Abstract
MiR-21 has been identified as one of the most common proto-oncogenes. It is hypothesized that up-regulated miR-21 could be served as a potential biomarker for human cancer diagnosis. However, inconsistencies or discrepancies about diagnostic accuracy of circulating miR-21 still remain. In this sense, miR-21′s diagnostic value needs to be fully validated. In this study, we performed an update meta-analysis to estimate the diagnostic value of circulating miR-21 in various human cancers. Additionally, we conducted a validation test on 50 endometrial cancer patients, 50 benign lesion patients and 50 healthy controls. A systematical literature search for relevant articles was performed in Pubmed, Embase and Cochrane Library. A total of 48 studies from 39 articles, involving 3,568 cancer patients and 2,248 controls, were included in this meta-analysis. The overall sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and area under the curve (AUC) were 0.76 (0.71-0.80), 0.82 (0.79-0.85), 4.3 (3.6-5.1), 0.29 (0.24-0.35), 15 (11-20) and 0.86 (0.83-0.89), respectively. In the validation test, the expression levels of serum miR-21 were significantly higher in benign lesion patients (p = 0.003) and endometrial cancer patients (p = 0.000) compared with that of healthy controls. Endometrial cancer patients showed higher miR-21 expression levels (p = 0.000) compared with benign lesion patients. In conclusion, the meta-analysis shows that circulating miR-21 has excellent performance on the diagnosis for various cancers and the validation test demonstrates that serum miR-21 could be served as a novel biomarker for endometrial carcinoma.
Keywords: miR-21, cancers, meta-analysis, diagnosis, endometrial carcinoma
INTRODUCTION
Early surveillance and diagnosis can potentially reduce the mortality of human cancers. Conventional diagnostic methods such as biopsy and imageological examination are invasive or harmful. Meanwhile, the diagnostic capability of common serum-based cancer biomarkers, such as carcinoembryonic antigen (CEA), alpha-fetalprotein (AFP) or carbohydrate antigen (CA), are also restricted due to the low sensitivity and specificity of these biomarkers. Consequently, a non-invasive and convenient diagnostic method with high sensitivity and specificity for human cancers is urgently needed.
MicroRNAs (miRNAs) are a class of small (19–25 nucleotides), non-coding and single-stranded RNAs that enhance or inhibit mRNAs expression at the post-transcriptional level [1, 2]. Over the past years, an increasing number of miRNAs, as oncogenes or tumor suppressor genes, have been proved differential expressions in a variety of cancers [3]. Studies also show that miRNAs are chemically stable and detectable in tissue, serum, plasma, urine, cerebrospinal fluid, feces as well as other body fluids [4]. The above findings demonstrate that circulating miRNAs could be used as novel biomarkers for human cancer screening and diagnosis for its superiorities of being non-invasive and convenient with high sensitivity and specificity [5].
MiR-21 has been identified as one of the most prominent oncogenic miRNAs and has been proved up-regulated in various human cancers [6, 7], which regulates the expression of multiple cancer-associated target genes [8–10]. Accumulating evidence strongly supports the role of miR-21 as oncogene in human cancers [11–19]. Therefore, it is hypothesized that up-regulated miR-21 could be employed as a potential biomarker for human cancer diagnosis. To date, numerous studies have testified the diagnostic value of circulating miR-21 in various human cancers [7, 20]. However, inconsistencies or heterogeneities about diagnostic accuracy of circulating miR-21 still remain and its diagnostic value in human cancers needs to be confirmed. In this study, we performed an update meta-analysis to assess the diagnostic value of circulating miR-21 in various human cancers.
As we all know, endometrial cancer has become one of dominant female cancers. But according to our meta-analysis, we only found one study focusing on the diagnostic value of circulating miR-21 in patients with endometrial cancer [21] and this study were excluded from our meta-analysis for the following reasons: (1) insufficient data to execute a two-by-two table; (2) only 12 cancer samples included which was less than 20. Given this, a validation test about serum miR-21 expression levels was also conducted among 50 endometrial cancer patients, 50 benign lesion patients and 50 healthy controls to make up the deficiency in diagnostic studies on circulating miR-21 of endometrial cancer.
RESULTS
Study selection and characteristics
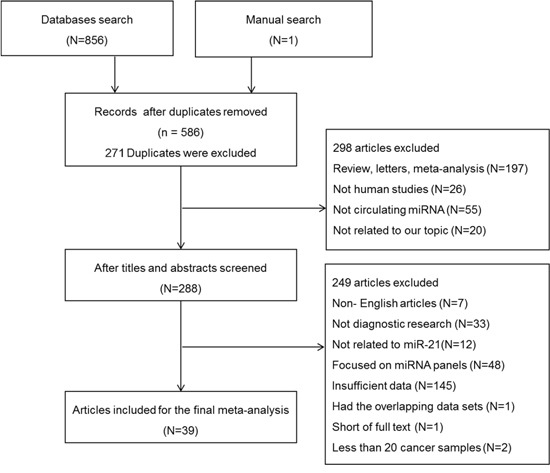
A total of 857 articles were identified by database search or manual search, among which 271 articles were excluded for duplicated data. After screening the titles and abstracts, 298 articles were excluded because they were review articles, letters, meta-analyses, non-human studies, studies on non-circulating miRNA or irrelevant to our topic. After reading and evaluating the full texts of the remaining 288 articles carefully, 249 articles were excluded in line with the exclusion criteria (7 articles were non-English articles; 33 articles were not diagnostic research; 12 articles were not related to miR-21; 48 articles focused on miRNA panels; 145 articles did not provide sufficient data; 1 article had the overlapping data sets; 1 article was short of full text; 2 articles collected less than 20 cancer samples). Finally, 48 studies from 39 articles were included in this meta-analysis [9, 22–59]. The flow diagram of study selection was shown in Figure 1.
Figure 1. Flow diagram of study selection process based on inclusion and exclusion criteria.
On the whole, 48 studies from 39 articles published as of 3 March 2016, involving 3,568 cancer patients and 2,248 controls, were included in this meta-analysis. The sample types included serum (n = 27), plasma (n = 19) and peripheral blood mononuclear cell (PBMC) (n = 2). The cancer types included lung cancer (n = 10), colorectal cancer (n = 7), gastric cancer (n = 7), hepatocellular cancer (n = 6), breast cancer (n = 7), esophageal cancer (n = 2), pancreatic cancer (n = 1), head and neck squamous cell cancer (n = 1), nasopharyngeal cancer (n = 1), lymphoma (n = 1), biliary tract cancer (n = 2), laryngeal squamous cell cancer (n = 1), prostatecancer (n = 1) and retinoblastoma (n = 1). The miR-21 expression levels were detected through quantitative reverse transcription polymerase chain reaction (qRT-PCR) in all the studies, including 25 studies with Taqman probe and 23 studies with SYBR dye. The basic characteristics of the included articles were listed in Table 1.
Table 1. Characteristics and quality assessment of 48 studies included in meta-analysis.
| First author | Year | Country | Ethnicity | Patients (controls) |
Cancer | Sample | method | ECon | Sen | Spe | Tp | Fp | Fn | Tn | AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wei J[23] | 2011 | China | Asian | 77(36) | LC | Plasma | SYBR | miR-16 | 61.04 | 83.33 | 47 | 6 | 30 | 30 | 0.729 |
| Shen J[24] | 2011 | USA | Caucasian African | 58(29) | LC | Plasma | SYBR | miR-16 | 79.31 | 65.52 | 46 | 10 | 12 | 19 | 0.816 |
| Li Y[25] | 2011 | China | Asian | 20(10) | LC | Serum | SYBR | mimics | 78.80 | 100.00 | 16 | 0 | 4 | 10 | 0.912 |
| Le HB[26] | 2012 | China | Asian | 82(50) | LC | Serum | Taqman | miR-16 | 46.30 | 92.00 | 38 | 4 | 44 | 46 | 0.686 |
| Wang B[9] | 2012 | China | Asian | 31(39) | LC | Serum | SYBR | miR-16 | 87.10 | 74.40 | 27 | 10 | 4 | 29 | 0.880 |
| Tang D[27] | 2013 | China | Asian | 62(60) | LC | Plasma | Taqman | RNU6 | 48.40 | 78.30 | 30 | 13 | 32 | 47 | 0.715 |
| Tang D[27] | 2013 | China | Asian | 34(32) | LC | Plasma | Taqman | RNU6 | 52.90 | 71.90 | 18 | 9 | 16 | 23 | 0.709 |
| Abd-EI-Fattah AA[28] | 2013 | Egypt | African | 65(37) | LC | Serum | SYBR | RNU48 | 85.70 | 86.50 | 56 | 5 | 9 | 32 | 0.850 |
| Mozzoni P[29] | 2013 | Italy | Caucasian | 54(46) | LC | Plasma | Taqman | miR-16 | 50.00 | 92.30 | 27 | 4 | 27 | 42 | 0.740 |
| Yang JS[56] | 2014 | China | Asian | 300(152) | LC | Serum | Taqman | RNU6 | 69.00 | 71.00 | 207 | 45 | 93 | 107 | 0.810 |
| Kanaan Z[57] | 2012 | USA | Caucasian | 50(50) | CC | Plasma | Taqman | RNU6 | 90.00 | 90.00 | 45 | 5 | 5 | 45 | 0.820 |
| Wang B[9] | 2012 | China | Asian | 32(39) | CC | Serum | SYBR | miR-16 | 87.50 | 74.40 | 28 | 10 | 4 | 29 | 0.850 |
| Toiyama Y[30] | 2013 | Japan | Asian | 186(53) | CC | Serum | Taqman | cel-miR-39 | 82.80 | 90.60 | 154 | 5 | 32 | 48 | 0.919 |
| Liu GH[31] | 2013 | China | Asian | 200(80) | CC | Serum | Taqman | miR-16 | 65.00 | 85.00 | 130 | 12 | 70 | 68 | 0.802 |
| Luo X[32] | 2013 | Germany | Caucasian | 80(144) | CC | Plasma | Taqman | miR-16 | 51.70 | 80.70 | 41 | 28 | 39 | 116 | 0.653 |
| Basati G[33] | 2014 | Iran | Caucasian | 40(40) | CC | Serum | SYBR | RNU6 | 77.00 | 78.00 | 31 | 9 | 9 | 31 | 0.870 |
| Ogata-kawata H[34] | 2014 | Japan | Asian | 88(11) | CC | Serum | Taqman | miR-451 | 61.40 | 90.90 | 54 | 1 | 34 | 10 | 0.798 |
| Tsujiura M[35] | 2010 | Japan | Asian | 69(30) | GC | Plasma | Taqman | RNU6 | 60.90 | 63.33 | 42 | 11 | 27 | 19 | 0.673 |
| Zheng Y[36] | 2011 | China | Asian | 53(20) | GC | Plasma | SYBR | RNU6 | 83.77 | 80.53 | 44 | 4 | 9 | 16 | 0.853 |
| Li BS[37] | 2012 | China | Asian | 60(60) | GC | Plasma | Taqman | cel-miR-39 | 74.29 | 75.71 | 45 | 15 | 15 | 45 | 0.794 |
| Wang B[9] | 2012 | China | Asian | 30(39) | GC | Serum | SYBR | miR-16 | 56.70 | 94.90 | 17 | 2 | 13 | 37 | 0.810 |
| Shiotani A[38] | 2013 | Japan | Asian | 64(64) | GC | Serum | Taqman | miR-16 | 58.60 | 86.10 | 38 | 9 | 26 | 55 | 0.720 |
| Wu J[22] | 2015 | China | Asian | 50(50) | GC | Serum | SYBR | RNU6 | 83.77 | 79.60 | 44 | 10 | 6 | 40 | 0.912 |
| Wu J[22] | 2015 | China | Asian | 50(50) | GC | PBMC | SYBR | RNU6 | 74.29 | 73.40 | 41 | 13 | 9 | 37 | 0.898 |
| Xu J[39] | 2011 | China | Asian | 101(89) | HCC | Serum | SYBR | miR-181 | 56.70 | 73.50 | 85 | 24 | 16 | 65 | 0.870 |
| Tomimaru Y[40] | 2012 | Japan | Asian | 126(50) | HCC | Plasma | Taqman | miR-16 | 87.30 | 92.00 | 110 | 4 | 16 | 46 | 0.953 |
| Tomimaru Y[40] | 2012 | Japan | Asian | 126(30) | HCC | Plasma | Taqman | miR-16 | 61.10 | 83.30 | 77 | 5 | 49 | 25 | 0.773 |
| Liu AM[41] | 2012 | China | Asian | 57(59) | HCC | Serum | Taqman | NA | 89.47 | 71.19 | 51 | 17 | 6 | 42 | 0.865 |
| Amr KS[42] | 2015 | Egypt | African | 23(17) | HCC | Serum | Taqman | RNU48 | 100.00 | 81.20 | 23 | 3 | 0 | 14 | 0.943 |
| Zhuang C[59] | 2015 | China | Asian | 52(43) | HCC | Serum | SYBR | cel-miR-39 RNU6 |
67.40 | 55.80 | 35 | 19 | 17 | 24 | 0.621 |
| Asaga S[43] | 2011 | USA | Caucasian | 79(20) | BC | Serum | SYBR | miR-16 | 67.00 | 75.00 | 53 | 5 | 26 | 15 | 0.721 |
| Wang B[9] | 2012 | China | Asian | 50(39) | BC | Serum | SYBR | miR-16 | 80.00 | 87.70 | 40 | 5 | 10 | 34 | 0.880 |
| Mar-Aguilar F[44] | 2013 | Mexico | Caucasian | 60(10) | BC | Serum | Taqman | 18s RNA | 94.40 | 80.00 | 57 | 2 | 3 | 8 | 0.950 |
| Gao J[45] | 2013 | China | Asian | 89(55) | BC | Serum | SYBR | miR-16 | 87.60 | 87.30 | 78 | 7 | 11 | 48 | 0.929 |
| Toraih EA[46] | 2015 | Egypt | African | 30(60) | BC | Serum | Taqman | RNU6 | 66.70 | 86.70 | 20 | 8 | 10 | 52 | 0.800 |
| Motawi TM[58] | 2016 | Egypt | African | 50(25) | BC | Serum | SYBR | RNU48 | 96.00 | 92.00 | 48 | 2 | 2 | 23 | 0.984 |
| Motawi TM[58] | 2016 | Egypt | African | 50(25) | BC | Serum | SYBR | RNU48 | 82.00 | 76.00 | 41 | 6 | 9 | 19 | 0.855 |
| Kurashige J[47] | 2012 | Japan | Asian | 71(39) | EC | Serum | Taqman | miR-16 | 46.50 | 100.00 | 33 | 0 | 38 | 39 | NA |
| Wang B[9] | 2012 | China | Asian | 31(39) | EC | Serum | SYBR | miR-16 | 71.00 | 69.20 | 22 | 12 | 9 | 27 | 0.740 |
| Wang J[48] | 2009 | USA | Caucasian | 49(36) | PC | Plasma | Taqman | miR-16 | 46.00 | 89.00 | 23 | 4 | 26 | 32 | 0.620 |
| Hsu CM[49] | 2012 | China | Asian | 50(36) | HNSCC | Plasma | Taqman | cel-miR-39 | 83.30 | 51.10 | 42 | 18 | 8 | 18 | 0.741 |
| Liu X[50] | 2013 | China | Asian | 217(73) | NPC | Plasma | SYBR | RNU6 | 76.00 | 69.90 | 165 | 22 | 52 | 51 | 0.792 |
| Kishimoto T[51] | 2013 | Japan | Asian | 94(50) | BTC | Plasma | Taqman | miR-16 | 85.10 | 100.00 | 80 | 0 | 14 | 50 | 0.930 |
| Kishimoto T[51] | 2013 | Japan | Asian | 94(23) | BTC | Plasma | Taqman | miR-16 | 72.30 | 91.30 | 68 | 2 | 26 | 21 | 0.830 |
| Jones K[52] | 2014 | Australia | Caucasian | 42(20) | Lym | Plasma | SYBR | cel-miR-39 | 95.00 | 86.00 | 40 | 3 | 2 | 17 | 0.920 |
| Wang J[53] | 2014 | China | Asian | 52(49) | LSCC | Serum | SYBR | RNU6 | 69.20 | 81.60 | 36 | 9 | 16 | 40 | 0.801 |
| Huang W[54] | 2015 | China | Asian | 75(75) | PCa | PBMC | Taqman | RNU6 | 87.50 | 85.70 | 66 | 11 | 9 | 64 | 0.833 |
| Liu SS[55] | 2014 | China | Asian | 65(65) | RB | Plasma | SYBR | RNU6 | 46.00 | 72.00 | 30 | 18 | 35 | 47 | 0.548 |
Sen: sensitivity, Spe: specificity, Econ: endogenous control, Tp: true positive, Fp: false positive, Fn: false negative, Tn: true negative, AUC: area under ROC curve, LC: lung cancer, CC: colorectal cancer, GC: gastric cancer, HHC: hepatocellular cancer, BC: breast cancer, EC: esophageal cancer, PC: pancreatic cancer, HNSCC: head and neck squamous cell cancer, NPC: nasopharyngeal cancer, BTC: biliary tract cancer, Lym: Lymphoma, LSCC: laryngeal squamous cell cancer, PCa: prostate cancer, RB: retinoblastoma, PBMC: peripheral blood mononuclear cell, NA: not available
The qualities of included articles were assessed by the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) (Table 2) [60]. Item 1 was scored as “no” because samples with the known disease were recruited in each study. Item 2 was scored as “yes” because selection criteria were clearly described in each study. Item 3 was scored as “yes” because the reference standard could classify the target condition in each study. No studies stated the interval time between reference standard and index test so Item 4 was scored as “unclear”. Item 5 was scored as “yes” because reference standard was used in each study. Item 6 was scored as “yes” when all patients receive verification of the reference standard, or it would be scored as “unclear” when this information is not reported. Item 7 was scored as “yes” because the reference standard was independent of the index test in each study. For all the studies, the index test and the reference standard were described in detail so Item 8 and 9 were scored as “yes”. For Item 10, it was unclear if the index test was conducted without knowledge of the results from the reference standard. Item 11 was scored as “yes” because the reference standard results were defined without knowledge of the results from the index test. Item 12 was scored as “yes” because clinical data was available. Item 13 was scored as “yes” because uninterpretable test results were reported. Item 14 was scored as “yes” because withdrawals from the study were explained. With these references, the qualities of included articles were scored from 10 to 11 in this meta-analysis, which indicated that the quality of the included studies was satisfactory.
Table 2. QUADAS assessment for the studies included in meta-analysis for diagnosis.
| First author | Item 1 | Item 2 | Item 3 | Item 4 | Item 5 | Item 6 | Item 7 | Item 8 | Item 9 | Item 10 | Item 11 | Item 12 | Item 13 | Item 14 | Q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wei J[23] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Shen J[24] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Li Y[25] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Le HB[26] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Wang B[9] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Tang D[27] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Abd-EI-Fattah AA[28] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Mozzoni P[29] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Yang JS[56] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Kanaan Z[57] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Toiyama Y[30] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Liu GH[31] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Luo X[32] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Basati G[33] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Ogata-kawata H[34] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Tsujiura M[35] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Zheng Y[36] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Li BS[37] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Shiotani A[38] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Wu J[22] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Xu J[39] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Tomimaru Y[40] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Liu AM[41] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Amr KS[42] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Asaga S[43] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Mar-Aguilar F[44] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Gao J[45] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Toraih EA[46] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Kurashige J[47] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Wang J[48] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Hsu CM[49] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Liu X[50] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Kishimoto T[51] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Jones K[52] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Wang J[53] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Huang W[54] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Liu SS[55] | N | Y | Y | U | Y | U | Y | Y | Y | U | Y | Y | Y | Y | 10 |
| Zhuang C[59] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
| Motawi TM[58] | N | Y | Y | U | Y | Y | Y | Y | Y | U | Y | Y | Y | Y | 11 |
Item 1: Was the spectrum of patients representative of the patients? Item 2: Were selection criteria clearly described? Item 3: Is the reference standard likely to classify the target condition? Item 4: Is the time period between reference standard and index test short enough? Item 5: Did the whole sample use a reference standard of diagnosis? Item 6: Did patients receive the same reference standard regardless of the index test result? Item 7: Was the reference standard independent of the index test? Item 8: Was the index test described in sufficient detail? Item 9: Was the reference standard described in sufficient detail? Item 10: Were the index test results interpreted without knowledge of the results of the reference standard? Item 11: Were the reference standard results interpreted without knowledge of the results of the index test? Item 12: Were the same clinical data available when test results were interpreted as would be available when the test is used in practice? Item 13: Were uninterpretable/intermediate test results reported? Item 14: Were withdrawals from the study explained? [60] N: No, Y: Yes, N: unclear, Q: quadas
Overall diagnostic accuracy and subgroup analyses
Heterogeneity across studies was determined and forest plots of sensitivity and specificity were shown in Figure 2. There appeared to be significant heterogeneity (for sensitivity, Q = 368.44, p = 0.00, I2 = 87.24; for specificity, Q = 162.29, p = 0.00, I2 = 71.04). Therefore, the random effects model was selected. The pooled results for diagnostic accuracy were shown in Table 3. The overall sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and area under the curve (AUC) were 0.76 (0.71-0.80), 0.82 (0.79-0.85), 4.3 (3.6-5.1), 0.29 (0.24-0.35), 15 (11-20) and 0.86 (0.83-0.89), respectively. The summary receiver operator characteristic (SROC) curve was illustrated in Figure 3. The above results indicated that miR-21 had high diagnostic accuracy for various cancers.
Figure 2. Forest plots for miR-21 in various cancers.
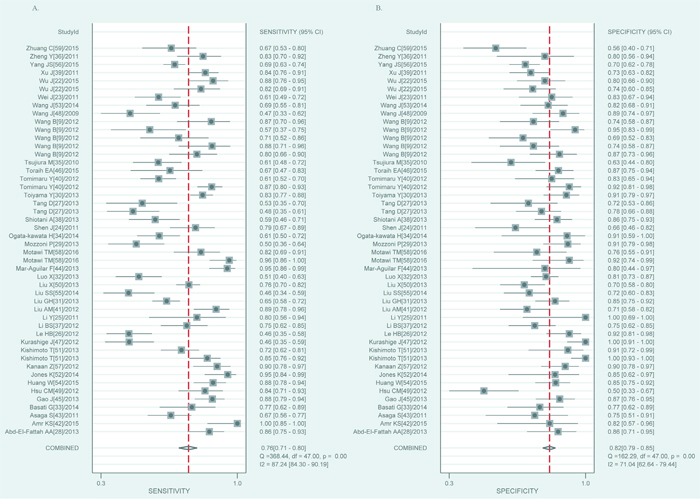
A. The pooled sensitivity. B. The pooled specificity.
Table 3. Summary results for diagnostic accuracy and their 95% confidence interval.
| Subgroup | Studies | Sensitivity (95%CI) | Specificity (95%CI) | PLR (95%CI) | NLR(95%CI) | DOR (95%CI) | AUC(95%CI) |
|---|---|---|---|---|---|---|---|
| Ethnicity | |||||||
| Asian | 34 | 0.74(0.68-0.78) | 0.82(0.77-0.86) | 4.1(3.2-5.2) | 0.32(0.27-0.39) | 13(9-18) | 0.85(0.81-0.88) |
| Caucasian | 8 | 0.77(0.59-0.89) | 0.84(0.79-0.88) | 4.8(3.3-6.8) | 0.28(0.14-0.53) | 17(7-44) | 0.85(0.81-0.88) |
| African | 5 | 0.89(0.75-0.96) | 0.86(0.79-0.90) | 6.2(4.0-9.5) | 0.13(0.05-0.32) | 49(15-154) | 0.87(0.84-0.90) |
| Sample | |||||||
| Serum | 27 | 0.78(0.72-0.83) | 0.83(0.78-0.86) | 4.5(3.6-5.7) | 0.26(0.21-0.34) | 17(12-25) | 0.88(0.84-0.90) |
| Plasma | 19 | 0.71(0.63-0.78) | 0.82(0.75-0.87) | 3.9(2.8-5.5) | 0.35(0.26-0.47) | 11(6-20) | 0.84(0.80-0.87) |
| Cancer | |||||||
| LC | 10 | 0.67(0.56-0.76) | 0.81(0.74-0.86) | 3.5(2.5-4.8) | 0.41(0.31-0.55) | 9(5-14) | 0.82(0.79-0.85) |
| CC | 7 | 0.75(0.63-0.83) | 0.84(0.79-0.87) | 4.6(3.4-6.3) | 0.30(0.20-0.46) | 15(8-30) | 0.86(0.83-0.89) |
| GC | 7 | 0.73(0.63-0.81) | 0.80(0.73-0.86) | 3.7(2.7-5.0) | 0.33(0.24-0.46) | 11(6-19) | 0.84(0.80-0.87) |
| HCC | 6 | 0.83(0.70-0.92) | 0.77(0.66-0.86) | 3.7(2.3-6.0) | 0.21(0.11-0.42) | 17(6-50) | 0.87(0.83-0.89) |
| BC | 7 | 0.85(0.75-0.91) | 0.85(0.80-0.90) | 5.8(4.0-8.5) | 0.18(0.10-0.31) | 33(14-76) | 0.89(0.86-0.91) |
| Others | 11 | 0.74(0.62-0.83) | 0.86(0.74-0.93) | 5.2(2.7-10.3) | 0.31(0.21-0.45) | 17(7-42) | 0.86(0.83-0.89) |
| Method | |||||||
| SYBR | 23 | 0.80(0.74-0.84) | 0.79(0.74-0.82) | 3.7(3.0-4.6) | 0.26(0.20-0.34) | 14(9-22) | 0.85(0.82-0.88) |
| Taqman | 25 | 0.72(0.64-0.79) | 0.85(0.80-0.89) | 4.8(3.6-6.6) | 0.33(0.25-0.42) | 15(9-24) | 0.87(0.84-0.90) |
| Endogenous control | |||||||
| MiR-16 | 20 | 0.69(0.62-0.76) | 0.87(0.82-0.91) | 5.3(3.9-7.2) | 0.35(0.28-0.44) | 15(10-23) | 0.87(0.83-0.89) |
| RNU6 | 14 | 0.74(0.65-0.81) | 0.78(0.73-0.81) | 3.3(2.6-4.2) | 0.34(0.25-0.47) | 10(6-17) | 0.82(0.78-0.85) |
| cel-miR-39 | 4 | 0.84(0.676-0.89) | 0.78(0.59-0.90) | 3.8(1.9-7.7) | 0.21(0.13-0.34) | 18(6-52) | 0.87(0.84-0.90) |
| Overall | 48 | 0.76(0.71-0.80) | 0.82(0.79-0.85) | 4.3(3.6-5.1) | 0.29(0.24-0.35) | 15(11-20) | 0.86(0.83-0.89) |
CI: confidence interval, PLR: positive likelihood ratio, NLR: negative likelihood ratio, DOR: diagnostic odds ratio, AUC: area under ROC curve
Figure 3. SROC curve of miR-21 for diagnostic value in various cancers.
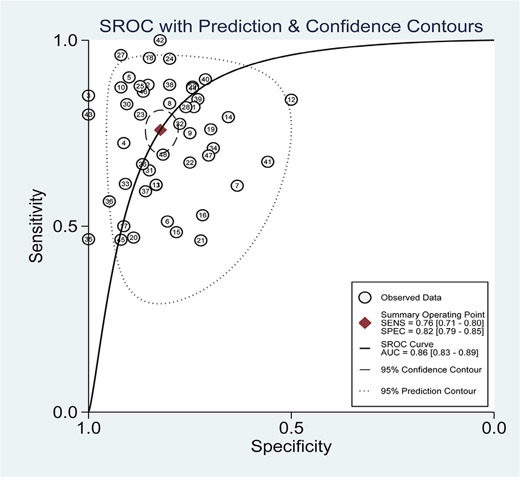
In order to identify potential sources of heterogeneity, subgroup analyses were performed based on ethnic groups, sample types, cancer types, test methods and endogenous controls. Based on the results of subgroup analyses, the pooled sensitivity, specificity, PLR, DOR and AUC of Caucasian-based studies were 0.77 (0.59-0.89), 0.84 (0.79-0.88), 4.8 (3.3-6.8), and 17 (7-44) respectively, which were respectively higher than that of Asian-based studies (0.74 (0.68-0.78), 0.82 (0.77-0.86), 4.1 (3.2-5.2) and 13(9-18)). Subgroup analyses by sample types showed that the pooled sensitivity, specificity, PLR, NLR, DOR and AUC were 0.78 (0.72-0.83), 0.83 (0.78-0.86), 4.5 (3.6-5.7), 0.26 (0.21-0.34), 17 (12-25) and 0.88 (0.84-0.90) respectively for serum-based miR-21 assays, and 0.71 (0.63-0.78), 0.82 (0.75-0.87), 3.9 (2.8-5.5), 0.35 (0.26-0.47), 11 (6-20) and 0.84 (0.80-0.87) respectively for plasma-based miR-21 assays, showing that serum-based miR-21 assays had a higher performance than plasma-based miR-21 assays. The pooled sensitivity of SYBR-based studies was 0.80 (0.74-0.84), which was higher than that of Taqman-based studies (0.72 (0.64-0.79)). On the contrary, for Taqman-based studies, the pooled specificity, PLR, NLR, DOR and AUC (0.85 (0.80-0.89), 4.8 (3.6-6.6), 0.33 (0.25-0.42), 15 (9-24) and 0.87 (0.84-0.90)) were respectively higher than that for SYBR-based studies (0.79 (0.74-0.82), 3.7 (3.0-4.6), 0.26 (0.20-0.34), 14 (9-22) and 0.85 (0.82-0.88)). The pooled results for diagnostic accuracy were also shown in Table 3.
Publication bias
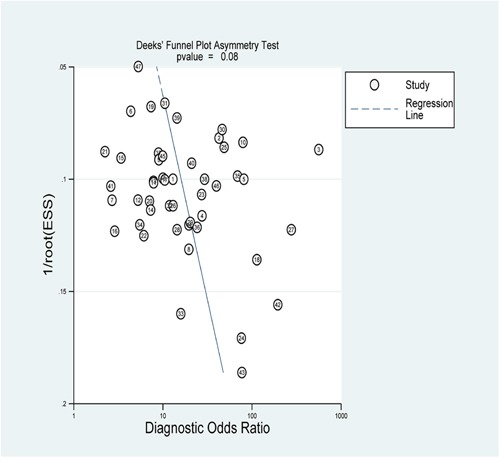
To assess the publication bias of the included studies, Deek's funnel plot was used in the meta-analysis. For our meta-analysis, the funnel plot was symmetric and the p value were 0.08 (Figure 4), indicating no significant publication bias occurred.
Figure 4. The Deek's test plot of the diagnostic meta-analysis.
Validation test on endometrial cancer patients
A total of 150 serum samples were detected, including those from endometrial cancer patients (n = 50), benign lesion patients (n = 50) and healthy controls (n = 50). No statistical differences in age were observed between healthy controls and benign lesion patients (p = 0.967), healthy controls and endometrial cancer patients (p = 0.992) or benign lesion patients and endometrial cancer patients (p = 0.983). The expression levels of serum miR-21 were significantly higher in benign lesion patients (p = 0.003) and endometrial cancer patients (p = 0.000) than that in healthy controls. Endometrial cancer patients showed higher miR-21 expression level (p = 0.000) compared with benign lesion patients (Figure 5). Receiver operating characteristic (ROC) curve was employed to assess the diagnostic value of serum miR-21 in benign lesion patients and endometrial cancer patients. Serum miR-21 showed an AUC value of 0.670 (0.562-0.777) in discriminating benign lesion patients from healthy controls. With a cutoff value of 1.502, the sensitivity and specificity were 56% and 76% respectively (Figure 6A). Serum miR-21 showed an AUC value of 0.831 (0.746-0.916) in discriminating endometrial cancer patients from healthy controls. With a cutoff value of 2.937, the sensitivity and specificity were 70% and 92% respectively (Figure 6B). Serum miR-21 showed an AUC value of 0.710 (0.608-0.813) in discriminating endometrial cancer patients from benign lesion patients. With a cutoff value of 3.457, the sensitivity and specificity were 64% and 76% respectively (Figure 6C).
Figure 5. Relative fold change of serum miR-21 in endometrial cancer patients (n = 50), benign lesion patients (n = 50) and healthy controls (n = 50).
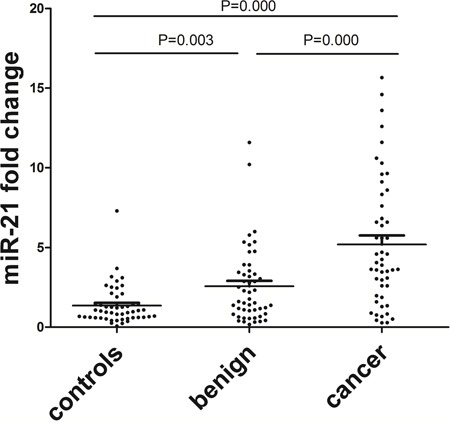
Benign lesion patients vs. healthy controls, p = 0.003; endometrial cancer patients vs. healthy controls, p = 0.000 and benign lesion patients vs. endometrial cancer patients, p = 0.000.
Figure 6. ROC curve analysis for evaluating serum miR-21 diagnostic performance.
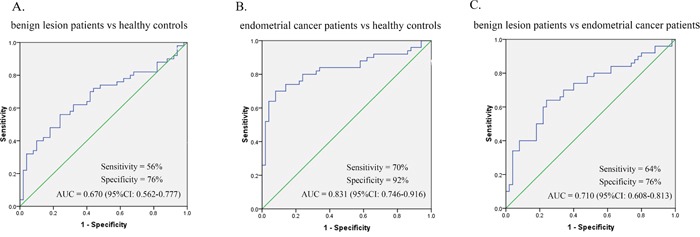
A. The performance in differentiating benign lesion patients from healthy controls. B. The performance in differentiating endometrial cancer patients from healthy controls. C. The performance in differentiating benign lesion patients from endometrial cancer patients.
DISCUSSION
In this meta-analysis, a total of 48 studies from 39 articles were included, involving 3,568 cancer patients and 2,248 controls. The overall sensitivity and specificity for identifying various cancers were 0.76 and 0.82 respectively, which were much higher than that of traditional serum-based cancer biomarkers. In this meta-analysis, the overall PLR was 4.3, which suggests that the possibility of developing cancer for potential patients is 4.3 times higher than that of healthy controls when circulating miR-21 levels are elevated; the overall NLR was 0.29, which indicates that the possibility of developing cancer is 29% when circulating miR-21 assay is normal. Moreover, in order to evaluate the diagnostic performance of circulating miR-21 in human cancers, SROC curve was plotted and the AUC was 0.86, suggesting the diagnosis performance of circulating miR-21 was excellent. All in all, the above results indicated that circulating miR-21 had high diagnostic accuracy for various human cancers. In order to trace the sources of heterogeneity, subgroup analyses were performed based on ethnic groups, specimen types, cancer types, test methods and endogenous controls which might have potential to cause variable results.
Based on the results of subgroup analyses, the diagnostic accuracy of Caucasian-based studies was higher than that of Asian-based studies. As for the selection of blood types, collection methods can significantly influence the miRNAs concentration. In this meta-analysis, serum samples have better performance than plasma which suggests that serum may be suitable in the detection of circulating miR-21. SYBR and Taqman probe are the most frequently used methods of real-time quantitative PCR. To compare the characteristic of these two methods, the sensitivity of SYBR was higher than that of Taqman probe while the specificity showed reverse result. The choice of endogenous controls is a contentious issue in the detection of circulating miRNAs. At present, miR-16, RNU6 and caenorhabditis elegans miRNA (cel-miR-39) are the most commonly used endogenous controls for circulating miR-21. In this meta-analysis, the performance of these three endogenous controls appeared to be equally matched.
Although some meta-analyses related to diagnostic value of circulating miR-21 in human cancers have been previously reported, there are some following advantages in this meta-analysis: (I) 14 kinds of cancers are included in this meta-analysis; (II) compared with previous reported meta-analysis, much more studies and samples are included; (III) more comprehensive subgroup analyses than any other reported meta-analysis are conducted, including ethnic groups, sample types, cancer types, test methods and endogenous controls; (IV) a relatively systematic analysis about pre-analysis variables is performed in this meta-analysis. In conclusion, this study is the first systematic review and comprehensive diagnostic meta-analysis to evaluate the overall diagnostic accuracy of circulating miR-21 for various cancers.
There are still several limitations in this meta-analysis despite the advantages mentioned above. First, there are only eight Caucasian-based studies and five African-based studies, which may lead to statistical insufficiency. Second, the heterogeneity exists in this meta-analysis. Subgroup analyses are applied, but the results could not fully explain the observed heterogeneity.
Except for the meta-analysis, we also identified the diagnostic value of circulating miR-21 in patients with endometrial cancer or benign lesion in this study, which can make up the deficiency in diagnostic studies on circulating miR-21 of endometrial cancer. Compared with the previous study [21], our validation test is more convincing. First, our validation test could provide more comprehensive data for the diagnosis research, such as cutoff value, the sensitivity and the specificity. Second, our validation test has larger sample size. Third, our validation test is the first study to detect serum miR-21 expression levels of benign lesion patients. In our validation test, serum miR-21 showed excellent performance on the diagnosis of endometrial cancer (p = 0.000), which was consistent with the results of our meta-analysis. The ROC curve analysis revealed robust levels of serum miR-21 in discriminating endometrial cancer patients from control subjects (sensitivity = 70%, specificity = 92%, AUC = 0.831). We also observed significance by comparing data collected from benign lesion patients with that of healthy controls (p = 0.003) or endometrial cancer patients (p = 0.000). In conclusion, serum miR-21 expression levels were statistically up-regulated in patients with benign lesion and endometrial cancer. In other words, the serum miR-21 expression levels of benign lesion patients were higher than that of healthy controls but lower than that of endometrial cancer patients.
This study assessed the role of circulating miR-21 as a biomarker for the diagnosis of various cancers by meta-analysis and further validated the role of circulating miR-21 in endometrial cancer. In conclusion, the meta-analysis shows that circulating miR-21 has excellent performance on the diagnosis for various cancers and the validation test demonstrates that serum miR-21 could be served as a novel biomarker for endometrial carcinoma.
MATERIALS AND METHODS
Search strategy
A systematical literature search for relevant articles was performed in PubMed, Embase and Cochrane Library as of 3 March, 2016. Our search was performed based on the following key terms: (miR-21 or microRNA-21 or miRNA-21) and (cancer or tumor or carcinoma or malignancy or neoplasm) and (circulating or serum or sera or plasma or blood) and (diagnostic or diagnosis or sensitivity or specificity). In addition, the reference lists of related review articles were scanned for further screening.
Inclusion and exclusion criteria
All articles included must be in accordance with the following criteria: (I) researches on patients with any type of human cancers; (II) cancers diagnosed by golden standard; (III) the relationship investigated between circulating miR-21 expression levels and cancer diagnosis; (IV) patients with benign diseases or healthy individuals as the control group; (V) sufficient data provided to execute a two-by-two table. Exclusion criteria were as follows: (I) review article, letter or meta-analysis; (II) non-English; (III) irrelevant to our topic; (IV) lack of key information; (V) less than 20 cancer samples. When the same patient groups were enrolled from more than one article, the largest and most comprehensive one was selected in this meta-analysis to avoid overlapping.
Data extraction and quality assessment
The whole process of this meta-analysis was executed and assessed by two independent reviewers (Yun Gao and Meiyu Dai). When inconsistencies occurred, consensuses were achieved through detailed discussions. The following data were extracted for each eligible study: the first author, year of publication, country, ethnicity, sample size, sample type, cancer type, detecting method, endogenous control and diagnostic results including sensitivity, specificity, true positive, false positive, false negative, true negative and AUC. The QUADAS [60] was used to systematically evaluate the quality of included articles. The QUADAS checklist includes 14 items which are listed in the notes of Table 2. Each item is assessed by “yes”, “no” or “unclear”. The answer “yes” scores a point, whereas “no” or “unclear” scores zero.
Validation test on endometrial cancer patients
In this study, we recruited 50 endometrial cancer patients, 50 benign lesion patients (uterine myoma and endometrial polyp) and 50 healthy controls matched in sex and age from the Fourth Affiliated Hospital of Guangxi Medical University. Endometrial cancer patients were confirmed by pathological diagnosis and the serum samples were collected before any treatment. Patients who had received chemotherapy, radiotherapy or operation were excluded from this study. This study was approved by institutional review board of the Fourth Affiliated Hospital of Guangxi Medical University.
MiRNA was extracted from 200μl serum using the miRNeasy Serum/Plasma Kit (Qiagen, CA, USA) according to the manufacturer's instructions. Cel-miR-39 was used as the reference gene. The reverse transcription reaction and real-time quantitative PCR were performed using miScript II RT Kit (Qiagen, CA, USA) and miScript SYBR Green PCR Kit (Qiagen, CA, USA) according to the manufacturer's instructions. The relative expression levels of miR-21 were calculated using 2−ΔΔCt method.
Statistical analysis
Meta-analysis was performed using the Stata 12.0 (Stata Corporation, College Station, TX, USA). The meta-analysis models were used to calculate pooled sensitivity, pooled specificity, PLR, NLR and DOR. The SROC curve was plotted and AUC was calculated to evaluate the performance of diagnostic studies. Heterogeneity was evaluated by the Q test and I2. When homogeneity is achieved (p ≥ 0.10, I2 ≤ 50%), a fixed-effect model is used for secondary analysis. If not (p < 0.10, I2 > 50%), a random-effect model is used to perform subgroup analyses for exploring sources of heterogeneity. Deek's funnel plot was performed to evaluate the publication bias. If p < 0.10, publication bias exists. In the validation test, statistical analyses were performed with SPSS 16.0 (SPSS, Inc., Chicago, IL). Statistical differences of miR-21 relative expression levels were evaluated by Mann-Whitney test. All tests were two sided and p < 0.05 was considered statistically significant. The ROC curve and AUC were performed to analyze the diagnostic value of miR-21. The cutoff value was obtained by Youden index.
Acknowledgments
This work was supported by grants from Key Laboratory Construction of Tumor Diseases Prevention in Liuzhou, Guangxi (No.2014G020403).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nature reviews Genetics. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nature reviews Clinical oncology. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 3.Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. MicroRNA: function, detection, and bioanalysis. Chemical reviews. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 4.Xiao YF, Yong X, Fan YH, Lu MH, Yang SM, Hu CJ. microRNA detection in feces, sputum, pleural effusion and urine: novel tools for cancer screening (Review) Oncology reports. 2013;30:535–544. doi: 10.3892/or.2013.2525. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Zhang KY, Liu SM, Sen S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules. 2014;19:1912–1938. doi: 10.3390/molecules19021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA biology. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karimi Kurdistani Z, Saberi S, Tsai KW, Mohammadi M. MicroRNA-21: mechanisms of oncogenesis and its application in diagnosis and prognosis of gastric cancer. Archives of Iranian medicine. 2015;18:524–536. [PubMed] [Google Scholar]
- 8.Chen J, Wang X. MicroRNA-21 in breast cancer: diagnostic and prognostic potential. Clinical & translational oncology. 2014;16:225–233. doi: 10.1007/s12094-013-1132-z. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. Journal of cancer research and clinical oncology. 2012;138:1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Wang JJ, Tang HM, To SS. Targeting strategies on miRNA-21 and PDCD4 for glioblastoma. Archives of biochemistry and biophysics. 2015;580:64–74. doi: 10.1016/j.abb.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Shen L, Wan Z, Ma Y, Wu L, Liu F, Zang H, Xin S. The clinical utility of microRNA-21 as novel biomarker for diagnosing human cancers. Tumour biology. 2015;36:1993–2005. doi: 10.1007/s13277-014-2806-z. [DOI] [PubMed] [Google Scholar]
- 12.Meng X, Xiao C, Zhao Y, Jia L, Tang Y, Li D. Meta-analysis of microarrays: diagnostic value of microRNA-21 as a biomarker for lung cancer. The International journal of biological markers. 2015;30:e282–285. doi: 10.5301/jbm.5000153. [DOI] [PubMed] [Google Scholar]
- 13.Wu R, Jiang Y, Wu Q, Li Q, Cheng D, Xu L, Zhang C, Zhang M, Ye L. Diagnostic value of microRNA-21 in the diagnosis of lung cancer: evidence from a meta-analysis involving 11 studies. Tumour biology. 2014;35:8829–8836. doi: 10.1007/s13277-014-2106-7. [DOI] [PubMed] [Google Scholar]
- 14.Yu W, Wang Z, Shen LI, Wei Q. Circulating microRNA-21 as a potential diagnostic marker for colorectal cancer: A meta-analysis. Molecular and clinical oncology. 2016;4:237–244. doi: 10.3892/mco.2015.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Yang X, Yang J, Zhen J, Zhang D. Serum microRNA-21 as a potential diagnostic biomarker for breast cancer: a systematic review and meta-analysis. Clinical and experimental medicine. 2016;16:29–35. doi: 10.1007/s10238-014-0332-3. [DOI] [PubMed] [Google Scholar]
- 16.Huang JT, Liu SM, Ma H, Yang Y, Zhang X, Sun H, Zhang X, Xu J, Wang J. Systematic review and meta-analysis: circulating mirnas for diagnosis of hepatocellular carcinoma. Journal of cellular physiology. 2016;231:328–335. doi: 10.1002/jcp.25135. [DOI] [PubMed] [Google Scholar]
- 17.Li G, Shen Q, Li C, Li D, Chen J, He M. Identification of circulating MicroRNAs as novel potential biomarkers for hepatocellular carcinoma detection: a systematic review and meta-analysis. Clinical & translational oncology. 2015;17:684–693. doi: 10.1007/s12094-015-1294-y. [DOI] [PubMed] [Google Scholar]
- 18.Li MY, Hu XX. Meta-analysis of microRNA expression profiling studies in human cervical cancer. Medical oncology. 2015;32:510. doi: 10.1007/s12032-015-0510-5. [DOI] [PubMed] [Google Scholar]
- 19.Shan L, Ji Q, Cheng G, Xia J, Liu D, Wu C, Zhu B, Ding Y. Diagnostic value of circulating miR-21 for colorectal cancer: a meta-analysis. Cancer biomarkers. 2015;15:47–56. doi: 10.3233/CBM-140437. [DOI] [PubMed] [Google Scholar]
- 20.Clancy C, Joyce MR, Kerin MJ. The use of circulating microRNAs as diagnostic biomarkers in colorectal cancer. Cancer biomarkers. 2015;15:103–113. doi: 10.3233/CBM-140456. [DOI] [PubMed] [Google Scholar]
- 21.Tsukamoto O, Miura K, Mishima H, Abe S, Kaneuchi M, Higashijima A, Miura S, Kinoshita A, Yoshiura K, Masuzaki H. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecologic oncology. 2014;132:715–721. doi: 10.1016/j.ygyno.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, Li G, Wang Z, Yao Y, Chen R, Pu X, Wang J. Circulating microRNA-21 is a potential diagnostic biomarker in gastric cancer. Disease markers. 2015;2015:435–656. doi: 10.1155/2015/435656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei J, Gao W, Zhu CJ, Liu YQ, Mei Z, Cheng T, Shu YQ. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chinese journal of cancer. 2011;30:407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, Jiang Z, Fang H, Katz RL, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Laboratory investigation. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Li W, Ouyang Q, Hu S, Tang J. Detection of lung cancer with blood microRNA-21 expression levels in Chinese population. Oncology letters. 2011;2:991–994. doi: 10.3892/ol.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le HB, Zhu WY, Chen DD, He JY, Huang YY, Liu XG, Zhang YK. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Medical oncology. 2012;29:3190–3197. doi: 10.1007/s12032-012-0303-z. [DOI] [PubMed] [Google Scholar]
- 27.Tang D, Shen Y, Wang M, Yang R, Wang Z, Sui A, Jiao W, Wang Y. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. European journal of cancer prevention. 2013;22:540–548. doi: 10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 28.Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell biochemistry and biophysics. 2013;67:875–884. doi: 10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 29.Mozzoni P, Banda I, Goldoni M, Corradi M, Tiseo M, Acampa O, Balestra V, Ampollini L, Casalini A, Carbognani P, Mutti A. Plasma and EBC microRNAs as early biomarkers of non-small-cell lung cancer. Biomarkers. 2013;18:679–686. doi: 10.3109/1354750X.2013.845610. [DOI] [PubMed] [Google Scholar]
- 30.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. Journal of the National Cancer Institute. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, Sun XF. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour biology. 2013;34:2175–2181. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 32.Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PloS one. 2013;8:e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basati G, Emami Razavi A, Abdi S, Mirzaei A. Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Medical oncology. 2014;31:205. doi: 10.1007/s12032-014-0205-3. [DOI] [PubMed] [Google Scholar]
- 34.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PloS one. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. British journal of cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, Cui L, Sun W, Zhou H, Yuan X, Huo M, Chen J, Lou Y, Guo J. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer biomarkers. 2011;10:71–77. doi: 10.3233/CBM-2011-0231. [DOI] [PubMed] [Google Scholar]
- 37.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF, Li N, Tang B, Liu KY, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PloS one. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, Inoue K, Uedo N, Iishi H, Haruma K. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. British journal of cancer. 2013;109:2323–2330. doi: 10.1038/bjc.2013.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Molecular carcinogenesis. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 40.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K, Kanto T, Doki Y, Mori M. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. Journal of hepatology. 2012;56:167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 41.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, Poon RT, Gao C, Luk JM. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amr KS, Ezzat WM, Elhosary YA, Hegazy AE, Fahim HH, Kamel RR. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene. 2016;575:66–70. doi: 10.1016/j.gene.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 43.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clinical chemistry. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 44.Mar-Aguilar F, Mendoza-Ramirez JA, Malagon-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodriguez-Padilla C, Resendez-Perez D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Disease markers. 2013;34:163–169. doi: 10.3233/DMA-120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J, Zhang Q, Xu J, Guo L, Li X. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chinese journal of cancer research. 2013;25:743–748. doi: 10.3978/j.issn.1000-9604.2013.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toraih EA, Mohammed EA, Farrag S, Ramsis N, Hosny S. Pilot study of serum microRNA-21 as a diagnostic and prognostic biomarker in Egyptian breast cancer patients. Molecular diagnosis & therapy. 2015;19:179–190. doi: 10.1007/s40291-015-0143-6. [DOI] [PubMed] [Google Scholar]
- 47.Kurashige J, Kamohara H, Watanabe M, Tanaka Y, Kinoshita K, Saito S, Hiyoshi Y, Iwatsuki M, Baba Y, Baba H. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. Journal of surgical oncology. 2012;106:188–192. doi: 10.1002/jso.23064. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer prevention research. 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu CM, Lin PM, Wang YM, Chen ZJ, Lin SF, Yang MY. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour biology. 2012;33:1933–1942. doi: 10.1007/s13277-012-0454-8. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Luo HN, Tian WD, Lu J, Li G, Wang L, Zhang B, Liang BJ, Peng XH, Lin SX, Peng Y, Li XP. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer biology & therapy. 2013;14:1133–1142. doi: 10.4161/cbt.26170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kishimoto T, Eguchi H, Nagano H, Kobayashi S, Akita H, Hama N, Wada H, Kawamoto K, Tomokuni A, Tomimaru Y, Umeshita K, Doki Y, Mori M. Plasma miR-21 is a novel diagnostic biomarker for biliary tract cancer. Cancer science. 2013;104:1626–1631. doi: 10.1111/cas.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones K, Nourse JP, Keane C, Bhatnagar A, Gandhi MK. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clinical cancer research. 2014;20:253–264. doi: 10.1158/1078-0432.CCR-13-1024. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M, Tian L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Medical oncology. 2014;31:148. doi: 10.1007/s12032-014-0148-8. [DOI] [PubMed] [Google Scholar]
- 54.Huang W, Kang XL, Cen S, Wang Y, Chen X. High-Level expression of microRNA-21 in peripheral blood mononuclear cells is a diagnostic and prognostic marker in prostate cancer. Genetic testing and molecular biomarkers. 2015;19:469–475. doi: 10.1089/gtmb.2015.0088. [DOI] [PubMed] [Google Scholar]
- 55.Liu SS, Wang YS, Sun YF, Miao LX, Wang J, Li YS, Liu HY, Liu QL. Plasma microRNA-320, microRNA-let-7e and microRNA-21 as novel potential biomarkers for the detection of retinoblastoma. Biomedical reports. 2014;2:424–428. doi: 10.3892/br.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang JS, Li BJ, Lu HW, Chen Y, Lu C, Zhu RX, Liu SH, Yi QT, Li J, Song CH. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour biology. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 57.Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Annals of surgery. 2012;256:544–551. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 58.Motawi TM, Sadik NA, Shaker OG, El Masry MR, Mohareb F. Study of microRNAs-21/221 as potential breast cancer biomarkers in Egyptian women. Gene. 2016 doi: 10.1016/j.gene.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 59.Zhuang C, Jiang W, Huang D, Xu L, Yang Q, Zheng L, Wang X, Hu L. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clinics and research in hepatology and gastroenterology. 2015 doi: 10.1016/j.clinre.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC medical research methodology. 2003;3:25. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]


