Abstract
Heat shock transcription factor 1 (HSF1), a key regulator of the heat-shock response, is deregulated in many cancers. HSF1 can mediate cancer cell survival and metastasis. High levels of HSF1 have been associated with poor prognosis in breast cancer. The nature of HSF1 upregulation needs to be validated in different cohorts to further validate its prognostic utility in breast cancer.
We first evaluated its expression in a cohort of breast cancer tissue microarrays with Oncotype DX recurrence scores available using immunohistochemistry. To further confirm the clinical relevance and prognostic impact, mutational and methylation status of the gene were also assessed in The Cancer Genome Atlas and publically available microarray datasets.
Immunohistochemical analysis showed that HSF1 expression is independent of Oncotype DX high recurrence score in ER-positive node-negative patients. Analysis of The Cancer Genome Atlas data revealed upregulation of HSF1 is not due to methylation or mutation. HSF1 copy number variations and amplifications (15%) were not associated with survival. In publicly available microarray datasets, a prognostic impact was observed in ER-positive tumors, but not in ER-negative tumors. Patients with ER-positive tumors with high HSF1 levels were associated with shorter overall survival (P = 0.00045) and relapse-free survival (P = 0.0057). In multivariable analysis, HSF1 remained a significant prognostic parameter.
The mRNA expression levels of HSF1 in ER-positive breast cancer are associated with both shorter relapse-free and overall survival. This prognostic impact is specific to mRNA expression, but stayed insignificant by protein expression or by analyzing amplification events.
Keywords: Breast cancer, HSF1, genomics, estrogen receptor, endocrine resistance
INTRODUCTION
Cells and organisms respond to stress by inducing heat shock proteins which act as molecular chaperones to restore protein homeostasis [1–4]. This adaptive mechanism is controlled by heat shock transcription factor (HSF1). When this transcription factor is activated, it gets phosphorylated, trimerized, and translocates to the nucleus. In the nucleus, it binds to specific DNA sequence motifs (known as heat shock elements) leading to the synthesis of heat shock proteins. In most experimental models, HSF1 enables adaptive changes in a diverse array of cellular processes including signal transduction, glucose metabolism and protein translation [5–10]. The binding of HSF1 to the DNA is dramatically different based on the phase of the cell cycle. HSF1 binds to only 35 target sites in mitotic chromatin, as opposed to 1242 target sites in freely cycling cells [11]. Its ability to activate transcription in mitosis is minimal. Consequently, mitotic cells are unable to induce expression of heat shock genes and are susceptible to protein damaging stress.
Cancer cells, being mutation prone and aneuploid, show a high activity of HSF1. Recently, Mendillo et al. identified genomewide target sites of HSF1 in breast cancer cell lines with different metastatic capacities [12]. They showed that HSF1 driven transcription is profoundly different in malignant cells compared with cells that are exposed to heat stress. Cancer cells seem to “hijack” HSF1 and utilize its transcriptional activity and central role in homeostasis to promote their growth and metastatic potential [13]. The importance of HSF1 in carcinogenesis is demonstrated by the dramatic reduction in susceptibility of HSF1-knockout mice to a wide spectrum of carcinogens [5, 14]. Similarly, depletion of HSF1 leads to marked decrease in proliferation and survival in established human cancer of cell lines [5, 9, 10, 14].
The role of HSF1 in breast cancer is not well established. Xi et al. have documented that deletion of HSF1 in mice overexpressing ERBB2 significantly reduces mammary tumorigenesis [15]. In addition the mice show a significant reduction in lung metastasis. Santagata et al. analyzed the expression of HSF1 in breast cancer samples from the Nurses' Health Study using immunohistochemistry [16]. They have documented an association of high HSF1 expression with increased mortality particularly in ER-positive patients (HR 2.1; P < 0.0001). They have postulated that targeting HSF1 might be a useful therapeutic strategy. In contrast Cheng et al. did not find HSF1 to be important in multivariate analysis in breast cancer [17]. In the current study, we focus on validating these findings using data from publicly available gene expression databases, including The Cancer Genome Atlas (TCGA), as well as by performing immunohistochemistry using a commercially available antibody. We confirmed that high expression of HSF1 mRNA, but not amplification, is associated with poor prognosis. However, we found only a weak association between protein expression and high Oncotype DX recurrence scores, a surrogate for adverse outcomes.
RESULTS
Upregulation HSF1 protein levels is independent of Oncotype DX high recurrence scores
Analysis of the National Surgical Adjuvant Breast and Bowel Project (B14 and B20) clinical trials has led to the development of the Oncotype DX recurrence score [20]. This score estimates the likelihood of disease recurrence in women with early-stage, ER-positive breast cancer and has been used as a surrogate for predicting outcomes. To validate the prognostic relevance of HSF1 observed by Santagata et al, we assessed the expression levels of HSF1 in a TMA cohort of patients with Oncotype DX scores using a commercially available antibody (see Materials & Methods) [16]. Immunohistochemistry results for HSF1 expression (Figure 1) were assessable for 161 (77.6% of 210) patients (87 low; 54 intermediate and 20 high Oncotype DX scores). As shown in Table 1, there was no association between Oncotype DX score and staining intensity (P = 0.23), percentage (less or more 10%; P = 0.17), or H-score (above or below 70; P = 0.08).
Figure 1. Expression of HSF1 in breast cancer.
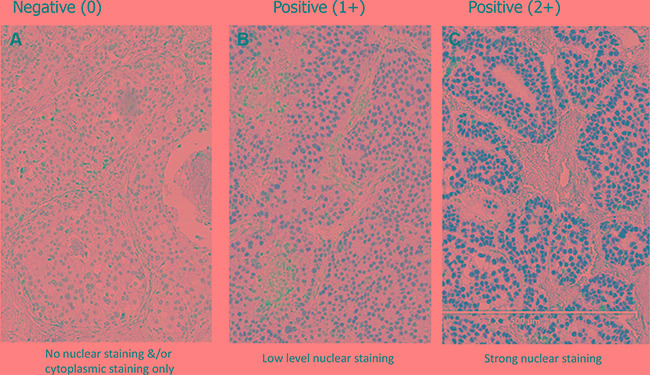
Representative immunohistochemical staining pattern of primary breast tumors for HSF1 (New England BioLabs, #4356). Nuclear staining intensity is categorized as the following; (A) no nuclear staining &/or cytoplasmic staining only (0), (B) low level nuclear staining (1+), (C) strong nuclear staining (2+).
Table 1. Correlation of the immunohistochemistry findings with Oncotype Dx scores.
| Table 1 | Nuclear Intensity | Percentage positivity | H-Score | ||||
|---|---|---|---|---|---|---|---|
| Oncotype DX score | 0 | 1 | 2 | < 10% | > 10% | H < 70 | H > 70 |
| High | 4 | 9 | 7 | 4 | 16 | 9 | 11 |
| Intermediate | 17 | 22 | 15 | 18 | 36 | 27 | 27 |
| Low | 36 | 36 | 15 | 38 | 49 | 57 | 30 |
| Grand Total | 57 | 67 | 37 | 60 | 101 | 93 | 68 |
HSF1 expression and copy number alterations in primary breast tumors
We next analyzed the genomic alterations in TCGA breast cancer dataset (cBio Cancer Genomics Portal). Copy number alterations (CNA) of HSF1 were observed in 146 (15%) out of 962 breast tumors (Figure 2A–2C). Of these, 14.8% were due to amplification and 0.2% due to homozygous deletion. In addition, only 0.2% patients had mutations which were not located in major domains (Figure 2D). Of the tumors with altered HSF1 expression, upregulation (25.6%) was more prevalent rather than downregulation (0.1%) (Figure 2E). These analyses confirm that amplification is the most common alteration, while mutations of HSF1 are not frequent in breast cancer. Survival analysis of cases with and without amplifications, in all 962 patients or ER+ and ER- subsets did not show an association with overall survival or disease-free survival in TCGA dataset (Supplementary Figure S1). We further analyzed 737 out of 1079 cases available with methylation data (HM450). Methylation was inversely associated with the expression of HSF1 but the correlation was weak (Spearman 0.45; Figure 2F). However, cases with alterations versus without alterations did not significantly correlate with the overall survival or disease-free survival in TCGA dataset (Supplementary Figure S1).
Figure 2. The cBio cancer genomics portal analysis.
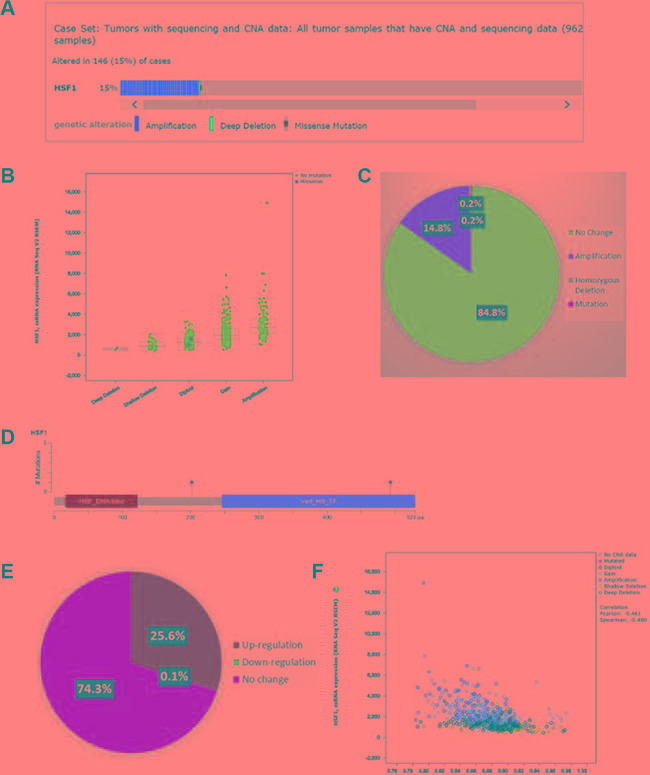
Copy number alterations (CNAs) and Mutations of HSF1 in TCGA breast cancer dataset. (A) individual genes are represented as rows, individual cases or patients are represented as columns, and glyphs and/ or color-coding is used to compactly summarize distinct genomic alterations, including somatic mutations and copy number alterations (CNAs), (B) correlation of CNAs with HSF1 expression (RNA-seq), (C) the pie chart showing the frequency of genomic alterations and mutations, (D) mutation details for HSF1, (E) the pie chart showing the frequency of upregulated or downregulated gene expression, (F) correlation of methylation status (HM450) with mRNA expression (RNA-seq) of HSF1.
High expression of HSF1 correlates with poor prognosis in patients with ER-positive breast cancer – The Affymetrix microarray datasets
The current study differed from the Santagata study in that a monoclonal antibody was used for the immunohistochemical analysis [16]. To further analyze whether the observed differences could be related to technical issues or biologic relevance, we next evaluated the prognostic value of HSF1 mRNA expression using overall survival and relapse-free survival as endpoints in subsets of breast cancer represented in 11 microarray datasets (GOBO tool). High expression of HSF1 was significantly associated with shorter overall survival (ANOVA; P = 0.00045), and relapse-free survival (ANOVA; P = 0.0057) in ER-positive breast cancer patients (Figure 3A, 3B), but not in ER-negative tumors (data not shown). In this patient group, high expression levels of HSF1 also correlated with worse overall survival in node-negative subset (ANOVA; P = 0.00022; Figure 3C), showing that cases with high HSF1 have a significant mortality risk when compared with low level of HSF1 cases. We also assessed the prognostic value of HSF1 patients in untreated versus tamoxifen-treated patients. High HSF1 expression is associated with shorter overall survival (ANOVA; P = 0.00049) in untreated population (Figure 3D) and with shorter relapse-free survival for tamoxifen-treated patients (Figure 3E). These results indicate that targeting HSF1 may help prevent development of metastases in ER-positive patients.
Figure 3. Kaplan-Meier analysis using the Affymetrix data sets (GOBO).
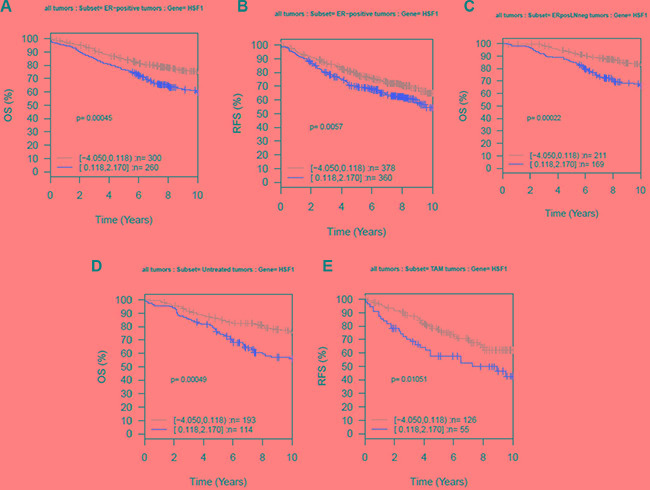
Ten-year (A) overall survival (OS) and (B) relapse-free survival (RFS) for ER-positive tumors, (C) ER-positive and lymph node (LN)-negative tumors, (D) OS for untreated tumors, and (E) RFS for tamoxifen (Tam)-treated tumors. The expression analysis was stratified into two quantiles based on HSF1 expression; low expression (gray) and high expression (red).
Multivariable Cox hazard analysis of HSF1 expression with clinical variables showed that low HSF1 expression was associated with good prognosis in all patients (overall survival (hazard ratio [HR] = 0.65; 95% CI = 0.47–0.89; P = 0.0083) and relapse-free survival (HR = 0.71; 95% CI = 0.51–1; P = 0.048) (Table 2). The other factors that retained significance in multivariable analysis were nodal status, tumor grade, age and tumor size (> 2 cm) for the overall survival. Tumor size (> 20 mm) was the only significant factor for relapse-free survival (HR = 1.98; 95% CI = 1.5–2.62; P = 1.58e–06). These results further validate high HSF1 is an independent poor prognostic marker in ER-positive tumors using large cohorts of breast cancer patients.
Table 2. Multivariable analysis in ER+ patients for overall survival and relapse-free survival (Affymetrix datasets).
| OS | RFS | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P value* | HR (95% CI) | P value* |
| low HSF1 versus high HSF1 | 0.65 (0.47–0.89) | 0.0083 | 0.71 (0.51–1) | 0.048 |
| LN- versus LN+ | 0.48 (0.35–0.67) | 1.93e-05 | 0.77 (0.57–1.03) | 0.079 |
| Grade 3 versus 1 + 2 | 1.5 (1.05–2.15) | 0.025 | 1.25 (0.92–1.7) | 0.148 |
| Age > 50yrs versus < 50yrs | 1.49 (1.05–2.13) | 0.025 | 0.82 (0.62–1.1) | 0.18 |
| Tumor size > 20 mm versus < 20 mm | 2.07 (1.48–2.9) | 2.54e-05 | 1.98 (1.5–2.62) | 1.58e–06 |
DISCUSSION
Santagata et al, in their study of the Nurses Health Cohort, demonstrated that HSF1 is an independent prognostic marker in breast cancer in a TMA-based analysis using immunohistochemical methods [16]. However, their study was performed using a cocktail of three different antibodies to detect HSF1 protein in TMAs. More recently, a monoclonal antibody directed against HSF1 has become commercially available. The differences in isotope specificity of this antibody and the antibody-cocktail to HSF1 is not known. Apart from the differences in reagents, our study employed a surrogate endpoint, Oncotype DX assay, for analyzing prognostic impact. In our study, upregulation of HSF1 protein was independent of Oncotype DX scores. In order to confirm that differences observed could be attributed to technical and tissue sampling issues and not related to biological relevance, we further evaluated the role of HSF1 in breast cancer using publically available datasets.
Analysis of the TCGA dataset showed that mutations were not common (0.2%); copy number alterations were noted in 14.8%, nearly 50% of which were amplification events. HSF1 amplification or upregulation of HSF1 was not associated with prognosis in all patients, or in ER-positive or ER-negative subsets. However, it is recognized that the TCGA dataset has limited followup/survival information, so we further analyzed the prognostic relevance of HSF1 in breast cancer in a publically available large (n = 1881) Affymetrix-based gene expression dataset. These analyses showed upregulation of HSF1 was associated with shorter overall survival and relapse-free survival in ER-positive, but not in ER-negative patients. HighHSF1 expression was significantly associated with worse outcome in treatment-naïve tumors and in node-negative tumors. Cheng et al. [17] have also analyzed the role of HSP90, and HSF1 in multiple data sets including many that are included in the current study. They failed to observe a significant impact of HSF1 in their analysis. The major differences in the two studies are the number of patients analyzed and analytical methods. They focused on subsets of patients with highest (top 10% or 25%) and lowest expression and did not find an impact once clinical confounding features were included. In contrast, our multivariable analysis that included age, tumor grade ER status, and treatment type confirmed that high HSF1 is an independent factor of poor clinical outcome in ER-positive breast cancer. Collectively, this data makes a strong case for a role of HSF1 in endocrine resistance and recurrence/metastasis.
HSF1 is a key transcription factor in the regulation of cellular homeostasis and modulates protein folding, stability, and protein-protein interactions. In response to a variety of stresses, HSF1 binds to the promoter regions of heat shock protein genes and drives transcription of these genes including HSP90 and HSP70. Although HSF1 supports the survival of normal cells under stress, aberrant upregulation of HSF1 has been shown to promote tumor cell survival and cancer progression. Therefore, HSP90 inhibitors can serve as potential target for therapeutics for patients with high HSF1 in ER-positive breast cancer. However, HSF1 can serve as a hub regulating a transcriptional program that can be distinct from heat shock protein network [12]. Therefore, better understanding of this program is necessary to target HSF1 therapeutically using agents other than HSP90 inhibitors.
In summary, high expression of HSF1 mRNA was associated with both shorter relapse-free survival and overall survival in patients with ER-positive breast cancer. Identification of high HSF1 could stratify patients at greater risk for recurrence/metastasis development independently of Oncotype DX. Therapies that can downregulate HSF1 need to be explored to prevent recurrence/metastasis development in ER-positive breast cancer.
MATERIALS AND METHODS
Tissue microarray patients
All protocols were reviewed and approved by the Institutional Review Board of Indiana University. All archival formalin-fixed, paraffin-embedded tumor blocks in this study were from patients with ER-positive (greater than 1% expression as per ASCO-CAP guidelines) breast carcinomas at the Indiana University Health Pathology Lab (IUHPL). The tissue samples consisted of a tissue microarray (TMA) containing duplicate (1mm) cores. On all of these 210 tumors, Oncotype DX recurrence score was available. The distribution of the Oncotype DX scores in the TMA series was 120 low; 65 intermediate; and 25 high scores (Supplementary Table S1).
Immunohistochemistry
Archival formalin-fixed, paraffin-embedded TMA blocks were cut at 4μ thickness. After deparaffinization and hydration, sections were incubated with the primary antibody. The HSF1 rabbit polyclonal antibody (New England BioLabs #4356) was diluted to 1/100 stained using the BOND-III (Leica Biosystems, Melbourne) automated stainer with Bond Polymer Refine Detection system (Leica Biosystems Newcastle, DS9800), Bond Diluent (Leica Biosystems Newcastle, AR9352) and ER2 reagent (Leica Biosystems Newcastle, AR9640). The ER2 solution was incubated for 20 minutes followed by a 15minute incubation with the diluted antibody. HSF1 scoring was based on H-Score method combining percentage and intensity.
Analysis of the cancer genome atlas (TCGA)
Somatic mutation rate, DNA copy number alterations (CNAs), mRNA, and methylation status for HSF1 were analyzed using the cBioPortal for Cancer Genomics (http://cbioportal.org). The portal is a Web resource to analyze complex cancer genomics data including genetic, epigenetic, gene expression and proteomic events [17, 18]. Tumors with CNA and RNA-sequencing data available were analyzed. For the survival analysis, cases with and without amplifications were identified and their survival data downloaded. Log-rank analysis was performed using Kaplan Meier method using the R statistical package.
Analysis of publicly available datasets
Expression of HSF1 was analyzed based on ER status, molecular subtypes, and other clinicopathological parameters using the datasets from the gene expression-based outcome for breast cancer online algorithm (GOBO) [19]. GOBO is a web-based analysis tool that utilizes 11 publicly available Affymetrix U133A gene expression data curated from 1881 breast cancer patients with associated stage, grade, nodal status, and intrinsic molecular classification [19]. Of all 1881 tumors, the groups were distributed as follows: a) ER-positive patients (n = 1225) of which 326 patients treated with tamoxifen alone while the remainder 927 did not receive systemic therapy and b) ER-negative patients (n = 395). Clinical characteristics of individual datasets were described previously [19]. Association of outcome was stratified into the two quantiles based on HSF1 gene expression level for each patient cohort with overall survival or relapse-free survival as endpoints and 10-year censoring in the above groups. The Kaplan-Meier survival analysis was calculated using Cox proportional hazard model, and the score test of the proportional hazard model was equivalent to the log-rank test.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
Sunil Badve is a Komen Scholar. Studies were supported by funds from the Susan G Komen for the Cure to Sunil Badve. Authors acknowledge the bioinformatics support provided by Matthew Palakal PhD., and Meeta Pradhan PhD. as well as Statistical support provided by Susan Perkins PhD from the IUSCC Statistical Core.
Footnotes
CONFLICTS OF INTEREST
This manuscript has not been and will not be submitted to any other journal while it is under your consideration. Each author listed on the manuscript has seen and approved the submission of this version of the manuscript and takes full responsibility for the manuscript. The authors have no conflicts of interest to disclose. No form of payment was given to anyone to produce the manuscript.
GRANT SUPPORT
This work was by research support provided by Leica. Sunil Badve is supported by the Susan G Komen for the Cure Scholar Award.
REFERENCES
- 1.Shamovsky I, Gershon D. Novel regulatory factors of HSF-1 activation: facts and perspectives regarding their involvement in the age-associated attenuation of the heat shock response. Mechanisms of ageing and development. 2004;125:767–75. doi: 10.1016/j.mad.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–60. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 3.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert opinion on therapeutic targets. 2009;13:469–78. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 4.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nature reviews Cancer. 2005;5:761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 5.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khaleque MA, Bharti A, Gong J, et al. Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene. 2008;27:1886–93. doi: 10.1038/sj.onc.1210834. [DOI] [PubMed] [Google Scholar]
- 7.Lee JH, Schoffl F. An Hsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana. Molecular & general genetics: MGG. 1996;252:11–9. doi: 10.1007/s004389670002. [DOI] [PubMed] [Google Scholar]
- 8.Lee YJ, Lee HJ, Lee JS, et al. A novel function for HSF1-induced mitotic exit failure and genomic instability through direct interaction between HSF1 and Cdc20. Oncogene. 2008;27:2999–3009. doi: 10.1038/sj.onc.1210966. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Shi H, Sevilimedu A, Liachko N, Nelson HC, Lis JT. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic acids research. 2006;34:3755–61. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Liu H, Liu Z, et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer research. 2011;71:4585–97. doi: 10.1158/0008-5472.CAN-11-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vihervaara A, Sergelius C, Vasara J, et al. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3388–97. doi: 10.1073/pnas.1305275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendillo ML, Santagata S, Koeva M, et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–62. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vihervaara A, Sistonen L. HSF1 at a glance. Journal of cell science. 2014;127:261–6. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 14.Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007;26:5086–97. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- 15.Xi C, Hu Y, Buckhaults P, Moskophidis D, Mivechi NF. Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. The Journal of biological chemistry. 2012;287:35646–57. doi: 10.1074/jbc.M112.377481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santagata S, Hu R, Lin NU, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18378–83. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PloS one. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


