Abstract
The cloning and expression of the CviPII DNA nicking and modification system encoded by chlorella virus NYs-1 is described. The system consists of a co-linear MTase encoding gene (cviPIIM) and a nicking endonuclease encoding gene (cviPIINt) separated by 12 nt. M.CviPII possesses eight conserved amino acid motifs (I to VIII) typical of C5 MTases, but, like another chlorella virus MTase M.CviJI, lacks conserved motifs IX and X. In addition to modification of the first cytosine in CCD (D = A, G or T) sequences, M.CviPII modifies both the first two cytosines in CCAA and CCCG sites as well. Nt.CviPII has significant amino acid sequence similarity to Type II restriction endonuclease CviJI that recognizes an overlapping sequence (RG^CY). Nt.CviPII was expressed in Escherichia coli with or without a His-tag in a host pre-modified by M.CviPII. Recombinant Nt.CviPII recognizes the DNA sequence CCD and cleaves the phosphodiester bond 5′ of the first cytosine while the other strand of DNA at this site is not affected. Nt.CviPII displays site preferences with CCR (R = A or G) sites preferred over CCT sites. Nt.CviPII is active from 16 to 65°C with a temperature optimum of 30–45°C. Nt.CviPII can be used to generate single-stranded DNAs (ssDNAs) for isothermal strand-displacement amplification. Nt.CviPII was used in combination with Bst DNA polymerase I large fragment to rapidly amplify anonymous DNA from genomic DNA or from a single bacterial colony.
INTRODUCTION
DNA nicking endonucleases (NEases) cleave one strand of DNA in a sequence-specific and strand-specific manner. Although there are over 240 Type II restriction endonucleases (REases) with unique specificities isolated from bacterial and viral sources, only a few site-specific NEases are currently commercially available (1). Efforts to develop more NEases consist of both genetic engineering of existing REases and screening of bacterial and viral sources for new enzymes. The NEases N.BstNBI and N.BstSEI (5′-GAGTCN4^-3′) were found in Bacillus stearothermophilus (2,3) and the bottom-strand NEase Nb.BsrDI (5′-^CATTGC-3′), a large subunit of BsrDI, was found in another isolate of B.stearothermophilus D70 (unpublished results). Two natural NEases Nt.CviPII (^CCD) (4) and Nt.CviQXI (R^AG) (5) are coded by chlorella viruses.
Plaque-forming chlorella viruses are ubiquitous in freshwater environments throughout the world and titers as high as 100 000 infectious particles per ml of native water have been reported (6). The viruses have many unusual properties including double-stranded DNA (dsDNA) genomes as large as 380 kb. Chlorella viruses can be distinguished from one another by the amount and site-specificity of genomic DNA methylation (7). DNA from each chlorella virus contains C5-methylcytosine (m5C) in amounts ranging from 0.1 to 47% of the total cytosines. Many viruses also contain N6-methyladenine (m6A) in amounts ranging from 1.5 to 37% of the total adenines. These properties led to the discovery that the chlorella viruses encode multiple DNA MTases and site-specific NEases/REases that cleave either one or both strands of dsDNA (8,9).
The virus NYs-1 genome contains a high level of m5C (47%) as well as 11% m6A (10). About half of the m5C methylation can be attributed to the NYs-1 encoded dinucleotide DNA MTase M.CviPI, which modifies GC sequences (11). NYs-1 encodes a site-specific NEase (now called Nt.CviPII) reported to recognize the sequence CCD and cleave 5′ to the first cytosine (4). The enzyme cleaves CmCD sequences but not mCCD sequences. Nt.CviPII creates breaks in dsDNA whenever two CCD sequences on opposite strands are close enough for the strands to separate; when the CCD sequences on opposite strands are further apart only a portion of the strands separate.
The low quantities of Nt.CviPII produced in a NYs-1 infected lysate limits its potential use in manipulating DNA. To overcome this limitation, we cloned and expressed this nicking–modification (N–M) system in Escherichia coli. Here, we describe the characterization of the CviPII N–M system, and Nt.CviPII's use in generating DNA oligonucleotides for random DNA amplification. Cloning Nt.CviPII required overcoming the toxicity problem caused by the frequent DNA nicking activity of the enzyme so as to produce a sufficient amount of recombinant Nt.CviPII. We also describe a novel method for isothermal DNA amplification without addition of exogenous primers.
MATERIALS AND METHODS
Molecular cloning of CviPII MTase and NEase genes
Procedures used to produce and purify chlorella virus NYs-1 and isolate NYs-1 DNA were described previously (12). NYs-1 DNA was partially digested by 2-fold serial dilutions of Sau3AI and ligated into BamHI-digested and CIP-treated pUCAC (a derivative of pUC19 with a chloramphenicol-resistant gene inserted into the AflIII site). The ligated products were used to transform E.coli ER1992 (13). Clones that expressed M.CviPII were selected by digesting pooled ampicillin (Amp) and chloramphenicol (Cam) resistant plasmids with MspI (cleaves CCGG and CmCGG sequences but not mCCGG sequence) prior to a second round of E.coli ER1992 transformation. Clones partially resistant to MspI were selected, which could contain the cviPIIM gene. Inserts from the plasmids were sequenced using pUC universal forward and reverse primers as well as custom-designed primers. The sequences were subjected to a BLAST search against REBASE (1) and GenBank (14).
DNA sequence adjacent to the cviPIIM gene was obtained by directly sequencing the NYs-1 genomic DNA using custom primers. To clone the cviPIIM and cviPIINt genes for expression, PCR primers were designed to amplify the genes from NYs-1 genomic DNA (Table 1). The MTase gene was cloned into the SphI and SalI sites of pUC19 where it is under the control of the lac promoter and the plasmid used to transform E.coli ER2502. The resultant pUC-cviPIIM was transferred into E.coli expression host ER2683. An N-terminal [His]6-tag coding sequence was added to the cviPIINt gene during PCR. The amplified product was ligated downstream of the tac promoter (15) sequence of plasmid pR976 (obtained from I. Hall and P. Riggs, NEB) such that the start codon was situated 18 nt downstream from the ribosome-binding site. The ligated DNA was used to transform the pre-modified ER2683 containing pUC-cviPIIM. All bacterial strains were obtained from M. Sibley and E. Raleigh of NEB.
Table 1. Oligos for expression and construction of nicking substrates.
| Name | Sequence (5′ to 3′) | Remarks |
|---|---|---|
| Cloning of cviPIIM | ||
| MP-SphI-F | 5′-GGTGGTGCATGCAAATAAATGCGTACAAAGTGATCGTATTTTTATC-3′ | SphI site added for cloning |
| MP-SalI-R | 5′-GGTGGTGTCGACTTAATAATGCATAAGATCACCCAAATATTC-3′ | SalI site added for cloning |
| Cloning of cviPIINt | ||
| NP-Nhis-PstI-F | 5′-GTGCTGCAGATGCATCATCATCATCATCATTATATATATATGTCTACTCCGCAGGCAAAGACCAAATAT-3′ | N-terminal His-tag and PstI site added for cloning |
| NYS1-2490-KpnI-R | 5′-CGGGGTACCTTAGTCGATCTCCAATCGGCCATTACGATATCGATG-3′ | KpnI site added for cloning |
| Construction of Nt.CviPII substrate | ||
| NP-sub-F2 | 5′-GTGAATTCACAAGTTAATACGACTCACTATAGCTGAAGTCGCGACTGCGTCAGCGAACAGCAG-3′ | EcoRI site added for cloning |
| NP-sub-F1 | 5′CGCGACTGCGTCAGCGAACAGCAGCTGTGCAGACGTTACGAGCACGACTAGCACAAGTGCTGA 3′ | |
| NP-sub-R1 | 5′-GTATCGCTCGAGCAGCTGCTGCTCGCTGATCACGTTACGACTTCAGCACTTGTGCTAGTCGTGCTC-3′ | |
| NP-sub-R2-CCA | 5′-AGTAAGCAAGCTTATCGCTTGCATTCGCCACAAGTATCTTACGACGTATCGCTCGAGCAGCTGCTGCTC-3′ | HindIII site added for cloning |
| NP-sub-R2-CCG | 5′-AGTAAGCAAGCTTATCGCTTGCATTCGCCGCAAGTATCTTACGACGTATCGCTCGAGCAGCTGCTGCTC-3′ | |
| NP-sub-R2-CCC | 5′-AGTAAGCAAGCTTATCGCTTGCATTCGCCCCAAGTATCTTACGACGTATCGCTCGAGCAGCTGCTGCTC-3′ | |
| NP-sub-R2-CCT | 5′-AGTAAGCAAGCTTATCGCTTGCATTCGCCTCAAGTATCTTACGACGTATCGCTCGAGCAGCTGCTGCTC-3′ |
Bold sequences CCN sites.
MTase protection assay
Modified plasmid pUC-cviPIIM was isolated from ER2502 [pUC-cviPIIM] cells after growing in medium containing 0.25 mM IPTG. The plasmid DNA was incubated with MspI (C^CGG) or ScrFI (CC^NGG) at 37°C for 1 h in NEBuffer 2 (10 mM Tris–HCl, 50 mM NaCl, 10 mM MgCl2 and 1 mM DTT, pH 7.9). The digested DNA was analyzed by agarose gel electrophoresis.
Determination of methylation site
Plasmid pUC-cviPIIM used in the MTase protection assay was treated with sodium bisulfite (EZ DNA Methylation Kit, Zymo Research) and the treated DNA was purified on a spin-column. The cviPIIM gene was amplified by PCR using primers MP-SphI-F and MP-SalI-F (Table 1). Untreated plasmid, amplified by the same pair of primers, served as a control. The PCR products were gel-purified and sequenced using the same PCR primers. Methylated cytosines are protected from sodium bisulfite, which converts un-modified cytosines to uracils; the uracils are amplified as thymidines in PCR. Thus, comparison of sodium bisulfite treated DNA sequences with control sequences identifies the sites containing m5C: cytosines-turned-thymidines were un-modified cytosines while those that remained cytosines were methylated.
Expression and purification of recombinant Nt.CviPII
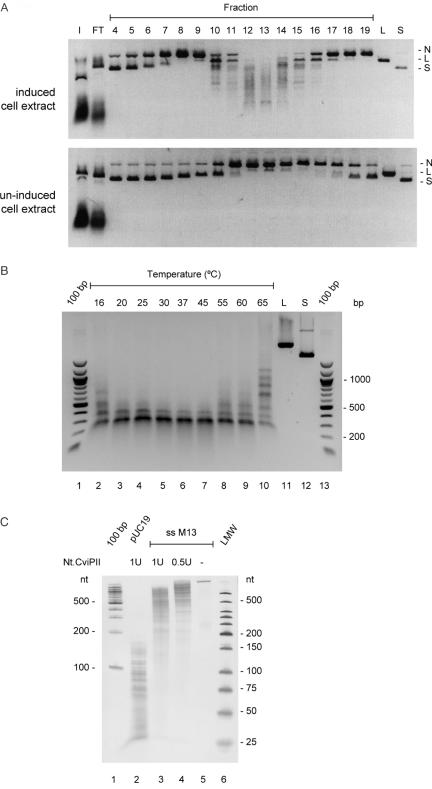
A single colony of ER2683, transformed with both pUC-cviPIIM and pR976-cviPIINt, was grown to mid-log phase in 2 l of rich media (10 g/l of Tryptone, 5 g/l of yeast extract and 5 g/l NaCl, pH adjusted to 7.2 with NaOH) containing Ampicillin, (Amp, 0.1 mg/ml), kanamycin (Km, 0.05 mg/ml) and tetracycline (Tc, 0.01 mg/ml) at 30°C at 280 r.p.m. One liter of culture was induced with 0.25 mM IPTG and the other was not. Both cultures were incubated at 16°C for 18 h and cells were harvested by centrifugation (3550 g). The cell pellets (wet weight of 3.9 g for the induced culture and 5.0 g for the uninduced culture) were sonicated in 100 ml of 20 mM NaHPO4 and 0.1 M NaCl, pH 7.4. After cleared lysates were obtained by centrifugation (26 700 g), the soluble fractions were loaded on an SP FF column (25 ml bed volume, Amersham Biosciences) and eluted with a linear gradient of 0.1–1 M NaCl. Two-microliter fractions were incubated with 0.5 μg of pUC19 at 37°C for 1 h. Reactions were stopped by adding 25 mM EDTA and analyzed by agarose gel electrophoresis.
To purify recombinant His-tagged Nt.CviPII, the cell extracts derived from 6 l of cell cultures were loaded on a Nickel-charged HisTrap column (HisTrap HP, 5 ml bed volume, Amersham Biosciences) and protein was eluted with a step gradient of 50, 100, 200, 300, 400 and 500 mM imidazole in 20 mM NaHPO4 and 0.5 M NaCl, pH 7.4. DNA nicking activity was detected in fractions eluting in 50 and 300 mM imidazole. The 50 mM imidazole fraction was further purified on a heparin FF column (25 ml bed volume, Amersham Biosciences) and an SP FF column (25 ml bed volume, Amersham Biosciences) (this preparation is referred to as His−-Nt.CviPII). The 300 mM imidazole fraction was concentrated and assayed (referred to as His+-Nt.CviPII). All purified protein preparations were concentrated by VIVASPIN 20 (10 000 MWCO, VIVASCIENCE) and stored in 10 mM NaHPO4 and 250 mM NaCl, pH 7.0 with 50% glycerol at −20°C.
DNA nicking activity assay
Nicking activity was assayed with 0.5 μg of pUC19 in NEBuffer 4 (20 mM Tris–acetate, 10 mM magnesium acetate, 50 mM potassium acetate and 1 mM DTT, pH 7.9) with 0.1 mg/ml BSA for 1 h at 37°C. Nt.CviPII nicking activity was distinguished from non-specific host nucleases by examining the banding pattern of the digested DNA. To check for non-specific exonuclease activity, reactions were incubated at 37°C for 16 h. For Nt.CviPII unit calculations, 1 μg of pUC19 was exposed to the same conditions. Protein concentrations were determined by the Bradford assay (BioRad Protein Assay) using BSA as a standard. To determine the temperature-dependence of the activity, 1 U of Nt.CviPII was incubated with 0.5 μg of pUC19 at various temperatures for 1 h. For single-stranded DNAs (ssDNA) cleavage activity determination, 250 ng of ss-M13 phage DNA was incubated with 0.5 or 1 U of Nt.CviPII at 37°C for 1 h. Nt.CviPII-cleaved DNA was subjected to electrophoresis in either 1.5% agarose gels, or 6% polyacrylamide gel (PAG) containing 7 M urea in 1× TBE buffer.
Determination of Nt.CviPII cleavage site
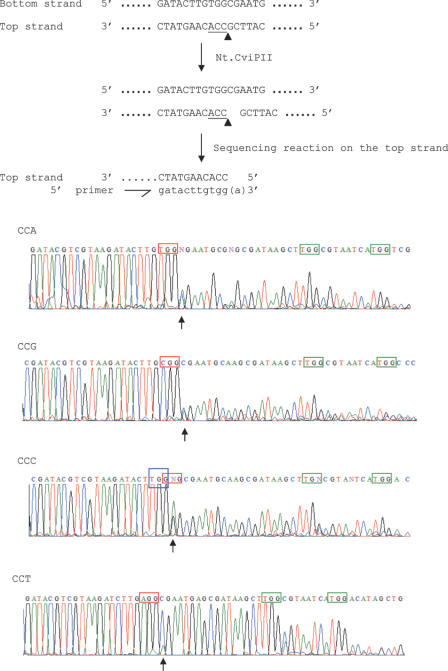
Double-stranded DNA substrates of 189 bp containing single CCA, CCT, CCC or CCG sites at 160–162 nt (Table 1) were constructed by PCR using the ‘inside-out’ method (16). The substrates contain an internal XhoI site (C^TCGAG) as a control to monitor cleavage. They also have EcoRI and HindIII sites at the 5′ and 3′ ends, respectively, for ligation into pUC19. The insert DNA sequence was confirmed by sequencing. Two micrograms of pUC19-substrate was incubated with Nt.CviPII at 37°C for 1 h. Reactions were terminated with 25 mM EDTA and DNA samples were purified by QIAprep spin columns (Qiagen). One-eighth dilutions of the purified cleaved products were sequenced with custom primers that anneal at the 5′ end of the substrate DNA (run-off sequencing).
Amplification of genomic DNA
Two hundred nanograms of Thermus thermophilus genomic DNA were incubated with 1 U of Nt.CviPII and either 16 U of Bst DNA polymerase large fragment, or 8 U of Taq DNA polymerase or 4 U of Vent DNA polymerase in ThermoPol reaction buffer at 55°C (20 mM Tris–acetate, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4 and 0.1% Triton X-100, pH 8.5), or 10 U of Klenow fragment of E.coli DNA polymerase I in EcoPol buffer (10 mM Tris–HCl, 5 mM MgCl2 and 7.5 mM DTT, pH 7.5) at 37°C for 30 min supplemented with 0.1 mM dNTPs. For DNA amplification from E.coli, single colonies were suspended in 50 μl of water, heated at 94°C for 8 min, centrifuged, and 20 μl of the supernatant was used for amplification. Amplified DNAs were subjected to electrophoresis on either 1.5–2% agarose gels or 6% PAG containing 7 M urea in 1× TBE buffer.
RESULTS
Cloning and identification of cviPIIM
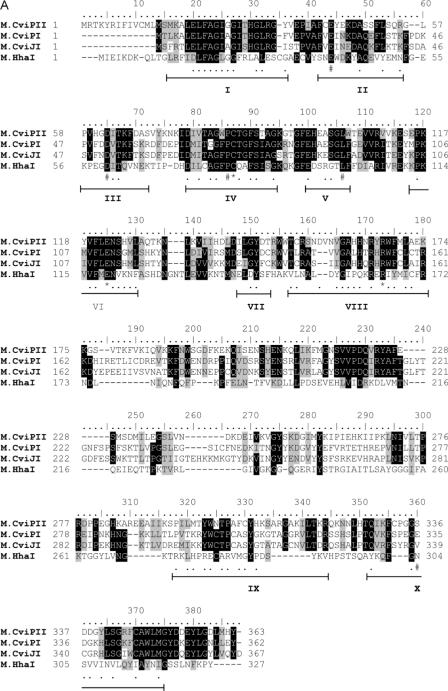
Plasmid DNA from E.coli cells that express M.CviPII (modifies the underlined C in CCD sequence) should be resistant to MspI digestion (modified mCCGG sequence is resistant to MspI cleavage). Eighteen plasmids were screened for resistance to MspI digestion after MspI challenge of the Sau3AI partial genomic DNA library. All 18 plasmids were partially resistant and insert DNAs from six isolates were sequenced. All six sequences had an identical open reading frame (ORF) (364 codons) that had 45.2% amino acid identity to NYs-1 encoded M.CviPI (recognition sequence GC) (11) and 41% amino acid identity to chlorella virus IL-3A encoded M.CviJI (recognition sequence RGCY) (17) (Figure 1A).
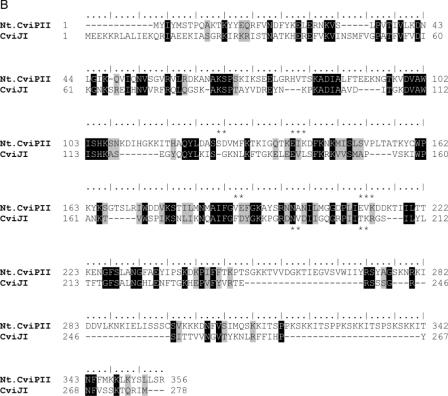
Figure 1.
(A) Alignment of M.CviPII, M.CviPI, M.CviJI and M.HhaI. Motifs I through X are indicated at the bottom of the alignment and conserved residues are indicated by dots. Sequences that are identical or similar are shown in black or grey boxes, respectively. Asterisks indicate catalytic residues and hashes indicate S-adenosyl-l-methionine binding residues. (B) Alignment of Nt.CviPII and CviJI amino acid sequences. Identical residues are shown in black boxes and similar residues are shaded in grey. Two Type II restriction REase active site motifs (P-D/E-Xn-D/E/S-Z-K/E; Z = hydrophobic residue) are found in Nt.CviPII sequences: Ser126-Asp127-X12-Glu139-Ile140-Lys141 and V189-Glu190-X21-Glu211-Val212-Lys213. The latter motif can be partially aligned to the proposed active site of CviJI. Asterisks indicate conserved residues of the active site motif.
Amino acid sequence alignment of M.CviPII with M.CviPI, M.CviJI and M.HhaI supports its identification as an m5C MTase (Figure 1A). M.HhaI is a prototypic m5C MTase that possesses all 10 motifs characteristic of m5C MTase motifs and for which structures at atomic resolution were reported (18,19). Alignment with M.HhaI showed that of the six most highly conserved motifs in other m5C MTases (motifs I, IV, VI, VIII, XI and X) (20,21), high conservation of motifs I to IV is found in all three chlorella virus enzymes. Motifs VI and VIII are identified in both M.CviPII and M.CviPI, albeit with lesser amino acid conservation. In M.HhaI and most m5C MTases, motifs I through VIII and the C-terminal part of motif X constitutes the large domain of the structure, whereas motifs IX and the N-terminal part of motif X make up the small domain. A variable target recognition domain (TRD), which is responsible for sequence recognition, links motif VIII to motif IX (22). Alignment of M.CviPII and M.CviPI against M.HhaI also indicated that the invariable active site residues Cys81, Glu119 and Arg165 of M.HhaI are found in all three chlorella virus enzymes. Residues that interact with S-adenosyl-l-methionine (AdoMet), namely Phe18, Gly20, Gly22 of motif I (FXGXGX), Glu40 of motif II, Asp60 of motif III, Pro80 of motif IV and Leu100 of motif V are also conserved. Motifs IX and X, however, do not appear to be present in these virus-encoded MTases. The absence of motifs IX and X in M.CviJI was reported previously (17,20). Nevertheless, the putative variable regions (TRD) following domain VIII of M.CviPII, M.CviPI, and M.CviJI share significant similarity. Sequence identities in this region are 32% between M.CviPII and M.CviPI, 26% between M.CviPII and M.CviJI, but only 8.3% between M.CviPII and M.HhaI. The high sequence identity in the predicted TRDs of M.CviPII and M.CviPI is unexpected because M.CviPII and M.CviPI modify CCD and GC sites, respectively, although both recognize short target sequences.
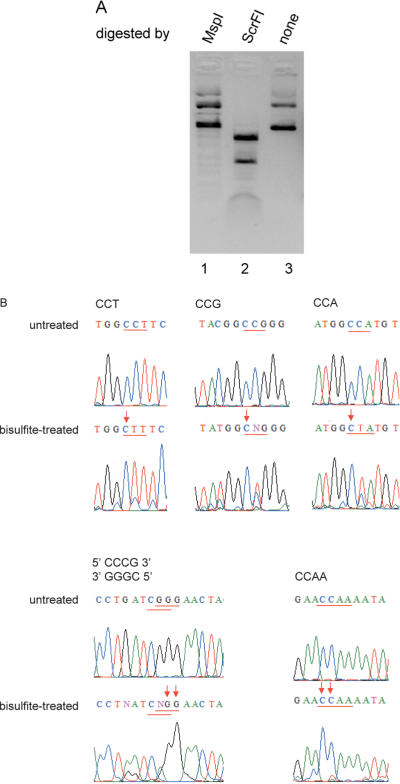
The putative cviPIIM gene was amplified by PCR, ligated into pUC19 and transferred into E.coli ER2502. The resultant plasmid, pUC-cviPIIM, was partially resistant to MspI but not to ScrF1 (Figure 2A). MspI cleaves CmCGG (23,24) but not mCCGG (25), whereas ScrFI cleaves mCCNGG (Nelson M., unpublished data) but not CmCNGG (25). This indirect evidence (resistant to MspI digestion) indicates that M.CviPII modifies the first cytosine of the CCGG sequence, which agrees with results obtained with the native enzyme (4). In addition, pre-modification of the E.coli expression host by the MTase allowed expression of Nt.CviPII (see below). Sequencing the PCR derived product from the sodium bisulfite-treated pUC-cviPIIM indicated that most of the first but not the second cytosines in CCG, CCA and CCT sequences were modified (Figure 2B). Surprisingly, the first two cytosines in the sequences CCCG and CCAA were also modified (Figure 2B, Table 2). Within CCCG sequence, there are overlapping CCC and CCG sites. Modification of the first cytosine in the CCC sites showed that M.CviPII also modifies CCC site (CCC is not cleaved by its cognate Nt.CviPII NEase). Methylation of the second cytosine in CCCG corresponds to the first cytosine of the CCG site. However, modification of both the first and second cytosines in CCAA sequences suggests that M.CviPII displays a relaxed recognition mode under over-expression conditions (the cviPIIM gene was cloned in a high-copy-number plasmid and IPTG-induced). We do not know if this is also a property of the native enzyme when expressed by the virus. We also found that M.CviQXI from chlorella virus NY-2A displayed a more relaxed specificity than the cognate nicking endonuclease when it is cloned in pUC19: the specificities of Nt.CviQXI and M.CviQXI are R^AG and AG, respectively (unpublished results). It is not clear whether this is a general property of viral DNA modification and protection systems.
Figure 2.
(A) Partial resistance to MspI challenge. pUC-cviPIIM was incubated with MspI or ScrFI. MspI does not cleave mCCGG whereas ScrFI cleaves mCCNGG. (B) Identification of methylation site. DNA sequences of PCR products derived from sodium bisulfite-treated pUC-cviPIIM were compared with those from untreated pUC-cviPIIM. The change of cytosine to thymidine corresponds to unmodified cytosine, whereas cytosine in both sodium bisulfite-treated and un-treated plasmids indicates C5-methylcytosine. The recognition sites stated on top of each panel are underlined in red and modified nucleotides are indicated by down arrows.
Table 2. Sites modified by M.CviPII in the ORF of M.CviPII in pUC-cviPIIM.
| Modified sites (mC) | Tally |
|---|---|
| CCG | 7 |
| CCA | 2 |
| CCT | 4 |
| CCCG | 1 |
| CCAA | 4 |
Cloning and Identification of cviPIINt
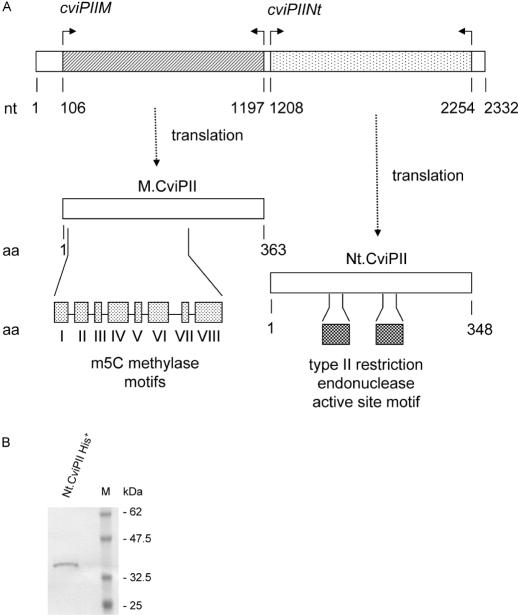
In chlorella viruses, like bacteria, REase genes are always located near their cognate DNA MTase gene, although the exact spacing and relative orientation of the two genes can vary (26). Therefore, we sequenced NYs-1 DNA adjacent to cviPIIM and identified an ORF of 349 codons (in the same orientation) that began 12 nucleotides downstream of the cviPIIM stop codon (Figure 3A). The ORF had low but significant amino acid sequence identity to chlorella virus IL-3A encoded REase CviJI (RG^CY, 23.5% identity) (Figure 1B) (17,27). The half site of CviJI (CY, Y = C or T) is similar to the Nt.CviPII recognition sequence CCD. Nt.CviPII does not resemble any other REases or NEases in GenBank.
Figure 3.
(A) The CviPII N-M system contains 2332 nt and two complete ORFs. The cviPIIM gene contains m5C DNA MTase motifs I through VIII as indicated (see also Figure 2). The cviPIINt gene contains two Type II REase active site motifs (see also Figure 2B). (B) Purified recombinant Nt.CviPII was analyzed on a 4–10% SDS–PAGE.
Due to the frequent number of Nt.CviPII nicking sites, we anticipated that it would be difficult to clone the cviPIINt gene in E.coli. Therefore, we initially tried to express Nt.CviPII in an in vitro transcription and translation system. A low level of nicking activity was detected; however, it was difficult to achieve a clear digestion pattern (data not shown). To achieve sufficient enzyme for purification, we tried to express the recombinant Nt.CviPII in E.coli. ER2683 whose genome was pre-modified by expression of M.CviPII from the plasmid pUC-cviPIIM. Additional measures were taken to construct a stable expression clone: (i) a low copy number plasmid pR976 with Ptac and p15A replication origin was used as the cloning vector for the cviPIINt gene; (ii) the cviPIINt gene was inserted 18 nt downstream of the ribosome-binding site to reduce Nt.CviPII expression. Efforts to express M.CviPII in pACYC184 (pACYC-cviPIIM gave only marginal modification of the host genomic DNA) and Nt.CviPII in pET21a vectors failed to generate a stable expression clone (data not shown).
Expression and purification of Nt.CviPII
Expression strain ER2683 (pUC-cviPIIM, pR976-cviPIINt) was successfully constructed. The Nt.CviPII expression plasmid alone could not transform E.coli cells, indicating that residual expression of Nt.CviPII in the absence of the cognate methylase is lethal to the host. To find out the basal level of expression, Nt.CviPII activity was compared with, and without IPTG induction. In the presence of IPTG, most fractions eluted from an SP FF column exhibited significantly higher DNA nicking activity (Figure 4A, top panel) than fractions from an uninduced culture (Figure 4A, bottom panel). IPTG induction of cells grown at 16°C produced more Nt.CviPII activity than cells grown at higher temperatures (data not shown). The pI value of Nt.CviPII was calculated to be 9.82 using ProtParam (28). For purification of the His-tagged Nt.CviPII, lysate was applied to a HisTrap column and protein eluted with an imidazole step gradient. Unexpectedly, nicking activity eluted at both low (50 mM) and high (300 mM) imidazole concentrations. The low imidazole-eluted activity was further purified through heparin FF and SP columns. N-terminal sequencing of the purified protein revealed the sequence MSTPQAKTKYY, which corresponds to amino acids 5–15 of Nt.CviPII. Thus this fraction contains a protein initiated at the cviPIINt fifth codon, which is an ATG. The mass of this protein is 34 110 Da, compared with a predicted value of 40 069 Da. This preparation was designated His−-Nt.CviPII.
Figure 4.
(A) Control of Nt.CviPII expression. Soluble extracts from equivalent induced (upper panel) and un-induced cultures (lower panel) were loaded on an SP FF column, eluted with a linear gradient of 0.1–1 M NaCl, and fractions were assayed for activity. (B) About 0.5 μg of pUC19 was incubated with 1 U of Nt.CviPII at the designated temperature for 1 h. Reactions were stopped and analyzed by electrophoresis on a 1.5% agarose gel. (C) Nt.CviPII-cleaved pUC19 and ss-M13 phage DNAs were analyzed by electrophoresis on a 6% PAG with 7 M urea. I, input; FT, flow-through; N, nicked pUC19; L, linearized pUC19; S, supercoiled pUC19; 100 bp, 100 bp DNA size marker; and LMW, low molecular weight DNA size marker.
Protein that eluted at 300 mM imidazole was concentrated and two bands of ∼34–36 kDa were observed by SDS–PAGE (data not shown). N-terminal sequencing established that the upper band was Nt.CviPII with a His6 tag while the lower band was an E.coli 2-dehydro-3-deoxyphosphoheptonate aldolase contaminant. The two proteins separated on an SP column, resulting in pure Nt.CviPII, as judged by SDS–PAGE (Figure 3B). This protein was designated His+-Nt.CviPII. Because this latter preparation was cleaner than His−-Nt.CviPII, it was used in all subsequent experiments.
Nt.CviPII nicking activity
Nt.CviPII activity was measured at a range of temperatures with a pUC19 substrate (Figure 4B). DNA nicking activity is highest at 30–45°C; nicking activity at 20–25°C is higher than that at 55–60°C. Nt.CviPII showed lowest activity at 65°C probably due to partial thermal denaturation. At 30–45°C, the cleavage products of pUC19 appear to be ∼200 bp in 1.5% agarose gel electrophoresis (Figure 4B). A time course experiment at 37°C established that a stable cleavage pattern was reached after 1 h incubation (results not shown). Longer incubation times or addition of more enzyme did not alter the pattern. Moreover, the pattern is not the result of Nt.CviPII inactivation at 37°C because pre-incubation of Nt.CviPII at 37°C for 90 min did not alter subsequent enzyme activity (data not shown). Extended incubation of pUC19 with two batches of Nt.CviPII (His+ and His−) for 16 h indicated no significant contamination with non-specific nucleases (data not shown). One unit of Nt.CviPII activity is defined as the amount of enzyme needed to cleave pUC19 into ∼200 bp products in 1 h at 37°C, as judged by electrophoresis in 1.5% agarose gels. Using this criterion, the specific activity of His+-Nt.CviPII is 9410 U/mg of protein.
Analyzing the pUC19 cleavage products by electrophoresis on a urea PAG indicated that the ssDNA fragments were smaller than 150 nt (Figure 4C). NEBcutter (Roberts R. and Vincze T., www.neb.com) identified 252 CCD sites distributed on both strands of pUC19. Upon complete digestion, the longest and the shortest fragments are expected to be ∼50 and ∼20 bp, respectively. However, the end products of pUC19 are ssDNAs that ranged from 25 to 150 nt (Figure 4C), suggesting that some sites are not nicked efficiently. Therefore, some Nt.CviPII cleavage sites in pUC19 are apparently less susceptible to cleavage than others (see below). To determine if Nt.CviPII is active toward ssDNA, ss-M13 DNA was digested by Nt.CviPII. However, only partial digestion was achieved when compared with digestion of pUC19 (Figure 4C). It is most likely that Nt.CviPII cleaved transient duplex DNA formed by the ss-M13 DNA.
Nt.CviPII cleavage specificity
To construct CCD sites with the same flanking sequence, synthetic dsDNA oligonucleotides (180 nt) that contain single CCA, CCG, CCT and CCC sites were ligated into pUC19. The plasmids were incubated with Nt.CviPII, analyzed on agarose gels, and the cleavage sites determined by ‘run-off’ sequencing. When a nick occurs in a dsDNA, the sequencing reaction stops after the nicked base and the peaks in the sequencing chromatogram diminish sharply. An extra adenine is added to the 3′ end of the newly synthesized ssDNA due to the template-independent polymerase activity of Taq DNA polymerase used in the sequencing reaction. The adenine peak also helps identify the cleavage site. Consistent with a previous report (4), run-off sequencing showed that the DNA was cleaved 5′ of the first cytosine in CCA, CCG and CCT sequences, but not at the CCC site (Figure 5). However, cleavage is less favorable at the CCT site than the CCA and CCG sites as only a small amount of the CCT substrate was cleaved (Figure 5, bottom panel). A small peak of adenine following the AGG triplet also indicates that some cleavage occurred at the CCT site. For the CCC site, there are always overlapping CCC and CCN sites because the base following CCC might not be a cytosine. In the synthetic substrate, the CCC site is followed by an adenine. Cleavage took place 5′ to the second cytosine in the CCCA sequence, indicating that Nt.CviPII recognizes the overlapping CCA site (boxed in blue, panel CCC, Figure 5) but not the CCC site. Therefore, CCA, CCG and CCT but not CCC sites are recognized and cleaved by Nt.CviPII.
Figure 5.
Run-off sequencing of Nt.CviPII-cleaved DNA. The sequences were read as reverse-complement by the sequencing primer. Therefore, TGG corresponds to CCA, CGG to CCG, GGG to CCC and AGG to CCT. Triplet sequences in red boxes are CCN sites that are added to the substrate DNA. Those in green boxes are native CCA sequences of pUC19. Arrows under the chromatographs indicate the nt adenine added by the template-independent activity of Taq DNA polymerase used in the sequencing reactions. Arrow heads also indicate the nicking site (complement).
Isothermal amplification of DNA
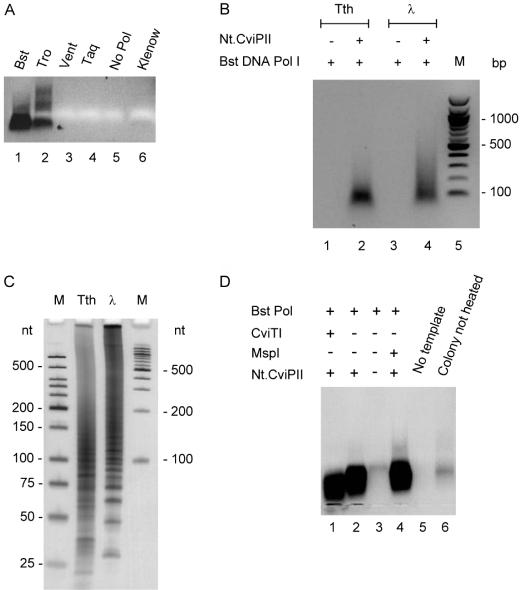
Because of the high frequency of Nt.CviPII cleavage sites and its single-stranded cleavage product, Nt.CviPII was used in conjunction with several DNA polymerases in isothermal random DNA amplification. E.coli DNA was incubated with Nt.CviPII and various DNA polymerases at various temperatures for 30 min. Bst DNA polymerase I large fragment generated the highest yield of DNA. The large fragment of DNA polymerase I from Thermomicrobium roseum (unpublished data) also synthesized significant amounts of DNA while Vent DNA polymerase and Taq DNA polymerase did not synthesize any DNA (Figure 6A). E.coli DNA polymerase Klenow fragment generated a small amount of amplified DNA. Random DNA amplification can be achieved from T.thermophilus HB27 and λ DNA in the presence of Nt.CviPII/Bst DNA polymerase I large fragment but not with Bst DNA polymerase large fragment alone (Figure 6B). As shown in urea PAGE, the amplified DNA ranged from 25 to 200 nt from T.thermophilus genomic DNA and some larger background DNA were also detected from λ DNA (Figure 6C). The DNA mass can be amplified approximately 50-fold with 10 ng input DNA generating 500 ng of amplified small DNA fragments. The Nt.CviPII/Bst DNA pol amplified products can be diluted and further amplified by addition of [N]6 or [N]9 random primers and Bst DNA pol large fragment by incubation at 50°C for 60 min; the second rounds of random amplification generated major products in the range of 1 to 2 kb for E.coli DNA (data not shown).
Figure 6.
Isothermal random DNA amplification. (A) E.coli DNA was amplified using Nt.CviPII in combination with Bst DNA polymerase I large fragment (Bst), T.roseum (Tro) DNA polymerase large fragment, Vent DNA polymerase (Vent), Taq DNA polymerase (Taq) or Klenow fragment of E.coli DNA polymerase I (Klenow). (B) T.thermophilus and λ DNAs were amplified with Nt.CviPII and Bst DNA polymerase I large fragment. The amplified products were analyzed by 1.5% agarose gel electrophoresis. (C) The same cleavage reaction products as in (B) were analyzed on a 6% PAG with 7 M urea. (D) DNA was amplified from single colonies of E.coli with or without 4-base cutters CviTI (RG^CY) or MspI (C^CGG).
Efforts were also made to amplify DNA from a single E.coli colony using Nt.CviPII and Bst DNA polymerase I large fragment (Figure 6D, lane 2). Heating the colony at 94°C for 8 min was necessary to release the DNA for optimal amplification (Figure 6D, lane 6). Inclusion of the 4-base cutting REase CviTI (RG^CY) or MspI (C^CGG) did not significantly alter the size of the amplified products (Figure 6D, lanes 1 and 4).
DISCUSSION
One unusual property of the chlorella viruses is that they encode multiple Type II DNA restriction and modification systems (8,9). This manuscript describes the identification and characterization of two genes from chlorella virus NYs-1 that encode a DNA NEase (Nt.CviPII) and its companion m5C MTase (M.CviPII). This is the first report characterizing recombinant M.CviPII and Nt.CviPII. Another nicking enzyme, Nt.CviQXI from chlorella virus NY-2A, has been synthesized in vitro (unpublished results). Nt.CviQXI nicks at R^AG sites (5).
NYs-1 has one of the most heavily methylated chlorella virus genomes known: 48% of the cytosines are m5C and 11% of the adenines are m6A (10). Previously, an NYs-1-encoded orphan m5C MTase M.CviPI was cloned and characterized (11); M.CviPI methylates GC sequences. In the current paper, we show that M.CviPII methylates not only CCD sequences but also the first and second cytosines in CCCG sequences (overlapping CCC and CCG sites) and both cytosines at CCAA sites. Because both NYs-1 MTases modify such short sequences, M.CviPI and M.CviPII could account for most, if not all, of the 48% m5C in NYs-1 DNA. None of the NYs-1-encoded m6A DNA MTases has been characterized.
M.CviPII is one of the few cloned DNA MTases that catalyzes m5C methylation of a trinucleotide sequence de novo. The other two frequent m5C MTases are orphan MTases: M.CviPI and M.SssI from Spiroplasma sp. strain MQ1, which methylate GC and CG sites (29), respectively. DNA MTases that methylate DNA frequently have been used to study yeast chromatin structure in vivo (11,30,31) and M.CviPII may be useful in similar studies.
The companion to M.CviPII is a nicking endonuclease Nt.CviPII, which recognizes the short sequence CCD (D = A, G, or T) and cleaves the phosphodiester bond 5′ to the first cytosine. However, not all CCD sequences are cleaved with equal efficiency: CCA and CCG sites are preferred over CCT sites. From sequence analysis, Nt.CviPII contains two putative Type II REase active site motifs (P-D/E-Xn-D/E/S-Z-K/E; Z = hydrophobic residue; Figures 1B and 3A). As a NEase, it is most likely that one of the active sites is functional. Mutagenesis studies should be carried out to determine the catalytic residues in the enzyme. In addition, biochemical studies are needed to determine the mechanism of nicking by Nt.CviPII, e.g. does the enzyme function as a monomer or a dimer? A previous study indicates that loss of the dimerization site of MlyI could generate a NEase (32). Alternatively, loss of one catalytic site could produce either a top strand or a bottom strand NEase (e.g. Nt.BsaI and Nb.BsaI) (33).
Restriction and modification genes are widespread in the chlorella viruses and their presence leads to the question: what is the origin of the genes? Several observations suggest that bacterial and chlorella virus restriction and modification enzymes have evolved from a common progenitor: (i) amino acid sequence motifs characteristic of bacterial m5C and m6A DNA MTases also exist in the chlorella virus DNA MTases. However, the three characterized chlorella virus m5C MTases lack motifs IX and X that are present in bacterial m5C MTases. (ii) some chlorella virus MTases have up to 37% amino acid identity with bacterial MTases. (iii) Chlorella virus REase genes (5,27,34,35), like bacterial REase genes (26), are adjacent to their companion DNA MTase gene. (iv) Chlorella virus MTase genes have upstream regions that function as promoters in E.coli (36,37).
Although evidence suggests that bacterial and chlorella virus DNA MTases probably have common evolutionary origins, amino acid and promoter sequence data suggest that the MTase genes have been associated with chlorella viruses for a very long time. The G+C content of the cviPIIM gene is 43%, which is close to the ∼40% G+C content of the NYs-1 genome (10). However, the G+C content of the cviPIINt gene is lower (32%), suggesting that this gene might have been acquired separately from the cviPIIM gene.
Several methods used for isothermal genome-wide amplification of DNA have been described recently. An early version of strand displacement amplification (SDA) depends on synthesis of hemi-phosphorothioated HincII or AvaI sites. where the appropriate REase nicks the template DNA on the un-modified strand to allow separation of the amplified strand (38–40). Rolling circle amplification requires a circular template (e.g. certain virus genomes, cloned plasmids or padlock-mediated circularized DNA) and random primers (41,42). More recently, specially designed DNA oligonucleotides have been used with Nt.BstNBI to conduct linear and exponential amplification of short oligonucleotides (43). Being a thermostable enzyme, Nt.BstNBI was incubated with template DNA, dNTPs and Vent exo- DNA polymerase. At 60°C, the nicked fragments (designed to be 12 nt) separated from the double strand template such that a free 3′ end was available for template-dependent amplification. To achieve exponential amplification, an oligonucleotide with sequence complementary to the target sequence is added as the ‘trigger’ oligonucleotide. On the other hand, using thermocycling, REases that cleave DNA frequently have been used to generate oligonucleotides for 2-step DNA amplification. CviJI*, which cleaves RG^CN and YG^CY sites in the presence of ATP, generates dsDNA fragments of 20–60 bp from Bacillus sp. (44). After heat inactivation, the cleavage products are mixed with Bacillus genomic DNA, a thermostable DNA polymerase, and dNTPs for amplification by thermocycling. The amplified DNA ranged from 0.1–10 kb in size, was independent of the size of the template DNA, and gave high sensitivity in Southern blots when biotinylated-dUTP was included in the amplification reaction.
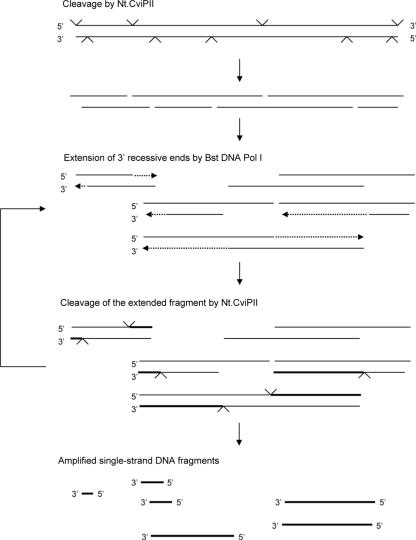
As demonstrated in this paper, Nt.CviPII and Bst DNA polymerase I large fragment can be used to amplify DNA in a single-step isothermal reaction. This process is termed nicking-endonuclease-mediated DNA amplification (NEMDA). Because Nt.CviPII is partially active at 55°C, the amplification reaction can be carried out at 55°C where denaturation and separation of nicked DNA (oligos) are favored. Nt.CviPII cleaves DNA frequently and produces single-stranded products or partial duplex DNAs with 5′ overhangs (Figure 7). Bst DNA polymerase I large fragment then fills in at the 3′ end until it reaches the end of the template. The extended DNA acts as a substrate for Nt.CviPII, which, in turn, provides new substrate for the polymerase, allowing linear amplification. The hallmark of NEMDA is that exogenous primers are not needed for DNA amplification, since partial duplex DNA is generated from the substrate DNA. From our collection of DNA polymerases, only Bst DNA polymerase I large fragment and Thermomicrobium roseum (Tro) DNA polymerase I large fragment produce significant amounts of amplified products. These polymerases possess relatively high strand displacement activity among the polymerases we tested. Strand displacement activity may be involved in removing the nicked fragment and revealing a recessive 3′ end for template-dependent amplification.
Figure 7.
Proposed mechanism of nicking-endonuclease-mediated DNA amplification (NEMDA) using Nt.CviPII and Bst DNA polymerase I large fragment. Note: partial nicking introduced by Nt.CviPII can generate products of varying length. See Discussion for description.
Unlike rolling circle amplification that can be used in whole genome amplification, NEMDA does not cover the entire genome. Nevertheless, the amplified DNA can be used as a probe to detect target DNA by Southern blotting (unpublished data). We have also demonstrated that incubation of random DNA oligonucleotides and fresh Bst DNA polymerase large fragment with the amplified product can further amplify the DNA. Alternatively, the amplified DNA can be purified and used as primers for direct amplification of genomic DNA through isothermal or thermocycling procedures.
Randomly amplified DNA has been used as a highly sensitive probe for arrays of DNA oligonucleotides carrying ‘signature sequences’ for biological agents of high concern such as E.coli O157:H7 (45,46). The DNA amplification method presented here does not require synthesis of primers and can generate large quantities of ssDNA from a single bacterial colony in a short time frame (e.g. 10 to 30 min). The procedure can be adapted for environmental or clinical samples and labels, such as biotin or fluorescein, can be incorporated into the amplified products by including modified dNTP. Development of timely, sensitive and specific detection methods to identify important pathogens is of tremendous interest to bio-defense and public health.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Richard Roberts for critical reading of the manuscript, Jim Gurnon for isolating NYs-1 DNA, Shelley Cushing and Jack Benner II for mass spectrometry and protein sequencing, Elisabeth Raleigh, Jim Samuelson, and Rebecca Kucera for discussion, Jim Samuelson for independent confirmation of NEMDA, Don Comb for support and encouragement. This work is partly supported by NIH STTR grant 1R41GM070212-01. The sequence has been deposited in GenBank with accession number of AY781802.
REFERENCES
- 1.Roberts R.J., Vincze,T., Posfai,J. and Macelis,D. (2003) REBASE: restriction enzymes and methyltransferases. Nucleic Acids Res., 31, 418–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan R.D., Calvet,C., Demeter,M., Agra,R. and Kong,H. (2000) Characterization of the specific DNA nicking activity of restriction endonuclease. N. BstNBI. Biol. Chem., 381, 1123–1125. [DOI] [PubMed] [Google Scholar]
- 3.Abdurashitov M.A., Belichenko,O.A., Shevchenko,A.V. and Degtyarev,S.K. (1996) N.BstSE—site-specific nuclease from Bacillus stearothermophilus SE-589—restriction endonuclease production. Molek. Biol., 30, 1261–1267. [PubMed] [Google Scholar]
- 4.Xia Y.N., Morgan,R., Schildkraut,I. and Van Etten,J.L. (1988) A site-specific single strand endonuclease activity induced by NYs-1 virus infection of a Chlorella-like green alga. Nucleic Acids Res., 16, 9477–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Nelson,M., Nietfeldt,J., Xia,Y., Burbank,D., Ropp,S. and Van Etten,J.L. (1998) Chlorella virus NY-2A encodes at least 12 DNA endonuclease/methyltransferase genes. Virology, 240, 366–375. [DOI] [PubMed] [Google Scholar]
- 6.Van Etten J.L. (2003) Unusual life style of giant chlorella viruses. Annu. Rev. Genet., 37, 153–195. [DOI] [PubMed] [Google Scholar]
- 7.Van Etten J.L., Lane,L.C. and Meints,R.H. (1991) Viruses and virus-like particles of eukaryotic algae. Microbiol. Rev., 55, 586–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson M., Burbank,D.E. and Van Etten,J.L. (1998) Chlorella viruses encode multiple DNA methyltransferases. Biol. Chem., 379, 423–428. [DOI] [PubMed] [Google Scholar]
- 9.Nelson M., Zhang,Y. and Van Etten,J.L. (1993) DNA methyltransferases and DNA site-specific endonucleases encoded by chlorella viruses. In Jost,P.J. Saluz,H.P., eds, DNA Methylation: Molecular Biology and Biological Significance, Birkhauser, Basel, pp. 186–211. [DOI] [PubMed] [Google Scholar]
- 10.Schuster A.M., Burbank,D.E., Meister,B., Skrdla,M.P., Meints,R.H., Hattman,S., Swinton,D. and Van Etten,J.L. (1986) Characterization of viruses infecting a eukaryotic Chlorella-like green alga. Virology, 150, 170–177. [DOI] [PubMed] [Google Scholar]
- 11.Xu M., Kladde,M.P., Van Etten,J.L. and Simpson,R.T. (1998) Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res., 26, 3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Etten J.L., Schuster,A.M., Girton,L., Burbank,D.E., Swinton,D. and Hattman,S. (1985) DNA methylation of viruses infecting a eukaryotic Chlorella-like green alga. Nucleic Acids Res., 13, 3471–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fomenkov A., Xiao,J.-P., Dila,D., Raleigh,E. and Xu,S.-Y. (1994) The ‘endo-blue method’ for direct cloning of restriction endonuclease genes in E.coli. Nucleic Acids Res., 22, 2399–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Boer H.A., Comstock,L.J. and Vasser,M. (1983) The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl Acad. Sci. USA, 80, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X., Yo,P., Keith,A., Ragan,T.J. and Harris,T.K. (2003) Thermodynamically balanced inside-out (TBIO) PCR-based gene synthesis: a novel method of primer design for high-fidelity assembly of longer gene sequences. Nucleic Acids Res., 31, e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shields S.L., Burbank,D.E., Grabherr,R. and Van Etten,J.L. (1990) Cloning and sequencing the cytosine methyltransferase gene M. CviJI from Chlorella virus IL-3A. Virology, 176, 16–24. [DOI] [PubMed] [Google Scholar]
- 18.Cheng X., Kumar,S., Posfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 19.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 20.Kumar S., Cheng,X., Klimasauskas,S., Mi,S., Posfai,J., Roberts,R.J. and Wilson,G.G. (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res., 22, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posfai J., Bhagwat,A.S., Posfai,G. and Roberts,R.J. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauster R., Trautner,T.A. and Noyer-Weidner,M. (1989) Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J. Mol. Biol., 206, 305–312. [DOI] [PubMed] [Google Scholar]
- 23.Hou P., Ji,M., He,N. and Lu,Z. (2004) Microarray-based method to evaluate the accuracy of restriction endonucleases HpaII and MspI. Biochem. Biophys. Res. Commun., 314, 110–117. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Hattar J. and Jiricny,J. (1988) Effect of cytosine methylation on the cleavage of oligonucleotide duplexes with restriction endonucleases HpaII and MspI. Nucleic Acids Res., 16, 4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson M., Christ,C. and Schildkraut,I. (1984) Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res., 12, 5165–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson G.G. and Murray,N.E. (1991) Restriction and modification systems. Annu. Rev. Genet., 25, 585–627. [DOI] [PubMed] [Google Scholar]
- 27.Swaminathan N., Mead,D.A., McMaster,K., George,D., Van Etten,J.L. and Skowron,P.M. (1996) Molecular cloning of the three base restriction endonuclease R.CviJI from eukaryotic Chlorella virus IL-3A. Nucleic Acids Res., 24, 2463–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasteiger E., Gattiker,A., Hoogland,C., Ivanyi,I., Appel,R.D. and Bairoch,A. (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res., 31, 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renbaum P., Abrahamove,D., Fainsod,A., Wilson,G.G., Rottem,S. and Razin,A. (1990) Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res., 18, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kladde M.P. and Simpson,R.T. (1994) Positioned nucleosomes inhibit Dam methylation in vivo. Proc. Natl Acad. Sci. USA, 91, 1361–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kladde M.P., Xu,M. and Simpson,R.T. (1996) Direct study of DNA–protein interactions in repressed and active chromatin in living cells. EMBO J, 15, 6290–6300. [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y., Lunnen,K.D. and Kong,H. (2001) Engineering a nicking endonuclease N.AlwI by domain swapping. Proc. Natl Acad. Sci. USA, 98, 12990–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Z., Samuelson,J.C., Zhou,J., Dore,A. and Xu,S.Y. (2004) Engineering strand-specific DNA nicking enzymes from the type IIS restriction endonucleases BsaI, BsmBI, and BsmAI. J. Mol. Biol., 337, 573–583. [DOI] [PubMed] [Google Scholar]
- 34.Skowron P.M., Swaminathan,N., McMaster,K., George,D., Van Etten,J.L. and Mead,D.A. (1995) Cloning and applications of the two/three-base restriction endonuclease R.CviJI from IL-3A virus-infected Chlorella. Gene, 157, 37–41. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Lu,Z., Sun,L., Ropp,S., Kutish,G.F., Rock,D.L. and Van Etten,J.L. (1997) Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome. Virology, 237, 360–377. [DOI] [PubMed] [Google Scholar]
- 36.Narva K.E., Wendell,D.L., Skrdla,M.P. and Van Etten,J.L. (1987) Molecular cloning and characterization of the gene encoding the DNA methyltransferase,M.CviBIII, from Chlorella virus NC-1A. Nucleic Acids Res., 15, 9807–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefan C., Xia,Y.N. and Van Etten,J.L. (1991) Molecular cloning and characterization of the gene encoding the adenine methyltransferase M.CviRI from Chlorella virus XZ-6E. Nucleic Acids Res., 19, 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milla M.A., Spears,P.A., Pearson,R.E. and Walker,G.T. (1998) Use of the restriction enzyme AvaI and exo-Bst polymerase in strand displacement amplification. Biotechniques, 24, 392–396. [DOI] [PubMed] [Google Scholar]
- 39.Walker G.T., Little,M.C., Nadeau,J.G. and Shank,D.D. (1992) Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc. Natl Acad. Sci. USA, 89, 392–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker G.T., Nadeau,J.G., Spears,P.A., Schram,J.L., Nycz,C.M. and Shank,D.D. (1994) Multiplex strand displacement amplification (SDA) and detection of DNA sequences from Mycobacterium tuberculosis and other mycobacteria. Nucleic Acids Res., 22, 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rector A., Tachezy,R. and Van Ranst,M. (2004) A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol., 78, 4993–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smirnov D.A., Burdick,J.T., Morley,M. and Cheung,V.G. (2004) Method for manufacturing whole-genome microarrays by rolling circle amplification. Genes Chromosomes Cancer, 40, 72–77. [DOI] [PubMed] [Google Scholar]
- 43.Van Ness J., Van Ness,L.K. and Galas,D.J. (2003) Isothermal reactions for the amplification of oligonucleotides. Proc. Natl Acad. Sci. USA, 100, 4504–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swaminathan N., McMaster,K., Skowron,P.M. and Mead,D.A. (1998) Thermal cycle labeling: zeptomole detection sensitivity and microgram probe amplification using CviJl* restriction-generated oligonucleotides. Anal. Biochem., 255, 133–141. [DOI] [PubMed] [Google Scholar]
- 45.Vora G.J. M., C.E., Stenger,D.A. and Andreadis,J.D. (2004) Nucleic acid amplification stratagies for DNA microarray-based pathogen detection. Appl. Environ. Microbiol., 70, 3047–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J.M., Bell,J., Huang,Y., Tirado,M., Thomas,D., Forster,A.H., Haigis,R.W., Swanson,P.D., Wallace,R.B., Martinsons,B. et al. (2002) An integrated, stacked microlaboratory for biological agent detection with DNA and immunoassays. Biosens. Bioelectron., 17, 605–618. [DOI] [PubMed] [Google Scholar]