Abstract
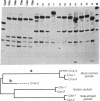
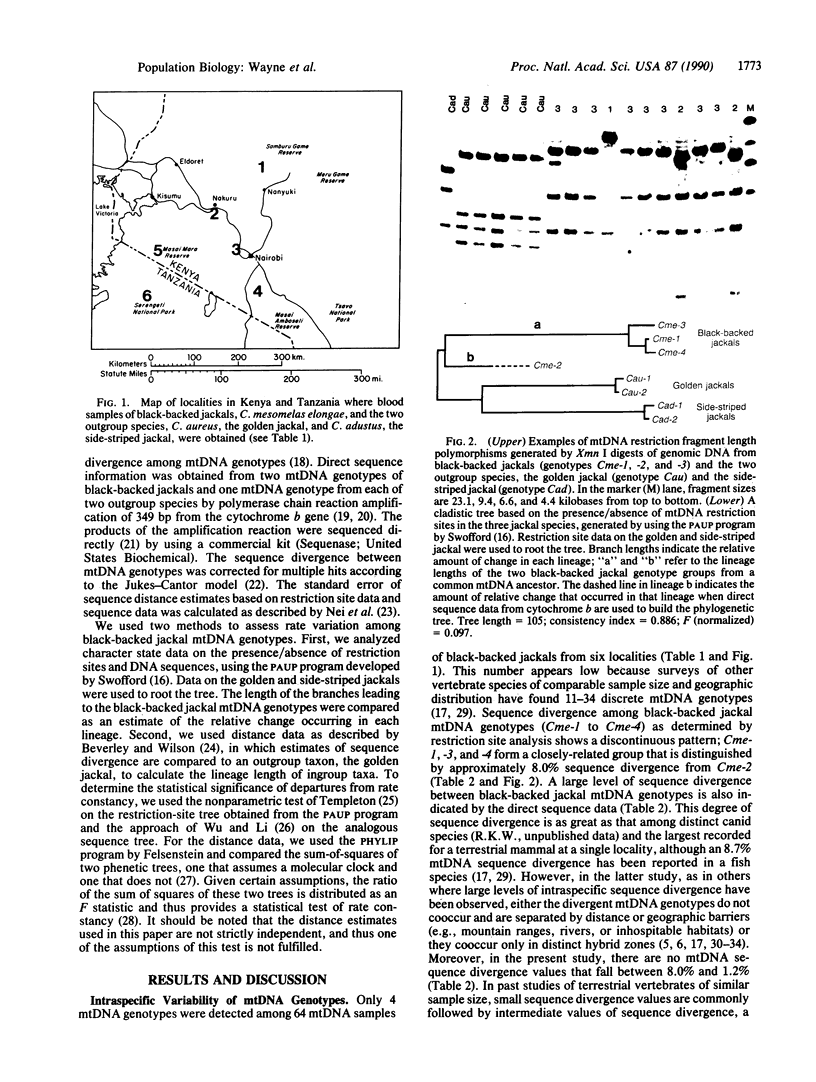
In discussions about the relative rate of molecular evolution, intraspecific variability in rate is rarely considered. An underlying assumption is that intraspecific sequence differences are small, and thus variations in rate would be difficult to detect or would not affect comparisons among distantly related taxa. However, several studies on mammalian mitochondrial DNA (mtDNA) have revealed considerable intraspecific sequence divergence. In this report, we test for differences in the rate of intraspecific evolution by comparing mtDNA sequences, as inferred from restriction site polymorphisms and direct sequencing, between mtDNA genotypes of the eastern African black-backed jackal, Canis mesomelas elongae, and those of two other sympatric jackal species. Our results are unusual for several reasons. First, mtDNA sequence divergence within several contiguous black-backed jackal populations is large (8.0%). Previous intraspecific studies of terrestrial mammals have generally found values of less than 5% within a single population, with larger divergence values most often occurring among mtDNA genotypes from geographically distant or isolated localities. Second, only 4 mtDNA genotypes were present in our sample of 64 jackals. The large sequence divergence observed among these mtDNA genotypes suggests there should be many more genotypes of intermediate sequence divergence if they had evolved in sympatry. Finally, estimates of the rate of mtDNA sequence evolution differ by approximately 2- to 4-fold among black-backed jackal mtDNA genotypes, thus indicating a substantial heterogeneity in the rate of sequence evolution. The results are difficult to reconcile with ideas of a constant molecular clock based on random fixation of selectively neutral or nearly neutral mtDNA sequence mutations.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Ball R. M., Arnold J. Current versus historical population sizes in vertebrate species with high gene flow: a comparison based on mitochondrial DNA lineages and inbreeding theory for neutral mutations. Mol Biol Evol. 1988 Jul;5(4):331–344. doi: 10.1093/oxfordjournals.molbev.a040504. [DOI] [PubMed] [Google Scholar]
- Avise J. C. Mitochondrial DNA and the evolutionary genetics of higher animals. Philos Trans R Soc Lond B Biol Sci. 1986 Jan 29;312(1154):325–342. doi: 10.1098/rstb.1986.0011. [DOI] [PubMed] [Google Scholar]
- Avise J. C. Mitochondrial DNA and the evolutionary genetics of higher animals. Philos Trans R Soc Lond B Biol Sci. 1986 Jan 29;312(1154):325–342. doi: 10.1098/rstb.1986.0011. [DOI] [PubMed] [Google Scholar]
- Avise J. C., Neigel J. E., Arnold J. Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J Mol Evol. 1984;20(2):99–105. doi: 10.1007/BF02257369. [DOI] [PubMed] [Google Scholar]
- Ball R. M., Freeman S., James F. C., Bermingham E., Avise J. C. Phylogeographic population structure of Red-winged Blackbirds assessed by mitochondrial DNA. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1558–1562. doi: 10.1073/pnas.85.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham E., Avise J. C. Molecular zoogeography of freshwater fishes in the southeastern United States. Genetics. 1986 Aug;113(4):939–965. doi: 10.1093/genetics/113.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverley S. M., Wilson A. C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21(1):1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Carr S. M., Ballinger S. W., Derr J. N., Blankenship L. H., Bickham J. W. Mitochondrial DNA analysis of hybridization between sympatric white-tailed deer and mule deer in west Texas. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9576–9580. doi: 10.1073/pnas.83.24.9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris S. D., Sage R. D., Huang C. M., Nielsen J. T., Ritte U., Wilson A. C. Flow of mitochondrial DNA across a species boundary. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2290–2294. doi: 10.1073/pnas.80.8.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich P. D. Temporal scaling of molecular evolution in primates and other mammals. Mol Biol Evol. 1986 May;3(3):205–221. doi: 10.1093/oxfordjournals.molbev.a040391. [DOI] [PubMed] [Google Scholar]
- Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Päbo S., Villablanca F. X., Wilson A. C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb T., Avise J. C. Directional introgression of mitochondrial DNA in a hybrid population of tree frogs: The influence of mating behavior. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2526–2530. doi: 10.1073/pnas.83.8.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Stephens J. C., Saitou N. Methods for computing the standard errors of branching points in an evolutionary tree and their application to molecular data from humans and apes. Mol Biol Evol. 1985 Jan;2(1):66–85. doi: 10.1093/oxfordjournals.molbev.a040333. [DOI] [PubMed] [Google Scholar]
- Ochman H., Wilson A. C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26(1-2):74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- Ohta T. Role of very slightly deleterious mutations in molecular evolution and polymorphism. Theor Popul Biol. 1976 Dec;10(3):254–275. doi: 10.1016/0040-5809(76)90019-8. [DOI] [PubMed] [Google Scholar]
- Pamilo P., Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988 Sep;5(5):568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Powell J. R. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: evidence from Drosophila. Proc Natl Acad Sci U S A. 1983 Jan;80(2):492–495. doi: 10.1073/pnas.80.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Spolsky C., Uzzell T. Evolutionary history of the hybridogenetic hybrid frog Rana esculenta as deduced from mtDNA analyses. Mol Biol Evol. 1986 Jan;3(1):44–56. doi: 10.1093/oxfordjournals.molbev.a040376. [DOI] [PubMed] [Google Scholar]
- Vawter L., Brown W. M. Nuclear and mitochondrial DNA comparisons reveal extreme rate variation in the molecular clock. Science. 1986 Oct 10;234(4773):194–196. doi: 10.1126/science.3018931. [DOI] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Clark A. G., Stoneking M., Cann R. L., Wilson A. C. Allelic variation in human mitochondrial genes based on patterns of restriction site polymorphism. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9611–9615. doi: 10.1073/pnas.83.24.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrischnik L. A., Higuchi R. G., Stoneking M., Erlich H. A., Arnheim N., Wilson A. C. Length mutations in human mitochondrial DNA: direct sequencing of enzymatically amplified DNA. Nucleic Acids Res. 1987 Jan 26;15(2):529–542. doi: 10.1093/nar/15.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. I., Li W. H. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]