Abstract
We previously observed reduced YWHAE (14-3-3ε) protein expression in a small set of gastric cancer samples. YWHAE may act as a negative regulator of the cyclin CDC25B, which is a transcriptional target of MYC oncogene. The understanding of YWHAE role and its targets is important for the better knowledge of gastric carcinogenesis. Thus, we aimed to evaluate the relationship among YWHAE, CDC25B, and MYC in vitro and in vivo. For this, we analyzed the YWHAE, CDC25B, and MYC expression in YWHA-silenced, CDC25B-silenced, and MYC-silenced gastric cancer cell lines, as well as in gastric cancer and non-neoplastic gastric samples. In gastric cancer cell lines, YWHAE was able to inhibit the cell proliferation, invasion and migration through the reduction of MYC and CDC25B expression. Conversely, MYC induced the cell proliferation, invasion and migration through the induction of CDC25B and the reduction of YWHAE. Most of the tumors presented reduced YWHAE and increased CDC25B expression, which seems to be important for tumor development. Increased MYC expression was a common finding in gastric cancer and has a role in poor prognosis. In the tumor initiation, the opposite role of YWHAE and CDC25B in gastric carcinogenesis seems to be independent of MYC expression. However, the inversely correlation between YWHAE and MYC expression seems to be important for gastric cancer cells invasion and migration. The interaction between YWHAE and MYC and the activation of the pathways related to this interaction play a role in the metastasis process.
Keywords: gastric cancer, cancer development, YWHAE, CDC25B, MYC
INTRODUCTION
Gastric cancer (GC) is one of the most common causes of cancer death in the World [1]. Advanced GC presents few treatment options and a poor prognosis, which is in part due to the tumor recurrence, invasion or metastasis. The relative five-year survival rate is below than 20% [2]. It is still necessary to determine the key molecular factors involved in GC initiation and progression.
In eukaryotes, the 14-3-3s are part of a highly conserved protein family. Seven 14-3-3 genes encode nine protein isoforms, including two phosphorylated forms (α and δ) [3, 4]. The 14-3-3 proteins are mainly dimeric within the cell and are able to bind several sites within a target or act as a bridge between proteins [5–7]. 14-3-3 proteins can interact with hundreds of proteins, including cdc25 phosphatase [4, 5, 7, 8]. The precise function of 14-3-3 proteins is not fully understand. However, these proteins seem to play a role as molecular scaffolds [4] and regulate different biologic processes, including apoptosis, mitogenic signal transduction, and cell cycle (for reviews, see references [5, 9, 10]).
Deregulated expression of 14-3-3 proteins has been detected in some GC proteomic studies [11–14]. We previously observed reduced YWHAE, also called 14-3-3ε, protein expression in a small set of GC specimens [15]. Reduced YWHAE expression has also been described in other cancers [16–18], suggesting that this protein may play a role as a tumor suppressor.
YWHAE acts as a negative regulator of CDC25 [19, 20]. CDC25 phosphatases play a key role in cell cycle proliferation. CDC25B seems to present oncogenetic properties [21] and its overexpression was described previously in GC [22–25]. The subcellular localization of CDC25B can be controlled by its association with 14-3-3 proteins. CDC25B subcellular location might contribute to stall the cell cycle at the G2 phase following DNA damage [26–29].
At the transcription level, CDC25B is also a target of MYC and they may mediate MYC-induced cell cycle activation and/or apoptosis [30]. A correlation between CDC25B and MYC immunoreactivity was earlier described in GC [25]. MYC, located at 8q24, is a key oncogene in gastric carcinogenesis [31]. We previously demonstrated that MYC mRNA and protein increased expression is a common finding in GC samples [32–35] and some preneoplastic gastric lesions [36, 37] from a Brazilian population. Our research group also showed MYC expression increases during gastric carcinogenesis in a nonhuman primate model [38]. Moreover, we described several genetic and epigenetic alterations involving MYC gene in GC samples or GC cell lines, including chromosome 8 trisomy [32, 39–43], gene or 8q24 amplification [32–36, 39, 44–46], gene insertion [47], promoter hypomethylation [34] and point mutations [34]. However, the understanding of MYC targets is important for the better knowledge of its role in gastric carcinogenesis and may help in the development of new anticancer therapies.
Based on our previous findings, we hypothesized that MYC or CDC25B up-regulation may induce YWHAE down-regulation in GC or YWHAE down-regulation would induce CDC25B up-regulation in this neoplasia, which would also contribute to MYC overexpression. In this study, we aimed to better understand the relationship of the expression of these genes in vivo and in vitro. For this, we simultaneously evaluated the YWHAE, CDC25B, MYC and mRNA and protein expression in GC cell lines and in a large set of GC and paired non-neoplastic gastric samples. Additionally, we investigated the possible associations between gene/protein expression and clinical variables.
RESULTS
mRNA and protein expression in gastric cell lines
We firstly accessed the mRNA and protein expression of YWHAE, CDC25B and MYC in GC cell lines in relation to the non-neoplastic MNP01 cells (Figure 1). GC cell lines presented a reduced YWHAE mRNA and protein expression in relation to MNP01 cells [mRNA median (interquartile range, IQR): 0.71 (0.31); protein median (IQR): 0.52 (0.40); respectively]. On the other hand, the GC cell lines presented an increased CDC25B [mRNA median (IQR): 1.79 (1.15); protein median (IQR): 1.45 (1.24); respectively] and MYC [mRNA median (IQR): 2.98 (1.13); protein median (IQR): 2.48 (0.66); respectively] expression.
Figure 1. YHWAE, CDC25B and MYC mRNA and protein expression in gastric cancer cell lines in relation to non-neoplastic cells.
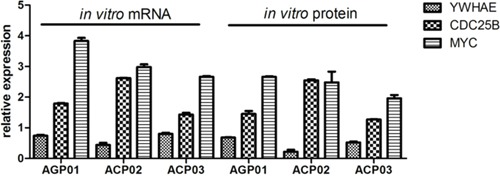
MNP01 non-neoplastic cells were used as a calibrator. Values of median and IQR are shown.
YWHAE silencing induces GC cell proliferation, invasion and migration
siRNA decresead YWHAE expression in more thand 80% in ACP03 and in more than 90% in AGP01 and ACP02 cell lines (Figure 2A–2B). Furthermore, YWHAE silencing induced cell proliferation (p<0.05, for all comparisions; Figure 3A–3C) and induces cell arrest by increase of G1/G0 cells and decrease in cells in S and G2/M (p<0.05, for all comparisions; Figure 4A–4C).
Figure 2. Effect of gene silencing in gene and protein expression in gastric cancer cell lines.
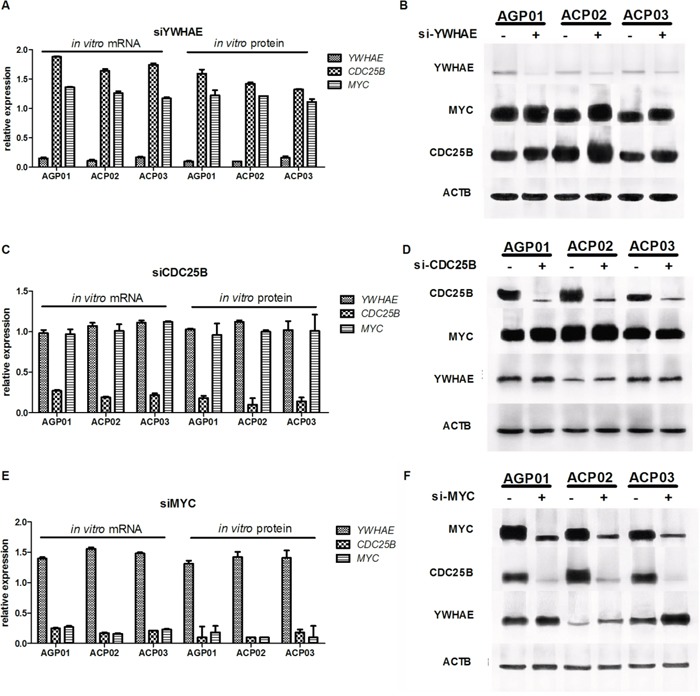
A. YWHAE silencing induced MYC and CDC25B increased expression in GC cell lines. B. GC cells with (+) or without (-) YWHAE silencing, equal amounts of whole cell extracts were analyzed by western blot with the indicated antibodies. C. CDCD25B silencing did not alter MYC and YHWAE expression in GC cell lines. D. GC cells with (+) or without (-) CDC25B silencing; Equal amounts of whole cell extracts were analyzed by western blot with the indicated antibodies. E. MYC silencing induced the reduction of CDC25B expression and increasing of YHWAE expression in GC cell lines. F. GC cells with (+) or without (-) MYC silencing; equal amounts of whole cell extracts were analyzed by western blot with the indicated antibodies. siRNA control-transfected cells were used as a calibrator. Values of median and IQR are shown.
Figure 3. Effect of gene silencing in gastric cancer cell proliferation.
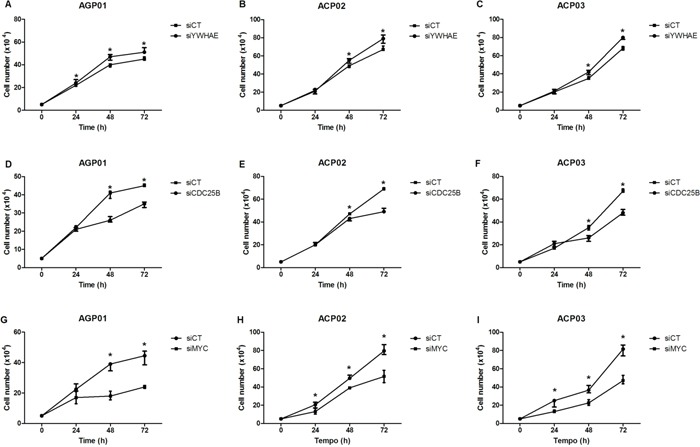
A. Effect of YWHAE silencing in AGP01 cell line. B. Effect of YWHAE silencing in ACP02 cell line. C. Effect of YWHAE silencing in ACP03 cell line. D. Effect of CDC25B silencing in AGP01 cell line. E. Effect of CDC25B silencing in ACP02 cell line. F. Effect of CDC25B silencing in ACP03 cell line. G. Effect of MYC silencing in AGP01 cell line. H. Effect of MYC silencing in ACP02 cell line. I. Effect of MYC silencing in ACP03 cell line. Cell counting was measured after 24, 48, and 72 h of silencing. *p<0.05, significant difference between controls and silenced cells by Mann-Whitney test. Values of median and IQR are shown. siCT: controls cells; siYWHAE: cells with YWHAE silencing; siCDC25B: cells with CDC25B silencing; siMYC: cells with MYC silencing.
Figure 4. Effect of si-RNA silencing on cell cycle progression of gastric cancer cell lines.
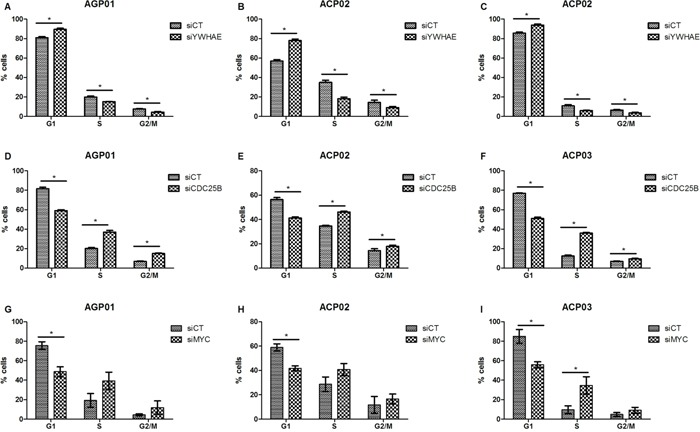
A. Effect of YWHAE silencing in AGP01 cell line. B. Effect of YWHAE silencing in ACP02 cell line. C. Effect of YWHAE silencing in ACP03 cell line. D. Effect of CDC25B silencing in AGP01 cell line. E. Effect of CDC25B silencing in ACP02 cell line. F. Effect of CDC25B silencing in ACP03 cell line. G. Effect of MYC silencing in AGP01 cell line. H. Effect of MYC silencing in ACP02 cell line. I. Effect of MYC silencing in ACP03 cell line. The cell cycle analysis was performed at 72 h after transfection. *p<0.05, significant difference between controls and silenced cells by Mann-Whitney test. Values of median and IQR are shown. siCT: controls cells; siYWHAE: cells with YWHAE silencing; siCDC25B: cells with CDC25B silencing; siMYC: cells with MYC silencing.
YWHAE silencing induced cell invasion and migration in all gastric cancer cell lines (p<0.05, for all comparisions; Figure 5A).
Figure 5. Effect of si-RNA silencing on gastric cancer cell lines invasion and migration.
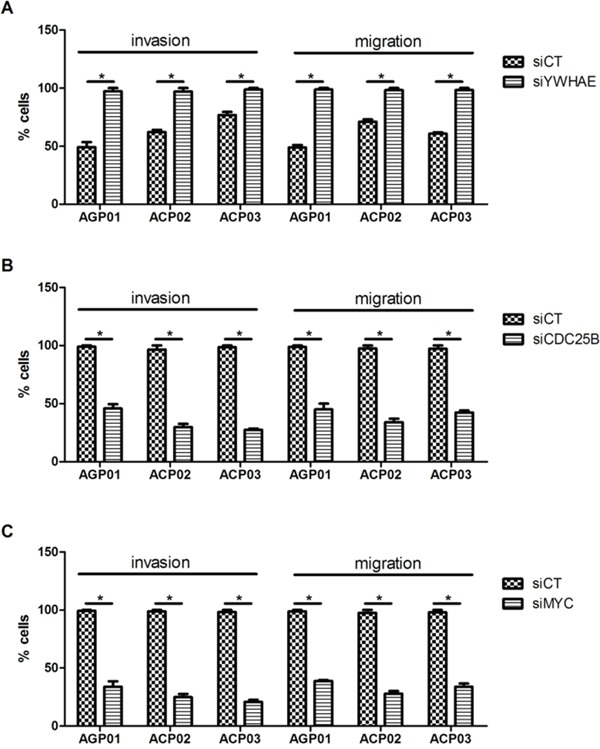
A. Effect of YWHAE silencing in GC cell line. B. Effect of CDC25B silencing in GC cell line. C. Effect of MYC silencing in GC cell line. *p<0.05, significant difference between controls and silenced cells by Mann-Whitney test. Values of median and IQR are shown. siCT: controls cells; siYWHAE: cells with YWHAE silencing; siCDC25B: cells with CDC25B silencing; siMYC: cells with MYC silencing.
YWHAE regulates CDC25B and MYC in GC cells
YWHAE silencing induced CDC25B up-regulation by at least 1.6-fold in all GC cell lines (Figure 2A) and a slight MYC up-regulation by approximately 1.25-fold in all GC cell lines (Figure 2A). At protein level, YWHAE expression was inversely correlated with MYC (ρ=-0.697; p=0.037; Figure 6A) and CDC25B expression (ρ=-0.676; p=0.046; Figure 6B). MYC and CDC25B protein expression was also correlated in YWHAE silencing cells (ρ=0.854; p=0.003; Figure 6C).
Figure 6. Correlation between protein expression in YWHAE-silenced AGP01, ACP02 and ACP03 gastric cancer cell lines.
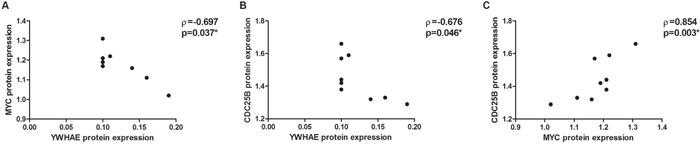
A. YWHAE and MYC. B. YWHAE and CDC25B. C. MYC and CDC25B. Correlation coefficients and p-values of Spearman correlation test are shown. *p<0.05, significant correlation by Spearman correlation test.
Then, we silenced CDC25B and MYC in GC cell to evaluate the effect of these genes in GC cells, as well as the effect on YWHAE expression. siRNA caused a reduction in CDC25B expression of more than 80% in all three GC cell lines (Figure 2C–2D). Although CDC25B silencing induced a significant decrease in cell proliferation (Figure 3D–3F), control cells were accumulated in G1 in relation to si-CDC25B cells (p<0.05, for all comparisions; Figure 4D–4F). CDC25B silencing inhibits GC cells invasion and migration (Figure 5B). However, CDC25B silencing did not induced alteration in MYC or YWHAE mRNA and protein expression (Figure 2B).
Conversely, siRNA caused a reduction in MYC expression of more than 70% in all three studied GC cell lines (Figure 2E–2F). The cell lines also presented significant alterations concerning cell proliferation. All cell lines presented a significant decrease in proliferation after 48 h and 72 h of silencing MYC (p<0.05, for all comparisons; Figure 3G–3I). Moreover, ACP02 and ACP03 also showed a reduction in cell proliferation after 24 h of MYC silencing (p<0.05, for all comparisons; Figure 3G–3I). After 72 h, MYC silencing leads to alterations in the GC cell cycle. Slight variations were observed among GC cell lines (Figure 4G–4I). Control cells were accumulated in G1, and there was a statistical significance when comparing control cells to siRNA-MYC cells (p<0.05, for all comparisions), as detected in si-CDC25B cells. Furthermore, MYC silencing inhibed GC cells invasion and migration (p<0.05, for all comparisions; Figure 5C).
We next evaluate whether MYC may regulate YWHAE and CDC25B in gastric cells. MYC silencing induced CDC25B down-regulation by at least 4-fold in all GC cell lines (Figure 2E). Conversely, MYC silencing up-regulated YWHAE by approximately 1.5-fold in all GC cell lines (Figure 2E). Considering all cell lines, we observed that MYC expression was directly correlated with CDC25B expression (ρ=0.996; p<0.001; Figure 7A) and inversely correlated with YWHAE expression (ρ=-0.996; p<0.001; Figure 7B). Moreover, a strong inverse correlation was detected between YWHAE and CDC25B expression (ρ=-0.992; p<0.001; Figure 7C). MYC and YWHAE protein expression was also inversely correlated (ρ=-0.703; p<0.033).
Figure 7. Correlation between the studied genes mRNA expression in MYC-silenced AGP01, ACP02 and ACP03 gastric cancer cell lines.
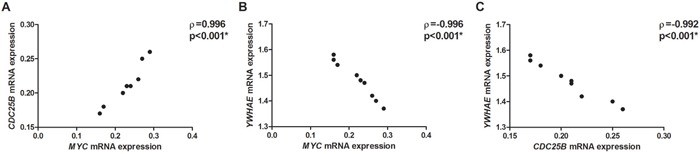
A. MYC and CDC25B. B. MYC and YWHAE. C. CDC25B and YWHAE. Correlation coefficients and p-values of Spearman correlation test are shown. *p<0.05, significant correlation by Spearman correlation test.
YWHAE, CDC25B, and MYC expression in gastric samples
YWHAE immunoreactivity was detected in the cytoplasm of lymphocytes and non-neoplastic gastric cells (Figure 8A). YWHAE immunoreactivity was observed in only 6 (4.7%) of the gastric tumor tissue samples (Figure 8B). CDC25B was detected in the nuclei and cytoplasm of 128 (99.2%) GC samples (Figure 8D). Nuclear immunoreactivity for MYC was detected in 117 (90.7%) GC samples (Figure 8F). Conversely, nonatypical gastric cells did not present CDC25B and MYC immunoreactivity (Figure 8C and 8E).
Figure 8. YWHAE, CDC25B and MYC immunoreactivity in gastric tissue samples.
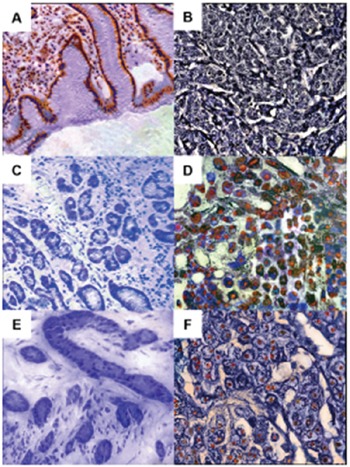
A. Non-neoplastic gastric mucosa showing cytoplasmic YWHAE staining. B. intestinal-type gastric cancer cells without YWHAE immunoreactivity. C. Non-neoplastic gastric tissue without CDC25B immunoreactivity. D. Diffuse-type gastric cancer presenting nuclear and cytoplasmic CDC25B immunoreactivity. E. Gastric mucosa without MYC immunoreactivity. F. Intestinal-type gastric cancer presenting nuclear immunoreactivity of MYC.
Down-regulation (at least 50% decrease in expression) of YWHAE protein and mRNA was detected in 89 (69%) and 61 (47.3%) GC samples, respectively (Figure 9A–9B, G).
Figure 9. Protein and gene expression in gastric cancer. A. YWHAE protein expression.
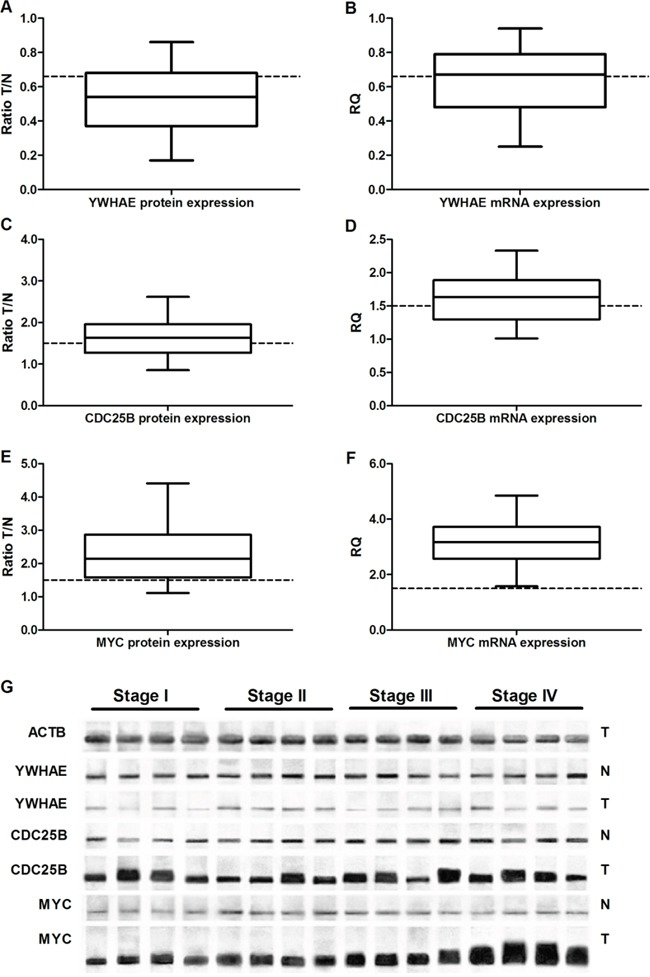
B. YHWAE mRNA expression. C. CDC25B protein expression. D. CDC25B mRNA expression. E. MYC protein expression. F. MYC mRNA expression. G. Representative image of Western-blot. In all graphs, the expression in gastric tumors was normalized by matched non-neoplastic gastric tissue. T: tumor sample; N: normal mucosa sample; RQ: relative quantification. The whiskers indicate the minimum and maximum values. The dotted lines representes the 1.5 fold-change.
Conversely, protein and mRNA levels of CDC25B were increased more than 1.5-fold (increment of at least 50% in expression) in 72 (55.8%) and 77 (59.7%) GC samples, respectively (Figure 9C–9D, G). In addition, MYC protein and mRNA expression increased more than 1.5-fold in 101 (78.3%) and 129 (100%) GC samples, respectively, in comparison to paired non-neoplastic gastric specimens (Figure 9E–9F, G).
MYC (p=0.002 and 0.001, respectively) and YWHAE (p=0.007 and <0.001, respectively) immunoreactivity was associated with higher protein and mRNA levels in GC samples. A strong correlation was observed between mRNA and protein expression for YWHAE (ρ=0.759; p<0.001; Figure 10A), CDC25B (ρ=0.972; p<0.001; Figure 10B), and MYC (ρ=0.968; p<0.001; Figure 10C).
Figure 10. Correlation between protein and mRNA expression in gastric cancer samples.
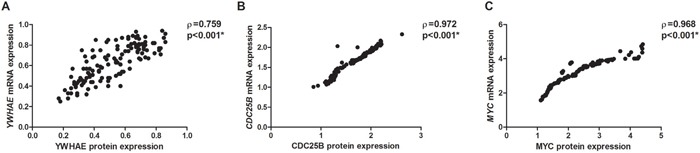
A. YWHAE. B. CDC25B. C. MYC. Correlation coefficients and p-values of Spearman correlation test are shown. *p<0.05, significant correlation by Spearman correlation test.
An inverse correlation was detected between YWHAE and CDC25B protein (ρ=-0.692; p<0.001; Figure 11A) and mRNA (ρ=-0.857; p<0.001; Figure 11B) expression. Moreover, 122 (94.6%) of tumors presented CDC25B immunoreactivity and lack of YWHAE immunostaining.
Figure 11. Correlation between YWHAE and CDC25B in all the studied gastric cancer samples.
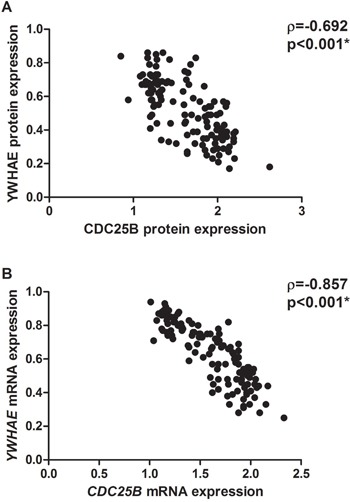
A. protein expression. B. mRNA expression. Correlation coefficients and p-values of Spearman correlation test are shown. *p<0.05, significant correlation by Spearman correlation test.
MYC protein and mRNA expression was not correlated with CDC25B or YWHAE expression (p>0.05 for all analyses). However, 117 (90.7%) of the tumors presented both MYC and CDC25B immunoreactivity, and 111 (86%) of the tumors presented MYC immunoreactivity and lack of YWHAE immunostaining.
We used the K-means clustering method to group samples based on their gene expression similarities. In the K-means clustering method, we observed the presence of two clusters. In both clusters, a strong inverse correlation was also detected between YWHAE and CDC25B mRNA (ρ=-0.844, p<0.001, Figure 12A for “Cluster 1” and ρ=-0.868, p<0.001, Figure 12G for “Cluster 2”) and protein (ρ=-0.737, p<0.001, Figure 12D for “Cluster 1” and ρ=-0.657, p<0.001, Figure 12J for “Cluster 2”) expression. In one of the clusters (named “Cluster 2”), MYC mRNA and protein expression was directly correlated with CDC25B expression (ρ=0.409, p=0.001, Figure 12I; ρ=0.400, p=0.001, Figure 12L; respectively) and inversely correlated with YWHAE mRNA expression (ρ=-0.340, p=0.007, Figure 12H). Interestingly, “Cluster 2” was composed of samples with the highest MYC expression, in which the mRNA level was increased by at least 3.2-fold and protein level was increased by at least 1.8-fold.
Figure 12. Correlation between mRNA and protein expression in two cluster of gastric cancer samples.
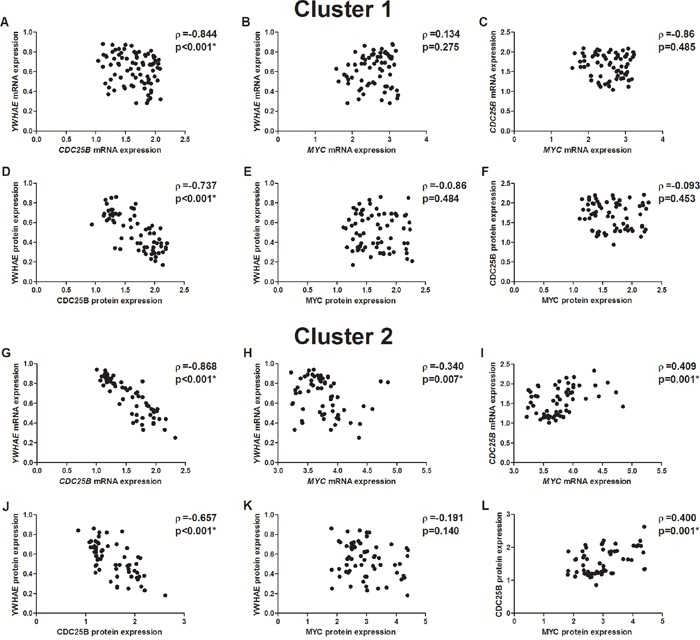
A. YWHAE and CDC25B mRNA expression in the cluster 1. B. YWHAE and MYC mRNA expression in the cluster 1. C. CDC25B and MYC mRNA expression in the cluster 1. D. YWHAE and CDC25B protein expression in the cluster 1. E. YWHAE and MYC protein expression in the cluster 1. F. CDC25B and MYC protein expression in the cluster 1. G. YWHAE and CDC25B mRNA expression in the cluster 2. H. YWHAE and MYC mRNA expression in the cluster 2. I. CDC25B and MYC mRNA expression in the cluster 2. J. YWHAE and CDC25B protein expression in the cluster 2. K. YWHAE and MYC protein expression in the cluster 2. L. CDC25B and MYC protein expression in the cluster 2. Correlation coefficients and p-values of Spearman correlation test are shown. *p<0.05, significant correlation by S Spearman correlation test.
Association between YWHAE, CDC25B, and MYC expression with clinicopathological features in GC
Clinicopathological variables and YWHAE, CDC25B, and MYC expression are shown in Table 1. YWHAE protein expression was reduced in tumors of male in relation to tumors of females (p=0.046; Table 1).
Table 1. Clinicopathological variables and gene expression in GC.
| YWHAE immunoreactivity | YWHAE protein | YWHAE mRNA | CDC25B immunoreactivity | CDC25B protein | CDC25B mRNA | MYC immunoreactivity | MYC protein | MYC mRNA | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | N | N (%) of positive cases | p valuea | Ratio T/N [median (IQR)] | p valueb | RQ [median(IQR)] | p valueb | N (%) of positive cases | P valuea | Ratio T/N [median(IQR)] | p valueb | RQ [median(IQR)] | P valueb | N (%) of positive cases | p valuea | Ratio T/N [median (IQR)] | p valueb | RQ [median (IQR)] | p valueb | |||||||||||||||||||
| Gender | ||||||||||||||||||||||||||||||||||||||
| Female | 45 | 2 (4.4) | 0.652 | 0.57 (0.26) | 0.046* | 0.64 (0.32) | 0.592 | 45 (100) | 0.651 | 1.71 (0.73) | 0.349 | 1.68 (0.61) | 0.376 | 41 (91.1) | 0.589 | 2.11 (1.55) | 0.687 | 3.22 (1.38) | 0.953 | |||||||||||||||||||
| Male | 84 | 4 (4.8) | 0.49 (0.33) | 0.69 (0.30) | 83 (98.8) | 1.61 (0.66) | 1.61 (0.60) | 76 (90.5) | 2.16 (1.18) | 3.11 (1.09) | ||||||||||||||||||||||||||||
| Onset | ||||||||||||||||||||||||||||||||||||||
| < 45 years | 34 | 3 (8.8) | 0.187 | 0.41 (0.27) | 0.006* | 0.49 (0.21) | <0.001* | 34 (100) | 0.736 | 1.96 (0.50) | 0.003* | 1.88 (0.40) | 0.005* | 2 (94.1) | 0.339 | 1.80 (0.92) | 0.013* | 3.04 (1.27) | 0.048* | |||||||||||||||||||
| ≥ 45 years | 95 | 3 (3.2) | 0.57 (0.29) | 0.71 (0.23) | 94 (99.2) | 1.45 (0.61) | 1.57 (0.52) | 85 (89.5) | 2.23 (1.27) | 3.28 (1.01) | ||||||||||||||||||||||||||||
| Tumor location | ||||||||||||||||||||||||||||||||||||||
| Cardia | 50 | 1 (2) | 0.247 | 0.57 (0.25) | 0.379 | 0.68 (0.22) | 0.489 | 50 (100) | 0.612 | 1.64 (0.64) | 0.625 | 1.61 (0.53) | 0.383 | 43 (86) | 0.126 | 1.87 (1.27) | 0.088 | 3.06 (1.32) | 0.197 | |||||||||||||||||||
| Non-cardia | 79 | 5 (6.3) | 0.49 (0.34) | 0.66 (0.35) | 78 (98.7) | 1.62 (0.72) | 1.68 (0.64) | 74 (93.7) | 2.20 (1.16) | 3.23 (0.93) | ||||||||||||||||||||||||||||
| Histological type | ||||||||||||||||||||||||||||||||||||||
| Diffuse | 62 | 3 (4.8) | 0.622 | 0.39 (0.25) | <0.001* | 0.58 (0.33) | 0.019* | 62 (100) | 0.519 | 1.84 (0.72) | 0.041* | 1.78 (0.62) | 0.027* | 52 (83.9) | 0.010* | 2.20 (1.13) | 0.786 | 3.21 (0.92) | 0.854 | |||||||||||||||||||
| Intestinal | 67 | 3 (4.5) | 0.63 (0.22) | 0.72 (0.26) | 66 (98.5) | 1.43 (0.57) | 1.52 (0.50) | 65 (97) | 2.11 (1.40) | 3.09 (1.22) | ||||||||||||||||||||||||||||
| Stage | ||||||||||||||||||||||||||||||||||||||
| Early | 12 | 0 (0) | 0.550 | 0.36 (0.28) | 0.027* | 0.48 (0.19) | 0.003* | 12 (100) | 0.907 | 1.83 (0.45) | 0.102 | 1.80 (0.33) | 0.046* | 10 (83.3) | 0.309 | 1.37 (0.60) | <0.001* | 2.25 (1.25) | 0.003* | |||||||||||||||||||
| Advanced | 117 | 6 (5.1) | 0.55 | 0.68 | 116 | 1.61 | 1.62 | 107 | 2.19 | 3.23 | (0.29) | (0.29) | (99.1) | (0.68) | (0.60) | (91.5) | (1.24) | (1.03) | ||||||||||||||||||||
| Tumor invasion | ||||||||||||||||||||||||||||||||||||||
| T1/T2 | 42 | 0 (0) | 0.089 | 0.42 (0.28) | 0.006* | 0.57 (0.23) | 0.001* | 42 (100) | 0.674 | 1.79 (0.42) | 0.004* | 1.76 (0.33) | 0.004* | 36 (85.7) | 0.151 | 1.63 (0.77) | <0.001* | 2.59 (1.12) | <0.001* | |||||||||||||||||||
| T3/T4 | 87 | 6 (6.9) | 0.58 (0.28) | 0.72 (0.29) | 86 (98.9) | 1.35 (0.67) | 1.48 (0.62) | 81 (93.1) | 2.38 (1.20) | 3.40 (0.95) | ||||||||||||||||||||||||||||
| Lymph node metastasis | ||||||||||||||||||||||||||||||||||||||
| Absent | 16 | 1 (6.3) | 0.556 | 0.52 (0.27) | 0.379 | 0.62 (0.27) | 0.438 | 15 (93.8) | 0.124 | 1.93 (0.65) | 0.092 | 1.86 (0.58) | 0.123 | 11 (68.8) | 0.007* | 1.42 (0.25) | <0.001* | 2.36 (0.48) | <0.001* | |||||||||||||||||||
| Present | 113 | 5 (4.4) | 0.55 (0.31) | 0.67 (0.32) | 113 (100) | 1.61 (0.65) | 1.62 (0.59) | 106 (93.8) | 2.23 (1.21) | 3.29 (0.97) | ||||||||||||||||||||||||||||
| Distant metastasis | ||||||||||||||||||||||||||||||||||||||
| Absent | 70 | 3 (4.3) | 0.576 | 0.57 (0.30) | 0.134 | 0.67 (0.27) | 0.970 | 69 (98.6) | 0.543 | 1.63 (0.69) | 0.620 | 1.63 (0.59) | 0.712 | 58 (82.9) | <0.001* | 1.67 (0.61) | <0.001* | 2.63 (0.76) | <0.001* | |||||||||||||||||||
| Present | 59 | 3 (5.1) | 0.49 (0.32) | 0.66 (0.36) | 59 (100) | 1.63 (0.68) | 1.67 (0.64) | 59 (100) | 2.93 (0.96) | 3.74 (0.44) | ||||||||||||||||||||||||||||
| H. pylori | ||||||||||||||||||||||||||||||||||||||
| Negative | 13 | 0 (0) | 0.522 | 0.39 (0.31) | 0.109 | 0.57 (0.35) | 0.315 | 13 (100) | 0.899 | 1.76 (0.82) | 0.134 | 1.72 (0.68) | 0.127 | 12 (92.3) | 0.654 | 2.17 (1.41) | 0.879 | 3.02 (1.27) | 0.656 | |||||||||||||||||||
| Positive | 116 | 6 (5.2) | 0.55 (0.31) | 0.67 (0.30) | 115 (99.1) | 1.62 (0.68) | 1.63 (0.60) | 105(90.5) | 2.14 (1.30) | 3.21 (1.16) | ||||||||||||||||||||||||||||
| CagA | ||||||||||||||||||||||||||||||||||||||
| Negative | 46 | 0 (0) | 0.066 | 0.54 (0.32) | 0.611 | 0.66 (0.30) | 0.508 | 45 (97.8) | 0.357 | 1.63 (0.71) | 0.526 | 1.63 (0.55) | 0.665 | 41 (89.1) | 0.435 | 2.08 (1.15) | 0.789 | 3.17 (1.05) | 0.904 | |||||||||||||||||||
| Positive | 83 | 6 (7.2) | 0.56 (0.31) | 0.70 (0.33) | 83 (100) | 1.64 (0.69) | 1.65 (0.66) | 76 (91.6) | 2.18 (1.33) | 3.17 (1.14) | ||||||||||||||||||||||||||||
| EBV | ||||||||||||||||||||||||||||||||||||||
| Negative | 108 | 4 (3.7) | 0.252 | 0.54 (0.30) | 0.742 | 0.67 (0.30) | 0.723 | 107 (99.1) | 0.837 | 1.64 (0.69) | 0.730 | 1.64 (0.57) | 0.990 | 96 (88.9) | 0.107 | 2.04 (1.14) | 0.024* | 3.08 (1.10) | 0.025* | |||||||||||||||||||
| Positive | 21 | 2 (9.5) | 0.58 (0.36) | 0.64 (0.34) | 21 (100) | 1.39 (0.68) | 1.60 (0.66) | 21 (100) | 2.57 (1.71) | 3.69 (1.19) |
p value by ÷2 test;
p value by Mann-Whitney test.
p<0.05, significantly difference between groups.
A tendency for different expression between groups.
N: number of samples; T/N: ratio of protein expression between neoplastic and matched non-neoplastic samples; RQ: relative quantification, in which the matched non-neoplastic sample was designated as a calibrator from each neoplastic samples; IQR: interquartile range; EBV: Epstein-Barr virus.
The cancer samples of patients with late-onset tumors presented higher YWHAE and MYC expression and lower CDC25B expression in relation to to early-onset GC samples (p<0.05 for all analyses; Table 1).
YWHAE protein and mRNA expression was reduced in diffuse-type GC (p<0.001 and 0.019, respectively; Table 1). Conversely, CDC25B protein and mRNA expression was increased in diffuse-type GC (p=0.041 and 0.027, respectively; Table 1). MYC immunoreactivity was more frequent in intestinal-type GC (p=0.010; Table 1).
Decreased YWHAE protein and mRNA expression was also associated with early-stage and T1/T2 tumors (p<0.05 for all comparisons; Table 1). In contrast, CDC25B protein and mRNA expression was higher in T1/T2 tumors in relation to T3/T4 tumors (p<0.05 for all analyses; Table 1). CDC25B protein expression was also increased in early-stage tumors in comparison to advanced-stage tumors (p=0.046; Table 1).
Increased MYC expression was detected in tumors with advanced stage, deeper invasion, lymph node and distant metastases and with EBV infection (p<0.05 for all analyses; Table 1). Moreover, MYC immunoreactivity was more frequent in tumors of patients with lymph node or distant metastases (p<0.05 for all analyses; Table 1).
YWHAE expression seems to increase and CDC25B expression seems to reduce between stages I and III; however, we did not detect a significant change among tumor stages after Bonferroni adjustment (p>0.008; Figure 13A–13D). A gradual increase of MYC expression was detected in GC samples through tumor stages I to IV (p<0.008 for all analyses; Mann-Whitney test followed by Bonferroni correction; Figure 13E–13F).
Figure 13. Protein and mRNA expression by tumor stage. A. YWHAE protein expression.
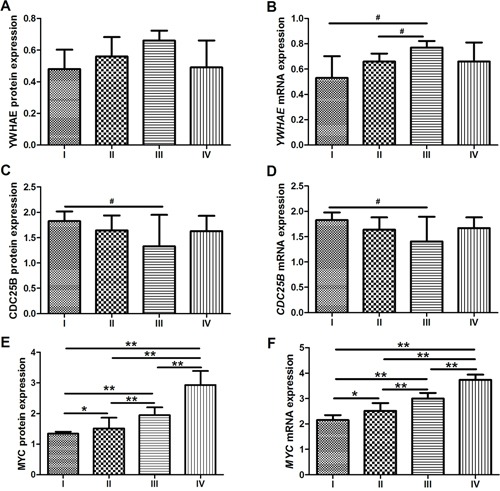
B. YWHAE mRNA expression. C. CDC25B protein expression. D. CDC25B mRNA expression. E. MYC protein expression. F. MYC mRNA expression. *p<0.008, significant difference between groups (Mann-Whitney test followed by Bonferroni corrections); **p<0.001, significant difference between groups (Mann-Whitney test followed by Bonferroni corrections); #p<0.05, difference between groups but not statistically significant after Bonferroni adjustment. Values of median and IQR are shown.
The inverse correlation between YWHAE and CDC25 mRNA and protein expression was independent of the tumor stage, tumor invasion or ocurrence of lymph node and distant metastases (p<0.05; for all comparisons). Interestingly, YWHAE and MYC expression was directly correlated (ρ=0.361; p=0.002) and CDC25B and MYC expression was inversely correlated (ρ=-0.271; p=0.023) in tumors of patients without distant metastasis. However, in tumors of patients with distant metastasis or in stage IV GC, MYC and CDC25B mRNA expression was positively correlated (ρ=0.269; p<0.039) and MYC and CDC25B protein expression also tended to be correlated (ρ=0.248; p=0.058).
Moreover, the “Cluster 2” described above was associated with advanced GC stage, deeper tumor invasion (T stage), the occurrence of lymph node metastasis and the occurrence of distant metastases (p<0.05 for all analyses; Supplementary Table S1).
DISCUSSION
Reduced YWHAE expression was described in different types of cancer, including lung [16], larynx [17], and brain [18] cancers. Our group also detected reduced expression of YWHAE in a smaller GC samples cohort [15]. Although these findings provide evidences to support classification of YWHAE as a tumor suppressor gene, the precise role of this gene in gastric carcinogenes was still unknown.
In vitro, we demonstrated that YWHAE acts as a tumor suppressor in GC inducing cell arrest and inhibiting cell invasion and migration through the down-regulation of MYC and CDC25B. On the other hand, MYC oncogene is able to induces GC cell proliferation, invasion and migration through the up-regulation of CDC25B and down-regulation of YWHAE. Our findings suggest that YWHAE and MYC may have a opposite role in GC cells. A previous study showed that the knockdown of another member of 14-3-3 family, the YWHAS, leads to increased expression of MYC protein [48]. Thus, other 14-3-3 family member can regulates MYC during gastric carcinogenesis.
In tissue samples, YWHAE immunoreactivity was detected in only approximately 5% of tumors and its 1.5-fold reduced protein and mRNA expression was observed in aproximately 70% and 50% of the studied tumors, respectively. Moreover, none of the tumors presented the YWHAE expression higher than the matched non-neoplastic gastric sample. This findings confirm that YWHAE may act as a tumor suppressor as observed in vitro and as described previously by our group in a smaller GC samples cohort [15]. On the contrary, CDCD25B immunoreactivity was observed in almost all tumors and its mRNA and protein expression was increased by at least 1.5-fold in more than 50% of the cases in relation to non-neoplastic samples. However, it is important to highlight that only two samples presented the ratio of CDC25B protein expression between tumor and corresponding non-neoplastic specimens below 1 in the studied cohort. This finding is in agreement with the oncogenic role of CDC25B in GC samples from our admixed population (mainly composed by European, Africans, and Amerindians [49]) as already described in GC samples from patients from East Asian [22–25].
Although direct silencing of CDC25B did not altered YWHAE expression, CDC25B and YWHAE expression were inversely correlated in MYC or YWHAE silenced cells, as well as in GC samples. YWHAE negatively regulates CDC25 [19, 20]. The hypothesis of CDC25B negative regulation by YWHAE in gastric carcinogenesis is also supported by our findings concerning the association of these genes or proteins expression and clinicopathological features. Reduced YWHAE and increased CDC25B expression was associated with early-onset GC, diffuse-type, T1/T2 stage, and early-stage tumors.
Our group previously described the association or tendency to association of reduced YWHAE expression with early-onset GC, diffuse-type, T1/T2 stage, and early-stage GC [15]. Thus, our findings reinforce that decreasing of YWHAE expression may be important for tumor initiation, especially in diffuse-type tumors an of early-onset. Early-onset GC presents distinct molecular and clinicopathological pattern in relation to late-onset tumors, which suggest that they are a two subsets of GC [50–52]. Furthermore, our results also supports that the intestinal and diffuse histological GC types follow different molecular pathways and may be two separate entities [53].
Although our results suggest that the highest level of CDC25B expression seems be important for GC initiation in our population, previous studies described its increased expression associated with advanced tumor stage, deeper invasion, and metastasis in East Asian population [22–24]. As described above, the expression of CDC25B was up-regulated in most of the studied GC samples, despite the invasion or metastasis. Further studies are still required to improve the knowledge about CDC25B function during GC progression; however, it is important to highlight that this is the largest cohort of GC and paired non-neoplastic gastric specimens in which CDC25B expression was evaluated.
Interestingly, even although not statistically significant, YWHAE expression was increased and CDC25B expression was reduced continuously between stages I to and III. However, in the stage IV, YWHAE expression returns to decrease and CDC25B expression returns to increase. In this stage, the highest MYC expression was detected. Here, we observed that only in GC samples with highest MYC expression, MYC expression was directly correlated with CDC25B expression and inversely correlated with YWHAE expression, such as observed in GC cell lines.
In GC samples from a Chinese population, a correlation between MYC and CDC25B immunoreactivity was described [25]; however, the effect of tumor stage in the immunoreactivity of these proteins was not accessed. MYC is able to regulate the transcription and hyperactivate cyclin/CDK complexes by the activation of CDC25 phosphatases and CDK kinases [54]. It has been proposed that the synthetic lethality based on the inhibition of CDK may be an interesting method for the treatment of tumors with MYC up-regulation [54]. However, we hypothesized that this approach is promising only in a subset of GC with highest MYC expression.
MYC is able to regulate several processes, including apoptosis, proliferation, cell growth, differentiation, angiogenesis, and cell metabolism [55]. Here, MYC silencing is able to reduce cancer cell proliferation, invasion and migraion in vitro in agreement with previous studies [56–59]. Although we observed that MYC induces cell proliferation, GC cells without MYC silencing presented more cells at G1 phase than MYC-silenced cells after 72 h of culture. This finding is probably due to the rapid accumulation in G1 phase just before confluence observed in controls cells, which therefore were not able to progress through the cell cycle even with active MYC.
All studied tumors presented increased MYC mRNA expression, and most of the tumors presented increased protein expression in relation to paired non-neoplastic specimens. As described by our group in a smaller cohort [34], MYC increased expression was associated with intestinal-type, deeper tumor extension and the presence of metastasis. Elevated expression of MYC was also associated with late-onset and advanced stage. In a previous study of our group, we reported that MYC amplification, which was associated with its immunoreactivity, was associated with these clinicopathological characteristics [34]. Moreover, MYC expression continuously increased during GC progression, with the highest expression detect in stage IV tumors. Thus, MYC deregulated expression is a common finding in GC, especially in intestinal-type and late-onset tumors, and has a role in poor prognosis.
In conclusion, decreasing YWHAE and increasing CDC25B expression seems to be important for tumor development, especially in diffuse-type tumors of early-onset. Conversely, increased MYC expression is a common finding in GC, especially in intestinal-type and late-onset tumors, and has a role in poor prognosis. In GC cell lines, YWHAE is able to regulate the GC cell proliferation, invasion and migration through the reduction of MYC and CDC25B expression. On the other hand, MYC also regulates the GC cell proliferation, invasion and migration through the induction of CDC25B and the reduction of YWHAE. In the tumor initiation, the opposite role of the possible tumor suppressor YWHAE and oncogene CDC25B in gastric carcinogenesis seems to be independent of MYC expression. However, the inversely correlation between YWHAE and MYC expression seems to be important for GC cells invasion and migration. The inverse correlation between these genes was only detected in a subset of GC, including GC samples at stage IV. Thus, the interaction between YWHAE and MYC and the activation of the pathways related to this interaction may be restricted to a subset of GC and may play a role in the metastasis process.
MATERIALS AND METHODS
Cell lines and culture
Three GC cell lines previously established and characterized by our group were used: AGP01, ACP02, and ACP03 [43]. The three cell lines present chromosome 8 trisomy and MYC amplification [43, 46].
A cell culture of non-neoplastic gastric mucosa cells (Normal Gastric Mucosa Cell Line 01, MNP01) pooled from 10 patients without gastric cancer was also used to initially evaluate the gene and protein expression of YWHAE, CDC25B, and MYC.
Cell were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco/Invitrogen, Germany) supplemented with 10% fetal bovine serum (Gibco/Invitrogen, Germany), 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B. All cultures were maintained in a 5% CO2 air-humidified atmosphere at 37°C.
Gene expression depletion by small interfering RNA (siRNA) transfection
An amount of 3×105 cells were seeded into 6 cm2 plates for each cell line before transfection. Cells were cultured for 24 h until cell density was approximately 50%. For YWHAE and CDC25B silencing, the cells were transfected into AGP01, ACP02, and ACP03 cell lines using either Silencer Select siRNA specific for YWHAE (s16; #4390824; Ambion, USA) and CDC25B (s2753; #4390824; Ambion, USA), respectively, or the Silencer select negative control #1 (#4390843; Ambion, USA). For MYC silencing, a pool of four different double-stranded siRNAs targeting MYC (20 μM; SMARTpool ON-TARGETplus MYC siRNA, L-003282-02-0020; GE Healthcare Dharmacon, USA) or scrambled control siRNAs (ON-TARGETplus Nontargeting Pool, D-001810-10-05; GE Healthcare Dharmacon, USA) were transfected into AGP01, ACP02, and ACP03 cell lines using Lipofectamine RNAiMAX Transfection Reagent (Life Technologies, USA).
All siRNA experiments were performed three times.
Cell proliferation by direct counting
After 24, 48, and 72 h of siRNA transfection, AGP01, ACP02, and ACP03 cells were harvested and directly counted in Neubauer chambers. The total number of cells was estimated and used to determine cell proliferation. Each sample was repeated three times and all experiments were carried in triplicates.
Cell cycle analysis by flow cytometry
For this analysis, siRNA transfection as carried for 72 h. Cells were treated with 10 μM BrdU for 60 min and then trypsinized and fixed in 80% ethanol at -20 °C overnight. The cell pellet was then treated with 2 M HCl/0.5% Triton X-100 for 30 min at room temperature, neutralized with 0.1 M Na2B4O7, and stained with FITC-anti BrdU antibodies. After centrifugation at 1000 rpm for 5 to 7 min, 400 μL propidium iodide (PI)-RNase solution (final concentrations: 38 mM Na3C6H5O7+69 μM PI+1 μL of 10 mg/mL RNase A) was added to the pellet and mixed well. Samples were incubated at room temperature in the dark for 30 min at 37 °C before analysis by BD FACSCanto™ II (BD Biosciences, USA) flow cytometer. Each sample was repeated three times. The forward light scatter (FSC) of nonfixed cells was used as a relative measure of cell size.
Invasion and migration analysis
For these analyses, siRNA transfection as carried for 24 h. Invasion assay was done in a 24-well transwell chamber. Cells were added to coated filters in 100 μL of serum-free medium. In the lower compartments of the chambers, 600 μL of human fibroblast serum-free-conditioned media was used as chemo attractant. After 18 h at 37 °C in a 5% CO2 incubator, the Matrigel coating on the upper surface of the filter was wiped off using a cotton swab. Cells that migrated through the filters were fixed, stained with crystal violet, photographed, and counted.
For the migration analysis, cells were loaded on transwell polycarbonate membrane inserts. The plates were incubated for 18 h at 37 °C in a 5% CO2 incubator, the cells in the lower wells were fixed, stained with crystal violet, and counted. The cells that had migrated to the lower compartment of the chambers were trypsinized and counted.
Each experiment was carried out in triplicate.
Patients and tissue specimens
We enrolled 129 patients with GC who underwent surgical resection with curative intent from Northern Brazil. All patients had negative histories of exposure to either chemotherapy or radiotherapy before surgery. Patients with co-occurrence of other diagnosed cancers were excluded from this study. Signed informed consent was obtained from all patients before sample collection. The study protocol was approved by the Ethics Committee of the Hospital Universitário João de Barros Barreto (Protocol #316737).
Part of each dissected tumor sample was formalin fixed and paraffin embedded (FFPE). Sections of the FFPE tissue were stained with hematoxylin and eosin for histological evaluation or used for immunohistochemical (IHC) analysis. Additional portions of each tumor and paired non-neoplastic tissue specimens were snap frozen in liquid nitrogen and stored at -80 °C until protein and nucleic acid purification.
All samples were classified according to Laurén [60] and the tumors were staged according to the tumor-node-metastasis (TNM) staging criteria [61]. The presence of Helicobacter pylori in gastric samples was detected by the rapid urease test, and its virulence factor cytotoxicity-associated gene A (CagA gene) was detect by polymerase chain reaction (PCR) using DNA purified simultaneously with proteins and mRNA, as previously performed by our group [62]. Epstein-Barr virus (EBV) was detected by RNA in situ hybridization [62].
DNA/RNA/protein purification
Total RNA and proteins were extracted with TRIzol reagent from GC cell lines after 48 h of transfection. Total protein, mRNA, and DNA were simultaneously isolated from gastric tissue samples using the AllPrep DNA/RNA/Protein Kit (Qiagen, Germany) according to the manufacturer's instructions. The protein pellet was dissolved in a buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 50 mM DTT, 1% Protease Inhibitor Cocktail (Sigma-Aldrich, USA), and 0.5% each of Phosphatase Inhibitor Cocktail 1 and 2 (Sigma-Aldrich, USA), as previously performed by our group [15]. The protein concentrations were determined by the method of Bradford (Sigma-Aldrich, USA). The RNA concentration and quality were determined using a NanoDrop spectrophotometer (Kisker, Germany) and 1% agarose gels, respectively. Samples were stored at -80 °C until use.
mRNA expression
RNA was reverse transcribed using the Reverse Transcription System according to the manufacturer's protocol (A3500; Promega, USA). Complementary DNA was then amplified by real-time reverse transcription quantitative PCR (RT-qPCR) using TaqMan probes purchased as Assays-on-Demand Products for Gene Expression (Life Technologies, USA) and a 7500 Fast Real-Time PCR instrument (Life Technologies, USA). The ACTB gene was selected as an internal control [63]. All RT-qPCRs were performed in triplicate for both the target genes (YWHAE: Hs00356749_g1; CDC25B: Hs00244740_m1; MYC: Hs00153408_m1) and the internal control (ACTB: 4333762F).
The relative quantification (RQ) of gene expression was calculated according to Livak and Schmittgen [64]. In tissue sample analyses, the corresponding control sample was designated as a calibrator from each tumor. In the cell line analysis, the siRNA control-transfected cells were used as a calibrator. The gene expression in the MNP01 was also designated as a calibrator from all GC cell lines.
Western blotting
Western blot analysis was performed as described previously [15]. Reduced protein (25 μg) from each sample was separated by 12.5% homogeneous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto a polyvinylidene fluoride (PVDF) membrane (Hybond-P; GE Healthcare, USA). The PVDF membrane was blocked with phosphate-buffered saline containing 0.1% Tween 20 and 5% low fat milk and incubated overnight at 4 °C with the corresponding primary antibodies: anti-YWHAE (dilution 1:1000; PA5-29773; Life Technologies, USA), anti-CDC25B (dilution 1:1000; PA5-14100; Life Technologies, USA), anti-MYC (dilution 1:50; MA5-12080; Life Technologies, USA), and anti-ACTB (dilution 1:250; Ac-15; Life Technologies, USA). After extensive washing, a peroxidase-conjugated secondary antibody was added for 1 h at room temperature. Immunoreactive bands were visualized using the Western blotting Luminol reagent, and the images were acquired using an ImageQuant 350 digital image system (GE Healthcare, Sweden). ACTB was used as a loading reference control.
Immunohistochemical staining and scoring
Tumor tissue sections (3 or 4 mm thick) were deparaffinized in xylene and rehydrated in a graded series of ethanol. After heat-induced epitope retrieval, the tissue sections were incubated with primary mouse monoclonal antibodies against YWHAE (dilution 1:100; PA5-29773; Life Technologies, USA), CDC25B (dilution 1:10; PA5-14100; Life Technologies, USA) or MYC (dilution 1:100; MA5-12080; Life Technologies, USA). A universal peroxidase-conjugated secondary antibody kit (LSAB System; DakoCytomation, USA) was used for detection. We used 3,3’-diaminobenzidine/H2O2 (DakoCytomation, Denmark) as the chromogen and hematoxylin as the counterstain. A protein immunoreactivity-positive sample was defined as one having 10% or more neoplastic cells that were positive for the protein.
Statistical analysis
The data are shown as the frequency, median, and IQR. The Shapiro-Wilk test was used to evaluate the distribution of the age, mRNA, and protein expression data and to determine the appropriate subsequent test for statistical comparisons. The Mann-Whitney test was used to investigate the possible associations between gene mRNA or protein expression and categorical variables, such as immunoreactivity and clinicopathological features. An association between categorical variables was analyzed using the χ2 test. The K-means clustering method was used to group samples based on their gene expression similarities. A Spearman correlation test was used to evaluate the possible correlation between mRNA and protein expression. p≤0.05 was considered significant. The Bonferroni adjustment of the p value was applied when multiple comparisons were performed, with the α level being divided by the number of comparisons.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest
FUNDING
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; grants #471072/2012-5 and 402283/2013-9; fellowship to MCS and RRB), Fundação de Amparo à Pesquisa do Pará (FAPESPA; grant #ICAAF 123/2014 to RRB) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; grant #2009/07145-9; fellowship to MFL) as grants and fellowship awards.
REFERENCES
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Kimura Y, Noguchi T, Kawahara K, Kashima K, Daa T, Yokoyama S. Genetic alterations in 102 primary gastric cancers by comparative genomic hybridization: gain of 20q and loss of 18q are associated with tumor progression. Mod Pathol. 2004;17:1328–37. doi: 10.1038/modpathol.3800180. [DOI] [PubMed] [Google Scholar]
- 3.Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–72. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 5.van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays. 2001;23:936–46. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- 6.Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satoh J, Nanri Y, Yamamura T. Rapid identification of 14-3-3-binding proteins by protein microarray analysis. J Neurosci Methods. 2006;152:278–88. doi: 10.1016/j.jneumeth.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan S, Sade RS, Townsley FM, Fersht AR. Mechanistic differences in the transcriptional activation of p53 by 14-3-3 isoforms. Nucleic Acids Res. 2010;38:893–906. doi: 10.1093/nar/gkp1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzivion G, Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–4. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 10.Yaffe MB. How do 14-3-3 proteins work? — Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–7. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 11.Leal MF, Chung J, Calcagno DQ, Assumpcao PP, Demachki S, da Silva ID, Chammas R, Burbano RR, de Arruda Cardoso Smith M. Differential proteomic analysis of noncardia gastric cancer from individuals of northern Brazil. PLoS One. 2012;7:e42255. doi: 10.1371/journal.pone.0042255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang JS, Cho HY, Lee YJ, Ha WS, Kim HW. The differential proteome profile of stomach cancer: identification of the biomarker candidates. Oncol Res. 2004;14:491–9. doi: 10.3727/0965040042380441. [DOI] [PubMed] [Google Scholar]
- 13.Chen YR, Juan HF, Huang HC, Huang HH, Lee YJ, Liao MY, Tseng CW, Lin LL, Chen JY, Wang MJ, Chen JH, Chen YJ. Quantitative proteomic and genomic profiling reveals metastasis-related protein expression patterns in gastric cancer cells. J Proteome Res. 2006;5:2727–42. doi: 10.1021/pr060212g. [DOI] [PubMed] [Google Scholar]
- 14.Tseng CW, Yang JC, Chen CN, Huang HC, Chuang KN, Lin CC, Lai HS, Lee PH, Chang KJ, Juan HF. Identification of 14-3-3beta in human gastric cancer cells and its potency as a diagnostic and prognostic biomarker. Proteomics. 2011;11:2423–39. doi: 10.1002/pmic.201000449. [DOI] [PubMed] [Google Scholar]
- 15.Leal MF, Calcagno DQ, Demachki S, Assumpcao PP, Chammas R, Burbano RR, Smith Mde A. Clinical implication of 14-3-3 epsilon expression in gastric cancer. World J Gastroenterol. 2012;18:1531–7. doi: 10.3748/wjg.v18.i13.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konishi H, Nakagawa T, Harano T, Mizuno K, Saito H, Masuda A, Matsuda H, Osada H, Takahashi T. Identification of frequent G(2) checkpoint impairment and a homozygous deletion of 14-3-3epsilon at 17p13. 3 in small cell lung cancers. Cancer Res. 2002;62:271–6. [PubMed] [Google Scholar]
- 17.Che XH, Chen H, Xu ZM, Shang C, Sun KL, Fu WN. 14-3-3epsilon contributes to tumour suppression in laryngeal carcinoma by affecting apoptosis and invasion. BMC Cancer. 2010;10:306. doi: 10.1186/1471-2407-10-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cvekl A, Jr, Zavadil J, Birshtein BK, Grotzer MA, Cvekl A. Analysis of transcripts from 17p13. 3 in medulloblastoma suggests ROX/MNT as a potential tumour suppressor gene. Eur J Cancer. 2004;40:2525–32. doi: 10.1016/j.ejca.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Kumagai A, Dunphy WG. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–72. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumagai A, Yakowec PS, Dunphy WG. 14-3-3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol Biol Cell. 1998;9:345–54. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galaktionov K, Jessus C, Beach D. Raf1 interaction with Cdc25 phosphatase ties mitogenic signal transduction to cell cycle activation. Genes Dev. 1995;9:1046–58. doi: 10.1101/gad.9.9.1046. [DOI] [PubMed] [Google Scholar]
- 22.Kudo Y, Yasui W, Ue T, Yamamoto S, Yokozaki H, Nikai H, Tahara E. Overexpression of cyclin-dependent kinase-activating CDC25B phosphatase in human gastric carcinomas. Jpn J Cancer Res. 1997;88:947–52. doi: 10.1111/j.1349-7006.1997.tb00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang XM, Yao GY, Zhang BY, Wang LL, Zhao M. [Study on the expression and significance of Galectin-3 and CDC25B mRNA in human gastric carcinoma] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26:288–92. doi: 10.3760/cma.j.issn.1003-9406.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi H, Murai Y, Tsuneyama K, Nomoto K, Okada E, Fujita H, Takano Y. High labeling indices of cdc25B is linked to progression of gastric cancers and associated with a poor prognosis. Appl Immunohistochem Mol Morphol. 2007;15:267–72. doi: 10.1097/01.pai.0000213120.58472.57. [DOI] [PubMed] [Google Scholar]
- 25.Xing X, Chen J, Chen M. Expression of CDC25 phosphatases in human gastric cancer. Dig Dis Sci. 2008;53:949–53. doi: 10.1007/s10620-007-9964-4. [DOI] [PubMed] [Google Scholar]
- 26.Uchida S, Kuma A, Ohtsubo M, Shimura M, Hirata M, Nakagama H, Matsunaga T, Ishizaka Y, Yamashita K. Binding of 14-3-3beta but not 14-3-3sigma controls the cytoplasmic localization of CDC25B: binding site preferences of 14-3-3 subtypes and the subcellular localization of CDC25B. J Cell Sci. 2004;117:3011–20. doi: 10.1242/jcs.01086. [DOI] [PubMed] [Google Scholar]
- 27.Davezac N, Baldin V, Gabrielli B, Forrest A, Theis-Febvre N, Yashida M, Ducommun B. Regulation of CDC25B phosphatases subcellular localization. Oncogene. 2000;19:2179–85. doi: 10.1038/sj.onc.1203545. [DOI] [PubMed] [Google Scholar]
- 28.Forrest A, Gabrielli B. Cdc25B activity is regulated by 14-3-3. Oncogene. 2001;20:4393–401. doi: 10.1038/sj.onc.1204574. [DOI] [PubMed] [Google Scholar]
- 29.Mils V, Baldin V, Goubin F, Pinta I, Papin C, Waye M, Eychene A, Ducommun B. Specific interaction between 14-3-3 isoforms and the human CDC25B phosphatase. Oncogene. 2000;19:1257–65. doi: 10.1038/sj.onc.1203419. [DOI] [PubMed] [Google Scholar]
- 30.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–7. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 31.Calcagno DQ, Leal MF, Assumpcao PP, Smith MA, Burbano RR. MYC and gastric adenocarcinoma carcinogenesis. World J Gastroenterol. 2008;14:5962–8. doi: 10.3748/wjg.14.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calcagno DQ, Leal MF, Seabra AD, Khayat AS, Chen ES, Demachki S, Assumpcao PP, Faria MH, Rabenhorst SH, Ferreira MV, de Arruda Cardoso Smith M, Burbano RR. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J Gastroenterol. 2006;12:6207–11. doi: 10.3748/wjg.v12.i38.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa Raiol LC, Figueira Silva EC, Mendes da Fonseca D, Leal MF, Guimaraes AC, Calcagno DQ, Khayat AS, Assumpcao PP, de Arruda Cardoso Smith M, Burbano RR. Interrelationship between MYC gene numerical aberrations and protein expression in individuals from northern Brazil with early gastric adenocarcinoma. Cancer Genet Cytogenet. 2008;181:31–5. doi: 10.1016/j.cancergencyto.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 34.de Souza CR, Leal MF, Calcagno DQ, Costa Sozinho EK, Borges Bdo N, Montenegro RC, Dos Santos AK, Dos Santos SE, Ribeiro HF, Assumpcao PP, de Arruda Cardoso Smith M, Burbano RR. MYC Deregulation in Gastric Cancer and Its Clinicopathological Implications. PLoS One. 2013;8:e64420. doi: 10.1371/journal.pone.0064420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calcagno DQ, Freitas VM, Leal MF, de Souza CR, Demachki S, Montenegro R, Assumpcao PP, Khayat AS, Smith Mde A, dos Santos AK, Burbano RR. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer. BMC Gastroenterol. 2013;13:141. doi: 10.1186/1471-230X-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calcagno DQ, Leal MF, Demachki S, Araujo MT, Freitas FW, Oliveira e Souza D, Assumpcao PP, Ishak G, de Arruda Cardoso Smith M, Burbano RR. MYC in gastric carcinoma and intestinal metaplasia of young adults. Cancer Genet Cytogenet. 2010;202:63–6. doi: 10.1016/j.cancergencyto.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Silva TC, Leal MF, Calcagno DQ, de Souza CR, Khayat AS, dos Santos NP, Montenegro RC, Rabenhorst SH, Nascimento MQ, Assumpcao PP, de Arruda Cardoso Smith M, Burbano RR. hTERT, MYC and TP53 deregulation in gastric preneoplastic lesions. BMC Gastroenterol. 2012;12:85. doi: 10.1186/1471-230X-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.da Costa Jde F, Leal MF, Silva TC, Andrade Junior EF, Rezende AP, Muniz JA, Lacreta Junior AC, Assumpcao PP, Calcagno DQ, Demachki S, Rabenhorst SH, Smith Mde A, Burbano RR. Experimental gastric carcinogenesis in Cebus apella nonhuman primates. PLoS One. 2011;6:e21988. doi: 10.1371/journal.pone.0021988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calcagno DQ, Leal MF, Taken SS, Assumpcao PP, Demachki S, Smith Mde A, Burbano RR. Aneuploidy of chromosome 8 and C-MYC amplification in individuals from northern Brazil with gastric adenocarcinoma. Anticancer Res. 2005;25:4069–74. [PubMed] [Google Scholar]
- 40.Assumpcao PP, Ishak G, Chen ES, Takeno SS, Leal MF, Guimaraes AC, Calcagno DQ, Khayat AS, Demachki S, Smith Mde A, Burbano RR. Numerical aberrations of chromosome 8 detected by conventional cytogenetics and fluorescence in situ hybridization in individuals from northern Brazil with gastric adenocarcinoma. Cancer Genet Cytogenet. 2006;169:45–9. doi: 10.1016/j.cancergencyto.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 41.Costa Guimaraes A, Goncalves Quintana L, Ferreira Leal M, Satomi Takeno S, Pimentel Assumpcao P, Moura Lima E, Salim Khayat A, Suchi Chen E, de Arruda Cardoso Smith M, Rodriguez Burbano R. Aneuploidy of chromosome 8 detected by fluorescence in situ hybridisation in ACP01 cell line gastric adenocarcinoma. Clin Exp Med. 2006;6:129–33. doi: 10.1007/s10238-006-0108-5. [DOI] [PubMed] [Google Scholar]
- 42.Lima EM, Rissino JD, Harada ML, Assumpcao PP, Demachki S, Guimaraes AC, Casartelli C, Smith MA, Burbano RR. Conventional cytogenetic characterization of a new cell line, ACP01, established from a primary human gastric tumor. Braz J Med Biol Res. 2004;37:1831–8. doi: 10.1590/s0100-879x2004001200008. [DOI] [PubMed] [Google Scholar]
- 43.Leal MF, Martins do Nascimento JL, da Silva CE, Vita Lamarao MF, Calcagno DQ, Khayat AS, Assumpcao PP, Cabral IR, de Arruda Cardoso Smith M, Burbano RR. Establishment and conventional cytogenetic characterization of three gastric cancer cell lines. Cancer Genet Cytogenet. 2009;195:85–91. doi: 10.1016/j.cancergencyto.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 44.Burbano RR, Assumpcao PP, Leal MF, Calcagno DQ, Guimaraes AC, Khayat AS, Takeno SS, Chen ES, De Arruda Cardoso Smith M. C-MYC locus amplification as metastasis predictor in intestinal-type gastric adenocarcinomas: CGH study in Brazil. Anticancer Res. 2006;26:2909–14. [PubMed] [Google Scholar]
- 45.Ribeiro HF, Alcantara DF, Matos LA, Sousa JM, Leal MF, Smith MA, Burbano RR, Bahia MO. Cytogenetic characterization and evaluation of c-MYC gene amplification in PG100, a new Brazilian gastric cancer cell line. Braz J Med Biol Res. 2010;43:717–21. doi: 10.1590/s0100-879x2010007500068. [DOI] [PubMed] [Google Scholar]
- 46.Leal MF, Calcagno DQ, Costa JFFB, Silva TCR, Khayat AS, Chen ES, Assumpção PP, Smith MAC, Burbano RR. MYC, TP53, and chromosome 17 copy-number alterations in multiple gastric cancer cell lines and in their parental primary tumors. J Biomed Biotechnol. 2011. p. 2011. [DOI] [PMC free article] [PubMed]
- 47.Calcagno DQ, Guimaraes AC, Leal MF, Seabra AD, Khayat AS, Pontes TB, Assumpcao PP, De Arruda Cardoso Smith M, Burbano RR. MYC insertions in diffuse-type gastric adenocarcinoma. Anticancer Res. 2009;29:2479–83. [PubMed] [Google Scholar]
- 48.Phan L, Chou PC, Velazquez-Torres G, Samudio I, Parreno K, Huang Y, Tseng C, Vu T, Gully C, Su CH, Wang E, Chen J, Choi HH, et al. The cell cycle regulator 14-3-3sigma opposes and reverses cancer metabolic reprogramming. Nat Commun. 2015;6:7530. doi: 10.1038/ncomms8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Batista dos Santos SE, Rodrigues JD, Ribeiro-dos-Santos AK, Zago MA. Differential contribution of indigenous men and women to the formation of an urban population in the Amazon region as revealed by mtDNA and Y-DNA. Am J Phys Anthropol. 1999;109:175–80. doi: 10.1002/(SICI)1096-8644(199906)109:2<175::AID-AJPA3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 50.Carvalho R, Milne AN, van Rees BP, Caspers E, Cirnes L, Figueiredo C, Offerhaus GJ, Weterman MA. Early-onset gastric carcinomas display molecular characteristics distinct from gastric carcinomas occurring at a later age. J Pathol. 2004;204:75–83. doi: 10.1002/path.1602. [DOI] [PubMed] [Google Scholar]
- 51.Milne AN, Carvalho R, Morsink FM, Musler AR, de Leng WW, Ristimaki A, Offerhaus GJ. Early-onset gastric cancers have a different molecular expression profile than conventional gastric cancers. Mod Pathol. 2006;19:564–72. doi: 10.1038/modpathol.3800563. [DOI] [PubMed] [Google Scholar]
- 52.Milne AN, Sitarz R, Carvalho R, Carneiro F, Offerhaus GJ. Early onset gastric cancer: on the road to unraveling gastric carcinogenesis. Curr Mol Med. 2007;7:15–28. doi: 10.2174/156652407779940503. [DOI] [PubMed] [Google Scholar]
- 53.Tahara E. Genetic pathways of two types of gastric cancer. IARC Sci Publ. 2004. pp. 327–49. [PubMed]
- 54.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–16. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 55.Pelengaris S, Khan M. The many faces of c-MYC. Arch Biochem Biophys. 2003;416:129–36. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 56.Khanna A, Bockelman C, Hemmes A, Junttila MR, Wiksten JP, Lundin M, Junnila S, Murphy DJ, Evan GI, Haglund C, Westermarck J, Ristimaki A. MYC-dependent regulation and prognostic role of CIP2A in gastric cancer. J Natl Cancer Inst. 2009;101:793–805. doi: 10.1093/jnci/djp103. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Q, Yang Y, Yu J, You QD, Zeng S, Gu HY, Lu N, Qi Q, Liu W, Wang XT, Guo QL. Posttranscriptional regulation of the telomerase hTERT by gambogic acid in human gastric carcinoma 823 cells. Cancer Lett. 2008;262:223–31. doi: 10.1016/j.canlet.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Hou Y, Ashktorab H, Gao L, Xu Y, Wu K, Zhai J. The impact of C-MYC gene expression on gastric cancer cell. Mol Cell Biochem. 2010;344:125–35. doi: 10.1007/s11010-010-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Jian W, Gao W, Zheng YX, Wang YK, Zhou ZQ, Zhang H, Wang CJ. RNAi silencing of c-Myc inhibits cell migration, invasion, and proliferation in HepG2 human hepatocellular carcinoma cell line: c-Myc silencing in hepatocellular carcinoma cell. Cancer Cell Int. 2013;13:23. doi: 10.1186/1475-2867-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauren P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. an Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 61.Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17:3077–9. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 62.de Souza CR, de Oliveira KS, Ferraz JJ, Leal MF, Calcagno DQ, Seabra AD, Khayat AS, Montenegro RC, Alves AP, Assumpcao PP, Smith MC, Burbano RR. Occurrence of Helicobacter pylori and Epstein-Barr virus infection in endoscopic and gastric cancer patients from Northern Brazil. BMC Gastroenterol. 2014;14:179. doi: 10.1186/1471-230X-14-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wisnieski F, Calcagno DQ, Leal MF, dos Santos LC, Gigek Cde O, Chen ES, Pontes TB, Assumpcao PP, de Assumpcao MB, Demachki S, Burbano RR, Smith Mde A. Reference genes for quantitative RT-PCR data in gastric tissues and cell lines. World J Gastroenterol. 2013;19:7121–8. doi: 10.3748/wjg.v19.i41.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


