Abstract
The solution structures of two 27 nt RNA hairpins and their complexes with cobalt(III)-hexammine [Co(NH3)63+] were determined by NMR spectroscopy. The RNA hairpins are variants of the P4 region from Escherichia coli RNase P RNA: a U-to-A mutant changing the identity of the bulged nucleotide, and a U-to-C, C-to-U double mutant changing only the bulge position. Structures calculated from NMR constraints show that the RNA hairpins adopt different conformations. In the U-to-C, C-to-U double mutant, the conserved bulged uridine in the P4 wild-type stem is found to be shifted in the 3′-direction by one nucleotide when compared with the wild-type structure. Co(NH3)63+ is used as a spectroscopic probe for Mg(H2O)62+ binding sites because both complexes have octahedral symmetry and have similar radii. Intermolecular NOE crosspeaks between Co(NH3)63+ and RNA protons were used to locate the site of Co(NH3)63+ binding to both RNA hairpins. The metal ion binds in the major groove near a bulge loop in both mutants, but is shifted 3′ by about one base pair in the double mutant. The change of the metal ion binding site is compared with results obtained on corresponding mutant RNase P RNA molecules as reported by Harris and co-workers (RNA, 1, 210–218).
INTRODUCTION
RNase P
RNase P is a ribonucleoprotein enzyme that catalyzes the processing of the 5′ end of tRNA in prokaryotes and eukaryotes (1,2). The RNA component of RNase P is essential for binding and positioning the precursor tRNA; the bacterial RNase P RNA is catalytically competent without the protein component of the enzyme (3). Catalysis is dependent on specific divalent metal ions (4); the protein subunit in Bacillus subtilis has been shown to enhance binding of several metal ions (5). In the case of eucaryal and archeal RNase P, one or more protein components are required for catalysis (1).
RNase P structure model
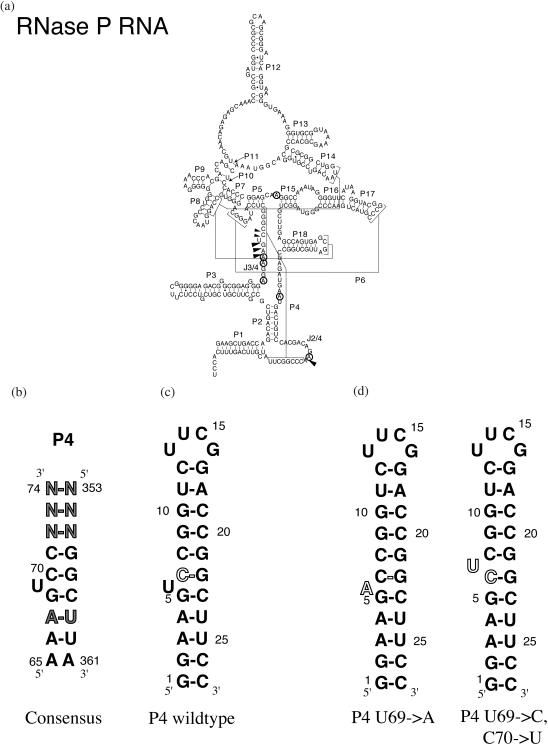
Based on phylogenetic comparison of a large number of RNase P RNA sequences, chemical and enzymatic structure probing, and site-specific UV crosslinking data, the secondary structure of RNase P RNA has been established and tertiary structure models have been proposed for the bacterial RNase P RNA (6–8). Crystal structures of the specificity domain of the B.subtilis RNase P RNA and the protein components of Thermotoga maritima and Escherichia coli have been reported (9,10), and a model of the complex of RNase P RNA with the E.coli C5 protein has been derived (11). The RNase P RNA adopts a highly base paired structure shown in Figure 1a (12). Several long-range interactions, including the P4 pseudoknot, result in a compact tertiary fold.
Figure 1.
Secondary structure of E.coli RNase P RNA (a) and hairpin P4 variants (b–d) used in this study. Arrows indicate sites of phosphorothioate modification interference, circles indicate sites of 7-deazaadenine interference (12). The long range interactions forming P4, P6 and two tetraloop–stem interactions are indicated by square brackets. Figure modified after Kanzatsev and Pace (12). (b) Secondary structure of the P4 element of E.coli RNase P RNA. The nucleotide numbering is adapted from the bacterial consensus structure. Nucleotides not strictly conserved are shown shaded. (c) Secondary structure of the wild-type P4 element. The nucleotide C70 next to the conserved bulge uridine is shown in outline. (d) Secondary structures of the U69→A and U69→C, C70→U mutant P4 elements. The sites of mutation near the conserved bulge uridine are shown in outline.
P4 element
One element of the RNase P RNA secondary structure (Figure 1a) was found to be strictly conserved among all bacterial RNase P RNAs. This so-called P4 element with the flanking J3/4 and J2/4 single-stranded regions is part of a pseudoknot structure and is positioned at the center of the catalytic domain immediately adjacent to the site of the pre-tRNA cleavage in the tertiary structure models. Binding of magnesium ions to phosphates at both ends of the P4 helix as well as to phosphates in the adjacent J2/4 and J3/4 regions has been concluded earlier from phosphorothioate modification interference studies (13–15). A single base change in P4 revealed changes in metal utilization as a result of changes in P4 structure (16–18).
Further mutagenesis on the P4 element was carried out (19) to investigate the effect of P4 structure on overall RNase P structure, metal binding sites and catalysis. Change of the conserved bulged nucleotide to adenosine (mutant U69A) had only a modest effect on catalysis, whereas change of the bulge position (mutant U69ΔU70, further denoted U70b in this study) reduced the rate of catalysis 70-fold. Intrigued by this apparent sensitivity to bulge position rather than sequence, these mutants were selected for further analysis in this study.
We investigated the structures using NMR spectroscopic techniques, and the cobalt(III)-hexammine [Co(NH3)63+] binding to both A bulge mutant (P4 U69A, Figure 1d) and bulge shift mutant P4 element (P4 U70b, Figure 1d). Co(NH3)63+ was used as an analog for magnesium hexahydrate (20) to locate Mg(H2O)62+ binding sites (18,21–23) The RNA molecules used in this study are derived from the E.coli RNase P RNA sequence and comprise nucleotides 66–73 and 354–360 in the E.coli sequence.
MATERIALS AND METHODS
RNA synthesis
P4 U69A and U70b RNAs were enzymatically synthesized and purified as described earlier (18,24). Typical sample concentrations as measured by UV absorbance were 2.2–2.4 mM.
NMR spectroscopy
All NMR experiments were done on a Bruker DMX 600 spectrometer at temperatures between 4 and 35°C. Exchangeable proton spectra were acquired in 90%H2O/10%D2O, 100 mM NaCl and 10 mM Na-phosphate, pH 6.4. Non-exchangeable proton spectra were acquired in 99.96% D2O, 100 mM NaCl and 10 mM Na-phosphate, pH 6.4. Proton chemical shifts were internally referenced to the solvent resonance.
NOESY experiments in 90% H2O/10% D2O were performed at NOE mixing times of 150 and 300 ms and temperatures of 5, 10, 15 and 25°C using jump-return water suppression (25) and z-gradient pulses during the NOE mixing time. The excitation maximum was set to 11 p.p.m. NOESY experiments in D2O (26) were performed at NOE mixing times of 50, 100, 150, 200, 300 and 400 ms and a temperature of 15°C. A presaturation pulse on the HDO resonance was used for solvent suppression. Zero- and double quantum artifacts in the short mixing time experiments were suppressed by a z-gradient pulse during the mixing time and by incrementing the mixing time by 10 μs between t1 increments. Four-hundred to five-hundred and twelve FIDs of 2048 complex points and 24 or 32 scans were acquired.
Double quantum filtered COSY experiments (27,28) were performed at 15 and 30°C. Broad band decoupling of phosphorus using GARP1 (29) was used in high resolution COSY experiments.
Proton detected 31P-1H COSY experiments were acquired at 600.14 MHz with 1600 Hz spectral width in the 31P dimension and 2000 Hz spectral width in the 1H dimension. Forty-eight scans of 2048 complex points and 320 FIDs were acquired. A 31P-1H Hetero-TOCSY-NOESY experiment (30–32) was performed at 15°C using a 70 ms DIPSI2 mixing pulse (33) and 500 ms NOESY mixing time. Ninety-six FIDs of 2048 complex points from 240 scans were acquired.
13C-1H HMQC experiments were run on unlabeled samples at natural abundance of 13C in 100% D2O with a sweep width of 12 500 Hz in the 13C dimension and the 13C carrier at 110 p.p.m. Two-hundred and twenty FIDs of 1024 complex points with 256 scans were acquired.
NOE distance constraints
NOE crosspeak intensities at 50, 100 and 150 ms mixing time were estimated from NOESY experiments acquired at 15°C in D2O. Crosspeaks were classified as strong, medium, weak and very weak as described earlier (18). Intermolecular crosspeaks between RNA and Co(NH3)63+ were derived from NOESY experiments in H2O. These intermolecular crosspeaks were classified as strong (4.3–5.0 Å) and medium (4.3–7.0 Å), accounting for the distance of 2.5 Å between the central Co atom and the amine protons.
Hydrogen bonding constraints were used to define base pairing geometry for all base pairs for which imino protons were observed in the 150 ms H2O NOESY experiments. Hydrogen bonding constraints for the C7:G22 base pair were slightly relaxed because of the larger imino proton line width. The non Watson–Crick U13:G16 in the tetraloop was constrained based on the published tetraloop structure (34); constraints for this base pair were relaxed similar to the C7:G22 base pair.
Dihedral constraints
Dihedral constraints on the sugar and backbone dihedral angles were constrained based on estimates of 3JH1′H2′, 3JH4′H5′, 3JH4′H5″, 3JPH3′, 3JPH5′, 3JPH5′ as described previously (18). The glycosidic conformation angle χ was constrained to anti for all nucleotides except G16 based on the strong H8-H1′ NOE crosspeak observed for this nucleotide. The α and ζ conformation angles were constrained to gauche- for nucleotides with phosphorus chemical shift between −4 and −5 p.p.m. No constraints on α and ζ were used otherwise.
Structure calculations
Structures were calculated using X-PLOR 3.1 (35) for restrained molecular dynamics. Initial structures were generated using random torsion angles. The molecular dynamics protocol was as described previously (36). Complex structures were calculated by merging coordinates of the RNA random torsion angle starting structures and Co(NH3)63+ before the first stage. The same sets of intramolecular NOE and torsion angle constraints were used for the complex structures as the NOE spectra did not differ significantly.
RESULTS
Assignments of exchangeable protons
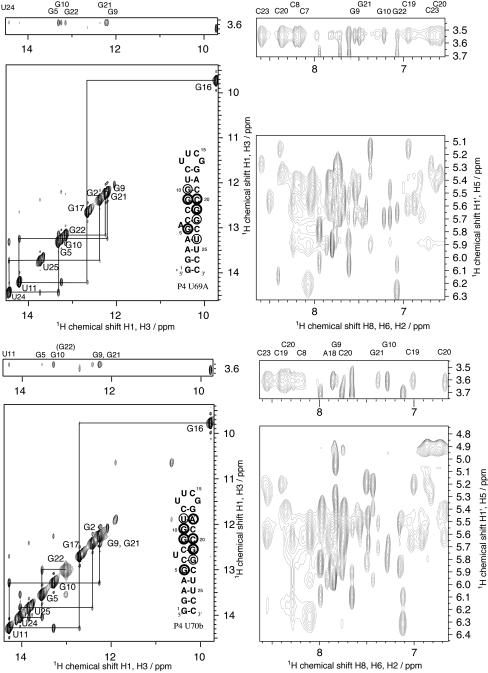
All imino proton resonances are assigned by standard procedures using data from H2O NOESY experiments (37,38) at 15°C and 150 ms mixing time (see Figure 3). The unusually shifted imino resonance at 9.8 p.p.m. in both the P4 U69A and P4 U70b spectrum is characteristic of the UUCG tetraloop structure (34,39), and confirms the formation of monomolecular stem–loop structures for both molecules. Sequential connectivity indicating close proximity of imino protons in stacked base pairs is observed from G2 through G5 and G22 through G16 in the P4 U69A molecule. A weaker crosspeak between G5 and G22 indicates continuation of base stacking across the bulge loop.
Figure 3.
H2O NOESY spectra of P4 U69A (top) and P4 U70b (bottom) acquired at 150 ms, 15°C in 6 mM Co(NH3)63+, 100 mM NaCl, 10 mM sodium phosphate, pH 6.4, 2.2 mM RNA. Left panels: imino-imino region (bottom) and imino-Co(NH3)63+ region (top) showing intermolecular crosspeaks between Co(NH3)63+ and RNA imino protons. The sequential connectivity between adjacent imino resonances is shown as a solid line. Right panel: aromatic-ribose region showing intermolecular crosspeaks between Co(NH3)63+ and aromatic and amino RNA protons. The inset on the left panel shows the localization of the intermolecular NOEs in the secondary structure. P4 U69A: Strong crosspeaks are observed between Co(NH3)63+ and RNA protons of G5, G9, G21, G10 and G22. Weaker crosspeaks to amino protons of C7, C8, C19, C20 and C23 and crosspeaks to aromatic protons of G9, G21, G10 and G22 are also observed. P4 U70b: Strong crosspeaks are observed between Co(NH3)63+ and RNA protons of G9, G10 and G21. Weaker crosspeaks to amino protons of C20 and C23 and crosspeaks to aromatic protons of A18, G10, C20 and G21 are also observed.
In the P4 U70b spectra, sequential crosspeaks are observed from G2 through G22 and G21 through G16. The G22-G21 crosspeak between the closing base pairs of the bulge loop is very weak. Thus, stacking across the bulge loop appears to be significantly less pronounced in the P4 U70b molecule.
Assignments of nonexchangeable protons
As evident from the aromatic-H1′ proton NOE spectra shown in Figure 2a, the U69A molecule displays sequential connectivities as expected from the proposed secondary structure. The sequential connectivities extend from G1 to G5 and C7 to U13 on the bulged strand, with very weak A-form like sequential crosspeaks observed across the bulge loop between G5 and C7, suggestive of stacking interaction between these two bases. These data require the bulge nucleotide to be in a non-stacked, extrahelical conformation. A-form connectivity is observed on the opposite strand through the bulge loop from G17 to C27, lending further support for stacking between the two stems.
Figure 2.
Aromatic-H1′ region of a 400 ms D2O NOESY spectrum of P4 U69A (a) and P4 U70b (b) at 15°C in 100 mM NaCl and 10 mM sodium phosphate, pH 6.4. Strong sequential H1′-aromatic connectivities indicative of regular A-form helical geometry are indicated by lines. P4 U69A (a): dashed line, G1-G5; dash-dotted line, C7-U13; solid line: G17-C27. The sequential walk is interrupted between G5 and C7, indicating a non stacking position of A6. Weak sequential crosspeaks between G5 and C7 suggest weaker stacking between these residues. Sequential connectivity is continuous between G17 and C27, indicating regular stacking geometry along the whole strand. P4 U70b (b): dashed line, G1-C6; dash-dotted line, C8-U13; solid line, G17-C27. The sequential walk is interrupted between C6 and C8, indicating a non stacking position of U7. Weak sequential cross peaks between C6 and C8 suggest weak stacking between those residues. Sequential connectivity is continuous between G17 and C27, indicating regular stacking geometry along the whole strand.
For the bulge position mutant P4 U70b molecule (Figure 2b), A-form sequential connectivity is observed from G1 to C6 and C8 to U13 on the bulged strand. Again, A-form sequential cross peaks are observed between C6 and C8, somewhat stronger than the G5-C7 connectivities observed in P4 U69A. This indicates that the U7 introduced by mutation is bulged out instead of the U6 in the wild-type molecule. On the opposite strand, A-form connectivity is again present from G17 to C27, indicating a stacked conformation of the base pairs closing the bulge loop.
The UUCG tetraloop in both molecules displays the characteristic spectral properties described earlier (34,39).
The sequential connectivity patterns demonstrated for the aromatic-H1′ spectral regions could be confirmed in the H1′-ribose, ribose-aromatic and aromatic-aromatic regions. Based on the sequential assignments of H1′ and aromatic protons, all H2′ protons and all but a few H3′ and H4′ protons could be unambiguously assigned from 1H-1H COSY, NOESY and 13C-1H HMQC experiments at natural abundance.
Intermolecular Co(NH3)63+–RNA NOE crosspeaks
The slowly exchanging Co(NH3)63+ ammine protons show a single resonance at 3.6 p.p.m. in H2O. Intermolecular NOE crosspeaks between this Co(NH3)63+ ammine proton resonance and RNA imino, amino and aromatic protons are observed in H2O NOESY experiments in the presence of 6 mM Co(NH3)63+ at 150 ms mixing time (Figure 3). The crosspeak line width generally reflects the RNA proton line width. The intermolecular crosspeaks to RNA imino protons are unambiguously assigned with the exception of the overlapped G9 and G21 imino crosspeaks in the P4 U70b spectra. The presence of intermolecular NOEs to both the G21 H8 and the C20 H6 and amino protons was used to confirm the existence of NOEs to both G9 and G21 imino protons in the P4 U70b RNA.
NOE crosspeak intensities for imino and aromatic protons are summarized in the inset secondary structures of P4 U69A and P4 U70b in Figure 3. The Co(NH3)63+ appears to be localized at the upper stem adjacent to the bulge loop in both molecules. The same region was found to bind Mg2+ ions in titration experiments (data not shown). A significant difference in binding position is seen from the distinctive crosspeak to U24 in P4 U69A and to U11 in P4 U70b. Distance constraints derived from these crosspeaks are used in structure calculations to locate the site of Co(NH3)63+ binding in the RNA structure.
Structure calculations
NOESY crosspeak intensities were estimated from a set of NOESY experiments at various mixing times, and distance constraints were obtained from classification of the crosspeak intensities into four distance categories as described in Materials and Methods. NOESY experiments in the presence of Mg2+ or Co(NH3)63+ did not show significant changes in crosspeak intensities or connectivities compared with experiments in the absence of divalent metal ions, suggesting little or no structural changes upon Co(NH3)63+ binding. The same set of distance constraints was therefore used in structure calculations for both free RNA and the RNA–Co(NH3)63+ complex. Dihedral constraints for ribose and backbone conformation were derived from high resolution, phosphorus-decoupled 1H-1H COSY experiments and 31P-1H correlation experiments as described in Materials and Methods. Based on the magnitude of 3JH1′H2′, sugar pucker was constrained to 3′-endo for all nucleotides except G5 and A6, which were constrained to range between 2′-endo and 3′-endo, and U14 and C15, which were constrained to be 2′-endo in P4 U69A. For P4 U70b, C6 was constrained to range between 2′-endo and 3′-endo and U7, U14 and C15 were constrained as 2′-endo, all other nucleotides were constrained 3′-endo. O4′-exo conformation for G5 and C6 could be excluded based on the intraresidue H1′-H4′ NOE intensity. Backbone dihedral angles that could not be directly measured were constrained to A-form ranges in the helical regions only. Conformation angles ε were found to be within A-form range on the non-bulged strand based on measurement of 3JPH3′.
Table 1 summarizes the statistics on distance and dihedral constraints used in the structure calculations for the P4 U69A and P4 U70b molecules. Of the 50 initial structures, 35 structures converged during the global fold stage as judged from NOE violation energies. During subsequent refinement, 36 (P4 U69A) and 39 (P4 U70b) structures converged to low NOE and dihedral angle violation energies. The converged structures of both P4 U69A and P4 U70b were analyzed and most of the structures were found to fall in a single structure family. Structures that differed from the major structure family for each RNA revealed proton–proton distances below 5 Å for which no NOE crosspeaks were observed even at long mixing time; six structures for P4 U96A were discarded from further analysis based on this criterion. Converged structures from the refinement stage that did not show NOE violations >0.1 Å were superimposed using X-PLOR (35) for calculation of RMS deviations and average structures. The average structure was energy minimized to resolve distortion of base geometries introduced by the averaging. All-atom average RMSD values of individual structures to the average structure are given in Table 1.
Table 1. Structure constraints and statistics.
| Molecule | P4 U69A | P4 U70B | P4 U69A complex | P4 U70b complex |
|---|---|---|---|---|
| NOE constraints | ||||
| Intraresidue | 96 | 74 | 96 | 74 |
| Interresidue | 194 | 187 | 194 | 187 |
| Base pairing | 48 | 48 | 48 | 48 |
| Intermolecular | NA | NA | 15 | 12 |
| Total | 338 | 309 | 353 | 321 |
| No. per nt. | 10.7 | 9.7 | 11.3 | 10.1 |
| Torsion constraints | ||||
| Global fold | 153 | 152 | 153 | 152 |
| Refinement | 245 | 246 | 245 | 246 |
| Structural statistics of individual structures | ||||
| NOE violations >0.1 Å | 0 | 0 | 0 | 0 |
| Dihedral violations >5° | 0 | 0 | 0 | 0 |
| Structural statistics of average structure | ||||
| NOE violations >0.1 Å | 0 | 0 | 0 | 0 |
| Dihedral violations >5° | 0 | 0 | 0 | 0 |
| RMS deviation to average | 1.78 | 1.77 | 1.85 | 1.87 |
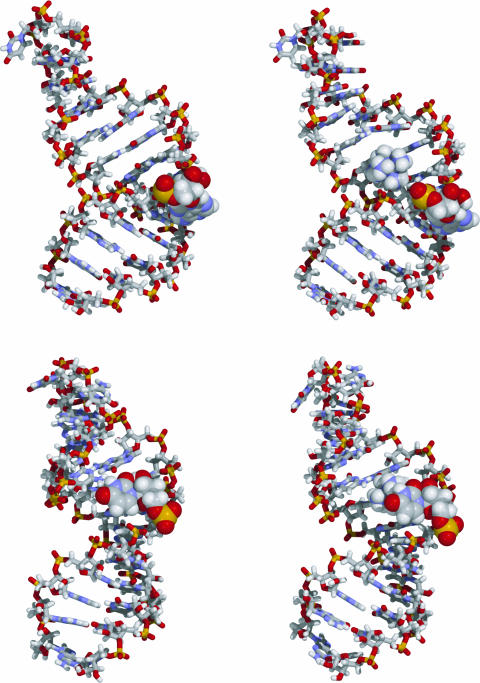
Both the P4 U96A and P4 U70b structures show a marked bend at the site of the bulge loop (Figure 4). The bulged nucleotide is found in the minor groove for P4 U69A, similar to the previously reported P4wt structure, whereas it is oriented towards the major groove in the P4 U70b structure, similar to the previously reported mutant structure. The location of the bulged nucleotide is determined by a number of unusual NOEs in both molecules. In P4 U69A, a stronger than usual NOE between A6 H2′ and C7 H5 combined with much weaker NOEs between A6 H2 and C7 H1′as well as C23 H1′ define the minor groove conformation of A6. A major groove position of A6 could be ruled out based on the absence of any A6 H2 to imino proton crosspeaks. For P4 U70b, stronger than usual NOEs from C6 H1′ and U7 H1′ to C8 H5 combined with a much weaker NOE between C6 H2′ and U7 H6 and the absence of other strong NOEs between U7 and C8 define the major groove position of U7.
Figure 4.
Side view of the average structures for the P4 U69A (upper panel) and P4 U70b (lower panel) RNA and RNA–Co(NH3)63+ complexes. The bulged nucleotides 6 (P4 U69A) and 7 (P4 U70b) are shown as space fill model, all other nucleotides are shown as stick drawing. The Co(NH3)63+ molecule is shown as spheres. The bulged adenosine occupies a position in the minor groove of the lower stem for P4 U69A, whereas the bulged uridine is located in the major groove of the upper stem for P4 U70bulge. The bound Co(NH3)63+ is located at the site of the bend slightly above the bulge loop in P4 U69A, and appears close to the G9:C20 and G10:G19 base pairs in the upper stem in P4 U70b. Comparing P4 U69A and P4 U70b, the position of the bound Co(NH3)63+ shifts up to the upper stem by about one base pair.
Structure calculations on the RNA–Co(NH3)63+ complex were done to locate the site of Co(NH3)63+ binding. After refinement, 14 structures for the P4 U69A–Co(NH3)63+ complex and 18 structures for the P4 U70b–Co(NH3)63+ complex were selected for averaging. The average structures are shown in Figure 4. The Co(NH3)63+ is located in the major groove at the bottom of the upper stem in the P4 U69A–Co(NH3)63+ complex structure. It is in close proximity to the G5-C23, C8-G21, and G9-C20 base pairs. In the P4 U70b–Co(NH3)63+ complex structure, the Co(NH3)63+ is located at the bottom of the upper stem, close to the G9-C20, G10-C19, and U11-A18 base pairs. The overall structural features of the RNA appear unchanged.
The coordinates for average structures of each molecule have been deposited in the PDB database under PDB ID 1XSG, 1XSH, 1XST and 1XSU.
DISCUSSION
Comparison of the structures of both P4 U69A and P4 U70b stems to the wild-type structure (18) reveals a significant difference between the two molecules. Change of the bulge nucleotide from uridine to adenosine does not change the secondary structure of the P4 element. The bulged nucleotide is found in the minor groove as observed in the wild-type structure, and the overall structure remains unchanged. The binding site of Co(NH3)63+ and Mg2+, not surprisingly, is preserved in the U69A mutant. Upon mutation of U69 to C and C70 to U, a GC base pair is introduced involving C69, thus extending the 5′ half of the P4 stem and shifting the bulge position by one nucleotide to U70. This difference in secondary structure is evident from the sequential NOESY crosspeaks shown in Figure 2. In addition to this change in bulge position, structure calculations based on NOE distance restraints and torsion angle restraints show that the bulged uridine changes its position from the minor groove to the major groove. Both the secondary structure and the change in bulged nucleotide position are similar to the situation in P4m. In contrast to P4m, however, no GU pair is introduced in P4 U70b. Comparing P4m and P4 U70b, the Mg2+ binding site is not preserved. Rather, a shift of the binding site to the 3′ direction is observed. From the comparison of metal ion binding positions, we can conclude that the introduction of a GU pair rather than the change in bulge position determines the site of binding in P4m. Without the GU pair, the metal ion is found to bind at the bottom of the upper stem adjacent to the bulge in both the wild-type structure and P4 U70b. Evidently, the expected widening of the major groove by the GU pair in P4m partly compensates the effect of the bulge shift, leading to a lesser change in positioning of the bound metal ion.
It is expected that the change in secondary structure and bulge nucleotide position found for the mutant P4 stems also occurs in the context of the full RNase P RNA. This is supported by changes in Tb3+ cleavage patterns consistent with the proposed secondary structure changes as reported by Kaye et al. (19).
The findings by Kaye et al. (19) reveal no large effect by local P4 structure changes on global structure as detected by hydroxyl radical experiments; any such global change would need to be rather subtle. Missing Tb3+ cleavage in the upper stem of P4 is probably due to a more stable helical structure in that region that prevents the necessary in-line backbone conformation required for cleavage, and should not be construed as evidence against the presence of a metal binding site in the upper stem. The proposed L8–P4 interaction (6) provides a possible mechanism for structural distortion beyond P4. In particular, the change in position of the universally conserved bulged uridine could have a profound effect if functional groups on the bulged nucleotide participate in tertiary contacts. The findings of Kaye et al. (19), however, indicate that bulge position, rather than identity, affects catalysis. The change in bulge position affects the riboyzme function by perturbation of a bulged-helix motif generally implicated in metal binding, according to these authors. The conserved uridine might be required for protein interaction rather than RNA tertiary contacts, since no profound change in RNA structure was found in the U69A mutant.
Binding of catalytic metal ions in the context of the complete RNase P sequence has been shown to involve the nucleotides A66 and A67, as well as single-stranded adenosines from the J3/4 junction 5′ to A66, and is mediated by inner sphere coordination. In the context of the isolated P4 stem, only A66 and A67 are present, and a different outer sphere binding site is observed instead. This difference both in context and metal binding properties limits functional interpretation of the metal binding site found in the major groove of P4. In particular, the Mg2+ binding site determined by this study may not correspond to a site of catalytic Mg2+ binding. On the other hand, tertiary structure models of RNase P RNA show both A66 and A67 to be quite removed from the scissile bond in the pre-tRNA. Functional groups implicated in metal coordination are spread along the whole of P4, with the A352 phosphate in J2/4 coming closest to the scissile bond. Hence, perturbation of an outer sphere metal binding site at the bulged-helix motif in P4 may well have indirect effects on overall metal binding to P4, including essential, inner-sphere coordinated metals.
Support for outer sphere coordinated metal ions in P4 arises from the observation by Kurz and Fierke (5) that Co(NH3)63+ competes for metal ions critical for catalysis. Up to four Mg2+ ions were found to bind cooperatively to the RNase P–pre-tRNA complex and stabilize substrate binding; the affinity for these ions was enhanced by the protein subunit. Co(NH3)63+ was found to inhibit catalytic activity by displacing a single essential outer sphere Mg2+ ion, as well as to compete with inhibitory, larger divalent ions. Taken together, these findings support an outer sphere Mg2+ binding site in P4. The role of this site remains unclear. It may serve to position an Mg2+ ion close to the site of inner sphere binding during folding, or it may be responsible for inhibition by larger divalent ions due to a displacement of the catalytic Mg2+ ion.
In addition to the perturbation of a bulged-helix motif, the precise location of the bulged nucleotide might also influence ribozyme function. In the wild-type and P4 U69A mutant, the minor groove placement of the bulged nucleotide positions it at the L8–P4 interface in the tertiary structure model, somewhat removed from the cleavage site. In the P4 U70 and U70b mutants, the major groove position of the bulged nucleotide is closer to the cleavage site but the roughly 13 Å distance to the scissile phosphate makes direct involvement in catalysis unlikely. Rather, the major groove placement of U70 might interfere with metal binding in the major groove of P4.
In summary, the position of helical structure distortion in P4 probably affects catalysis through a large change in ion binding position. This is an example of how a single base mutation can change the function of an RNA through metal interactions rather than effects on conformation.
Acknowledgments
ACKNOWLEDGEMENTS
The author is indebted to Dr D. Riesner for his support of this work. Acknowledgement is made to Dr G. Steger for critical reading of the manuscript.
REFERENCES
- 1.Pace N.R. and Brown,J.W. (1995) Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J. Bacteriol., 177, 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris M.E., Frank,D.N. and Pace,N.R. (1998) Structure and catalytic function of the bacterial ribonuclease P enzyme. In Simons, R.W. and Grunberg-Manago, M., (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 309–337. [Google Scholar]
- 3.Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N.R. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 4.Guerrier-Takada C., Haydock,K., Allen,L. and Altman,S. (1986) Metal ion requirements and other aspects of the reaction catalyzed by M1 RNA, the RNA subunit of ribonuclease P from Escherichia coli. Biochemistry, 25, 1509–1515. [DOI] [PubMed] [Google Scholar]
- 5.Kurz J.C. and Fierke,C.A. (2002) The affinity of magnesium binding sites in the Bacillus subtilis RNase P·pre-tRNA complex is enhanced by the protein subunit. Biochemistry, 41, 9545–9558. [DOI] [PubMed] [Google Scholar]
- 6.Harris M.E., Nolan,J.M., Malhotra,A., Brown,J.W., Harvey,S. and Pace,N.R. (1994) Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J., 13, 3953–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massire C., Jaeger,L. and Westhof,E. (1998) Derivation of the three dimensional architecture of bacterial ribonuclease P RNA from comparative sequence analysis. J. Mol. Biol., 279, 773–793. [DOI] [PubMed] [Google Scholar]
- 8.Chen J.L., Nolan,J.M., Harris,M.E. and Pace,N.R. (1998) Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J., 17, 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krasilnikov A.S., Jang,X., Pan,T. and Mondragon,A. (2003) Crystal structure of the specificity domain of ribonuclease P. Nature, 42, 760–764. [DOI] [PubMed] [Google Scholar]
- 10.Kanzatsev A.V., Krivenko,A.A., Harrington,D.J., Carter,R.J., Holbrook,S.R., Adams,P.D. and Pace,N.R. (2003) High-resolution structure of RNase P protein from Thermotoga maritima. Proc. Natl Acad. Sci. USA, 100, 7497–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai H.-Y., Masquida,B., Biswas,R., Westhof,E. and Gopalan,V. (2003) Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol., 325, 661–675. [DOI] [PubMed] [Google Scholar]
- 12.Kanzatsev A.V. and Pace,N.R. (1998) Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial ribonuclease P. RNA, 4, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardt W.-D., Erdmann,V.A. and Hartmann,R.K. (1996) Rp-deoxy-phosphorothioate modification interference experiments identify 2′-OH groups in RNase P RNA that are crucial to tRNA binding. RNA, 2, 1189–1198. [PMC free article] [PubMed] [Google Scholar]
- 14.Christian E.L., Kaye,N.M. and Harris,M.E. (2002) Evidence for a polynuclear metal ion binding site in the catalytic domain of ribonuclease P RNA. EMBO J., 21, 2253–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris M.E. and Pace,N.R. (1995) Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA, 1, 210–218. [PMC free article] [PubMed] [Google Scholar]
- 16.Frank D.N., Ellington,A.D. and Pace,N.R. (1996) In vitro selection of RNase P RNA reveals optimized catalytic activity in highly conserved structural domain. RNA, 2, 1179–1188. [PMC free article] [PubMed] [Google Scholar]
- 17.Frank D.N. and Pace,N.R. (1997) In vitro selection for altered divalent metal specificity in the RNase P RNA. Proc. Natl Acad. Sci. USA, 94, 14255–14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz M. and Tinoco,I.,Jr (2000) Solution structure and metal-ion binding of the P4 element from bacterial Rnase P RNA. RNA, 6, 1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaye N.M., Zahler,N.H., Christian,E.L. and Harris,M.E. (2002) Conservation of helical structure contributes to functional metal ion interactions in the catalytic domain of ribonuclease P RNA. J. Mol. Biol., 324, 429–442. [DOI] [PubMed] [Google Scholar]
- 20.Cowan J.A. (1993) Metallobiochemistry of RNA. Co(NH3)63+ as a probe for Mg2+(aq) binding sites. J. Inorg. Biochem., 49, 171–175. [DOI] [PubMed] [Google Scholar]
- 21.Kieft J.S. and Tinoco,I.,Jr (1997) Solution structure of a metal binding site in the major groove of RNA complexed with cobalt (III) hexammine. Structure, 5, 713–721. [DOI] [PubMed] [Google Scholar]
- 22.Colmenarejo G. and Tinoco,I.,Jr (1999) Structure and thermodynamics of metal binding in the P5 helix of a group I intron ribozyme. J. Mol. Biol., 290, 119–135. [DOI] [PubMed] [Google Scholar]
- 23.Rüdisser S. and Tinoco,I.,Jr (2000) Structure and thermodynamics of metal-ion binding to GA mismatches and to the GAAA tetraloop. J. Mol. Biol., 295, 1211–1223. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt J.R., Chastain,M. and Puglisi,J.D. (1991) Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques, 11, 764–769. [PubMed] [Google Scholar]
- 25.Sklenár V. and Bax,A. (1987) Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra. J. Magn. Reson., 74, 469–479. [Google Scholar]
- 26.Macura S., Wüthrich,K. and Ernst,R.R. (1982) Separation and suppression of coherent transfer effects in two-dimensional NOE and chemical exchange spectroscopy. J. Magn. Reson., 46, 269–282. [Google Scholar]
- 27.Shaka A.J. and Freeman,R. (1983) Simplification of NMR spectra by filtration through multiple-quantum coherence. J. Magn. Reson., 51, 169–173. [Google Scholar]
- 28.Müller N., Ernst,R.R. and Wüthrich,K. (1986) Multiple-quantum-filtered two-dimensional correlated NMR spectroscopy of proteins. J. Am. Chem. Soc., 108, 6482–6492. [Google Scholar]
- 29.Shaka A.J., Barker,P.B. and Freeman,R. (1985) Computer-optimized decoupling scheme for wideband applications and low-level operation. J. Magn. Reson., 64, 547–522. [Google Scholar]
- 30.Kellogg G.W. (1992) Proton-detected hetero-TOCSY experiments with application to nucleic acids. J. Magn. Reson., 98, 176–182. [Google Scholar]
- 31.Kellogg G.W. and Schweitzer,B.I. (1993) Two- and three-dimensional 31P-driven NMR procedures for complete assignment of backbone resonances in oligodeoxyribonucleotides. J. Biomol. NMR, 3, 577–595. [DOI] [PubMed] [Google Scholar]
- 32.Kellogg G.W., Szewczak,A.A. and Moore,P.B. (1992) Two-dimensional hetero-TOCSY-NOESY. Correlation of 31P resonances with anomeric and aromatic 1H resonances in RNA. J. Am. Chem. Soc., 114, 2727–2728. [Google Scholar]
- 33.Rucker S.P. and Shaka,A.J. (1989) Broadband homonuclear cross polarization in 2D N.M.R. using DIPSI-2. Mol. Phys., 68, 509–517. [Google Scholar]
- 34.Cheong C., Varani,G. and Tinoco,I.,Jr (1990) Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC. Nature, 346, 680–682. [DOI] [PubMed] [Google Scholar]
- 35.Brünger A.T. (1993) X-PLOR Version 3.1: A System for X-ray Crystallography and NMR, Yale University Press, New Haven, CT, USA. [Google Scholar]
- 36.Wimberly B., Varani,G. and Tinoco,I.,Jr (1993) The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry, 32, 1078–1087. [DOI] [PubMed] [Google Scholar]
- 37.Varani G., Aboul-ela,F. and Allain,F.H.-T. (1996) NMR investigation of RNA structure. Progr. Nucl. Magn. Reson. Spectrosc., 29, 51–127. [Google Scholar]
- 38.Varani G. and Tinoco,I.,Jr (1991) RNA structure and NMR spectroscopy. Quart. Revs. Biophysics, 24, 479–532. [DOI] [PubMed] [Google Scholar]
- 39.Allain F.H.T and Varani,G. (1995) Divalent metal ion binding to a conserved wobble pair defining the upstream site of cleavage of group I self-splicing introns. Nucleic Acids Res., 23, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]