Abstract
Human lactoferrin, a component of the innate immune system, kills a wide variety of microorganisms including the Gram positive bacteria Streptococcus pneumoniae. Pneumococcal surface protein A (PspA) efficiently inhibits this bactericidal action. The crystal structure of a complex of the lactoferrin-binding domain of PspA with the N-lobe of human lactoferrin reveals direct and specific interactions between the negatively charged surface of PspA helices and the highly cationic lactoferricin moiety of lactoferrin. Binding of PspA blocks surface accessibility of this bactericidal peptide preventing it from penetrating the bacterial membrane. Results of site-directed mutagenesis, in vitro protein binding assays and isothermal titration calorimetry measurements corroborate that the specific electrostatic interactions observed in the crystal structure represent major associations between PspA and lactoferrin. The structure provides a snapshot of the protective mechanism utilized by pathogens against the host’s first line of defense. PspA represents a major virulence factor and a promising vaccine candidate. Insights from the structure of the complex have implications for designing therapeutic strategies for treatment and prevention of pneumococcal diseases that remain a major public health problem worldwide.
Keywords: host–pathogen interaction, immunity, lactoferricin, lactoferrin
Introduction
Survival of a microorganism inside the host depends to a large extent on its ability to avoid, escape or counter the host’s defense mechanisms. The host’s first line of defense against microbial infection relies on the mechanisms of its innate immunity. As part of the innate immune system the body fluids and organized tissues of animals contain a number of naturally produced antimicrobial agents that kill various microbes or inhibit their growth. A major component of this regimen is a multifunctional iron-binding glycoprotein, lactoferrin (LF), found in milk and other exocrine secretions, which plays an active role in host defense. LF’s antimicrobial activity, both bacteriostatic and bactericidal, against various pathogens including bacteria, fungi and viruses has been clearly established.1–3
LF is secreted into the mucosal fluids.4 Therefore, bacteria that colonize mucosal surfaces are exposed to LF and must protect themselves from this antimicrobial agent. Various pathogenic species that live in the mucosal environment have developed different mechanisms for adaptation. For example, Gram negative bacteria of the Neisseriaceae family use specific receptor systems for acquiring iron from LF5,6 while various strains of the Gram positive bacterium Streptococcus pneumoniae bind LF via a surface protein called pneumococcal surface protein A (PspA).7,8
S. pneumoniae is one of the most important pathogens affecting human and is responsible for causing at least million deaths among children worldwide annually†. PspA is an important virulence factor expressed on the surface of all pathogenic strains of S. pneumoniae.9 PspA reduces the killing of pneumococci by LF.10 The protective role of PspA is apparent from the demonstration of increased susceptibility to LF in pneumococcal strains lacking PspA as compared to the wild-type bacteria.10 Furthermore, anti-PspA antibodies enhance the bactericidal effect of LF against wild-type pneumococci.10 Although LFs from various mammalian species are highly homologous in their amino acid sequence, and biochemical and structural properties, PspA exhibits preferential binding to human LF (hLF) as compared to the bovine homolog (bLF).8 Since S. pneumoniae is a human pathogen, the preference of PspA for hLF suggests a functional significance for this protein–protein interaction.
LF is a member of the transferrin family of proteins and, as in other transferrins, the N and C-terminal halves of LF form two homologous lobes referred to as the N and C-lobes. Each lobe contains one iron binding site situated in a deep cleft.11 The low iron saturation level of naturally occurring LF and its extremely high binding affinity for iron allows it to sequester free iron from body secretions and neutrophils depriving the pathogen of this essential metal, thus giving rise to the bacteiostatic effect.12–14 However, the bactericidal activity of LF is independent of iron, but for reasons not well understood is restricted only to its iron-free or apo form.3,10,15 This function of apo-LF presumably results from direct interaction with the bacterial surface.
Since Ellison et al. first demonstrated that LF damages the outer membrane of Gram negative bacteria16 it has been proposed that a highly cationic domain located at the N terminus of LF is responsible for its bactericidal activity.17 The N terminus of LF from human and other mammals contains positively charged bioactive peptides, collectively known as lactoferricins (Lfcn), that are released in the stomach and mucosal secretions through proteolytic digestion of LF.18,19 The composition of these peptides is characterized by a relatively large proportion of basic amino acids and a number of hydrophobic residues, especially tryptophan, that render them uniquely suitable for interacting with bacterial lipopolysaccharides and disrupting the membrane structure.20,21 Subsequent studies demonstrated that lactoferricin peptides, LfcnH and LfcnB, derived from hLF and bLF, respectively, exhibit potent antimicrobial activity against many bacteria, fungi and viruses22,23 although the exact mechanism of this action remains elusive. Recently we showed that a number of shorter peptides derived from LfcnH possess potent bactericidal activity against pneumococci.10 The central role of PspA in shielding the pneumococci from the lethal action of hLF is underscored by the observation that PspA, either as a component of the bacterial surface or as a recombinant protein, also protects the bacteria from killing by LfcnH peptides.10
PspA is a mutidomain protein which remains anchored to the pneumococcal cell wall through a C-terminal choline binding domain.24 The primary sequence of PspA shows no significant homology to any other protein in the database except a second choline binding surface protein of S. pneumoniae, PspC. The molecular mass of PspA ranges between 67 kDa and 98 kDa in various pneumococcal strains. The N-terminal half of mature PspA is predicted to be entirely α-helical.25 A lactoferrin-binding region of PspA has been localized within residues 168–288 of this α-helical domain.8 However, the nature of molecular interactions between PspA and LF remains unknown. Moreover, the PspA binding site on LF is not known. It is also not known if PspA binds to one or both lobes of LF.
Data presented here show that PspA binds to only the N-lobe of hLF. In order to gain a detailed understanding of the molecular interactions that protect S. pneumoniae and possibly other pathogens from the bactericidal effect of LF, we have determined the crystal structure of the complex of the LF-binding domain of PspA with the N-lobe of human LF. This is the first structure of LF (or either of its lobes) bound to a protein molecule. The structure of the complex reveals that through a set of specific interactions PspA binds to the lactoferricin domain. Results of this structure analysis contradict a previously proposed model.26 Using site-directed alanine mutations we confirmed that the protein–protein interactions identified in the crystal structure represent the major association between LF and PspA in solution and that the only binding site for PspA on hLF is located on the lactoferricin domain. The structure provides an insight into the mechanism by which this surface protein of pneumococci protects the bacteria from the host’s first line of defense.
Results
In vitro binding of PspA and LF
We cloned, expressed, and purified two truncated PspA fragments and two recombinant versions of human lactoferrin, the N-lobe (NLF) and full length (rhLF), as specified in Materials and Methods. PspA1 represents the complete N-terminal α-helical domain of mature PspA from strain Rx1 (residues 1–288). PspA2 contains the region (residues 168–288) that has been shown to be necessary for binding human LF.8 NLF consists of amino acid residues 1–343 of the mature hLF sequence, which include the N-lobe and the hinge domain between the N and C-lobes. Using an in vitro protein-binding assay with purified PspA1 and PspA2 and LF preparations, we conclude that both recombinant forms of PspA bind NLF as well as rhLF (Supplementary Data, Figure S1). Isothermal titration calorimetry (ITC) data for binding of PspA fragments with rhLF and NLF fit well to a single binding site model. The calculated stoichiometries for binding of PspA1: rhLF, PspA1:NLF and PspA2:rhLF are 1:1 (0.93, 0.88 and 1.14, respectively). The dissociation constants computed for these three complexes (4.8(±1.6), 10.3(±3.2) and 6.0(±2.3) nM, respectively) are consistent with that determined for binding of radio-labeled LF to pneumococci.7 Based on the results of size exclusion chromatography and ITC, PspA1 and PspA2 each form a 1:1 complex with NLF and with hLF. Since PspA2 and NLF represent the minimum binding domain of each component, a complex of PspA2 with NLF was subjected to structural investigation.
Structure of PspA2 in the complex with NLF
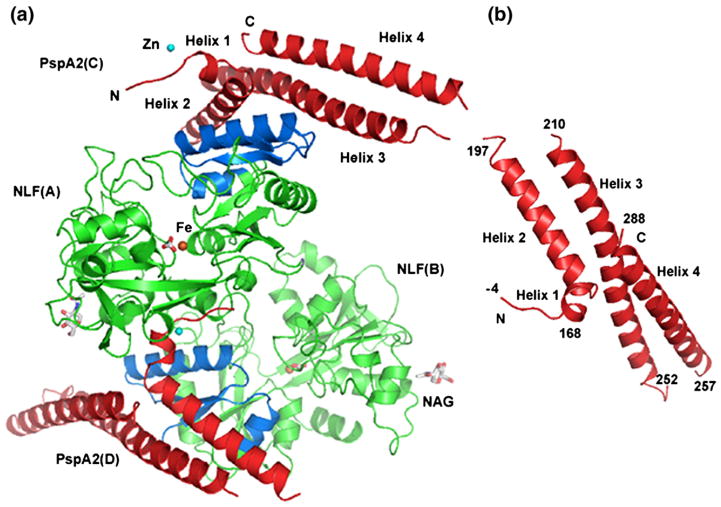
The PspA2:NLF complex crystallized in the space group P32. The asymmetric unit contains two copies of a 1:1 complex of NLF and PspA2 related by a non-crystallographic 2-fold symmetry (Figure 1(a)). The two molecules of each protein in the asymmetric unit are very similar to each other. The calculated r.m.s. deviations for all the Cα atoms of the NLF chains A and B, and of the PspA2 chains C and D are 0.1 Å and 0.2 Å, respectively. Data collection and refinement statistics are presented in Table 1.
Figure 1.
Structure of the PspA2:NLF complex. (a) Asymmetric unit content. LF N-lobes are shown in green (Lfcn peptide is colored blue), PspA2 molecules in red. One Fe ion (orange) is bound to each NLF. The coordinating carbonate ligand is shown in sticks. One Zn ion (cyan) is bound at the N-terminal end of each PspA2 molecule. The N-acetylglucosamine molecule is labeled NAG. (b) PspA2 structure. Residue numbering for PspA2 helices are shown.
Table 1.
Crystal parameters, X-ray data collection and refinement statistics
| Space group | P32 |
| Cell dimensions | |
| a, b, c (Å) | 130.18, 130.18, 80.80 |
| α, β, γ (°) | 90, 90, 120 |
| Data collection statistics | |
| Resolution (Å) | 15.0–2.91 (3.0–2.91)a |
| No. unique reflections (total set) | 33,481 (3355) |
| Rsym | 0.098 (0.36) |
| I/σ | 7.0 (3.0) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 2.9 (2.9) |
| Refinement and model statistics | |
| Resolution (Å) | 15.0–2.91 (3.0–2.91) |
| No. reflections used (working set) | 30, 285 |
| Rwork | 20.3 (26.2) |
| Rfree | 24.9 (33.3) |
| r.m.s.d. bond lengths (Å) | 0.009 |
| r.m.s.d. bond angles (°) | 1.16 |
| Average B-factor (Å2) (main-chain/side-chains/overall) | |
| Molecule A | 29.5/30.7/30.1 |
| Molecule B | 29.5/30.7/30.1 |
| Molecule C | 36.7/38.3/37.5 |
| Molecule D | 36.8/38.4/37.5 |
| Ramachandran statistics (%) | |
| Core region | 90.0 |
| Additional allowed region | 9.5 |
| Generously allowed | 0.3 |
| Disallowed | 0.3 |
Numbers in the parentheses are for the highest resolution bin.
LF structure
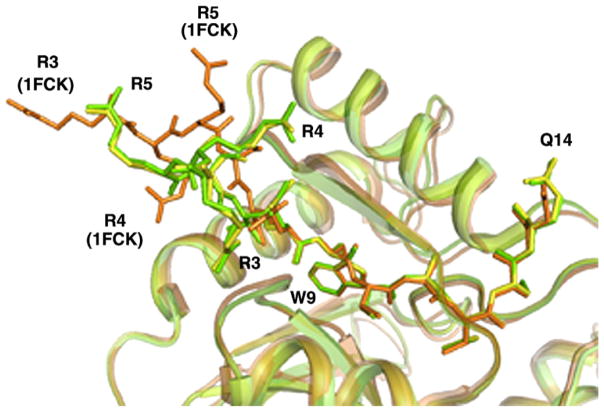
The structure of the LF N-lobe in the complex with PspA2 is very similar to the described structure of the iron-bound N-lobe27 (calculated r.m.s. deviation: 0.5 Å for all Cα atoms). There are two seven-strand β-sheets surrounded by a total of 11 α-helices. The structure of the N-lobe in the truncated form is almost identical to that in the full length LF.28–30 In the intact hLF, helix 11 of the N-lobe is joined by a loop to the short helix that links the two lobes. In the crystal structures of the truncated N-lobe the secondary structure at the C terminus varies depending on the stability of this region. In the present complex, although residues of the hinge domain are not visible in the electron density map, residues 317–331 form a helical structure similar to the one seen in the full length protein. The N-terminal four or five residues of hLF show some flexibility in various crystal structures. In the present complex the first three residues (Arg3, Arg4 and Arg5) in both NLF molecules (A and B) move noticeably away from the positions of the corresponding residues in other structures (Figure 2). As seen in Figure 2, both the main-chain and side-chains of these terminal residues of NLF deviate significantly in the complex although the rest of the structure remains unaffected by PspA2 binding. Compared to the previously reported hLF N-lobe structures,27,31 the only other significant difference is the conformation of His92, for which the side-chain assumes a different rotamer conformation. This new conformation allows the side-chain of His92 to form a hydrogen bond with the glycine residue (−4) in the N-terminal tag of a symmetry-related PspA2 molecule.
Figure 2.
Superposition of LF structures showing the movement of N-terminal residues in the PspA2: NLF complex. NLF chains (A and B, shown in yellow and green, respectively) are superposed on the full length hLF structure (PDB ID, 1FCK, shown in brown). The N-terminal 15 amino acid residues of each chain are shown in stick model. While terminal residues 3–5 of NLF in the complex shift considerably from the corresponding residues in free hLF, the remaining residues show no movement.
Although the complex was purified and crystallized without adding iron, electron density for one Fe3+ in the iron binding site of each NLF molecule (in the asymmetric unit) was clearly identifiable in the Fo–Fc map. Six chelating atoms are contributed by two carbonate oxygen atoms, two hydroxyl oxygen atoms from Tyr93 and Tyr193, one carboxyl oxygen atom from Asp61 and the NE2 from His254. The average Fe3+–O distance is 2.10 Å (range 1.74 Å–2.58 Å); the two Fe3+–N distances are 2.07 Å and 2.03 Å. There are three sulfate ions (possibly from the crystallization reagents) bound to the surface of each molecule of NLF. One N-acetyl glucosamine molecule has also been modeled into the electron density near the known glycosylation site, Asp138, in each N-lobe in the asymmetric unit.
PspA structure
The PspA2 molecule consists of four α-helices. Immediately following the vector-derived tetra-peptide (GSHM, numbered −4 to −1) at the N terminus there is a short helix (helix 1, residues 168–172) followed by three long helical segments (helix 2, residues 174–191; helix 3, 212–248; and helix 4, 258–288) connected by mobile loops (Figure 1(b)). Only the loop between the first two helices is visible in the structure; the other two are disordered. The three long helices are amphipathic with numerous hydrophobic interactions between neighboring antiparallel helices. The calculated inter-helix angle between helix 2 and helix 3 is −144° and between helix 3 and helix 4 is −158°‡. A plot of correlation coefficients of PspA2 residues is provided in Supplementary Data, Figure S2.
Structural comparison with the DALI database32 shows that the overall structure of PspA2 has similarity with the coiled-coil segments of many functionally unrelated proteins that are involved in various protein–protein interactions. In the SCOP database33 similar proteins containing at least two long antiparallel helices are classified as “antiparallel coiled coil”. The superfamilies of proteins in this class include among others the N-terminal domains of variant surface glycoproteins, colicins and some colicin receptors.
There was one unexplained peak (>5σ) in the electron density map near Asp168 in each PspA2 molecule at a site suitable for metal ion binding. After consideration of the coordination geometry and crystallographic parameters, we placed a Zn2+ at this site. It should be noted that LF is known to bind Zn2+ (among other metal ions)34 and the culture media from which recombinant NLF was purified contained Zn2+. Although zinc binding sites have been found in the C-lobe of bLF35 no such site has been identified in hLF. The quality of electron density at this site in both molecules is excellent (Supplementary Data, Figure S3). The octahedral coordination of each Zn2+ is satisfied by residues from PspA2 and NLF molecules (Supplementary Data, Figure S3). The biological relevance of this apparent zinc binding is not known.
Protein–protein interactions
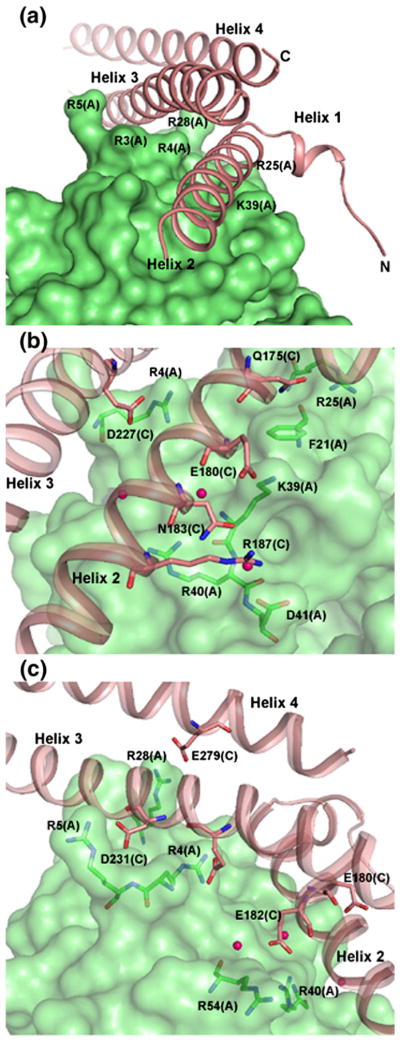
The PspA2 molecule binds to the surface of NLF. Helices 2 and 3 of PspA2 fit into two grooves on the N-lobe as shown in Figure 3(a). Side-chains from a number of NLF residues create the surface for binding the PspA2 helices; these residues include Arg3, Arg4, Arg28 and Arg31 for helix 3 and Ser36, Ile38, Lys39 and the aromatic ring of Phe21 for helix 2. Calculated buried surface areas for the two complexes (A:C and B:D) in the asymmetric unit are 1643 and 1650 Å2, respectively. The protein interface accessible surface area on the NLF molecules is ~818 Å2 while that on the PspA2 molecules is ~843 Å2. These represent approximately 5.35% and 9.99% of NLF and PspA2 surface, respectively. The protein–protein interface is formed by approximately 57% polar residues and 43% non-polar residues. The protein–protein interaction server§ identifies a total of 25 residues of PspA2 spanning the range 170–279 and 21 residues of NLF spanning the range of 3 to 54 as participating in the formation of the interface.
Figure 3.
Binding of PspA2 with the LF N-lobe. (a) Grooves on NLF for binding PspA2 helices. NLF is shown in surface drawing and PspA2 in ribbons. Several LF residues around the grooves are labeled. Helix 2 has the most interactions with NLF. (b) and (c) Close-up views of the intermolecular interactions between PspA2 and NLF. Amino acid residues for both PspA2 (salmon, labeled C) and NLF (green, labeled A) participating in binding are shown in sticks. Water molecules involved in bridging PspA2 and NLF are shown as red spheres.
As shown in Table 2 there are a total of 19 interactions between NLF(A) and PspA2(C) and 21 interactions between NLF(B) and PspA2(D). Residues from all three long helices of PspA2 contribute to binding (Figure 3(b) and (c)) with helix 2 providing six contacts (Table 2). The majority of the PspA residues involved in binding to hLF are acidic. However, three contacts are noticed between amide nitrogen atoms on PspA with oxygen atoms from residues on LfcnH domain (Table 2).
Table 2.
Intermolecular contacts between PspA and lactoferrin N-lobe
| Distance (Å)
|
||||
|---|---|---|---|---|
| PspA residue/atom | NLF residue/atom | Complex A:C | Complex B:D | |
| Glu171 OE2 | Gln14 NE2 | – | 3.15 | |
| Gln175 NE2 | Phe21 O | 3.04 | 3.25 | |
| Gln175 NE2 | Gln24 OE1 | 2.68 | 2.53 | |
| Gln175 OE1 | Arg25 NH1 | 3.14 | 3.06 | |
| Glu180 OE1 | Lys39 NZ | 3.05 | 2.92 | |
| Glu182 OE2 | Arg40 NH1 | 2.45 | 2.52 | |
| Asn183 ND2 | Lys39 O | 2.76 | 2.81 | |
| Glu227 OE1 | Arg4 NH2 | 3.04 | – | |
| Glu227 OE1 | Arg4 NH1 | 3.17 | 2.79 | |
| Glu227 OE | Arg4 NH1 | – | 3.17 | |
| Ser230 OG | Arg4 NH2 | 3.12 | 3.16 | |
| Asp231 OD1 | Arg5 NE | 3.23 | 3.14 | |
| Asp231 OD1 | Arg5 N | – | 3.15 | |
| Asp231 OD2 | Arg5 NE | 3.00 | 3.25 | |
| Asp231 OD2 | Arg5 NH2 | 3.27 | 2.91 | |
| Asp237 OD2 | Arg28 NE | 2.77 | 2.76 | |
| Glu279 OE1 | Arg28 NH2 | 2.97 | 3.10 | |
| Glu279 OE2 | Arg28 NH1 | 3.07 | 3.02 | |
| Gln 279 OE2 | Arg28 NH2 | – | 3.07 | |
| Contacts through bridging water molecules
| ||||
| Distance (Å)
|
||||
| PspA residue/atom | Water | NLF residue/atom | PspA2 … O | O … NLF |
|
| ||||
| Glu182 OE2(C) | 404 | Lys39 N(A) | 2.70 | 2.73 |
| Glu182 OE1(C) | 406 | Arg54 O(A) | 2.96 | 2.64 |
| Asn183 ND2(C) | 402 | Asp41 OD2(A) | 3.11 | 2.96 |
| Arg187 NH2(C) | 402 | 2.82 | ||
| Ala179 O(D) | 403 | Lys39 N(B) | 2.73 | 2.85 |
| Glu182 OE1(D) | 405 | Arg54 O(B) | 2.83 | 2.71 |
| Asn183 ND2(D) | 401 | Asp41 OD1(B) | 3.31 | 3.21 |
Interaction sites on the LF N-lobe are almost entirely confined to the lactoferricin region. Since the structure of the Lfcn domain remains unchanged in the apo-LF and holo-LF,36,37 similar protein–protein interactions are expected between PspA and both forms of LF. In the crystal structure of intact LF or its N-lobe, the Lfcn domain consists of two β-sheets flanked by two α-helices. There is no major change in the overall structure of N-lobe or Lfcn domain due to binding of PspA2 except for the first three N-terminal residues that move considerably with respect to their positions in uncomplexed LF (see Figure 2). Since resides Arg4 and Arg5 are involved in direct interactions with PspA2 this observed deviation may be induced by PspA2 binding. The side-chain of Arg4 is inserted between helix 2 and helix 3 to form two contacts with Ser230 and Glu227 in helix 3, while Arg5 of the Lfcn domain interacts with carboxyl group of Asp231 at the polar end of helix 3 of PspA2. It has been noted earlier that these three residues together with Arg25, Arg28, Arg29 and Arg31 from the C-terminal end of the first LF helix render this region a hot spot for binding to negatively charged surfaces of a variety of molecules including DNA and lipopolysaccharides.20,21
A total of six positively charged residues (Arg4, 5, 25, 28 and 40 and Lys39) of NLF directly interact with various residues in PspA2. In addition, the side-chain of Gln24 and the peptide oxygen atoms of Lys39 and Phe21 also interact with PspA2. Three water molecules are involved in bridging each NLF molecule with its binding partner PspA2 (Table 2). There are incidental contacts between the NLF(A) molecule and the N-terminal residues His-4, Ser-3 and Asp168 of PspA2(D) molecule. Two of these terminal residues of PspA2 (His-4 and Asp168) are also involved in coordination with the Zn2+.
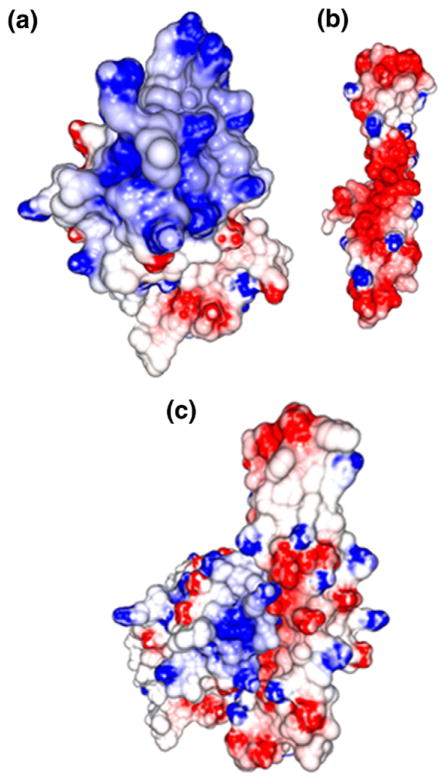
Results of our crystal structure analysis contradict a previously published model26 which proposed that a positive surface potential of PspA is responsible for LF binding and that the PspA binding grooves on LF are in the vicinity of the iron binding site. In fact, PspA binds far from the iron binding pocket (see Figure 1(a)) and its negatively charged surface interacts with LF thereby neutralizing the cationic Lfcn domain (Figure 4).
Figure 4.
Surface potentials: (a) NLF, (b) PspA2 and (c) PspA2:NLF complex. Surface potentials were calculated using the program GRASP2.38 Positive potential is depicted in blue and negative potential in red. Uneven distribution of positive charge is seen on NLF with a high concentration at the N-terminal end of the molecule. The PspA2 molecule shows a highly negatively charged surface.
Mutational analysis of PspA–LF interaction
In order to understand the relative contribution of different regions of PspA to the overall binding to LF, we conducted site-directed mutagenesis by selectively replacing one or more residues in each of the helices 2, 3 and 4 of PspA2 with alanine. Each residue selected for mutation was identified by the crystal structure as forming one or more contacts with NLF. Mutations were introduced in the DNA sequence for PspA1 (residues 1–288) and PspA2 (168–288) fragments. We expressed and purified mutant forms of both PspA1 and PspA2 and examined their ability to bind the N-lobe and full length rhLF.
After incubation of mutant proteins with purified NLF and rhLF, binding of PspA fragments with these proteins was monitored by electrophoresis on non-denaturing gels and Western blot analysis using an anti-PspA monoclonal antibody (Xi126).39 Due to high carbohydrate content and basic pI (8.4–9),2,11 LF does not migrate as a band on non-denaturing gel (Figure 5(a), lanes 7, N-lobe and 8, full length). However, when bound to PspA, both NLF and rhLF migrate as a single band representing the complex (Figure 5(a), lanes 2 and 3). A quadruple mutant of PspA1 (lane 4) containing two mutations in helix 2 (Glu180Ala, Glu182Ala) and one mutation in each of helix 3 (Asp237Ala) and helix 4 (Glu279Ala) does not form a complex with either NLF (lane 5) or intact rhLF (lane 6). The absence of binding interaction between PspA1 mutant and LF (both NLF and rhLF) was confirmed by Western blot (Figure 5(b)).
Figure 5.
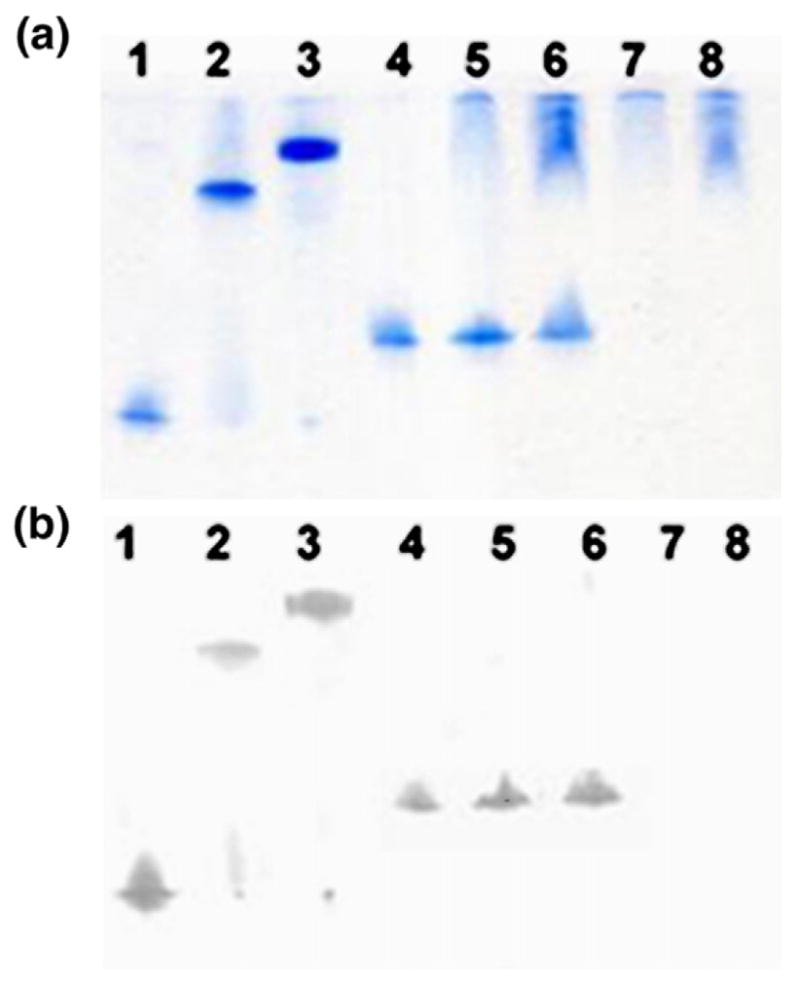
Analysis of binding of PspA1 mutant to LF. Various PspA1 residues (Glu180, Glu182, Asp237 and Glu279) involved in binding to NLF were mutated to alanine. The resulting mutant protein was expressed and used in binding studies. After incubation of PspA1 (wild-type and mutant) with LF (NLF and rhLF), the mixtures were subjected to gel electrophoresis on a non-denaturing gel. For Western blot analysis proteins were then transferred to a nitrocellulose membrane and detected using monoclonal mouse IgG Xi126 (as primary antibody) and alkaline phosphate-conjugated anti-mouse IgG antibody (as secondary ones). (a) Non-denaturing gel. (b) Western blot. 1, PspA1 (residues 1–288); 2, PspA1+ NLF; 3, PspA1+ rhLF; 4, PspA1 mutant; 5, PspA1 mutant+ NLF; 6, PspA1 mutant+ rhLF; 7, NLF; 8, rhLF.
In contrast, double mutants containing two mutations in helix 2 (Glu180Ala and Glu182Ala) or one mutation in helix 3 (Asp237Ala) and helix 4 (Glu279Ala), were still able to form a complex with rhLF and NLF (Supplementary Data, Figure S4). Three triple mutants (Glu180Ala, Glu182Ala, Asp237Ala), (Glu180Ala, Glu182Ala, Glu279Ala) and (Glu182Ala, Asp237Ala, Glu279Ala) lost the ability to form a complex, but another triple mutant (Glu180Ala, Asp237Ala, Glu279Ala) retained some binding capacity (Supplementary Data, Figure S4).
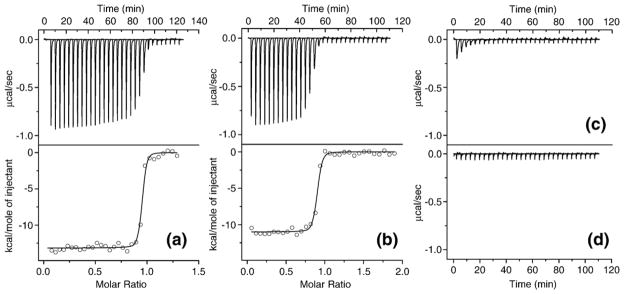
For quantitative assessment of binding interaction between rhLF or NLF with wild-type or mutant PspA fragments we performed ITC. There is no significant difference in thermodynamic parameters for binding of wild-type PspA1 with rhLF and NLF as shown in Figure 6(a) and (b), respectively. On the other hand, no interaction was observed when binding of rhLF with quadruple mutant of PspA1 (Glu180Ala, Glu182Ala, Asp237Ala, Glu279Ala) was measured under similar conditions. The titration curve in this case (Figure 6(c)) was comparable to that obtained for rhLF titration by buffer (Figure 6(d)). The titration curve for rhLF and PspA2 triple mutant (Glu180Ala, Asp237Ala, Glu279Ala) suggested some residual binding capacity though the interaction parameters could not be calculated accurately (data not shown). Therefore, ITC data are in good agreement with the results of gel shift experiments.
Figure 6.
Isothermal titration calorimetry data for the binding of different PspA and LF fragments. (a) rhLF (38 μM) was titrated by PspA1 (251 μM), (b) NLF (30 μM) was titrated by PspA1 (295 μM), (c) rhLF (29 μM) was titrated by PspA1 mutant (Glu180Ala, Glu182Ala, Asp237Ala, Glu279Ala) (233 μM), (d) rhLF (32 μM) was titrated by 50 mM sodium phosphate buffer (pH 7.5). The top panels of (a) and (b) and (c) and (d) show the heat released per second during the addition of 31×8 μl of PspA1 (native or mutant) or buffer to the cell containing rhLF or NLF. The bottom panels of (a) and (b) show the integrated binding isotherms with the continuous lines representing the fit of the data to a single site binding model. Binding enthalpy, ΔH: (a) −13.2(±0.1), (b) −11.0(±0.1) kcal/mol; binding entropy, ΔS: (a) −6.15, (b) −0.3 cal/(K mol); stoichiometry: (a) 0.932±0.002, (b) 0.875±0.003, and dissociation constant, KD: (a) 4.8(±1.6), (b) 10.3(±3.2) nM.
Results of our protein binding assays using various PspA mutants show that contributions from multiple helices are necessary for binding to LF and binding is not prevented by mutation of one binding residue in any helix of PspA. Moreover, mutation of the two binding residues on helix 2 (Glu180 and Glu182) or one binding residue on each of the PspA helices 3 (Asp237) and 4 (Glu279) to alanine, did not inhibit LF binding, showing that the protein–protein interactions between PspA and LF can withstand some degree of alterations on the PspA surface. However, replacing three binding residues (two in helix 2 and one in helix 3 or helix 4) with alanine was detrimental to binding. Similarly, mutation of Glu182 in helix 2 and one LF-binding residue in helices 3 and 4 inhibited complex formation between the triple mutant and LF. When Glu180 was mutated to alanine (instead of Glu182) the resulting triple mutant was still capable of binding LF, suggesting a relatively important contribution of Glu182, which is involved in multiple interactions with NLF (see Table 2).
All PspA1 and PspA2 mutants showed identical binding properties with full length rhLF and NLF, suggesting that LF C-lobe or PspA residues 1–167 did not contribute to binding. Results of binding assays were also similar when purified native human LF (Sigma) was used instead of recombinant protein, indicating that the difference in the carbohydrate composition of various LF preparations has no influence on binding to the PspA1 and PspA2 fragments (data not shown). Therefore, only specific protein–protein interactions between PspA2 and NLF domain are responsible for the binding affinity between these proteins.
Discussion
In spite of the availability of antibiotic treatment and a number of vaccines, pneumococcal infection still remains one of the main causes of morbidity and mortality especially among high risk groups.40,41 Emergence of antibiotic resistance42 and limitations of the currently available polysaccharide-based vaccines43,44 prompted a search for new therapeutic avenues, including identification of cross-reactive protein antigens conserved across capsular serotypes for vaccine preparation.45,46 PspA is the most intensively studied potential vaccine candidate.46–48 The multi-faceted biological activity of PspA includes inhibition of complement activation and deposition on the pneumococcal surface49 and retardation of clearance of pneumococci from the blood.48 However, the most intriguing property of PspA is its ability to protect pneumococci from the bactericidal action of LF and the Lfcn peptides.10
LF’s antibacterial activity involves complex interactions of LF, bacterial cells, bacterial products, and host cells.1,2 The stretch of N-terminal 49 residues of human LF which constitutes the LfcnH is crucial for the bactericidal functions.50 This effect is mediated through direct interaction with the bacterial surface, either as released peptide/peptides or while within the N-lobe of LF. Several lines of evidence established that PspA quite effectively inhibits the bactericidal action of apo-LF. LF is more lethal in the absence of PspA (in PspA– strain), and anti-PspA antibodies enhance the killing. Moreover, addition of recombinant PspA reduces killing of pneumococci by apo-LF and LfcnH peptides.10 These data combined with the fact that PspA is the only protein on the surface of S. pneumoniae which is known to bind LF7,8 emphasize the intimate relationship between PspA’s interaction with LF and its ability to protect the bacterium from attack by the latter. However, the exact mechanism of this defense is not known.
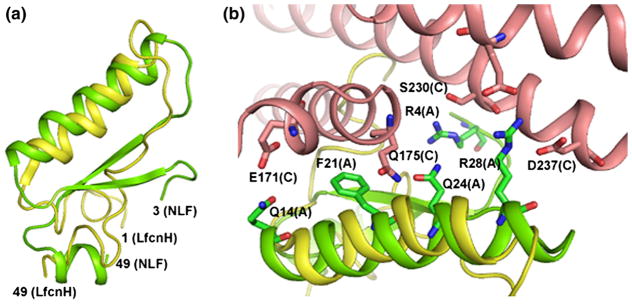
The crystal structure of PspA2 with NLF offers insight on how PspA’s ability to bind lactoferrin can be protective for the bacteria. First, the structure shows that PspA specifically binds to the Lfcn domain, thereby blocking its access to the bacterial surface. Second, although it is not known if in vivo the lactoferrin-mediated killing of S. pneumoniae is caused entirely by Lfcn peptides released from human LF, the crystal structure presented here shows that binding of PspA hinders the surface accessibility of the LF N-lobe. It is therefore reasonable to assume that PspA prevents protease access to the cleavage site and thereby inhibits the release of bactericidal peptides from LF. Finally, even in the incidence of proteolysis, PspA is likely to be able to scavenge cationic Lfcn peptide/peptides and thus inhibit their effect against pneumococci. The recently resolved NMR structure of LfcnH reveals that the helical structure of this domain resembles that in the intact LF.50 As shown in Figure 7(a), the first helix in the free LfcnH (yellow) and in the Lfcn domain (green) from the NLF:PspA2 complex superposes well while the β-sheets are completely unfolded in the solution structure. As this NLF helix contributes many interactions with PspA2 (Figure 7(b)), it is expected that PspA can efficiently bind released 49 residue LfcnH peptide. The unfolded regions of LfcnH may provide additional contacts to PspA via exposed residues. Shorter peptides generated by proteolysis from LfcnH may involve other binding interactions than those seen in the crystal structure.
Figure 7.
(a) Superposition of the NMR structure of intact LfcnH peptide (yellow) on the lactoferricin domain (residues 3–49) of the NLF (green) from the PspA2:NLF complex. The conformation of the first helix of the Lfcn domain is very similar in both structures although the β-sheet is unfolded in the LfcnH peptide. (b) Residues in the first helix of NLF (green) that interact with various residues of PspA2 (salmon) are shown in stick model. This helix of NLF provides many contacts with PspA. The corresponding section of LfcnH peptide is shown in yellow.
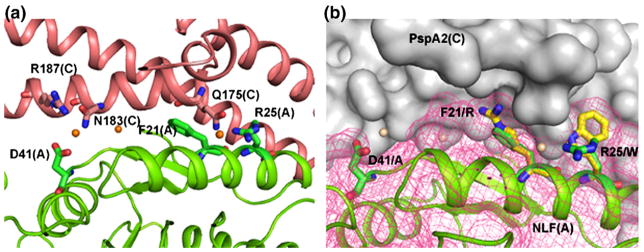
Bovine LF and the corresponding lactoferricin peptides also show strong bactericidal properties.23,51 bLF also binds to PspA but the binding efficiency is approximately 10–15-fold less than hLF.8 This may reflect the evolutionary history of S. pneumoniae, which is a human pathogen. Within the Lfcn domain there are seven differences between bovine and human proteins. Among these, the three most striking differences are Phe21 (human) Arg (bovine), Arg25Trp and Asp41Ala. All other differences between human and bovine LF are conservative changes (Arg5, 28, 54 of hLF to Lys residues in bLF and Lys39 to Arg). A careful look at the sites of the non-conservative mutations provides some clues to the observed difference in the binding efficiency of bovine and human LF to PspA. First, the oxygen atom Asp41OD2 of hLF forms contacts with PspA (Asn183 and Arg187) through a water molecule (see Figure 8(a)). Replacement of the negatively charged side-chain of this acidic residue with the aliphatic side-chain of alanine in bLF would abolish these interactions. Second, side-chain NH1 of Arg25 in hLF forms a hydrogen bond, though weak, with Gln175. This hydrogen bond would be absent in bLF where a bulky aromatic side-chain is present. Finally, replacing Phe21 in hLF with an Arg residue would alter the packing environment near the side-chain (Figure 8(b)). The crystal structure therefore provides clear and testable hypotheses to explain the decreased ability of bLF to bind PspA.
Figure 8.
Comparison of Lfcn domain of bLF and hLF. (a) There are three non-conservative differences in the Lfcn domains of human and bovine LF. These residues (Phe21, Asp41 and Arg48) of NLF (green), shown in stick model, provide important interactions for binding PspA (red). (b) Interactions between PspA2 (surface drawing, grey) and the LF N-lobe (ribbons, green). Sites for three non-conservative mutations are shown in stick representation (bovine residues in yellow, human residues in green). These residues are Asp41 (human) to Ala (bovine), Phe21 to Arg and Arg25 to Trp. Mesh (rose) was calculated after mutating the residues according to the bovine sequence. These mutations are expected to alter the nature of interactions considerably.
Although targeting protein–protein interactions is more challenging than traditional approaches of identification of small molecule inhibitors for protein targets, inhibitors of protein–protein interaction are becoming increasingly popular in the discovery of lead compounds.52,53 The crystal structure of PspA2: NLF offers an interesting possibility for screening small molecule inhibitors. Inhibitors blocking PspA–LF interactions may allow the natural bactericidal effect of lactoferrin to protect the host from colonization and infection by S. pneumoniae.
In summary, the crystal structure of the lactoferrin-binding domain of PspA in complex with LF N-lobe shows details of the specific interaction between human LF and PspA. The structure reveals how PspA acts as a barrier between the bacterial outer membrane and LF (or Lfcn) and thereby protects pneumococci from exposure to the highly cationic lactoferricin domain, and provides the first glimpse of the exposed surface epitopes of the PspA2 region. This domain is known to elicit the most cross-protective immunity54,55 and therefore knowledge of the three-dimensional structure will aid design of vaccines based on peptides within this region.
Materials and Methods
Cloning, expression, and purification of PspA fragments
The coding sequences of truncated S. pneumoniae surface protein A constructs PspA1 (amino acid residues 1–288 of mature sequence) and PspA2 (168–288) were amplified by polymerase chain reaction (PCR) from plasmid pUAB103 containing amino acid residues 1–370 of mature PspA sequence of the strain Rx1 using Triple Master polymerase mix (Eppendorf, Germany) according to the manufacturer’s protocol. Amplified DNAs were cloned into NdeI and XhoI sites of pET15b vector (Novagen). Recombinant PspAs were expressed in Escherichia coli strain BL21(DE3) (Stratagene) via induction of the culture with 0.4 mM isopropyl thio-β-D-galactoside when A600 =0.8 and subsequent overnight growth at room temperature. Initially, N-terminally His6-tagged proteins were purified from cell-free extracts by metal-chelate affinity chromatography on Talon column (Clontech) followed by removing the His-tag by proteolysis with thrombin. Additional purification steps included anion-exchange chromatography on a Q Sepharose column (GE Healthcare) and size exclusion chromatography on a Superdex 75 (16/60) column (GE Healthcare).
Point mutations in plasmids containing PspA1 and PspA2 were performed using QuikChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Mutated PspA fragments were expressed and purified by the same protocol as wild-type proteins.
Cloning, expression, and purification of LF and its N-lobe
The coding sequences of human LF and its N-lobe (NLF, amino acid residues 1–343 of mature hLF sequence) were amplified from plasmid pNUT:hLF by PCR using triple master polymerase mix. Amplified DNAs were subcloned into the BamHI site of baculovirus transfer vector pAcGP67A (BD Biosciences). Positive clones were checked for the correct orientation of inserts by restriction digestion of plasmid with SmaI enzyme. The transfer vector containing the rhLF or NLF sequence and linearized BD BaculoGold Bright Baculovirus DNA (BD Biosciences) were co-transfected into Spodoptera frugiperda insect cell line (Sf9) using cellfectin reagent (Invitrogen) according to the manufacturer’s instructions. For rhLF and NLF expression, insect cell line BTI-TN-5B1-4 (High Five cells) were infected with recombinant baculovirus and grown at 27 °C for four to five days. Recombinant hLF and NLF were purified from the supernatant of the insect cells using cation exchange chromatography on SP Sepharose.
Isothermal titration calorimetry measurements
Calorimetric titrations were carried out with VP-ITC microcalorimeter (MicroCal, Inc., Northamphon, MA). The solution of rhLF (Agennix Inc.) or NLF at concentration of 25–38 μM was loaded into the sample cell and the injection syringe contained PspA1 or PspA2 (wild-type or mutant) at concentration of 176–295 μM (all concentrations were calculated assuming that proteins are monomers). PspA was delivered to stirred cell (300 rpm) at 210–240 s intervals by 31 injections of 8 μl each. Experiments were conducted in 50 mM sodium phosphate buffer (pH 7.5) at 25(±0.02) °C. Data were fitted to a single binding site model56 using Origin software (version 5.0, MicroCal, Inc.). The binding enthalpy (ΔH), and entropy (ΔS), association constant (KA) and stoichiometry were obtained.
Complex preparation, crystallization, and structure solution
For complex preparation, purified PspA2 and NLF were mixed in ~1:1 molar ratio. The PspA2:NLF complex was separated from unbound proteins by size exclusion chromatography on a Superdex 75 column. Finally, complex was dialyzed against 50 mM Tris-HCl (pH7.5), 50 mM NaCl and concentrated to 45 mg/ml by ultrafiltration using a 10 kDa-cutoff membrane (Amicon). Crystals were grown using sitting drop vapor diffusion at 4 °C. The drops contained 1:1 mixture of the complex and the reservoir solution composed of 30% (v/v) PEG 400, 0.2 M lithium sulfate and 0.1 M sodium cacodylate (pH 6.5). For data collection a crystal was flash frozen in paraffin oil.57 X-ray data were collected from a single crystal at 100 K on a Mar165 image plate at IMCA-CAT beamline (Advanced Photon Source, Argonne National Laboratory). Crystallographic data processing was performed using HKL 2000 (Table 1). The crystal belongs to space group P32 with unit cell dimensions a=b=130.18 Å and c=80.80 Å. There are two 1:1 complexes per asymmetric unit, and the value of Vm is 3.79 Å3/Da, which corresponds to a solvent volume fraction of 68%.
Structure determination and refinement
The structure of the N-terminal lobe of LF was solved by molecular replacement, using the human N-terminal lobe as a search model (PDB entry 1H45). The two highest peaks in the cross-rotation function corresponded to the two molecules in the asymmetric unit. A difference electron density map calculated with phases from the partial structure showed three long stretches of α-helical density, and idealized polyalanine helices were built and fit to the density using Coot.58 Subsequent electron density maps calculated with phases from the partial structure allowed alignment of the sequence and fitting of the side-chains.
Refinement of the structure was carried out by simulated annealing using CNS.59 No sigma cutoff was applied to the data. Five percent of the data were randomly selected and removed prior to refinement for analysis of the free R factor. The two complexes were restrained by the non-crystallographic symmetry during the refinement. The progress of the refinement was guided by the decrease in both the conventional and free R factors. Individual B-factors were included in the final refinement. Two final rounds of TLS restrained refinement (without any non-crystallographic symmetry restraints) were carried out using REFMAC5.0.60 The refinement statistics are shown in Table 1. The final model includes residues 3–333 of NLF, residues 168–288 of PspA (plus the four residue N-terminal tag), two Fe ions, two carbonate ions, six sulfate ions, two molecules of N-acetylglucosamine, two Zn ions and 24 water molecules. Overall quality of the model is excellent with 99.4% residues in the allowed regions of Ramachandran plot, two NLF residues (Ser192 and Leu300) in each complex in the disallowed region. These residues are in the disallowed region in previously described structures.
Data deposition
Atomic coordinates have been deposited to the RCSB Protein Data Bank (PDB ID 2PMS).
Supplementary Material
Acknowledgments
We thank Kathryn Stowell of Massey University, New Zealand for the gift of pNUT:hLF plasmid and Agennix, Inc. for human lactoferrin expressed in Aspergillus awamori. This work was supported by a grant NIH 1R21 AI061155 (to D.C.). Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago.
Abbreviations used
- LF
lactoferrin
- hLF
human lactoferrin
- bLF
bovine lactoferrin
- rhLF
recombinant human lactoferrin (full length)
- NLF
recombinant human lactoferrin N-lobe
- Lfcn
lactoferricin
- LfcnH
human lactoferricin
- LfcnB
bovine lactoferricin
- PspA
pneumococcal surface protein A
- ITC
isothermal titration calorimetry
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2007.04.075
References
- 1.Valenti P, Antonini G. Lactoferrin: an important host defence against microbial and viral attack. Cell Mol Life Sci. 2005;62:2576–2587. doi: 10.1007/s00018-005-5372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farnaud S, Evans RW. Lactoferrin – a multifunctional protein with antimicrobial properties. Mol Immunol. 2003;40:395–405. doi: 10.1016/s0161-5890(03)00152-4. [DOI] [PubMed] [Google Scholar]
- 3.Arnold RR, Brewer M, Gauthier JJ. Bactericidal activity of human lactoferrin: sensitivity of a variety of microorganisms. Infect Immun. 1980;28:893–898. doi: 10.1128/iai.28.3.893-898.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward PP, Paz E, Conneely OM. Multifunctional roles of lactoferrin: a critical overview. Cell Mol Life Sci. 2005;62:2540–2548. doi: 10.1007/s00018-005-5369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling JML, Schryvers AB. Perspectives on interactions between lactoferrin and bacteria. Biochem Cell Biol. 2006;84:275–281. doi: 10.1139/o06-044. [DOI] [PubMed] [Google Scholar]
- 6.Yu RH, Schryvers AB. Bacterial lactoferrin receptors: insights from characterizing the Moraxella bovis receptors. Biochem Cell Biol. 2002;80:81–90. doi: 10.1139/o01-235. [DOI] [PubMed] [Google Scholar]
- 7.Hammerschmidt S, Bethe G, Remane PH, Chhatwal GS. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect Immun. 1999;67:1683–1687. doi: 10.1128/iai.67.4.1683-1687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakansson A, Roche H, Mirza S, McDaniel LS, Brooks-Walter A, Briles DE. Characterization of the binding of human lactoferrin to pneumococcal surface protein A (PspA) Infect Immun. 2001;69:3372–3381. doi: 10.1128/IAI.69.5.3372-3381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crain MJ, Waltman WD, 2nd, Turner JS, Yother J, Talkington DF, McDaniel LS, et al. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect Immun. 1990;58:3293–3299. doi: 10.1128/iai.58.10.3293-3299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaper M, Hollingshead SK, Benjamin WH, Jr, Briles DE. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin. Infect Immun. 2004;72:5031–5040. doi: 10.1128/IAI.72.9.5031-5040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker EN, Baker HM. Molecular structure, binding properties and dynamics of lactoferrin. Cell Mol Life Sci. 2005;62:2531–2539. doi: 10.1007/s00018-005-5368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullen JJ, Rogers HJ, Leigh L. Iron-binding proteins in milk and resistance to Escherichia coli infection in infants. Brit Med J. 1972;1:69–75. doi: 10.1136/bmj.1.5792.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rainard P. Bacteriostatic activity of bovine milk lactoferrin against mastitic bacteria. Vet Microbiol. 1986;11:387–392. doi: 10.1016/0378-1135(86)90068-4. [DOI] [PubMed] [Google Scholar]
- 14.Levay PF, Viljoen M. Lactoferrin: a general review. Haematologica. 1995;80:252–267. [PubMed] [Google Scholar]
- 15.Arnold RR, Russel JE, Champion WJ, Brewer M, Gauthier JJ. Bactericidal activity of human lactoferrin: differentiation from the status of iron deprivation. Infect Immun. 1982;35:792–799. doi: 10.1128/iai.35.3.792-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison RT, 3rd, Giehl TJ, LaForce FM. Damage of outer membrane of enteric gram-negative bacteria by lactoferrin and transferrin. Infect Immun. 1988;56:2774–2781. doi: 10.1128/iai.56.11.2774-2781.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellamy W, Takase M, Yamauchi H, Wakabayashi H, Kawase K, Tomita M. Identification of the bactericidal domain of lactoferrin. Biochim Biophys Acta. 1992;1121:130–136. doi: 10.1016/0167-4838(92)90346-f. [DOI] [PubMed] [Google Scholar]
- 18.Kuwata H, Yip T, Tomita M, Hutchens TW. Direct evidence of the generation in human stomach of an antimicrobial peptide domain (lactoferricin) from ingested lactoferrin. Biochim Biophys Acta. 1998;1429:129–141. doi: 10.1016/s0167-4838(98)00224-6. [DOI] [PubMed] [Google Scholar]
- 19.Britigan BE, Hayek MB, Doebbeling BN, Fick RB., Jr Transferrin and lactoferrin undergo proteolytic cleavage in the Pseudomonas aeruginosa-infected lungs of patients with cystic fibrosis. Infect Immun. 1993;61:5049–5055. doi: 10.1128/iai.61.12.5049-5055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnaud S, Spiller C, Moriarty LC, Patel A, Gant V, Odell EW, Evans RW. Interactions of lactoferricin-derived peptides with LPS and antimicrobial activity. FEMS Microbiol Letters. 2004;233:193–199. doi: 10.1016/j.femsle.2004.01.039. [DOI] [PubMed] [Google Scholar]
- 21.Geerts MEJ, van Heen HA, Mericskay M, de Boer HA, Nuijens JH. N-terminal stretch Arg2, Arg3, Arg4 and Arg5 of human lactoferrin is essential for binding to heparin, bacterial lipopolysaccharide, human lysozyme and DNA. Biochem J. 1997;328:145–151. doi: 10.1042/bj3280145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gifford JL, Hunter HN, Vogel HJ. Lactoferricin: a lactoferrin-derived peptide with antimicrobial, antiviral, antitumor and immunological properties. Cell Mol Life Sci. 2005;62:2588–2598. doi: 10.1007/s00018-005-5373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakabayashi H, Takase M, Tomita M. Lactoferricin derived from milk protein lactoferrin. Curr Pharm Des. 2003;9:1277–1287. doi: 10.2174/1381612033454829. [DOI] [PubMed] [Google Scholar]
- 24.Yother J, White JM. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J Bacteriol. 1994;176:2976–2985. doi: 10.1128/jb.176.10.2976-2985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yother J, Briles DE. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992;174:601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jedrzejas MJ. Unveiling molecular mechanism of pneumococcal surface protein A interactions with antibodies and lactoferrin. Clin Chim Acta. 2006;367:1–10. doi: 10.1016/j.cca.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 27.Day CL, Anderson BF, Tweedie JW, Baker EN. Structure of the recombinant N-terminal lobe of human lactoferrin at 2.0 Å resolution. J Mol Biol. 1993;232:1084–1100. doi: 10.1006/jmbi.1993.1462. [DOI] [PubMed] [Google Scholar]
- 28.Baker HM, Baker CJ, Smith CA, Baker EN. Metal substitution in transferrins: specific binding of cerium (IV) revealed by the crystal structure of cerium-substituted human lactoferrin. J Biol Inorg Chem. 2000;5:692–698. doi: 10.1007/s007750000157. [DOI] [PubMed] [Google Scholar]
- 29.Sun XL, Baker HM, Shewry SC, Jameson GB, Baker EN. Structure of recombinant human lactoferrin expressed in Aspergillus awamori. Acta Crystallog sect D. 1999;55:403–407. doi: 10.1107/s0907444998011226. [DOI] [PubMed] [Google Scholar]
- 30.Thomassen EAJ, Van Veen HA, Van Berkel PHC, Nuijens JH, Abrahams JP. The protein structure of recombinant human lactoferrin produced in the milk of transgenic cows closely matches the structure of human milk-derived lactoferrin. Transgen Res. 2005;14:397–405. doi: 10.1007/s11248-005-3233-0. [DOI] [PubMed] [Google Scholar]
- 31.Peterson N, Arcus V, Anderson B, Tweedie J, Jameson G, Baker E. “Dilysine trigger” in transferrins probed by mutagenesis of lactoferrin: crystal structures of the R210G, R210E, and R210L mutants of human lactoferrin. Biochemistry. 2002;41:14167–14175. doi: 10.1021/bi020443a. [DOI] [PubMed] [Google Scholar]
- 32.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 33.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 34.Vegarud GE, Langsrud T, Svenning C. Mineral-binding milk proteins and peptides; occurrence, biochemical and technological characteristics. Brit J Nutr. 2000;84:S91–S98. doi: 10.1017/s0007114500002300. [DOI] [PubMed] [Google Scholar]
- 35.Jabeen T, Sharma S, Singh N, Bhushan A, Singh TP. Structure of the zinc-saturated C-terminal lobe of bovine lactoferrin at 2.0 Å resolution. Acta Crystallog sect D. 2005;61:1107–1115. doi: 10.1107/S0907444905016069. [DOI] [PubMed] [Google Scholar]
- 36.Anderson BF, Norris GE, Rumball SV, Thomas DH, Baker EN. A comparison of the three-dimensional structures of human lactoferrin in its iron free and iron saturated forms. Adv Exp Med Biol. 1994;357:227–230. doi: 10.1007/978-1-4615-2548-6_22. [DOI] [PubMed] [Google Scholar]
- 37.Jameson GB, Anderson BF, Norris GE, Thomas DH, Baker EN. Structure of human apolactoferrin at 2.0 Å resolution. Refinement and analysis of ligand-induced conformational change. Acta Crystallog sect D. 1998;54:1319–1335. doi: 10.1107/s0907444998004417. [DOI] [PubMed] [Google Scholar]
- 38.Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structure and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 39.McDaniel LS, Scott G, Kearney JF, Briles DE. Monoclonal antibodies against protease-sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J Exp Med. 1984;160:386–397. doi: 10.1084/jem.160.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez R. Pneumococcus: the sugar-coated bacteria. Int Microbiol. 2006;9:179–190. [PubMed] [Google Scholar]
- 41.Schmidt-Ioanas M, Lode H. Treatment of pneumonia in elderly patients. Expert Opin Pharmacother. 2006;7:499–507. doi: 10.1517/14656566.7.5.499. [DOI] [PubMed] [Google Scholar]
- 42.Jacobs MR. Streptococcus pneumoniae: epidemiology and patterns of resistance. Am J Med. 2004;117(Suppl 3A):3S–15S. doi: 10.1016/j.amjmed.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Bogaert D, Hermans PWM, Adrian PV, Rumke HC, de Groot R. Pneumococcal vaccines: an update on current strategies. Vaccine. 2004;22:2209–2220. doi: 10.1016/j.vaccine.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 44.Pelton SI, Dagan R, Gaines BM, Klugman KP, Laufer D, O’Brien K, Schmitt HJ. Pneumococcal conjugate vaccines: Proceedings from an interactive symposium at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy. Vaccine. 2003;21:1562–1571. doi: 10.1016/s0264-410x(02)00681-3. [DOI] [PubMed] [Google Scholar]
- 45.Ogunniyi AD, Grabowicz M, Briles DE, Cook J, Paton JC. Development of a vaccine against invasive pneumococcal disease based on combination of proteins of Streptococcus pneumoniae. Infec Immun. 2007;75:350–357. doi: 10.1128/IAI.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tai SS. Streptococcus pneumoniae protein vaccine candidates: properties, activities and animal studies. Crit Rev Microbiol. 2006;32:139–153. doi: 10.1080/10408410600822942. [DOI] [PubMed] [Google Scholar]
- 47.Briles DE, Hollingshead SK, King J, Swift A, Braun PA, Park MK, et al. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis. 2000;182:1694–1701. doi: 10.1086/317602. [DOI] [PubMed] [Google Scholar]
- 48.Briles DE, Tart RC, Swiatlo E, Dillard JP, Smith P, Benton KA, et al. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA) Clin Microbiol Rev. 1998;11:645–657. doi: 10.1128/cmr.11.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren B, McCrory MA, Pass C, Bullard DC, Ballantyne CM, Xu Y, et al. The virulence function of Streptococcus pneumoniae surface protein A involves inhibition of complement activation and impairment of complement receptor-mediated protection. J Immunol. 2004;173:7506–7512. doi: 10.4049/jimmunol.173.12.7506. [DOI] [PubMed] [Google Scholar]
- 50.Hunter HN, Demcoe AR, Jenssen H, Gutteberg TJ, Vogel HJ. Human lactoferricin is partially folded in aqueous solution and is better stabilized in a membrane mimetic solvent. Antimicrob Agents Chemother. 2005;49:3387–3395. doi: 10.1128/AAC.49.8.3387-3395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamauchi K, Tomita M, Giehl TJ, Ellison RT., 3rd Antimicrobial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect Immun. 1993;61:719–728. doi: 10.1128/iai.61.2.719-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berg T. Modulation of protein–protein interactions with small organic molecules. Angew Chem Int Ed Engl. 2003;42:2462–2481. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 53.Toogood PL. Inhibition of protein-protein association by small molecules: approaches and progress. J Med Chem. 2002;45:1543–1558. doi: 10.1021/jm010468s. [DOI] [PubMed] [Google Scholar]
- 54.Roche H, Hakansson A, Hollingshead SK, Briles DE. Regions of PspA/EF3296 best able to elicit protection against Streptococcus pneumoniae in a murine infection model. Infect Immiun. 2003;71:1033–1041. doi: 10.1128/IAI.71.3.1033-1041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDaniel LS, Ralph BA, McDaniel DO, Briles DE. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microbial Pathogenesis. 1994;17:323–337. doi: 10.1006/mpat.1994.1078. [DOI] [PubMed] [Google Scholar]
- 56.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 57.Riboldi-Tunnicliffe A, Hilgenfeld R. Cryo-crystallography with oil – an old idea revived. J Appl Crystallog. 1999;32:1003–1005. [Google Scholar]
- 58.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallog sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallog sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 60.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallog sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.