoan species representative of all main metazoan lineages (Deuterostomia, Ecdysozoa, Lophotrochozoa, Ctenophora, Cnidaria and Porifera), but surveyed more extensively lophotrochozoan species as they have been undersampled in earlier studies due to a lack of available genomes. Data comparisons among lophotrochozoan, ecdysozoan and deuterostomian species are of special interest in order to reconstruct bilaterian ancestral states (figure 1a). We identified a single unambiguous Notch orthologue in all metazoans species investigated, except in vertebrates, in the enteropneust Saccoglossus kowalevskii, in the annelid H. robusta and in the planarian Schmidtea mediterranea. A more complicated situation emerged for the evolution of Delta and Jagged proteins. In all species but the ctenophore, several Delta-related proteins with various domain compositions are identified. One particular domain composition (MNLL-DSL-(9)xEGF-TM-ATEV), in which not only epidermal growth factor (EGF) motif numbers but also their specific spacings are conserved, presumably corresponds to an ancestral Delta protein that was already present in the bilaterian ancestor [36]. Our study supports this interpretation; indeed genes corresponding to this proposed ancestral organization are present in several bilaterian species we sampled. In addition, the EGF domains generally follow the pattern of repeat spacings and cysteine residue spacings noticed previously [36] (electronic supplementary material, figure S1C). As noted above, a conserved alternative splicing upstream of the ATEV peptide is found in both Platynereis and vertebrate proteins, which suggests that this particular splicing is as ancient as the bilaterian ancestor. In addition to these ancestral Delta proteins, a number of bilaterian genomes code for ‘Delta-like’ proteins (three in Platynereis, two in H. robusta, S. mediterranea and L. gigantea, one in I. scapularis, and the human Dll3), displaying domain variations, the most common being the loss of the ATEV motif and of a number of EGF repeats. There is no indication that orthology relationships can be traced back to the bilaterian ancestors for any of these Delta-like proteins. Jagged proteins are absent outside eumetazoans (alternative scenarios for their emergence have been already discussed [7]) and have been lost or diverged beyond recognition in some bilaterians, such I. scapularis and Aplysia californica (figure 1a).
Phylogenetic analyses of metazoan Delta and Jagged ligands (electronic supplementary material, figure S1A) are based on a very limited length of sequences: only the domains corresponding to the MNLL and Delta/Serrate/Lag (DSL). Yet, the limited resolution these trees show is compatible with the interpretations given above. Briefly, phylogenetic analyses show that Pdu-Delta and Pdu-Jagged are included in robust Delta and Jagged clades, respectively. Sponge Deltas are clustered and form the sister group of the eumetazoan Delta + Jagged groups (approximate likelihood-ratio test (aLRT) > 0.90). Inside the Delta and Jagged clades, relationships are not fully resolved but Platynereis and Capitella sequences always group together. We thus tried unsuccessfully to infer the evolutionary origin of the three Delta-like genes found in Platynereis (electronic supplementary material, figure S1A). The phylogenetic relationships between Platynereis Notch receptor and other metazoan receptors are provided in the electronic supplementary material, figure S1A. As previously noticed [7], the relationships among Notch receptors are not perfectly well resolved and do not follow the metazoan species tree. However the orthology of the Platynereis Notch is not questionable (electronic supplementary material, figure S1A).
To sum up, core components of the Notch pathway in the lophotrochozoan Platynereis appear to be very similar to the ancestral situation of bilaterians. Therefore, investigating its functions in Platynereis is of special relevance to draw evolutionary conclusions.
2.2. The components of the Platynereis Notch pathway are not generally expressed in neurogenic tissues but they are expressed in forming chaetal sacs
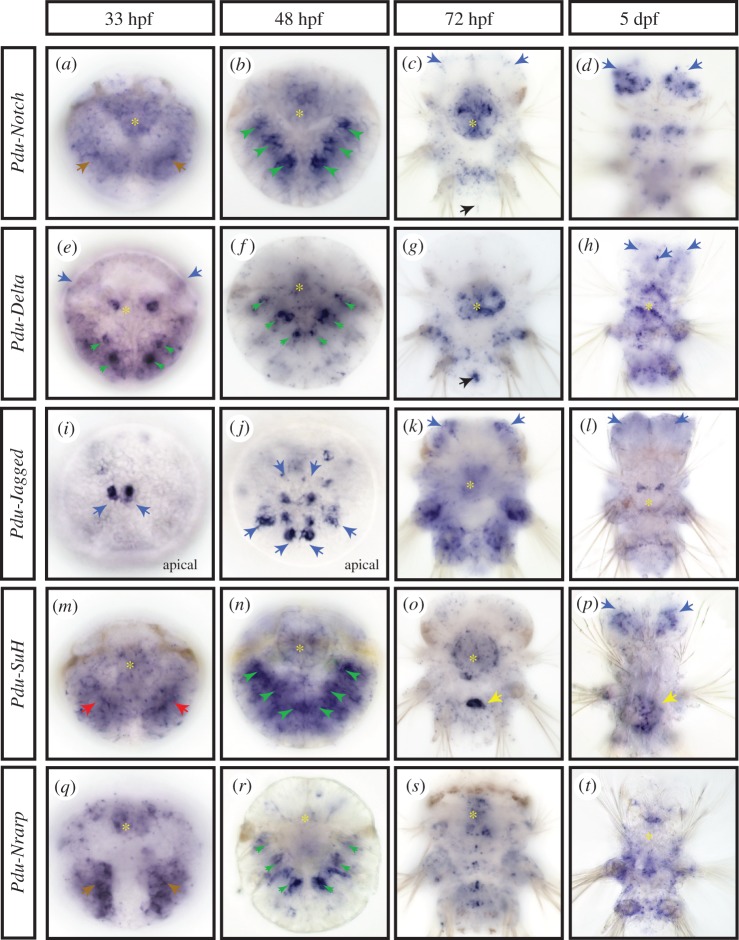
We analysed by whole-mount in situ hybridization (WMISH) the expression patterns of five genes, Pdu-Notch, Pdu-Delta, Pdu-Jagged, Pdu-SuH and Pdu-Nrarp (figure 2). We targeted developmental stages when mass adult neurogenesis is taking place: at 33 hpf (hours post-fertilization; mid trochophore), the anterior episphere (mostly the future brain of the worm) and the ventral ectoderm (the future nerve cord) start to thicken and numerous mitoses occur superficially; at 48 hpf (late trochophore), neuronal differentiation has started and cell proliferation becomes more localized along the midline; at 72 hpf (three segment nectochaete larva), many brain and nerve cord neurons have differentiated but cell proliferation and thus neurogenesis continue locally and superficially; at 5 dpf (days post-fertilization; late nectochaete larva), cell proliferation and differentiation continue in the brain [20,34,37,38]. The Notch core component expression patterns can be categorized in three main structures of the larva: the chaetal sacs, some brain cells including the apical organ and the stomodeum.
Figure 2.
Expression patterns of the main components of the Notch pathway during Platynereis larval development. WMISH for Pdu-Notch (a–d), Pdu-Delta (e–h), Pdu-Jagged (i–l), Pdu-Su(H) (m–p) and Pdu-Nrarp (q–t), are shown at four larval stages (33, 48, 72 hpf and 5 dpf). All panels are ventral views (anterior is up) except (i and j) that are apical (dorsal is up). Yellow asterisks mark expression in the stomodeum/forming pharynx. Brown arrows indicate an expression in the ectoderm, green arrowheads in the chaetal sacs, blue arrows in brain cells, red arrows in the mesoderm, black arrows in the putative mesoteloblasts and yellow arrows in a cluster of midgut cells.
All genes but Pdu-Jagged are expressed in specific structures called chaetal sacs. The chaetal sacs are responsible for producing chaetae, the retractile chitinous bristles displayed by the annelid leg-like appendages (parapodia) that help in locomotion. Chaetal sacs appear by invagination of ectodermal pockets [39]. In the late trochophore stage (48 hpf), when the chaetal sacs are fully formed and producing the first chaetae, Pdu-Notch, Pdu-Delta and Pdu-Nrarp are found in two bilateral ensembles of six patches of expression (figure 2b,f,n,r; green arrowheads). These lateral patches coincide with the 12 chaetal sacs of the larvae: two per future hemi-segment, six in the future neuropodia and six in the future notopodia (the ventral and dorsal moieties of the parapodia). All three gene expressions appear a little stronger in the ventral sacs (figure 2b,f,n,r; green arrowheads). Pdu-Delta expression is more restricted compared with Pdu-Notch and Pdu-Nrarp (figure 2f, green arrowheads) in a few deep cells of the chaetal sac and is observable early (at 33 hpf) in four to six bilateral and internalized patches of cells (figure 2e, green arrowheads). Pdu-Su(H) expression while being widespread is more intense in the chaetal sac areas at 48 hpf (figure 2n, green arrowheads).
Most of the genes studied (all but Pdu-Nrarp) are also expressed in different populations of brain cells and/or apical organ cells (a sensory structure composed of neurons and ciliary tuft [40] and present in many marine invertebrate larvae [32]), at several stages. Indeed, at 33 hpf, Pdu-Delta is expressed in few lateral brains cells (figure 2e, blue arrows), while Pdu-Jagged is found in two small groups of different brain cells (figure 2i, blue arrows, apical view). At 48 hpf, the Pdu-Jagged expression pattern expands in the brain, and forms several rows of cells inside and surrounding the apical organ (figure 2j, blue arrows, apical view; electronic supplementary material, figure S2A, internal dotted circle). Co-localization experiments reveal that some of those Pdu-Jagged + cells in the apical organ are serotoninergic, FMRFAmidergic and RYa peptidergic neurons (electronic supplementary material, figure S2A (v to x), internal dotted circle, purple arrow). Other brain cells expressing Pdu-Jagged are cholinergic neurons (electronic supplementary material, figure S2A (y), orange arrows). Later, at 72 hpf, the Notch receptor and the two ligands are expressed in several populations of brain cells (figure 2c,k, blue arrows; electronic supplementary material, figure S2A (b to d)). Co-localization with the c-amidated dipeptide RYa neuropeptide antibody [41] allows us to identify a small number of peptidergic neurons among the Pdu-Notch+ brain cells (electronic supplementary material, figure S2A (g to h), orange arrow). Similarly, Pdu-Delta + cells are most probably but not exclusively FMRFAmidergic, FLAmidergic and cholinergic neurons (electronic supplementary material, figure S2A (i to o), internal dotted circle, purple arrows). Pdu-Delta and Pdu-Notch are also co-expressed in two bilateral patches of brain cells of unknown identity (electronic supplementary material, figure S2A (e), orange arrows). At this stage, Pdu-Jagged expression is wider in the brain, where it is broadly co-expressed with Pdu-Notch but not Pdu-Delta (figure 2k, blue arrows; electronic supplementary material, figure S2A (d and f)). In addition, Pdu-Delta is found in pyramidal cells of the apical organ also expressing Pdu-Notch (electronic supplementary material, figure S2A (e), internal dotted circle, purple arrow). At 5 dpf, a long time after the main wave of neuronal differentiation occurred, widespread expression of Pdu-Notch appears in the brain (figure 2d, blue arrows). At this stage, Pdu-Delta, Pdu-Jagged and Pdu-Su(H) have also an expression in a few brain cells (figure 2h,l,p blue arrows).
Remarkably, all genes are expressed in the stomodeum and/or pharynx at several stages, either broadly (Pdu-Notch) or in few specific cells (Pdu-Delta; figure 2, yellow asterisks). Finally, at 72 hpf, Pdu-Notch and Pdu-Delta expressions are found in posterior internal cells that correspond to the mesoteloblasts (figure 2c,g black arrows), as evidenced by the coexpression with Vasa and Smb, two stem cell markers [42] (electronic supplementary material, figure S2B (b to j, yellow arrows)).
We thus find no general expression of Notch core components in tissues that are undergoing the main wave of neurogenesis between 33 and 55 hpf. Strikingly, we find no evidence of expression patterns resembling the grid-like expression of Delta genes in the ventral neural ectoderm of arthropods [43–46] or of the ‘salt and pepper’ expression in the neural tube of vertebrates [47]. Instead, Delta expression is limited at these stages and later ones to a few putative neuronal precursor cells, indicating a restricted role in the specification of a limited number of specific neurons. This surprisingly suggests that the Notch pathway may not be generally involved in Platynereis neurogenesis. Conversely, the prominent expression of Notch core components in chaetal sacs suggests a function in Platynereis chaetal sac patterning.
2.3. Notch pathway chemical disruption in Platynereis: initial characterization of effects
Although the core components of the Notch pathway apparently do not show general expression patterns associated with Platynereis larval neurogenesis, we cannot exclude that their expression levels are below the detection level of our WMISH experiments. To test the role of Notch in central nervous system (CNS) formation and chaetogenesis, therefore, we used three pharmacological agents: DAPT, LY-411575 and RO-4929097, all of which are inhibitors of the γ-secretase complex [48]. These drugs act by preventing the third cleavage of the Notch receptor, thereby blocking the nuclear relocalization and the activity of NICD as a transcriptional activator in the receiving cell. A large number of larvae (150 at least) were treated in each experiment. As adult neurogenesis and chaetogenesis overlap broadly in time, we applied drugs during a time window encompassing both processes: 24–48 hpf (Time window 1, table 1). During this time period, massive neural ectoderm proliferation occurs, the neural epithelium thickens considerably and neuronal differentiation starts, with the first elements of the adult neurite scaffold being put in place [34]. The chaetal sacs also start to form from 32 hpf onwards and the production of chaetae starts around 40 hpf [39].
Table 1.
Types of treatment performed to study specifically chaetogenesis and neurogenesis processes. Four time windows were chosen. Time window 1 encompasses both processes and gives a general view of the potential functions of the Notch pathway in the annelid Platynereis. Time window 2 allows targeting the cell fates of chaetal sacs. Time window 3 encompasses the larval neuron formation. Time window 4 encompasses ‘adult’ neurogenesis. For each time window and drug concentration, the phenotype and results observed at several stages are mentioned. VNC, ventral nerve cord; conc., concentration.
| drug conc. | Time window 1: 24–48 hpf |
Time window 2: 20–48 hpf |
Time window 3: 12/16–24 hpf |
Time window 4: 30–48 hpf |
||||
|---|---|---|---|---|---|---|---|---|
| stages | phenotypes | stages | phenotypes | stages | phenotypes | stages | phenotypes | |
| LY-411575 1 µM |
48 hpf | follicle cells markers expression decreased; neurogenesis markers expression maintained | 48 hpf | overexpression of chaetoblast marker; presence of abortive chaetoblasts | 24 hpf | Elav expression in maintained in 9 neurons | 72 hpf | normal number of brain cells/ number of VNC neurons slightly enhanced |
| 72 hpf | absence of chaetae/normal chaetoblasts; normal VNC |
72 hpf | absence of chaetae; development slightly delayed |
72 hpf | absence of chaetae | |||
| 6 dpf | absence of chaetae/normal VNC | |||||||
| RO-4929097 2 µM |
48 hpf | follicle cells markers expression decreased; neurogenesis markers expression maintained | 48 hpf | absence of chaetae/very few abortive chaetoblasts | ||||
| 72 hpf | absence of chaetae/normal chaetoblasts; normal VNC |
72 hpf | absence of chaetae | |||||
| 6 dpf | absence of chaetae/normal VNC | |||||||
| RO-4929097 30 µM |
48 hpf | absence of chaetae/development delayed | 48 hpf | abnormal expression of chaetoblast marker; presence of abortive supernumerary chaetoblasts | 24 hpf | Elav expression is maintained in 9 neurons | 72 hpf | normal number of brain cells; development delayed |
| 72 hpf | absence of chaetae/development delayed | 72 hpf | absence of chaetae; development delayed |
72 hpf | absence of chaetae; development delayed | |||
| later | dead | |||||||
| DAPT 40 µM |
48 hpf | follicle cells markers expression decreased; neurogenesis markers expression maintained | ||||||
| 72 hpf | absence of chaetae/normal chaetoblasts; normal VNC | |||||||
| 6 dpf | absence of chaetae/normal VNC | |||||||
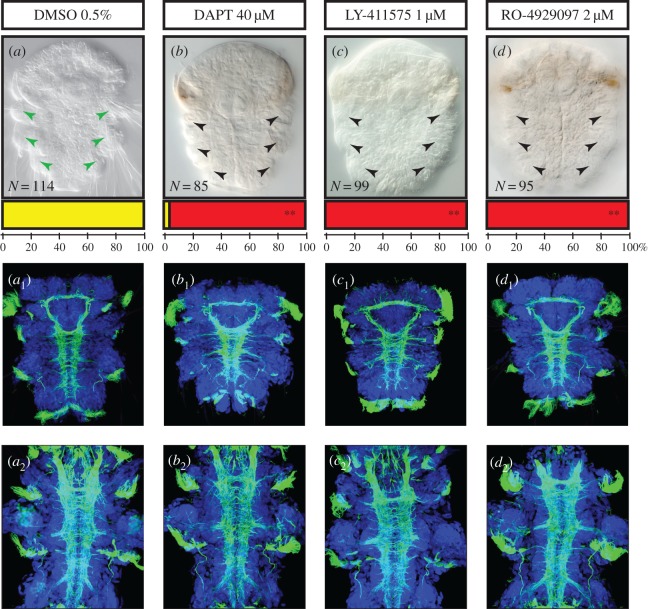
We first tested different concentrations of drugs to determine whether they cause characteristic defects. We considered the lowest concentrations (40 µM DAPT, 2 µM RO-4929097 and 1 µM LY-411575) causing these defects to minimize the potential off-target effects and toxicity of the drugs. Importantly, LY-411575 precipitates above 2 µM, RO-4929097 and DAPT above 40–50 µM in seawater (as already noticed for DAPT [49]). Only RO-4929097 thus causes defects at a concentration well below solubility problems. Morphological defects were then assessed at 72 hpf, when the three segments, the brain and the VNC are fully formed and all chaetae are externally visible [31], and at 6 days (figure 3).
Figure 3.
Inhibition of γ-secretase induces defects in bristle formation but no major nervous system phenotype. Ventral views of whole nectochaete larvae (72 hpf) and 6 dpf worms are shown (anterior is up). Larvae were incubated with DAPT (40 µM, b to b2), or LY-411575 (1 µM, c to c2), or RO-4929097 (2 µM, d to d2) in DMSO or in DMSO only (control group, a to a2) from 24 to 48 hpf. Larvae treated with the three drugs targeting the Notch pathway display a clear reduction of the bristles (a–d). Phenotypic classes were scored as ‘presence of chaetae’ (yellow) and ‘absence or abnormal chaetae pattern’ (red). Bars at the base of each image represent the percentages of larvae in each phenotypic class. Double asterisks indicate the highly significant differences (Student's test p < 0.01) between the mean numbers of affected versus unaffected larvae in the control and treated groups (sample sizes are indicated on the figure). Antibody labelling against acetylated tubulin (green) show the axon scaffold of the VNC at 72 hpf (a1 to d1) and 6 dpf (a2 to d2). Hoechst nuclear staining is in blue. Larvae treated with the three drugs display no gross defect in axon guidance, commissural projections or connectives in both stages.
Treated larvae showed limited but completely similar defects as far as gross morphology and behaviour are concerned. At 3 and 6 days, they are well elongated and segmented (figure 3) and show normal muscular contractions, swimming behaviour and phototactism. The swimming speed is faster than normal because treated larvae display very few or completely absent protruding chaetae (in a highly significant way, Student's test p < 0.01; figure 3a–d). Labellings of the neurite scaffold with an anti-acetylated alpha-tubulin antibody show no gross defects in the head and trunk nervous systems as connectives and commissures appear to form normally at 72 hpf and 6 days (figure 3a1–d2). Consistent results for treated embryos obtained with all three drugs (with different molecular structures but targeting the same molecule) clearly suggest that their action is specific and that limited off-target effects appear at the chosen concentrations. Nevertheless, to determine whether drug-induced cell death occurred, we used the terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) method and found just a few apoptotic cells in the treated embryos, as for the control (electronic supplementary material, figure S2C (a to a4)).
As Notch plays a role in vertebrates in maintaining stocks of undifferentiated progenitors, it could have also a general role in Platynereis in regulating cell proliferation. To test that, we assessed proliferation at 48 hpf using 5-ethynyl-2′-deoxyuridine (EdU) labelling [34] and quantified EdU-labelled cells in structures related to the nervous system (i.e. in the ventral neurectoderm and the episphere), but also in the whole embryo, stomodeum and chaetal sacs (n = 8 for each condition; electronic supplementary material, figure S2C (b to d)). We found few significantly different percentages of EdU + cells (Student's test p < 0.05), in treated embryos compared to the control, when looking at the whole embryo, the ventral neurectoderm or the episphere (electronic supplementary material, figure S2C (d)). Only three comparisons (between dimethyl sulfoxide (DMSO) and DAPT and LY-411575 for the ventral neurectoderm and between DMSO and LY-411575 for the episphere) appeared to give slightly more mitoses in the treated larvae, in a significant way. However, we observed that the total numbers of cells in the whole embryo, the ventral neurectoderm and the episphere (defined by quantifying DAPI+ cells) are similar in treated embryos and controls (electronic supplementary material, figure S2C (d)).
Together those results tend to indicate that the Notch pathway disruption does not affect considerably either the general VNC or brain formation, nor does it affect in a major way cell proliferation profiles in the ventral and anterior neurectoderm. On the contrary, the disappearance of chaetae rather supports a role in chaetal sac patterning. Those two aspects are extensively studied using specific time windows for treatments (table 1) depending on the process in the following sections.
2.4. Chaetoblast addition is progressive and their number is finely regulated
To unravel the role of the Notch signalling pathway in the chaetogenesis process, we decided to investigate in depth the development of the chaetal sacs of Platynereis. Chaetal sacs are composed of several pockets called follicles, each of which produces a single bristle [50]. Each follicle consists of one chaetoblast that builds the bristle by proximal addition of material, mostly chitin and four surrounding follicle cells arranged on top of each other along the chaetae [44] (electronic supplementary material, figure S3A) that possibly add more material along the bristle length. The bristle grows along the canal formed by follicle cells and finally outside of the parapodium surface [51]. Morphological and ultrastructural studies revealed that new follicles emerge by internalization of epidermal surface cells. A central cell, the future chaetoblast, sinks down into the epithelium, surrounded by several cells that will become follicle cells (electronic supplementary material, figure S3A).
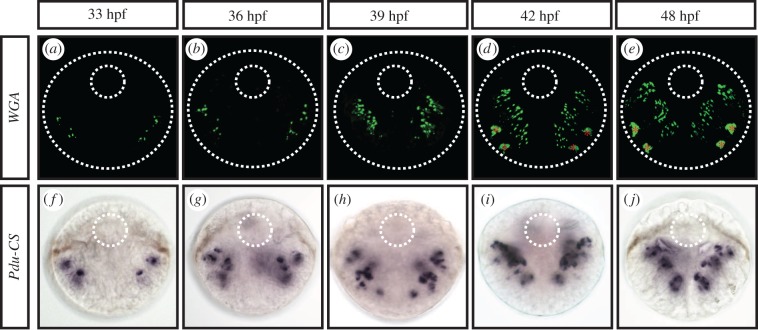
We used the properties of the wheat germ agglutinin (WGA), which binds strongly to the β-chitin [39], the main structural component of chitinous chaetae. We thus identified during larval development the position and number of chaetae and, at the proximal end of the bristle, chaetoblasts (figure 4a–e). We also investigated the expression pattern dynamic of Chitin synthase 1 (CS1), a conserved gene encoding an enzyme crucial for chitin polymerization [52] and expressed specifically in Platynereis chaetoblasts [44] (figure 4f–j). Both stainings give similar results: chaetoblasts and their corresponding chaetae are present in increasing numbers from 33 to 42 hpf (figure 4; electronic supplementary material, figure S3B), first in segments 1 and 2, and then in segment 3. Chaetoblasts first appear near the surface of the larval ectoderm and then sink deeper in the larval body as the chaetal sacs themselves internalize. Chaetoblast numbers are specific for each chaetal sac, depending on segment and dorsal–ventral position (figure 4; electronic supplementary material, figure S3B).
Figure 4.
Chaetogenesis dynamics during Platynereis embryonic development. WGA staining reveals the arrangement and number of chaetoblasts during the course of development (33, 36, 39, 42 and 48 hpf) (a–e). Expression patterns of the chaetoblast marker Pdu-CS1 show similar results (f–j). From 33 to 36 hpf, active chaetoblasts are present in only two segments (1 and 2) and then appears in segment 3 at 39 hpf. Around 42 hpf the chaetal sacs show a fixed number of follicles. All panels are ventral views (anterior is up). Red asterisks mark the artificial staining of glands at 42 and 48 hpf. Large dotted circles indicate the outline of the embryos, while the small dotted circles indicate the position of the stomodeum.
When are chaetal sac cell identities defined? As some chaetoblasts start being functional as early as 33 hpf, their specification and initial internalization must have taken place quite early in larval development. We complemented these stainings with live imaging experiments using the plasma membrane vital dye FM-464 to observe early chaetoblast formation (electronic supplementary material, figure S3C). We detected the presence of ‘rosettes’ of ectodermal cells at the level of the future parapodia, made of one or two central bottleneck-shaped cells that appears to be in the course of internalization, surrounded by six or seven petal-shaped cells, already present at the surface of larvae at 27 hpf (electronic supplementary material, figure S3C (a, a′ and a″)). Later in the development deeper roundish structures appear revealing the outlines of the maturing chaetal sacs (electronic supplementary material, figure S3C (b to d″)). We investigated the very early expressions of potential chaetoblast precursor markers (i.e. Pdu-Delta and Pdu-Hes12; figure 5) and follicle cells markers (not shown). We found that both types of markers are expressed as early as 20 hpf. Pdu-Delta and Pdu-Hes12 are expressed in scattered lateral superficial ectodermal cells (figure 5a,f, green arrowheads) in the parapodial field. Those cells are possibly the future chaetoblast cells, as suggested by their position and arrangement. Later, from 24 to 33 hpf, more cells express both genes in a ‘salt and pepper’ fashion in the whole parapodial field (figure 5b,c,g,e green arrowheads). While most of these cells are in a superficial position, some are located more internally, suggesting a possible mechanism of internalization. At 36–39 hpf, superficial expression decreases considerably while a few more internal cells continue to express both genes (figure 5d,e,i,j). As the fully formed chaetal sacs appear from 42 hpf on, Pdu-Delta is first restricted to a few deep cells in the chaetal sacs of segments 1 and 2 and later appears in segment 3 while Pdu-Hes12 expression is found in all chaetal sacs.
Figure 5.
Expression patterns of chaetoblast markers during Platynereis early embryonic development. WMISH for Pdu-Delta (a–e) and Pdu-Hes12 (f–j), at five developmental stages (20, 24, 33, 36 and 39 hpf) are shown. All panels are ventral views (anterior is up) except (c and h) that are lateral. Blue arrows indicate an expression in brain cells, green arrowheads in the presumptive chaetal sacs, presumably in chaetoblast cells. From 20 to 33 hpf, scattered superficial cells are observed (a–c, f–h). Those cells start to get internalized between 33 and 36 hpf and reach their final position after 39 hpf (d,e,i,j). (b) Inset details the ectodermal nature of Pdu-Delta+cells and (c) inset shows an apical view of Pdu-Delta + brain bilateral cells.
2.5. Molecular signature and arrangement of the chaetal sac cell types
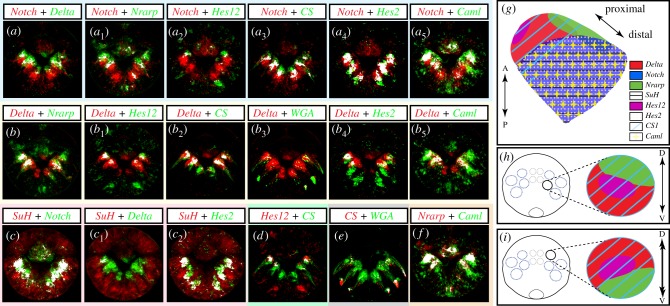
We then used the pattern registration technique recently developed for Platynereis larvae [52] to better unravel the mature follicle cell types' composition and arrangement. Pattern registration in the case of chaetal sacs does not allow a perfect one-cell resolution analysis because cells do not always display perfectly constant positions within the sacs. It is sufficient however to accurately visualize expression domains in three-dimensional views, to establish gross overlap of expression domains and to corroborate hypotheses of coexpression. Expression patterns of all Notch components expressed in chaetal sac cells (Pdu-Notch, Pdu-Delta, Pdu-SuH and Pdu-Nrarp) were thus scanned, aligned and averaged. We have shown previously [53] the expressions of two Hes (Hairy/enhancer of Split) genes (potential target genes of the Notch pathway in eumetazoans) in the chaetal sacs. Pdu-Hes2 and Pdu-Hes12 that are found in 12 patches corresponding to, respectively, a large proportion and a few internal cells of the chaetal sac, were also included in the database. As we wanted to compare those gene expressions with known markers of chaetal sacs cells, we also add the Pdu-CS1 (a chaetoblast marker) and the Pdu-Caml (a follicle cell marker [39]) genes to this analysis. Finally, to better understand the architecture of chaetal sacs in trochophore larvae, we also used Pdu-Twist, expression of which spans mesodermal territories surrounding the chaetal sacs [54]. We used again the WGA labelling to identify the position of the chaetoblasts, located in the proximal-most part of each chaetal sac [39].
In the electronic supplementary material, figure S3D, we provided three different views of three-dimensional surface labelling (either expression patterns or WGA staining) rendering possible the understanding of the complex organization of the 12 chaetal sacs (six in the neuropods and six in the notopods) surrounded by mesodermal tissues (Pdu-Twist) in the 48 hpf larvae. Pdu-Delta expression is localized in the internal-most part of the sac (electronic supplementary material, figure S3D (a4, b4, c4, d3, d4)) while Pdu-Notch expression is more widespread in a large medial area of the sac (electronic supplementary material, figure S3D (a3, b3, c3, d3, d4)).
We next tried to define the identity and molecular signature of the cells composing a chaetal sac by looking at the colocalization patterns of gene expression areas using registration (figure 6). With the WGA staining to localize the chaetoblasts, as well as the Pdu-CS1 expression (figure 6e), we identified Pdu-Delta as a marker of at least some cells in the chaetoblast region (figure 6b2 to b3). Pdu-Hes12 + cells fall also in the Pdu-CS1+domain and are thus chaetoblast cells too (figure 6b1 and d). Pdu-Notch and Pdu-Hes2 expression territories are broadly in the medial part of the sac, and probably correspond to several if not all follicle cells (figure 6a4). Their colocalization with the follicle cell marker Pdu-Caml supports this interpretation (figure 6a5). Pdu-Nrarp is co-localized partially with Pdu-Delta, Pdu-Notch and Pdu-Caml, and is expressed both in chaetoblasts and some follicle cells (figure 6a1, b and f). Altogether those different combinations of markers allowed us to identify two domains of expression patterns among the sac: one proximal area (being Pdu-Delta+, Pdu-Hes12+, Pdu-CS1+ and Pdu-Nrarp+) and one distal area (being Pdu-Notch+, Pdu-Hes2+, Pdu-Su(H)+, Pdu-Caml+ and Pdu-Nrarp+) (figure 6g). In addition, Pdu-Nrarp expression pattern is polarized along the dorsal–ventral axis, but in an inverse disposition in notopodial versus neuropodial chaetal sacs (figure 6h,i).
Figure 6.
Molecular fingerprint of Platynereis chaetal sac cell types at 48 hpf. All images are confocal maximum z-projection of averaged expression patterns registered on an average larva. Ventral surface views of the whole larvae, anterior side up, are shown. White patterns reveal the colocalization of green and red pixels. (a to a5) Pdu-Notch expression slightly overlaps with Pdu-Delta, Pdu-Nrarp and Pdu-CS1, is mutually exclusive with Pdu-Hes12 and is broadly co-expressed with Pdu-Hes2 and Pdu-Caml. (b to b5) Pdu-Delta expression overlaps partly with Pdu-Nrarp, broadly with Pdu-Hes12, Pdu-CS1 and WGA, and slightly with Pdu-Hes2 and Pdu-Caml. (c to c2) Pdu-Su(H) is largely co-expressed with Pdu-Notch and Pdu-Hes2 but not with Pdu-Delta. The chaetoblast marker, Pdu-CS1, is co-expressed with Pdu-Hes12, Pdu-Delta and WGA staining (d, b2 and e). Pdu-Caml and Pdu-Nrarp are broadly overlapping (f). The summary of expression patterns territories relative to a chaetal sac is provided in (g,h,i). (g) Schematic representation of a ventral chaetal sac with the gene expression territories mentioned, in a proximal–distal axis. See figure inset for the gene colour legends. The cartoons in (h,i) show apical view of a 48 hpf larva with the chaetal sacs represented by blue dashed circles. The black circles correspond to the location of the stomodeum. (h) Focus on an apical view of ventral chaetal sac gene expression territories. (i) Focus on a vegetal view of dorsal chaetal sac gene expression territories. Note the mirror-like expression patterns territories between the dorsal and chaetal sacs. A–P, antero-posterior axis; D–V, dorso-ventral axis.
2.6. Treatments with γ-secretase inhibitors suggest that Notch patterns Platynereis chaetal sac cell types
The morphological defects obtained through 24–48 hpf (time window 1, table 1) chemical disruptions (figure 3) as well as the continuous expressions of Notch pathway components, especially Pdu-Delta (figures 2 and 5), from the onset of chaetal sac formation to chaetae elongation strongly suggest an extensive role in chaetal sac pattern formation and chaetogenesis. We thus investigated the effects of Notch pathway chemical inhibition (Time window 1, table 1) on genes expressed in chaetal sacs to test whether Notch pathway component expressions themselves are affected (electronic supplementary material, figure S3E). Interestingly, Pdu-Notch is significantly downregulated (electronic supplementary material, figure S3E (a to a3); an expected effect, for example, see [55]), whereas Pdu-Delta seems not affected by any treatments (electronic supplementary material, figure S3E (b to b3)). Similarly, treatments also lead to drastic decreases of follicle cells marker expression (Pdu-Nrarp, Pdu-Hes2 and Pdu-Caml) while chaetoblast markers (Pdu-Hes12 and Pdu-CS1) are maintained (electronic supplementary material, figure S3E (c to g3)). As Notch pathway disruption does not affect cell proliferation profile and does not induce specific cell death in follicles and chaetal sacs (electronic supplementary material, figure S2C), the expression losses observed may be related to an abnormal differentiation of follicle cells.
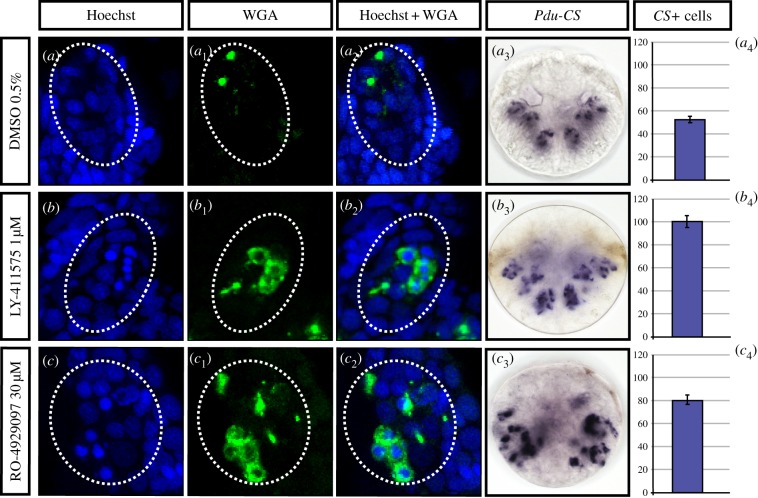
Notch pathway components (Pdu-Delta, Pdu-Hes12, Pdu-Nrarp and Pdu-Hes2) are expressed early in surface ectodermal cells, suggesting that they play a role in selecting cells that are going to differentiate into chaetoblasts or follicle cells, in a way that could involve lateral inhibition. We thus also performed early drug treatments (20–48 hpf, Time window 2, table 1) and assessed the resulting phenotypes on chaetoblast differentiation at 48 hpf, using WGA staining as well as the chaetoblast marker Pdu-CS1 (figure 7). LY-411575 was used at 1 µM while RO-4929097 had been raised to a higher concentration (30 µM) to obtain similar results. DAPT used at the limit of solubility (40 µM), while affecting chaetae production as the two other drugs do, gave no phenotype related to chaetoblast differentiation (not shown). In both efficient treatments, our experiments revealed the presence of roundish, intensely DAPI-stained cells surrounded by cytoplasmic WGA-reactive chitin (figure 7b to c2). Many of these cells do not show however a WGA-stained bristle elongating from it, suggesting that although they have some characteristics of chaetoblasts, they are not capable of producing bristles. We thus call them ‘abortive chaetoblasts’. Chaetoblasts and abortive chaetoblasts are more numerous in treated embryos than in the control (figure 7a to c4), as highlighted by Pdu-CS+ expression and cell counting (figure 7a4, b4 and c4). Abortive chaetoblasts are located at dispersed, abnormal positions in the sacs, including close to the surface and presumably at the ‘normal’ locations of follicle cells in a control embryo. No recognizable chaetal sacs form in these early treated larvae, suggesting that the patterning of these structures is completely compromised, in sharp contrast with later treatments. The internalization process, linked to the chaetal sac patterning, is consequently presumably affected.
Figure 7.
Early inhibitions of the Notch pathway induce supernumerary abnormal chaetoblasts within chaetal sacs. Embryos were incubated with LY-411575 (1 µM, b to b4), or RO-4929097 (30 µM, c to c4) in DMSO or in DMSO only (control group, a to a4) from 20 to 48 hpf. Treated larvae present several packed, intensely DAPI-stained nuclei surrounded by WGA-stained cytoplasm (b to b2, c to c2) instead of WGA spots neighbouring the chaetoblast nuclei (a to a2). The chaetoblast marker Pdu-CS1 is overexpressed within the whole sacs in treated larvae instead of a packed internal region (b to b2, c to c2, and compare to a3 to a4). All panels are ventral views (anterior is up).
As we are not able to establish unambiguously that the early Delta expressions are exclusively linked to chaetoblast differentiation, we also tested the possible involvement of the Notch pathway in the formation or selection of neural progenitors that will give rise to the few pioneer neurons of the 24 hpf larvae [31]. We thus treated larvae with the drugs (same concentrations as before) from 12 and 16 to 24 hpf (to be sure to encompass the very first neuron formation, Time window 3, table 1) and assessed the numbers of neurons (revealed by the postmitotic neuron marker Elav [33]) in both treated and control embryos at 24 hpf. At 24 hpf, the number of Elav+ cells in the trunk is very precise (nine) and is not at all affected by any early treatments (electronic supplementary material, figure S4A).
In conclusion, the inhibition of the Notch pathway in an early time window (Time window 2) results in missformed chaetal sacs and supernumerary chaetoblasts, consistent with a lateral inhibition mechanism, while additional experiments should be performed in the future to firmly prove it. By contrast, early larval neurogenesis is not affected. Later treatments (window 1), or with more limited drug doses, result in abnormal gene expressions and possibly abnormal differentiation of follicle cells.
2.7. No major involvement of the Notch pathway in ventral neurogenesis and brain patterning is evidenced
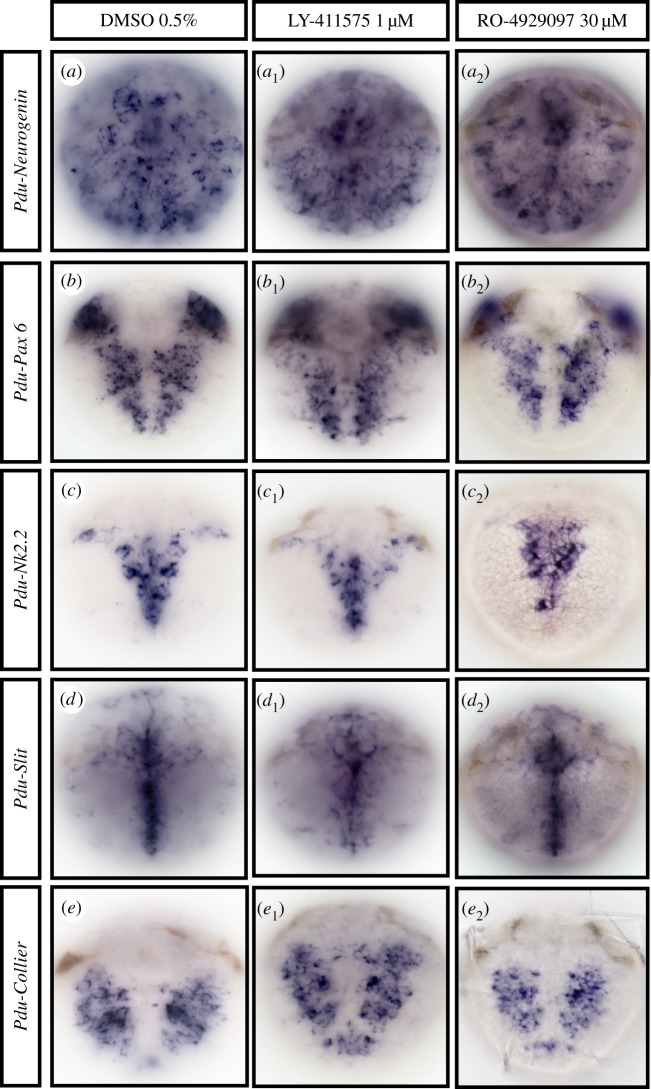
Although none of the components of the Notch pathway are obviously expressed in the forming VNC (figures 2 and 5) and the apparent morphology of the ventral nervous system does not appear much disturbed after γ-secretase inhibitor treatments (figure 3), we cannot exclude that Notch nevertheless could have a role in patterning and neuronal specification. We thus tested further the normal formation of the ventral nervous system by using several markers specific for different steps of neurogenesis and VNC patterning (Neurogenin, Pax6, Nk2.2, Slit and Collier) on 48 hpf-treated embryos for all drugs (Time window 1, table 1 and figure 8). The proneural gene Neurogenin (Pdu-Ngn) is exclusively expressed in neural progenitor cells (figure 8a) [33]. Pdu-Pax6 and Pdu-Nk2.2 are expressed in neurogenic columns and are key to medio-lateral patterning of neurons in vertebrates (figure 8b,c) [20]. Pdu-Slit is expressed in the ventral midline and is a key axon guidance factor in insects and vertebrates (figure 8d) [34]. Collier (Pdu-Coe), as a marker of neuronal differentiation, is expressed in differentiating and/or fully differentiated neurons (figure 8e) [56]. In concordance with the morphology and behaviour, we observed no expression pattern alteration in the forming CNS on treated larvae at concentrations (LY-411575 1 µM, RO-4929097, 30 µM, DAPT 40 µM; data not shown) that perturb chaetal sac patterning, either at gross levels or in the precise spatial patterns of these five genes (figure 8a to e2). In addition, no overlapping expression was observed between either Pdu-Notch or Pdu-Delta and Pdu-SoxB, Pdu-Ngn and Pdu-Pax6, central elements of the neurogenic network at eumetazoan scale [24] (electronic supplementary material, figure S4B).
Figure 8.
The Notch pathway does not regulate general ventral nerve chord patterning in 48 hpf Platynereis larvae. Ventral views of whole trochophore larvae (48 hpf) are shown (anterior is up). Larvae were incubated with LY-411575 (1 µM, a1 to e1), in DMSO or in DMSO only (control group, a–e) from 24 to 48 hpf, or RO-4929097 (30 µM, a2 to e2) from 30 to 48 hpf. WMISH, in control and treated larvae, reveals no gross defects in the expression of several genes involved in the process of neurogenesis after inhibition of the γ-secretase.
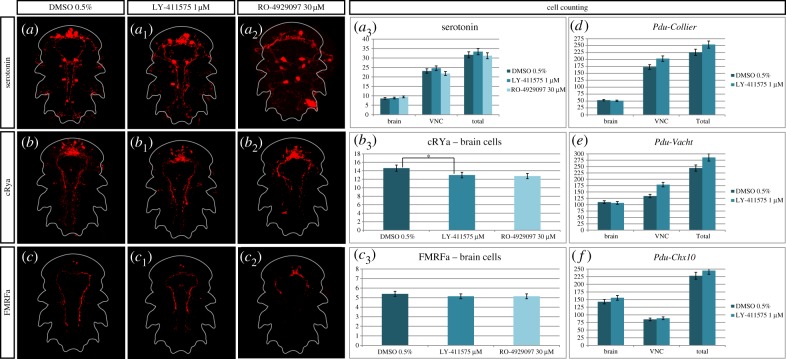
While inhibiting Notch clearly does not disrupt patterning of the VNC at 48 hpf, we then tested the possibility that a Notch activity reduction may enhance neurogenesis in older embryos. We thus treated larvae (LY-411575 (1 µM) and RO-4929097 (30 µM)) from 30 to 48 hpf (Time window 4, table 1), a time window that encompasses the massive neurogenesis process in Platynereis [34] but prevents too drastic phenotypes. We assessed the phenotype at 72 hpf, at the cellular level (figure 9). We used several markers and antibodies of differentiated neurons or specific types of neurons (serotoninergic, FMRFAmidergic, RYa peptidergic, cholinergic and interneurons), and investigated the number of cells that are positive for each of them, in both control and treated larvae. Serotonin, Crya [41] and FMRFa antibody stainings reveal no gross defaults between the control and LY-411575 treated larvae, while the RO-4929097 treated larvae appear to be delayed in their development (a to c2). Manual counting of serotoninergic (a3), RYa peptidergic (b3) and FMRFAmidergic (c3) neurons shows no significant differences in both brain and VNC between the control and treated larvae (even for the delayed ones), except for one case (RYa brain cells are significantly reduced in the RO-4929097 condition compared to control). We then used three other markers of neurogenesis: Pdu-Collier for all the differentiated neurons, Pdu-VAchT for the cholinergic neurons [20] and Pdu-Chx10 for a subset of interneurons [20]. Using registration, we obtained an averaged expression pattern for these genes in both control and LY-411575-treated larvae (not for RO-4929097 as the concentration used led to delayed larvae, preventing from a correct alignment during registration; electronic supplementary material, figure S4C). Using the Imaris software, we counted on these averages the number of positive cells for each gene in both the VNC and brain. This automatized approach for cell counting is highly linked to the in situ hybridization experiment efficiency and gives only a rough estimate of number of neurons. For all three genes, the number of neurons in the brain is almost equivalent between the control and treated larvae, while a few more interneurons are found after Notch inhibition (figure 9d–f). The number of interneurons is nevertheless constant in the VNC, while the cholinergic neurons and the overall differentiated neurons in general appear to be slightly more numerous in treated larvae. However, these numbers may be biased by the quality of the in situ hybridization and are in no way comparable to the drastic phenotypes observed in other models (in different contexts, see [57–60]).
Figure 9.
γ-Secretase inhibitor treatments, while possibly causing a slight increase in the number of neurons, do not change their overall differentiation patterns in 72 hpf Platynereis larvae. Ventral views of whole nectochaete larvae (72 hpf) are shown (anterior is up). Larvae were incubated with LY-411575 (1 µM, a1 to c1) or RO-4929097 (30 µM, a2 to c2) in DMSO or in DMSO only (control group, a–c) from 30 to 48 hpf and fixed at 72 hpf. Total number of neurons as well as specific populations of neurons (serotoninergic, FMRFAmidergic, RYa peptidergic, cholinergic and interneurons) in the VNC and the brain were counted in both control and treated larvae (a3 to c3, d–f). The serotoninergic, FMRFAmidergic and RYa peptidergic numbers of neurons are equivalent between control and treated larvae. The numbers of differentiated trunk neurons (Pdu-collier), as well as the cholinergic neurons (Pdu-Vacht) appear to be slightly increased after Notch inhibition (d–f) but a specific category of interneurons (Pdu-Chx10) remains stable.
3. Discussion
3.1. Evolution of the Notch pathway architecture: a lophotrochozoan focus
We investigated the evolution of the Notch pathway core components in metazoan genomes with a strong focus on lophotrochozoan species, and updated a previous analysis done at the eukaryotic level [7]. In a majority of species, a single Notch gene was found. The presence of supplementary copies of genes in H. sapiens is most probably due to the two whole-genome duplication events (2R) at the origin of vertebrates [61] while some lineage-specific duplications are also evidenced (for example, in H. robusta and S. mediterranea). The receptor Notch structure (domain composition and numbers of domain copies) is conserved in a large majority of cases, suggesting that the last common ancestor of bilaterians probably possessed a single Notch protein composed of 36 EGF repeats, three LNR repeats, NOD and NODP domains and seven ANK repeats. A more complex evolutionary history of the Delta/Delta-like proteins is suspected, but a single Delta gene was probably present in the urbilaterian ancestor with a structure (MNLL/DSL/9 EGF repeats/ATEV) that has been mostly conserved in extant bilaterians. The ATEV motif, which binds to PDZ domain proteins [62], is coded in most bilaterian Delta genes, while we identified in Platynereis a splice variant lacking this motif. Additional lophotrochozoan Delta-like proteins are found but their origin is uncertain. Drawing strong conclusions on the early metazoan history of the Delta-like ligand is difficult as non-bilaterian data are rather limited. At least one Delta-like gene was present in the last metazoan ancestor (with presumably multiple duplications events in the sponge lineages) but the structure of this ancestral gene cannot be firmly reconstructed from the available data (figure 1a; electronic supplementary material, figure S1D) [7]. In conclusion, the urbilaterian genome harboured well-defined Notch, Delta and Jagged genes. This situation is remarkably conserved in Platynereis, thus making it legitimate to discuss their functions in the worm in a bilaterian evolutionary context.
3.2. Is the Notch pathway ancestrally linked to central neurogenesis?
We think we have here gathered convergent arguments to rule out a major involvement of the Notch pathway in the early events of central neurogenesis in Platynereis, thereby contrasting with the situation known from insects and vertebrates:
-
—
There is no major expression of the receptor Pdu-Notch or the ligand Pdu-Delta at the time when mass adult neurogenesis is taking place in the episphere (future brain) and the ventral neuroectoderm. Both genes are found expressed in sub-populations of cells in the brain (notably in the apical organ) but only at very late larval stages (72 hpf onward) when the bulk of brain neurons have already differentiated.
-
—
Previously, we have shown the expression of two of the 13 Hes genes present in Platynereis (Hes12 and Hes13) in the presumptive VNC during larval neurogenesis [53]. As Hes genes are sometimes considered as ‘canonical Notch effector genes’, their expression could indicate a limited involvement of Notch in the events of central neurogenesis. Nevertheless, none of them is found in a very broad pattern that would suggest a role in general neurogenesis [53]. Finally, larvae treated with γ-secretase inhibitors do not show any Hes12 expression increase (electronic supplementary material, figure S3E). We thus feel confident that Notch does not act through any Hes gene (alone or in combination with another Hes) to pattern Platynereis VNC, while we cannot exclude that some Hes may play a role in more limited aspects such as the differentiation of specific neuron types. We also would like to stress the fact that the status of Hes genes as ‘core’ Notch pathway targets is the result of a historical over-simplification, based mostly on Drosophila studies. The Hes superfamily was shaped by many independent lineage-specific tandem duplication events leading to a high diversity of Hes members [53]. Consequently, a regulation of Hes by Notch should not be systematically expected.
-
—
Treatments with three drugs preventing the normal cleavage of the Notch receptor do not result in major defects in the nervous system, when applied during the two time windows of neurogenesis (i.e. during the formation of the specific larval and pioneer neurons, and during the later mass adult neurogenesis events in the VNC and the brain). The normal patterning of the CNS is preserved as indicated by gene expressions. Near normal proliferation of both anterior and ventral neural ectoderm is maintained. Near normal numbers of neurons are differentiating and form nerve connections that seem unperturbed. We nevertheless observed a slight increase of cholinergic neurons in the VNC after drug treatments. Pdu-VAchT (the cholinergic marker used) is known to be also expressed in the peripheral nervous system (PNS) of Platynereis (J Béhague and P Kerner 2017, unpublished data). We thus cannot exclude a limited role of the Notch pathway in peripheral neuron differentiation. However, the persistence of normal peripheral nerves in treated larvae does not suggest a predominant role in PNS formation either.
-
—
By contrast, the same three drugs are causing specific defects in organs where the core components of the Notch pathway are specifically expressed, the chaetal sacs. These defects seem to be compatible with the Notch pathway being involved in a process of lateral inhibition in the chaetal sacs.
The minor involvement of the Notch pathway in Platynereis generic neurogenesis comes as an important surprise and two alternative evolutionary scenarios can be proposed to interpret this striking fact:
(i) The Notch pathway may be ancestrally involved in neurogenesis and this role has been lost in ancestors of the Platynereis lineage. The ‘salt and pepper’ expression of proneuronal bHLH genes in Platynereis neurectoderm suggests that a process of selection of cells with neuronal fate occurs [33]. If the Notch pathway is not playing this role, one has to wonder how this process is performed and no alternate candidates are suspected so far.
(ii) The situation in Platynereis reflects the fact that the Notch pathway may not be ancestrally involved in neurogenesis. The Notch pathway could have been co-opted independently in ancestors of the arthropod and vertebrate lineages to perform analogous functions in the formation of nervous systems.
How plausible does each of these scenarios seem? The first scenario may be the most tempting as the view that the Notch pathway is probably ancestrally linked to neurogenesis is relatively widespread. However, careful examination of the available data leads to questioning the functional similarities that were initially discovered between insect and vertebrate central neurogenesis. In vertebrates, the Notch pathway plays a key role in maintaining a pool of neural stem cells during the progressive process of neurogenesis [63]. Therefore, Notch is not involved in directing neural progenitors towards one fate or another, this choice being regulated by different signalling pathways (FGF, Wnt and BMP [64]). By contrast, the Notch lateral inhibition process described in the fruit fly very precisely switches the central cell in proneural clusters towards a neural stem cell fate, while all the other cells in the cluster will become epidermal cells. Hence, in Drosophila, the first role of Notch is to choose between neural and non-neural fates, which is an earlier role compared to what the pathway does in vertebrates. Conversely, there does not seem to be a general role for Notch in the maintenance of the neural stem cell pools in the fruit fly although Notch plays a role locally in the maintenance of post-embryonic type II neuroblasts [65]. Consequently, the overlap between the insect and vertebrate functions of Notch in central neurogenesis could be seen as limited.
Do data coming from other model animals strategically placed in the metazoan tree support an independent recruitment of Notch in neurogenesis? The general picture is more ambiguous than usually acknowledged. In arthropods, expression data support a role of Notch similar to what is known from Drosophila (reviewed in [12]) but strong differences exist in neurogenetic processes between arthropod lineages. In particular, neuroblasts exist only in insects and crustaceans. In ecdysozoans, an ancestral role of Notch in neurogenesis comparable to arthropods is not warranted. In onychophorans [66,67], Delta does not show an expression similar to the arthropod situation. In nematodes, the derived components of the Notch machinery do not seem to have a general role in neurogenesis although they are necessary to specify the fate of a number of neural cells [68].
In lophotrochozoans, two annelid species other than Platynereis also do not suggest a general role of Notch in neurogenesis: in larvae of the worm C. teleta [27], both Notch and Delta are expressed in two longitudinal rows corresponding to chaetal sacs but show no prominent expression in the ventral neurogenic ectoderm. In the leech H. robusta, Notch inhibition does not lead to gross defect in the production of neurons per se [25,26]. In deuterostomes other than vertebrates, the existing data are insufficient to firmly conclude in favour of an ancestral role of Notch in neurogenesis, but in the sea urchin, a role for Notch in controlling the number of serotoninergic neurons in the larva is proposed while nothing is known on the formation of the adult nervous system [69]. In the chordates amphioxus [70] and ascidians [49,71], the expression of Delta homologues and the effects of Notch pathway perturbations are in good agreement with a role in the selection of neural cells.
Animals branching outside of bilaterians, namely cnidarians and sponges, are of special interest to potentially unravel the ancestral Notch role in neurogenesis. Cnidarians do not have a centralized nervous system but possess neural cells. In the evolutionarily derived Hydra, a proper Delta gene is missing and the small Notch receptor found in this species is involved in head and tentacle patterning [72]. In Nematostella (a sea anemone), a proper Notch protein and a short Delta ligand are present. An initial report [21] proposed an involvement of Notch in general neurogenesis. However, another article [22], while confirming a role of Notch in the production of at least a part of the anemone neurons, surprisingly showed that this role was not dependent on SuH and therefore not based on the ‘canonical’ Notch pathway. This point already pushes some authors to propose that Notch may have a more general role in cell differentiation at the metazoan scale and could have been coopted later in both cnidarians and bilaterians in a different manner in the neurogenic pathways [23]. A recent detailed study on Nematostella supports the idea that Notch signalling negatively regulates neurogenesis while its exact function remains controversial [24]. Last but importantly, the Notch pathway has been shown to exist in the sponge Amphimedon [73] with no less than six Delta-like ligands. The Notch pathway thus predates the appearance of a nervous system. Interestingly, some of the Delta-like genes are expressed in larval cells interpreted as sensory cells. Some of these cells might be derived from ancestral metazoan sensory cells that were the starting point for the first neurons [74,75].
All these data open the possibility that the roles of Notch in early neurogenesis may have evolved in parallel in several metazoan groups, while we cannot rule out now the possibility that the differing roles of this pathway in different species can also be derived from an ancestral involvement in neurogenesis, secondarily lost in some groups. Nevertheless, the sponge data support the view that the Notch pathway is a crucial ‘toolkit’ that was involved in cell type differentiation very early in metazoan history.
3.3. The Notch signalling pathway plays a central role in chaetal sacs morphogenesis in Platynereis
Annelid chaetae are extracellular locomotory structures, displayed on the worm appendages, that function in bundles alternatively deployed and retracted in epidermal pockets, the chaetal sacs. We show in this work that the number of chaetae produced in each chaetal sac is specific and strictly regulated. Chaetae are deciduous structures: they are shed progressively and new chaetae grow out of new follicle continuously during the worm's life. This process of replacement has been described as spatially polarized within the chaetal sac [50], chaetae being shed on one side and new follicles forming on the opposite side.
Before the formation and during the invagination of the chaetal sacs (20–36 hpf, figure 5), Delta is found in a dynamic ‘salt and pepper’ pattern in superficial or slightly more internal ectodermal cells. These cells may correspond to precursors being selected towards differentiating into chaetoblasts. Notch is expressed in broad lateral domains at these stages. Consistent with a role in the selection and strict control of the chaetoblast numbers, drug inhibitions of Notch signalling result in missformed chaetal sacs and the differentiation of abnormal and supernumerary chaetoblasts. We suggest that a process similar to lateral inhibition may be involved in the progressive selection of chaetoblast precursors from the surface ectoderm. We are not able, however, to demonstrate that Delta-expressing cells at these early stages are chaetoblast precursors, because of the absence of efficient lineage tracing in Platynereis embryos. But both Delta and Hes12 are expressed in the chaetoblasts later and Hes12 shows a ‘salt and pepper’ expression very similar to Delta. In addition, drug treatments do not affect the differentiation of larval and pioneer neurons, suggesting that Delta expression is not associated with differentiating neurons in embryogenesis. If Delta expressing cells indeed differentiate as chaetoblasts, surrounding cells that are laterally inhibited could give rise either to epidermal or to the various types of follicle cells.
After the complete formation of the chaetal sacs (42–48 hpf), Delta and Notch are expressed in specific regions of the chaetal sacs. With the help of pattern registration and WGA staining, we obtained a general picture of the gene combinations expressed by the cell populations that compose a chaetal sac (figure 6). Notch is found in at least part of the follicle cells, as well as Hes2, a potential target transcription factor, and Nrarp as shown by CamL (a follicle cell marker [39]) expression. Delta is found in few cells in the region where chaetoblasts are sitting, as shown by CS1 (a chaetoblast marker [39]) expression. A sub-population of the cells in the chaetoblast domain also expresses Pdu-Hes12.
Drug treatments during later time windows or with a lesser dose of drug (table 1) lead to mild phenotypes. Larvae do not show any clear defects but the absence of protruding bristles. Importantly, the chaetal sacs seem properly formed in late trochophore stage (48 hpf) and a normal number of chaetoblasts, as shown by chitin-synthase and WGA labellings, is found. Additionally, drug treatment is not accompanied by a modification of cell death and cell proliferation patterns in the sacs (electronic supplementary material, figure S2C). DIC microscopy reveals that short abnormal chaetae are present internally (not shown). Gene expressions reveal that Notch/Hes2/Nrarp and CamL expressing cells, corresponding to at least part of the follicle cells, are affected. We propose that follicle cells play an important role in the production of chaetae. Forming the canal through which the bristle is progressively built by basal addition, they have the capacity to secrete materials that could contribute to the bristle structure and rigidity, hence the abortive bristles seen in treated larvae. The Notch receptor expressed at the surface of these cells would play a role in maintaining their identity and normal function. What is the significance of the Delta and Hes12 expressing cells in the deep side of the chaetal sac? One speculative idea may be that the cells are new differentiating chaetoblasts that will eventually replace the larval ones, i.e. the start of the ‘treadmill’ process of replacement of chaetae. It is interesting to note in this context that Nrarp is expressed in a dorsoventrally polarized way in each chaetal sac and may play a role in the polarized process of chaetae replacement. Parapodia and their chaetal sacs are symmetrically organized respective to a medial longitudinal plane and Nrarp is expressed on the externally facing side of each sac respective to this plane (figure 6).
In summary, in late embryogenesis, Pdu-Notch and Pdu-Delta play a likely role in building chaetal sacs by selecting prospective chaetoblasts, and follicle cells, potentially through a process of lateral inhibition. Later during larval development, when chaetal sacs are in place, Notch probably plays a role in the maintenance and normal function of follicle cells.
3.4. Chaetogenesis in lophotrochozoans and the role of the Notch pathway
We can definitively assume that the Notch pathway plays an important role in chaetogenesis in the errantian annelid Platynereis dumerilii. In a previous study, Thamm & Seaver [27] proposed, based on expression patterns data, that Notch may be also involved in chaetal development in the sedentarian annelid Capitella teleta. As a majority of annelid species are encompassed into the errantia and the sedentaria [76], the requirement of Notch signalling in chaetae formation is probably an ancestral feature in annelids. Chaetogenesis is not restricted to annelids, but also occurs in other lophotrochozoan phyla: brachiopods and molluscs [39,50]. Interestingly, morphological and ultrastructural studies suggest that brachiopod chaetae-like structures are potentially similar to the annelid ones, but they have often been interpreted as non-homologous structures with an independent origin [39]. Recently, phylogenomic studies highlight the fact that brachiopods and annelids are much more closely related phyla than previously thought [77,78]. In this context, gaining insight into the molecular mechanisms underlying chaetogenesis in brachiopods by investigating the formation of the larval chaetae would fill a critical gap. In a comparative approach, several candidate genes that have been shown to be expressed in chaetal sac anlagen in annelids, such as Hox2, Post1, Nk3, Tlx, GATA456 and of course the Notch pathway core components could be tested [79–81]. The ultimate aim of such an evolutionarily comparative study would be to decipher whether the Notch pathway involvement in the chaetogenesis process is a specific developmental circuit co-opted in the lineage of annelids or whether it was ancestrally present in the common ancestor of both annelids and brachiopods to regulate chaetogenesis.
4. Conclusion
The Platynereis Notch pathway molecular architecture is well conserved and similar to the urbilaterian ancestral conformation. This pathway was certainly coopted, at least in the lineage of annelids, to play a role in bristle development, more precisely to pattern chaetal sac cells probably through a lateral inhibition process. Its absence of major function in annelid early neurogenesis could be seen as surprising when considering the general idea of a systematic involvement of this pathway to specify neuronal cell fate in bilaterians. We nevertheless highlight the fact that an ancestral link between the Notch pathway and neurogenesis is not as well supported as suggested before, and argue that independent recruitments of Notch in neurogenesis cannot be ruled out. Such a scenario of co-option of genetic networks involved in cell differentiation towards a role in early embryonic patterning has indeed already been advocated with several examples by Erwin & Davidson [82]. Exploring the Notch pathway in other lophotrochozoans and non-bilaterian species will undoubtedly help us to understand whether reasoning on homologies based on the comparison of Notch functions or modes of actions at very large scale is relevant or not.
5. Material and methods
5.1. Survey of Notch signalling pathway core components in Platynereis dumerilii: identification and cloning
Platynereis Notch pathway core components were identified by sequence similarity searches against large collections of expressed sequence tags (ESTs) and genomic sequences (Platynereis resources, 4dx.embl.de/platy/, D. Arendt, Jékely Lab, jekely-lab.tuebingen.mpg.de/blast/) [28] using the specific conserved domains diagnostic for each gene family in Drosophila and/or vertebrate genes. A Platynereis Notch gene had already been identified from a BAC sequence ([28], Genbank: AM114766). For Delta and Jagged family genes, a combination of the specific MNLL and DSL domains found in both type of proteins was used. Complete coding sequences for all genes were assembled from EST fragments using CodonCode Aligner (CodonCode Corporation, USA). Putative exons positions, for the Platynereis Delta gene, were mapped on genomic DNA by comparison with ESTs using Artemis [83]. Large gene fragments (1–3 kb) were subsequently cloned by PCR using sequence-specific primers using cDNAs from mixed larval stages as templates (primer sequences and PCR conditions are available upon request). PCR products were TA cloned into the PCR2.1 vector following the manufacturer's instructions (Invitrogen) and sequenced. Partial cDNAs obtained were then used as templates to produce RNA antisense probes for WMISH using Roche reagents. Orthology relationships were defined using as criteria sequence similarities, presence of specific domains and phylogenetic analyses (see below). The newly identified Platynereis gene sequences were deposited in Genbank with the following accession numbers: Pdu-Delta, KP293866; Pdu-Jagged, KP293873; Pdu-Su(H), KP293861; Pdu-psen, KP293862; Pdu-Nrarp, KP293864; Pdu-fng, KP293863; Pdu-numb, KP293865; Pdu-dl-like1, KP293867; Pdu-dl-like2, KP293868; Pdu-dl-like3, KP293869; Pdu-DSL-like1, KP293870; Pdu-DSL-like2, KP293871; Pdu-DSL-like3, KP293872.
5.2. Survey of Notch signalling pathway core components in a panel of metazoans and phylogenetic analyses
5.2.1. Data sources, sequence retrieving and domains composition
Notch pathway core component searches were carried out using the tblastn or blastp algorithms implemented in ngKlast (Korilog V 4.0, Questembert, France) with Drosophila, vertebrate and Nematostella proteins as query sequences, with the default Basic Local Alignment Search Tool (BLAST) parameters and a low cut-off E-value threshold of 0.1, against 15 genome datasets. Lists of BLAST hits were then reciprocally BLASTed against the human proteins dataset of the NCBI database (reciprocal best hits [69]). Those genomes correspond to 15 species representatives of the main lineages of animals: Porifera, Ctenophora, Cnidaria, Lophotrochozoa, Ecdysozoa and Deuterostomia, with a lophotrochozoan focus (plus one choanoflagellate). For each species, we screened the genome assembly, the predicted protein sequence dataset and transcriptomes when available. The presence of the specific protein domains (available upon request) was systematically checked by scanning sequences with both NCBI Conserved Domain search option V3.10 [84] and InterProscan v. 42 online software [85]. Orthology relationships were defined using as criteria sequence similarities, the presence of specific domains and phylogenetic analyses. The structural diversity of DSL proteins was very broad in metazoans. We had thus to define different classes of proteins to conduct a sensible phylogenetic analysis. We defined the organizations of Delta and Jagged proteins in metazoans (modified from a previous one made by Rassmussen et al. [36]) based on the presence of a minimum number and specific arrangement of domains. Based on its likely presence in at least the bilaterian ancestor, we defined as a ‘Delta’ protein a sequence that comprises all of the signal peptide (SP), an MNLL domain, a DSL domain, at least nine EGF repeats, a transmembrane domain (TM) and an intra-cellular domain (ICD). A number of proteins we found that displayed all these domains but with a number of EGF repeats smaller than seven were defined as ‘Delta-like’ proteins. All DSL-containing proteins that comprised at least one additional conserved domain not mentioned above were classified as ‘DSL-like’ proteins and not included in the scope of this phylogenetic analysis. For Jagged proteins, likewise, the minimal domain arrangement is considered to be an SP, an MNLL domain, a DSL domain, at least 16 EGF, a Von Willebrand factor C (VWC) domain, a TM and an ICD. Similarly, Notch proteins that do not possess the minimal domain arrangement of six EGF, three Lin12/Notch repeats (LNR), a TM and three ANK repeats were excluded.
5.2.2. Phylogenetic analyses
The predicted amino acid sequences of the identified Platynereis gene fragments were aligned with their presumptive orthologues from 13 metazoan species. These genes were either annotated as such in public databases, or found as predicted genes in whole-genome BLAST screening. Monosiga brevicolis sequences were selected as outgroup in order to root the tree, when possible. For Notch and Delta/Jagged proteins, the number of EGF domains is variable, preventing from adequate alignments; they were thus excluded. The Notch alignment includes a partial sequence that begins at the first LNR domain until the end of the protein. The Delta/Jagged alignment includes partial protein sequence from the signal peptide to the end of the DSL domain. Multiple alignments were then performed with MUSCLE 3.7 online [86,87] under default parameters and manually adjusted and improved in Bioedit [88]. Handling of the multiple alignments was also done using Bioedit. Maximum-likelihood analyses were performed using PHYML [89,90] with the Le and Gascuel amino acid substitution model [91] and six rate categories. Statistical support for the different internal branches was assessed by aLRT [92]. Trees were handled using FigTree and provided in the electronic supplementary material, figure S1A. The whole list of sequences used in this study and the multiple alignments are available in the electronic supplementary material, file S1.
5.3. Animal culture and collection
Platynereis embryos were obtained from an 18°C breeding culture established in the Institut Jacques Monod (Paris), according to the protocol of Dorresteijn et al. [93]. Staging of the embryos was done following Fischer et al. [31]. Embryos and larvae were fixed in 4% PFA, 1× phosphate-buffered saline (PBS), 0.1% Tween 20 and stored at −20°C in methanol 100% [94].
5.4. WMISH, immunohistochemistry, chitin and membrane labelling
NBT/BCIP whole-mount in situ hybridizations were performed as previously described for wild-type and treated embryos [34] with a modified protocol allowing for sharper signal as required for three-dimensional confocal imaging [52] (performed on two larval stages (48 and 72 hpf)). Cilia and neurite staining on 72 hpf treated larvae (see above) was done as previously described [34] using the mouse anti-acetylated tubulin antibody (Sigma T7451, 1 : 500), fluorescent secondary anti-mouse IgG Alexa Fluor 488 or 555 conjugate (Invitrogen, 1 : 500) and DAPI (1 : 1000). Neuronal antibodies were also used to label specific neuron populations as well as their axons and dendrites as previously described: antibodies against the neuropeptide RFamide [32], the monoamine transmitter serotonin [32] and the amidated neuropeptides RYamide, and FLamide [41]. The chitinous chaetae were stained with WGA FITC conjugate (Sigma L4895) at a concentration of 100 µg ml−1 for 12 h at 4°C. The staining was stopped with several washes in PBS/0.1% Tween 20. Larvae for three-dimensional imaging were counterstained with DAPI (Sigma), diluted at 1 µg ml−1 in PBS/0.1% Tween 20. Following WMISH or antibody labelling, larvae were transferred into 97% TDE diluted with PBS/0.1% Tween 20 [52]. Three-dimensional stacks of the nuclear DAPI labelling and of the reflected signal of NBT/BCIP labelling [95] were recorded and used for three-dimensional reconstructions and pattern registration. The FM4-64 dye (ThermoFisher Scientific T3166), as a membrane stain, was used to detect cellular outlines. 15 hpf embryos (before ciliae appearance) were embedded in 1% UltraPure™ Low Melting Point agarose containing FM4-64 (2 µl ml−1), in a glass bottom Petri dish covered by seawater with FM4-64 (2 µl ml−1) before imaging.
5.5. Small molecule inhibition
Embryos were incubated in the chosen time window (see main text) in natural seawater containing anti-presenilin drugs: DAPT (Tocris Bioscience), LY-411575 (A. G. Scientific) or RO-4929097 (Stemgent). For all inhibitors, stock solutions were 10 mM in 100% DMSO and different final concentrations were used (consequently the DMSO concentrations too). Control embryos originating from the same batch were incubated in natural seawater containing 0.5% DMSO. After drug incubations, embryos were either (i) rinsed in seawater for 24 h (and so, until 72 hpf) to assess their phenotypes or (ii) fixed at 48 hpf to perform WMISH with several markers. Approximately 150 embryos per treatment were examined and counted.
5.6. Whole-body gene expression registration
Image registration of three-dimensional gene pattern scans based on the DAPI nuclear labelling were performed using the open-source software package ITK4.0 and a collection of specific scripts developed in the Jékely Lab [52]. All patterns were registered to average anatomical reference templates (48 and 72 hpf). Both of these reference templates were calculated using a total of 40 DAPI-labelled larva three-dimensional stacks. The complete protocol for unbiased anatomical reference calculation and the reference files themselves are available upon request. Average patterns for a given gene were calculated using at least five individual larvae and gene pattern comparisons were performed using ImageJ and Imaris.
5.7. Edu cell proliferation and TUNEL assay
Incorporation in EdU (200 µM) was done directly in seawater, for 30 min prior to tissue fixation. Labelling was done after rehydration, using the Click-iT® 488/555 EdU kit, following the manufacturer's instructions (Molecular Probes). Embryos were counterstained with Hoechst or DAPI (1 µg ml−1) for nuclear staining. For TUNEL assays, after embryo rehydration, cuticle digestion and post-fixation, the terminal deoxynucleotide transferase reaction was performed following the protocol of the Click-iT TUNEL kit (Molecular Probes), with previously described minor modifications [34]. Embryos used for positive control were incubated with DNAse I to generate DNA breaks (similar incubation, 1 h on ice followed by 1 h at 37°C). Labelling was then performed following the protocol of the Click-iT TUNEL kit (Molecular Probes). For cell counting of EdU experiments, confocal stacks were obtained for five control embryos and five embryos treated with each of the three different drugs. To enumerate precisely all cells in a given larval region of interest (ROI), all embryos were registered with ITK4.0 as described above to the 48 hpf anatomic reference template. ROIs corresponding to the neurectoderm, episphere, chaetal sacs and stomodeum were delimitated using the 48 hpf template and the three-dimensional surface tool of Imaris software (Bitplane). DAPI- and EdU-positive nuclei were then counted for each registered sample stack and each ROI using Imaris.
5.8. Imaging
Bright field images were taken on a Leica microscope. Classical confocal images were taken with a Leica SP5 confocal microscope. Adjustments of brightness, contrast and Z projections were performed using the ImageJ software. Confocal images for registration procedure were taken in a Zeiss LSM 710 confocal microscope. Three-dimensional surface of expression patterns and associated videos were performed using, respectively, the surface and animation tools of Imaris. Co-localization stacks of expression patterns were obtained with the Coloc tool of Imaris and co-localization images were then edited on ImageJ. Live imaging of the 24 hpf FM4-64 labelled embryos was performed on a Zeiss LSM 780 confocal microscope with a 63× water immersion objective. Z-stacks (z = 0.53 µM) of several whole embryos were performed every hour (using the multiposition option). The figure panels were compiled using Adobe Illustrator and Adobe Photoshop software.
5.9. Statistical analyses
To compare the number of embryos harbouring a normal versus abnormal chaetae arrangement (Y variable) in control and treated conditions (X variable), a Fisher's test (two sided) was performed to test the independency of X and Y. To compare the number of positive cells for several markers (CS, Crya, Fmrfa, serotonin and EdU) in control and treated embryos, a Welch's 2 sample t-test (two sided) was performed for each comparison, to test whether the means are equivalent between the two groups. Statistical tests were performed using the BioStaTGV online platform (http://marne.u707.jussieu.fr/biostatgv/?module=tests). All raw data are provided in the electronic supplementary material, table S1.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to the Genoscope and the EMBL for giving access to unpublished Platynereis ESTs and genomic sequences. We also acknowledge the platform ImagoSeine staff for help in confocal imagery and Imaris software use. The authors wish to especially thank Dr G. Jékely and Dr A. Asadulina for their help in adapting the registration tools in our laboratory. Dr G. Jékely is also thanked for sharing neuropeptide antibodies. We thank Dr P. Steinmetz and Dr N. Dray for sharing preliminary data. We also thank A. Guillou for his help with gene cloning at the beginning of this study, O. Tournière for her help with ISH of Hes2 and Vasa genes, Dr S. Schneider for the Chitin synthase clone, Dr A. Zakrzewski for her expertise on chaetogenesis and Dr D. Aurelle for his help with statistic tests. We express our gratitude to Prof. Michel Vervoort and Dr Pierre Kerner for support and valuable suggestions on the study/manuscript.
Data accessibility
The data from this study are available as the electronic supplementary material.
Authors' contributions
E.G. carried out most of the molecular experiments: gene cloning, in situ hybridizations, antibody staining, EdU, Tunel experiments and drug treatments. G.B. adapted the three-dimensional pattern registration computer tools and generated the average reference larvae. E.G. generated average expression data for image registration (excepted Neurogenin), confocal imaging, FM4-64 staining, cell counting and three-dimensional surface rendering. Q.I.B.L. performed Neurogenin image registration. G.B. performed genome research and genomic analyses. E.G. performed phylogenetic analyses. E.G. and G.B. conceived and designed the study, analysed data, wrote the manuscript and were involved in editing the final version of the manuscript. All authors read and approved the final manuscript.
Competing interests
The author(s) declare that they have no competing interests.
Funding
This work was funded by the CNRS, the Agence Nationale de la Recherche (France) (ANR grant no. BLAN-0294 and ANR BLAN-METAMERE), the Institut Universitaire de France and the Who am I? laboratory of excellence (No.ANR-11-LABX-0071) funded by the French Government through its ‘Investments for the Future’ program operated by ANR under grant No.ANR-11-IDEX-0005-01.
References
- 1.Gazave E, Renard E. 2010. Evolution of Notch transmembrane receptors. Encyclopedia of Life Sciences (ELS). Chichester, UK: John Wiley & Sons Ltd; (doi:10.1002/9780470015902.a0022893) [Google Scholar]
- 2.Bray SJ. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. 7, 678–689. (doi:10.1038/nrm2009) [DOI] [PubMed] [Google Scholar]
- 3.Fortini ME. 2009. Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633–647. (doi:10.1016/j.devcel.2009.03.010) [DOI] [PubMed] [Google Scholar]
- 4.Kopan R, Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233. (doi:10.1016/j.cell.2009.03.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon WR, Arnett KL, Blacklow SC. 2008. The molecular logic of Notch signaling--a structural and biochemical perspective. J. Cell Sci. 121, 3109–3119. (doi:10.1242/jcs.035683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. 2003. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proc. Natl Acad. Sci. USA 100, 7638–7643. (doi:10.1073/pnas.1230693100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. 2009. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol. Biol. 9, 249 (doi:10.1186/1471-2148-9-249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hori K, Sen A, Artavanis-Tsakonas S. 2013. Notch signaling at a glance. J. Cell Sci. 126, 2135–2140. (doi:10.1242/jcs.127308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. 2007. Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 19, 166–175. (doi:10.1016/j.ceb.2007.02.012) [DOI] [PubMed] [Google Scholar]
- 10.Schwanbeck R, Martini S, Bernoth K, Just U. 2011. The Notch signaling pathway: molecular basis of cell context dependency. Eur. J. Cell Biol. 90, 572–581. (doi:10.1016/j.ejcb.2010.10.004) [DOI] [PubMed] [Google Scholar]
- 11.Poellinger L, Lendahl U. 2008. Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Curr. Opin. Genet. Dev. 18, 449–454. (doi:10.1016/j.gde.2008.07.013) [DOI] [PubMed] [Google Scholar]
- 12.Hartenstein V, Stollewerk A. 2015. The evolution of early neurogenesis. Dev. Cell 32, 390–407. (doi:10.1016/j.devcel.2015.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egger B, Chell JM, Brand AH. 2008. Insights into neural stem cell biology from flies. Phil. Trans. R. Soc. B 363, 39–56. (doi:10.1098/rstb.2006.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarman A. 2013. Neurogenesis in Drosophila. Encyclopedia of Life Sciences (ELS). Chichester, UK: John Wiley & Sons Ltd: (doi:10.1002/9780470015902.a0000825.pub2) [Google Scholar]
- 15.Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. 2002. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol. Cell. Neurosci. 21, 285–300. (doi:10.1006/mcne.2002.1174) [DOI] [PubMed] [Google Scholar]
- 16.Latasa MJ, Cisneros E, Frade JM. 2009. Cell cycle control of Notch signaling and the functional regionalization of the neuroepithelium during vertebrate neurogenesis. Int. J. Dev. Biol. 53, 895–908. (doi:10.1387/ijdb.082721ml) [DOI] [PubMed] [Google Scholar]
- 17.Formosa-Jordan P, Ibanes M, Ares S, Frade JM. 2013. Lateral inhibition and neurogenesis: novel aspects in motion. Int. J. Dev. Biol. 57, 341–350. (doi:10.1387/ijdb.120259jf) [DOI] [PubMed] [Google Scholar]
- 18.Neves J, Abello G, Petrovic J, Giraldez F. 2013. Patterning and cell fate in the inner ear: a case for Notch in the chicken embryo. Dev. Growth Differ. 55, 96–112. (doi:10.1111/dgd.12016) [DOI] [PubMed] [Google Scholar]
- 19.Arendt D, Nubler-Jung K. 1999. Comparison of early nerve cord development in insects and vertebrates. Development 126, 2309–2325. [DOI] [PubMed] [Google Scholar]
- 20.Denes AS, Jekely G, Steinmetz PR, Raible F, Snyman H, Prud'homme B, Ferrier DE, Balavoine G, Arendt D. 2007. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129, 277–288. (doi:10.1016/j.cell.2007.02.040) [DOI] [PubMed] [Google Scholar]
- 21.Marlow H, Roettinger E, Boekhout M, Martindale MQ. 2012. Functional roles of Notch signaling in the cnidarian Nematostella vectensis. Dev. Biol. 362, 295–308. (doi:10.1016/j.ydbio.2011.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Layden MJ, Martindale MQ. 2014. Non-canonical Notch signaling represents an ancestral mechanism to regulate neural differentiation. Evodevo 5, 30 (doi:10.1186/2041-9139-5-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelava I, Rentzsch F, Technau U. 2015. Evolution of eumetazoan nervous systems: insights from cnidarians. Phil. Trans. R. Soc. B 370, 20150065 (doi:10.1098/rstb.2015.0065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards GS, Rentzsch F. 2015. Regulation of Nematostella neural progenitors by SoxB, Notch and bHLH genes. Development 142, 3332–3342. (doi:10.1242/dev.123745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera AS, Gonsalves FC, Song MH, Norris BJ, Weisblat DA. 2005. Characterization of Notch-class gene expression in segmentation stem cells and segment founder cells in Helobdella robusta (Lophotrochozoa; Annelida; Clitellata; Hirudinida; Glossiphoniidae). Evol. Dev. 7, 588–599. (doi:10.1111/j.1525-142X.2005.05062.x) [DOI] [PubMed] [Google Scholar]
- 26.Rivera AS, Weisblat DA. 2009. And Lophotrochozoa makes three: Notch/Hes signaling in annelid segmentation. Dev. Genes Evol. 219, 37–43. (doi:10.1007/s00427-008-0264-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thamm K, Seaver EC. 2008. Notch signaling during larval and juvenile development in the polychaete annelid Capitella sp. I. Dev. Biol. 320, 304–318. (doi:10.1016/j.ydbio.2008.04.015) [DOI] [PubMed] [Google Scholar]
- 28.Raible F, et al. 2005. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science 310, 1325–1326. (doi:10.1126/science.1119089) [DOI] [PubMed] [Google Scholar]
- 29.Dray N, et al. 2010. Hedgehog signaling regulates segment formation in the annelid Platynereis. Science 329, 339–342. (doi:10.1126/science.1188913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauri A, Brunet T, Handberg-Thorsager M, Fischer AH, Simakov O, Steinmetz PR, Tomer R, Keller PJ, Arendt D. 2014. Development of the annelid axochord: insights into notochord evolution. Science 345, 1365–1368. (doi:10.1126/science.1253396) [DOI] [PubMed] [Google Scholar]
- 31.Fischer AH, Henrich T, Arendt D. 2010. The normal development of Platynereis dumerilii (Nereididae, Annelida). Front. Zool. 7, 31 (doi:10.1186/1742-9994-7-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marlow H, Tosches MA, Tomer R, Steinmetz PR, Lauri A, Larsson T, Arendt D. 2014. Larval body patterning and apical organs are conserved in animal evolution. BMC Biol. 12, 7 (doi:10.1186/1741-7007-12-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simionato E, Kerner P, Dray N, Le Gouar M, Ledent V, Arendt D, Vervoort M. 2008. atonal- and achaete-scute-related genes in the annelid Platynereis dumerilii: insights into the evolution of neural basic-Helix-Loop-Helix genes. BMC Evol. Biol. 8, 170 (doi:10.1186/1471-2148-8-170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demilly A, Steinmetz P, Gazave E, Marchand L, Vervoort M. 2013. Involvement of the Wnt/beta-catenin pathway in neurectoderm architecture in Platynereis dumerilii. Nat. Commun. 4, 1915 (doi:10.1038/ncomms2915) [DOI] [PubMed] [Google Scholar]
- 35.Krebs LT, Deftos ML, Bevan MJ, Gridley T. 2001. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the Notch signaling pathway. Dev. Biol. 238, 110–119. (doi:10.1006/dbio.2001.0408) [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen SL, Holland LZ, Schubert M, Beaster-Jones L, Holland ND. 2007. Amphioxus AmphiDelta: evolution of Delta protein structure, segmentation, and neurogenesis. Genesis 45, 113–122. (doi:10.1002/dvg.20278) [DOI] [PubMed] [Google Scholar]
- 37.Kerner P, Simionato E, Le Gouar M, Vervoort M. 2009. Orthologs of key vertebrate neural genes are expressed during neurogenesis in the annelid Platynereis dumerilii. Evol. Dev. 11, 513–524. (doi:10.1111/j.1525-142X.2009.00359.x) [DOI] [PubMed] [Google Scholar]
- 38.Tomer R, Denes AS, Tessmar-Raible K, Arendt D. 2010. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 142, 800–809. (doi:10.1016/j.cell.2010.07.043) [DOI] [PubMed] [Google Scholar]
- 39.Zakrzewski AC. 2011. Molecular characterization of chaetae formation in annelida and other Lophotrochozoa. PhD thesis, Freien Universität Berlin, Berlin, Germany. [Google Scholar]
- 40.Richter S, et al. 2010. Invertebrate neurophylogeny: suggested terms and definitions for a neuroanatomical glossary. Front. Zool. 7, 29 (doi:10.1186/1742-9994-7-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conzelmann M, Jekely G. 2012. Antibodies against conserved amidated neuropeptide epitopes enrich the comparative neurobiology toolbox. Evodevo 3, 23 (doi:10.1186/2041-9139-3-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gazave E, Behague J, Laplane L, Guillou A, Preau L, Demilly A, Balavoine G, Vervoort M. 2013. Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularly related to primordial germ cells. Dev. Biol. 382, 246–267. (doi:10.1016/j.ydbio.2013.07.013) [DOI] [PubMed] [Google Scholar]
- 43.Dove H, Stollewerk A. 2003. Comparative analysis of neurogenesis in the myriapod Glomeris marginata (Diplopoda) suggests more similarities to chelicerates than to insects. Development 130, 2161–2171. (doi:10.1242/dev.00442) [DOI] [PubMed] [Google Scholar]
- 44.Stollewerk A, Schoppmeier M, Damen WG. 2003. Involvement of Notch and Delta genes in spider segmentation. Nature 423, 863–865. (doi:10.1038/nature01682) [DOI] [PubMed] [Google Scholar]
- 45.Eriksson BJ, Ungerer P, Stollewerk A. 2013. The function of Notch signalling in segment formation in the crustacean Daphnia magna (Branchiopoda). Dev. Biol. 383, 321–330. (doi:10.1016/j.ydbio.2013.09.021) [DOI] [PubMed] [Google Scholar]
- 46.Muskavitch MA. 1994. Delta–Notch signaling and Drosophila cell fate choice. Dev. Biol. 166, 415–430. (doi:10.1006/dbio.1994.1326) [DOI] [PubMed] [Google Scholar]
- 47.Louvi A, Artavanis-Tsakonas S. 2006. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93–102. (doi:10.1038/nrn1847) [DOI] [PubMed] [Google Scholar]
- 48.Golde TE, Koo EH, Felsenstein KM, Osborne BA, Miele L. 2013. γ-Secretase inhibitors and modulators. Biochim. Biophys. Acta 1828, 2898–2907. (doi:10.1016/j.bbamem.2013.06.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasini A, Amiel A, Rothbacher U, Roure A, Lemaire P, Darras S. 2006. Formation of the ascidian epidermal sensory neurons: insights into the origin of the chordate peripheral nervous system. PLoS Biol. 4, e225 (doi:10.1371/journal.pbio.0040225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hausen H. 2005. Chaetae and chaetogenesis in polychaetes (Annelida). Hydrobiologia 535–536, 37–52. (doi:10.1007/s10750-004-1836-8) [Google Scholar]
- 51.O'Clair RM, Cloney RA. 1974. Patterns of morphogenesis mediated by dynamic microvilli: Chaetogenesis in Nereis vexillosa. Cell. Tissue Res. 151, 141–157. (doi:10.1007/BF00222219) [DOI] [PubMed] [Google Scholar]
- 52.Asadulina A, Panzera A, Veraszto C, Liebig C, Jekely G. 2012. Whole-body gene expression pattern registration in Platynereis larvae. Evodevo 3, 27 (doi:10.1186/2041-9139-3-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gazave E, Guillou A, Balavoine G. 2014. History of a prolific family: the Hes/Hey-related genes of the annelid Platynereis. Evodevo 5, 29 (doi:10.1186/2041-9139-5-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfeifer K, Schaub C, Wolfstetter G, Dorresteijn A. 2013. Identification and characterization of a twist ortholog in the polychaete annelid Platynereis dumerilii reveals mesodermal expression of Pdu-twist. Dev. Genes Evol. 223, 319–328. (doi:10.1007/s00427-013-0448-6) [DOI] [PubMed] [Google Scholar]
- 55.Wang M, Wu L, Wang L, Xin X. 2010. Down-regulation of Notch1 by gamma-secretase inhibition contributes to cell growth inhibition and apoptosis in ovarian cancer cells A2780. Biochem. Biophys. Res. Commun. 393, 144–149. (doi:10.1016/j.bbrc.2010.01.103) [DOI] [PubMed] [Google Scholar]
- 56.Demilly A, Simionato E, Ohayon D, Kerner P, Garces A, Vervoort M. 2011. Coe genes are expressed in differentiating neurons in the central nervous system of protostomes. PLoS ONE 6, e21213 (doi:10.1371/journal.pone.0021213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikolaou N, Watanabe-Asaka T, Gerety S, Distel M, Koster RW, Wilkinson DG. 2009. Lunatic fringe promotes the lateral inhibition of neurogenesis. Development 136, 2523–2533. (doi:10.1242/dev.034736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizoguchi T, Togawa S, Kawakami K, Itoh M. 2011. Neuron and sensory epithelial cell fate is sequentially determined by Notch signaling in zebrafish lateral line development. J. Neurosci. 31, 15 522–15 530. (doi:10.1523/JNEUROSCI.3948-11.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGraw HF, Snelson CD, Prendergast A, Suli A, Raible DW. 2012. Postembryonic neuronal addition in zebrafish dorsal root ganglia is regulated by Notch signaling. Neural Dev. 7, 23 (doi:10.1186/1749-8104-7-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dyer C, Linker C, Graham A, Knight R. 2014. Specification of sensory neurons occurs through diverse developmental programs functioning in the brain and spinal cord. Dev. Dyn. 243, 1429–1439. (doi:10.1002/dvdy.24184) [DOI] [PubMed] [Google Scholar]
- 61.Putnam NH, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071. (doi:10.1038/nature06967) [DOI] [PubMed] [Google Scholar]
- 62.Mara A, Schroeder J, Holley SA. 2008. Two deltaC splice-variants have distinct signaling abilities during somitogenesis and midline patterning. Dev. Biol. 318, 126–132. (doi:10.1016/j.ydbio.2008.03.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cau E, Blader P. 2009. Notch activity in the nervous system: to switch or not switch? Neural Dev. 4, 36 (doi:10.1186/1749-8104-4-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Groves AK, LaBonne C. 2014. Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev. Biol. 389, 2–12. (doi:10.1016/j.ydbio.2013.11.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu S, Wildonger J, Barshow S, Younger S, Huang Y, Lee T. 2012. The bHLH repressor Deadpan regulates the self-renewal and specification of Drosophila larval neural stem cells independently of Notch. PLoS ONE 7, e46724 (doi:10.1371/journal.pone.0046724) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eriksson BJ, Stollewerk A. 2010. The morphological and molecular processes of onychophoran brain development show unique features that are neither comparable to insects nor to chelicerates. Arthropod Struct. Dev. 39, 478–490. (doi:10.1016/j.asd.2010.07.005) [DOI] [PubMed] [Google Scholar]
- 67.de Sena Oliveira I, Tait NN, Strubing I, Mayer G. 2013. The role of ventral and preventral organs as attachment sites for segmental limb muscles in Onychophora. Front. Zool. 10, 73 (doi:10.1186/1742-9994-10-73) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chao MY, Larkins-Ford J, Tucey TM, Hart AC. 2005. lin-12 Notch functions in the adult nervous system of C. elegans. BMC Neurosci. 6, 45 (doi:10.1186/1471-2202-6-45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei Z, Angerer RC, Angerer LM. 2011. Direct development of neurons within foregut endoderm of sea urchin embryos. Proc. Natl Acad. Sci. USA 108, 9143–9147. (doi:10.1073/pnas.1018513108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu TM, Luo YJ, Yu JK. 2012. BMP and Delta/Notch signaling control the development of amphioxus epidermal sensory neurons: insights into the evolution of the peripheral sensory system. Development 139, 2020–2030. (doi:10.1242/dev.073833) [DOI] [PubMed] [Google Scholar]
- 71.Akanuma T, Hori S, Darras S, Nishida H. 2002. Notch signaling is involved in nervous system formation in ascidian embryos. Dev. Genes Evol. 212, 459–472. (doi:10.1007/s00427-002-0264-x) [DOI] [PubMed] [Google Scholar]
- 72.Munder S, et al. 2013. Notch-signalling is required for head regeneration and tentacle patterning in Hydra. Dev. Biol. 383, 146–157. (doi:10.1016/j.ydbio.2013.08.022) [DOI] [PubMed] [Google Scholar]
- 73.Richards GS, Degnan BM. 2012. The expression of Delta ligands in the sponge Amphimedon queenslandica suggests an ancient role for Notch signaling in metazoan development. Evodevo 3, 15 (doi:10.1186/2041-9139-3-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakarya O, Armstrong KA, Adamska M, Adamski M, Wang IF, Tidor B, Degnan BM, Oakley TH, Kosik KS. 2007. A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE 2, e506 (doi:10.1371/journal.pone.0000506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Renard E, Vacelet J, Gazave E, Lapébie P, Borchiellini C, Ereskovsky AV. 2009. Origin of the neuro-sensory system: new and expected insights from sponges. Integr. Zool. 4, 294–308. (doi:10.1111/j.1749-4877.2009.00167.x) [DOI] [PubMed] [Google Scholar]
- 76.Struck TH, et al. 2011. Phylogenomic analyses unravel annelid evolution. Nature 471, 95–98. (doi:10.1038/nature09864) [DOI] [PubMed] [Google Scholar]
- 77.Hejnol A, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. R. Soc. B 276, 4261–4270. (doi:10.1098/rspb.2009.0896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Podsiadlowski L, Braband A, Struck TH, von Dohren J, Bartolomaeus T. 2009. Phylogeny and mitochondrial gene order variation in Lophotrochozoa in the light of new mitogenomic data from Nemertea. BMC Genomics 10, 364 (doi:10.1186/1471-2164-10-364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kulakova M, et al. 2007. Hox gene expression in larval development of the polychaetes Nereis virens and Platynereis dumerilii (Annelida, Lophotrochozoa). Dev. Genes Evol. 217, 39–54. (doi:10.1007/s00427-006-0119-y) [DOI] [PubMed] [Google Scholar]
- 80.Saudemont A, Dray N, Hudry B, Le Gouar M, Vervoort M, Balavoine G. 2008. Complementary striped expression patterns of NK homeobox genes during segment formation in the annelid Platynereis. Dev. Biol. 317, 430–443. (doi:10.1016/j.ydbio.2008.02.013) [DOI] [PubMed] [Google Scholar]
- 81.Gillis WJ, Bowerman B, Schneider SQ. 2007. Ectoderm- and endomesoderm-specific GATA transcription factors in the marine annelid Platynereis dumerilli. Evol. Dev. 9, 39–50. (doi:10.1111/j.1525-142X.2006.00136.x) [DOI] [PubMed] [Google Scholar]
- 82.Erwin DH, Davidson EH. 2002. The last common bilaterian ancestor. Development 129, 3021–3032. [DOI] [PubMed] [Google Scholar]
- 83.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. (doi:10.1093/bioinformatics/16.10.944) [DOI] [PubMed] [Google Scholar]
- 84.Marchler-Bauer A, et al. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35, D237–D240. (doi:10.1093/nar/gkl951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. (doi:10.1093/nar/gki442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5, 113 (doi:10.1186/1471-2105-5-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. (doi:10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hall TA. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 89.Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33, W557–W559. (doi:10.1093/nar/gki352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. (doi:10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 91.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. (doi:10.1093/molbev/msn067) [DOI] [PubMed] [Google Scholar]
- 92.Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. (doi:10.1080/10635150600755453) [DOI] [PubMed] [Google Scholar]
- 93.Dorresteijn AWC, O'Grady B, Fischer A, Porchet-Henere E, Boilly-Marer Y. 1993. Molecular specification of cell lines in the embryo of Platynereis (Annelida). Rouxs Arch. Dev. Biol. 202, 264–273. (doi:10.1007/BF00363215) [DOI] [PubMed] [Google Scholar]
- 94.Tessmar-Raible K, Steinmetz PR, Snyman H, Hassel M, Arendt D. 2005. Fluorescent two-color whole mount in situ hybridization in Platynereis dumerilii (Polychaeta, Annelida), an emerging marine molecular model for evolution and development. Biotechniques 39, 462–464. (doi:10.2144/000112023) [DOI] [PubMed] [Google Scholar]
- 95.Jekely G, Arendt D. 2007. Cellular resolution expression profiling using confocal detection of NBT/BCIP precipitate by reflection microscopy. Biotechniques 42, 751–755. (doi:10.2144/000112462) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from this study are available as the electronic supplementary material.