Abstract
Mitochondrial gene expression in yeast is believed to be regulated predominantly at the post-transcriptional level. However, the contribution of mitochondrial transcription and RNA-turnover rates to differential gene regulation is still largely unknown. Mitochondrial run-on transcription and hybrid selection assays showed that some of the multigenic transcription units of the mitochondrial genome are transcribed evenly, whereas others are transcribed asymmetrically, with higher transcription rates for promoter-proximal genes, than for promoter-distal genes. The tRNAE-cytochrome b (COB) operon was analyzed in detail to investigate the mechanisms underlying transcription rate asymmetries in yeast mitochondria. We showed that a drop in transcription rates occurs in a particular region between the coding sequences and is independent of the coding sequence of the downstream COB gene. Deletion of the region between tRNAE and COB coding sequences decreases the drop in transcription rates. Deletion of the nuclear gene encoding the Pet 127 protein, which is involved in mitochondrial RNA 5′ processing and degradation, also partially relieves transcriptional asymmetry. Therefore, asymmetry is probably due to a combination of attenuated transcription at specific sites between the coding sequences and very rapid RNA degradation.
INTRODUCTION
In the yeast Saccharomyces cerevisiae, a single circular mitochondrial chromosome contains the genes for the cytochrome oxidase subunits I, II and III (COX1, COX2 and COX3), ATPase subunits 6, 8 and 9 (ATP6, ATP8 and ATP9), apocytochrome b (COB), a ribosomal protein (VAR1) and several intron-related open reading frames (1). In addition, the 21S and 15S ribosomal RNAs, 24 transfer RNAs and the 9S RNA component of RNaseP are encoded by the mitochondrial DNA. Mitochondrial DNA is transcribed by a single RNA polymerase consisting of a catalytic subunit, Rpo41, and a specificity-conferring factor, Mtf1, which exhibits homology to bacterial sigma factors (2,3). Similar to other proteins involved in mitochondrial gene expression, both polymerase subunits are encoded by nuclear genes and are imported from the cytosol.
Mitochondrial gene expression is regulated primarily at the post-transcriptional level. Several proteins are involved in an mRNA-specific manner in the processing of polycistronic precursors, the stability of mature transcripts and the activation of translation (1). Examples of these processes are particularly well-studied for the tRNAE-COB operon. Following endonucleolytic cleavages of the primary transcript that release the tRNA, the 5′ end of the COB precursor mRNA is further shortened to yield the stable, mature, monocistronic transcript. The product of the nuclear gene CBP1 specifically stabilizes the mature COB mRNA (4,5). Another protein that affects the expression of the COB gene is the nuclear-encoded Pet127 protein. In pet127 strains, 5′-unprocessed precursor RNAs accumulate to levels equivalent to those of processed transcripts in wild-type strains, indicating that 5′ end processing is blocked (6). In contrast to Cbp1, Pet127 is not a COB-specific factor. A pet127 mutation also prevents the formation of mature ATP8/6, VAR1 and 15S rRNA transcripts from longer 5′ precursor forms (6,7).
The complete sequence analysis of the S.cerevisiae mitochondrial DNA identified 19 sites of transcriptional initiation (8), which implies that several genes are organized in polycistronic transcription units. One example is the COB gene, which is transcribed together with the upstream gene for tRNAE (9). A consensus sequence consisting of 8 bp is present immediately upstream of each transcription initiation site (10). Transcription has been studied by a variety of means, employing purified organellar RNA polymerase (11–13), organellar lysates (14), purified intact organelles (15–18) and pulse labeling of whole yeast cells with 32PO4 (19). Rates of transcriptional initiation reportedly vary up to 20-fold between different transcription units (19). Furthermore, Mueller and Getz (19) observed differences in transcription rates between genes of the same polygenic transcription unit. These differences were interpreted as evidence that transcription attenuation regulates mitochondrial gene expression.
In the present study, run-on transcription assays with isolated mitochondria were employed to study transcriptional profiles of the mitochondrial genome, in particular, the tRNAE-COB operon. The advantage of this in organello system over in vitro systems is that labeled run-on transcripts represent RNAs that were in the process of being transcribed at the time of organelle isolation. We found that some transcription units were evenly transcribed, whereas other operons showed a strong reduction in transcription rates of downstream genes. Using the tRNAE-COB transcription unit as a model operon, we observed a sharp drop in hybridization intensities within a 300 bp region between the two genes. When a 910 bp region between the two coding sequences was deleted, the hybridization with probes of the downstream gene was increased, suggesting that transcription is attenuated in this interval. Also, in a mutant strain lacking the Pet127 protein, the drop in transcriptional activity was reduced significantly. This result suggests an involvement of Pet127 in rapid degradation of COB RNA during the time that transcription rates are measured.
MATERIALS AND METHODS
Strains and media
The strains used in this study are listed in Table 1. Mutant mitochondria were introduced into the strain S150-Bio1/ρo by cytoduction (20). The matings were done as described by Dieckmann and Mittelmeier (21), except that the final cytoductants were selected on the media containing 5-fluoroorotic acid (FOA) (22). Other media used for this study were YPD (2% glucose, 2% peptone and 1% yeast extract), YEPG (3% glycerol, 2% peptone and 1% yeast extract) and WO (2% glucose, 0.67% yeast nitrogen base without amino acids). Amino acid supplements were added to WO at a final concentration of 20 μg/ml. Solid media contained 2% agar. For run-on transcription analysis, the strains were grown in 1 liter of YPD.
Table 1. Names and genotypes of yeast strains used in this study.
| Strain name | Genotype or description | Reference |
|---|---|---|
| S150-2B | a, [ρ+], ura3-52, his3, leu2-3,2-112 | (35) |
| S150-Bio1 | a, [ρ+], ura3-52, leu2-3,2-112 | (26) |
| 2/ΔCO | α, [ρΔCO mit−], met6 | (28) |
| TG955 | α, [ρTG955] in LL20 | (5) |
| S150TG955 | α, [ρTG955] in S150-Bio1 | This study |
| JC3M9410 | a, [ρ M9410 mit−] in JC3 | (4) |
| S150M9410 | a, [ρ M9410 mit−] in S150-Bio1 | This study |
| Δ pet127 | a, pet127::LEU2 in S150-2B | This study |
Isolation of intact mitochondria
Mitochondria were isolated following standard procedures (23). In brief, 1 liter of YPD was inoculated with 10 ml of an overnight culture and grown for 16 h at 30°C in a rotary shaker. Cells were harvested by centrifugation, washed with 1.2 M sorbitol and digested with 30 ml of buffer containing 30 mg of Zymolyase per 10 g cell wet weight. The digestion was carried out at 34°C until at least 90% of a sample of the cells lysed on dilution in water. Cells were washed twice with 1.2 M sorbitol, resuspended in wash buffer [0.5 M sorbitol, 0.06 M Tris–HCl (pH 7.5) and 1 mM EDTA] supplemented with 0.1% BSA and briefly mixed in a Waring blender. Mitochondria were separated from cell debris by differential centrifugation at 640 × g. The supernatant was collected and the mitochondria were pelleted at 30 000 × g. After several washes, the protein concentration of the mitochondrial suspension was determined and mitochondria were frozen and stored at −80°C as pellets containing 1 mg of protein each.
Run-on transcription with isolated mitochondria
Mitochondria were resuspended in ice-cold 330 mM sorbitol, 50 mM HEPES–KOH, pH 8.0 to yield a concentration of ∼10 mg protein/ml and the suspension was kept on ice. For run-on transcription, 10 μl of this suspension were used in a total volume of 100 μl. The standard reaction mixture contained 50 mM HEPES–KOH, pH 8.0, 10 mM MgCl2, 25 mM KOAc, 10 mM DTT, 0.2 U/μl RNasin, 0.5 μg/μl heparin, 125 μM of CTP, ATP and GTP each, and 100 μCi [α-32P]UTP. Transcription was allowed to proceed for 4 min at 30°C unless stated otherwise. Incorporation of [α-32P]UTP into elongating transcripts was determined as described by Hallick et al. (24) with aliquots of the reaction mixture spotted onto DE81 filters (Whatman, Maidstone, UK). The labeled RNA was extracted from the mitochondria in a volume of 250 μl Trizol reagent (GIBCO). After incubation for 5 min at room temperature, 50 μl chloroform/1% isoamylalcohol were added and the solution was mixed vigorously. Following centrifugation at 14 000 r.p.m. (for 15 min at 4°C), the aqueous phase containing the RNA was recovered. The RNA was precipitated using 0.7 vol of isopropanol, washed once with ice-cold 70% ethanol and resuspended in 50 μl of 50% deionized formamide.
Oligonucleotides
Oligonucleotides were obtained from Sigma Genosys and are listed in Table 2. The oligonucleotides were selected such that, those that were to be compared, had an equally high G+C content (30–35% for coding region-specific 40mer oligonucleotides and 8–10% for 50mer oligonucleotides localized in the non-coding intergenic regions, respectively).
Table 2. Oligonucleotides used for dot-blot hybridizations.
| Oligo name | 5′ to 3′ sequence |
|---|---|
| ATP6 5′ | CCAAATAATAGTCTAATCTCAAATTGATCTAATGGTGATG |
| ATP6 3′ | TGACCAGCTAAGATATTAGAACCTAATCTTAAACCTAATG |
| ATP9 | ACCATTAATTAAAGCTGCGAATACGATAGCAATACCAATA |
| COB-EX1 | CCCATATTTCATCAATAATTAATTGATGATGGTTGTGGTG |
| COB-EX6 | CATGGCATGCTCCAATTTGTCCTAATAATACGAAATTGAA |
| COX1-A4 | AGTTGATGTAACTAAACATACTAACCCCATAGGTAATACT |
| COX1-A5β | ATCCTAATAATCCAATTGAAGCCATAGCATATACCATTGA |
| COX1-A6 | TGATAATAGTGCAATGAATGAACCAATAGAAGCGACATAA |
| tRNAE | GATATTATCAACATGAAGAGGTGATGTCGTAACCATTAGA |
| tRNAS | GTAGCCAATAAAGACTAATGATAGCTCAAATATCACACTT |
| VAR1 | AAATGTTGTAATTTATTATTAATATTCCCCGCGGGACCAA |
| tRNAE-112 | AATTATTATTATTATTATTAAGTTTATTATTACTATTTATTAACCTAATT |
| COB −1079 | TATTAATTAATATAAAGAATATGTTTAATTATATATACT-TTTTTTACAAA |
| COB −949 | AAAATATATTATATATTTTATTATAGTATAATAACAATCGGTATTTATTA |
| COB −562 | TAATATTAATGATATCATTATTATTTAATAATAATTAATATTATGAAAGA |
| COB −319 | GTTAATAGATTTTATTAATATTATTATTATTATTTATTAAATATAAACCT |
| COB −29 | ATTTGATTTTCTAAATGCCATATTATTATTATTTATTTATAAAATATTAA |
| c1 | TTGGATGCATAGCTTGAGTATTCTATAGTGTCACCTAAAT |
| c2 | AACCTACGTATCGAACTCATAAGATATCACAGTGGATTTA |
Dot-blot filter preparation and hybridization
Gene-specific PCR-fragments of the yeast mitochondrial genome with an average size of 220 bp or, alternatively, 40 or 50mer oligonucleotides were diluted in 0.4 M NaOH, 10 mM EDTA, heat denatured and subsequently transferred onto a nylon filter (Hybond N+, Amersham), using a vacuum dot-blot apparatus (Schleicher&Schuell) essentially as described by the manufacturer. Bound nucleic acids were UV-crosslinked to the membrane with a Stratalinker (Stratagene). Hybridizations of run-on transcripts to immobilized PCR fragments were carried out at 65°C in 5× SSC, 5× Denhardt's solution, 0.5% SDS and 40 μg/ml of denatured, sonicated salmon sperm DNA. Oligonucleotide-containing filters were hybridized at 42°C in the same buffer.
RESULTS
Kinetics of run-on transcription
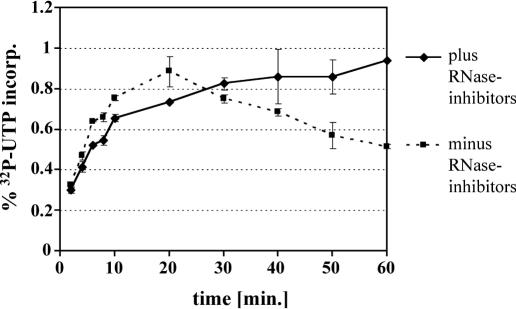
Reaction conditions, and rates of incorporation of radiolabeled nucleotides into isolated mitochondria of S.cerevisiae, were assayed and optimized prior to the analysis of transcription profiles. The composition of the reaction mixture was adapted from a protocol that has been employed successfully for chloroplasts (25). The pretreatment of the organelles, the reaction temperature and reaction times were varied. Overall incorporation of radiolabeled nucleotides into newly synthesized mitochondrial transcripts was assessed by a filter binding assay as described by Hallick et al. (24). Both, [α-32P]UTP and [α-32P]ATP were incorporated very efficiently into mitochondrial run-on transcripts. In double-labeling experiments in which both radiolabeled nucleotides were added to the reaction mixture, the incorporation rates were increased, but did not double. Several wild-type strains (e.g. S150, LL20 and LU20) and strain S150-Bio1 (26), which contains a peptide tag on the Cbp1 protein, were assayed for mitochondrial transcription rates. No differences in overall incorporation rates of isolated mitochondria from these strains were observed. Incorporation of [α-32P]UTP in the presence and absence of RNase inhibitors was linear for ∼10 min. After this time period, the increase in incorporation rates was markedly reduced in reactions containing RNasin (Roche) and heparin (Sigma) and even became negative after 20 min in the absence of these inhibitors (Figure 1). During the first 4 min of run-on transcription, incorporation rates of labeled nucleotides were similar both in the absence and presence of RNase inhibitors. Only after >4 min, a slight inhibition of transcription was detectable, which was presumably due to the DNA-binding activity of heparin (Figure 1 and data not shown). A treatment with RNase A during or after run-on transcription reactions strongly enhanced the decay of labeled run-on transcripts (data not shown). Based on the data described above, all of the following experiments were performed in the presence of RNase inhibitors with incubation times not exceeding 4 min.
Figure 1.
Kinetics of run-on transcription in isolated mitochondria. Transcription in isolated mitochondria was monitored by a transcription pulse with 32P-UTP in the presence (straight black line) or absence (broken black line) of the RNase inhibitors, RNasin and heparin. Aliquots were taken at 2, 4, 6, 8, 10, 20, 30, 40, 50 and 60 min and the overall amount of incorporated radiolabeled nucleotides was measured using DE81 filters (see Materials and Methods). The mean values and standard deviations are based on four individual measurements.
Several operons exhibit strong transcriptional polarity
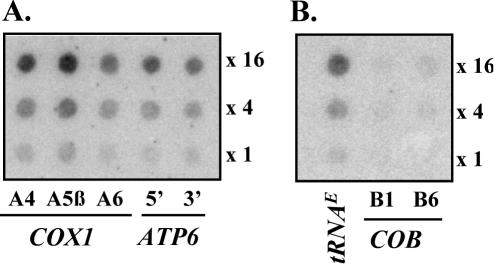
Transcription profiles of different operons were analyzed by hybridization of labeled run-on transcripts to gene-specific 40mer oligonucleotides dotted onto a nylon filter. To facilitate the comparison of hybridization intensities, three dilutions of each oligonucleotide were dotted onto the filters. All oligonucleotides possessed a G+C content of 30–35% to prevent differences in hybridization intensities due to different base compositions. Three oligonucleotides located in exons A4, A5β and A6 of COX1 all showed equal hybridization signals with run-on transcripts (Figure 2A), providing evidence that transcription rates can be faithfully assessed for an individual gene, independent of the location of the oligonucleotide probe and of the distance from the promoter. Oligonucleotides representing the 5′ and 3′ ends of the downstream ATP6 gene, which is co-transcribed with COX1 (27), showed equally strong hybridization signals (Figure 2A). The same was observed for the 21S rRNA–tRNAT transcription unit and for operons containing tRNA genes exclusively (data not shown). In contrast, several other operons did not display even transcription rates. For example, while tRNAE is transcribed at a high rate, transcription of the downstream COB gene is significantly reduced as deduced from the differences in hybridization intensities between a tRNAE-specific probe and two COB-specific oligonucleotides (Figure 2B). Other operons exhibiting this pattern of transcriptional asymmetry were the ATP9-tRNAS-VAR1 operon (see Figure 3) as well as the tRNAF-COX3 transcription unit (data not shown).
Figure 2.
Dot-blot analysis of run-on transcripts from two transcription units. 32P-UTP labeled yeast mitochondrial run-on transcripts were hybridized to gene-specific 40mer oligonucleotides representing the COX1-ATP8-ATP6 operon (A) and the tRNAE-COB operon (B), respectively. Each oligonucleotide was dotted onto the nylon filter in three dilutions corresponding to 2000, 8000 and 32 000 fmol per dot, respectively.
Figure 3.
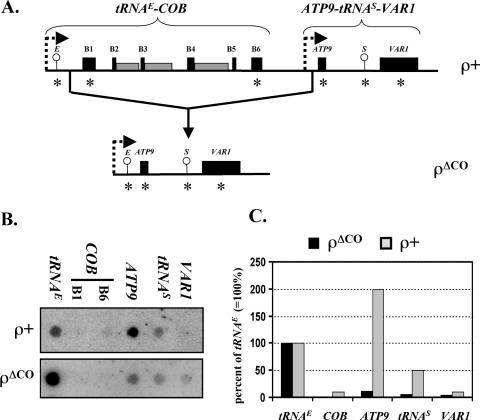
Run-on transcription analysis of the ρΔCO mutant. (A) Schematic depiction of a portion of the mitochondrial genome from wild-type yeast including the tRNAE-COB and ATP9-tRNAS-VAR1 transcription units (top) and the corresponding region of the ρΔCO mitochondrial genome (bottom). Circles indicate the location of tRNA genes, black boxes represent coding sequences while gray boxes represent introns. Promoters are symbolized by broken rightward arrows. The relative positions of the oligonucleotides used for dot-blot analysis as shown in (B) are indicated by asterisks. (B) Dot-blot analysis of run-on transcripts. The gene-specific oligonucleotides indicated in (A) were dotted onto a nylon filter and hybridized with 32P-labeled run-on transcripts from S150-2B (ρ+) and ρΔCO mitochondria (see Table 1). (C) Hybridization intensities of COB, ATP9, tRNAS and VAR1 in relation to tRNAE. Densitometric values of the hybridization signals were determined using a PhosphorImager (see Materials and Methods). The values obtained for tRNAE were set to 100% and used as a reference for the downstream genes.
To investigate whether sequences within the COB gene could be responsible for its weak transcription, mitochondrial run-on transcription was assayed with a mutant, ρΔCO (28), in which much of the COB gene along with the promoter of the downstream ATP9-tRNAS-VAR1 operon is deleted. This novel transcription unit consists of tRNAE, 143 bp between tRNAE and the COB 5′-untranslated region (5′-UTR), COB 5′-UTR sequence from positions −955 to −696, ATP9 sequence starting at position −355 of the 5′-UTR, tRNAS and VAR1 (see Figure 3A). Besides the deletion in the COB-ATP9 interval, there are no other alterations in this otherwise wild-type mitochondrial genome. In the wild-type strain (ρ+), ATP9 RNAs hybridized strongly, indicating a high transcription rate, whereas hybridization to the downstream tRNAS and VAR1 genes was low (Figure 3B). In the mutant strain ρΔCO, however, labeled run-on transcripts produced a comparatively weak hybridization signal for ATP9, although the upstream gene, tRNAE, was transcribed at a high rate (Figure 3B). As expected, the COB-specific oligonucleotides did not hybridize due to the deletion of the COB sequence in the mutant strain. Densitometric evaluation of the individual hybridization signals revealed that the drop in hybridization intensities between tRNAE and ATP9 was ∼90% (Figure 3C). A similar drop in hybridization intensities as seen in ρΔCO was observed in the ρ+ wild-type strain between the tRNAE and COB coding sequences (Figure 3C). The sequence responsible for the drop in apparent transcription rates appears to reside upstream of the gene fusion site at position −696 of the COB 5′-UTR.
Sequences between the tRNAE and COB coding regions influence transcription rates
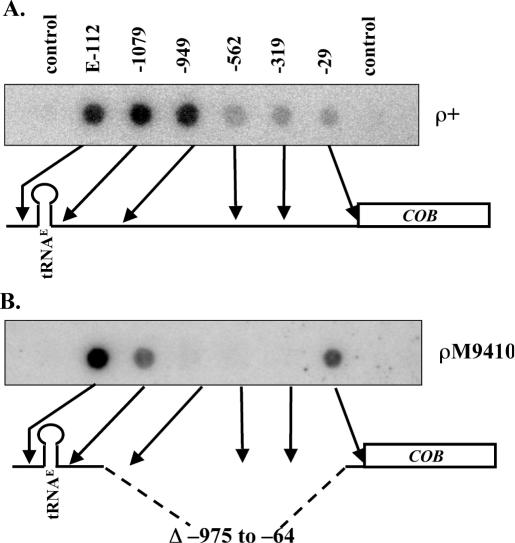
From the previous experiment, it could be concluded that the significant drop in transcription rates within the tRNAE-COB operon is not provoked by the coding sequence of the downstream COB gene. To check whether non-coding sequences between the two genes might have an influence on polymerase processivity along this operon, we used run-on transcription assays to analyze the transcription rates of both genes in two mutants bearing deletions between the coding sequences. While a deletion of 143 bp between tRNAE and COB 5′-UTR coding sequence in strain S150TG955 did not result in a change of the transcription pattern, a longer deletion of >900 bp in strain S150M9410, which affected most of the region between coding sequences, reduced the difference in hybridization intensities between tRNAE and COB by ∼3-fold (data not shown). Oligonucleotides, which were complementary to different sites between tRNAE and COB coding sequences revealed, in addition, that hybridization signals were equally strong at least for the first 150 nucleotides in ρ+ mitochondria but dropped significantly between nucleotides −949 and −562 relative to the COB translation start site (Figure 4A). This result is in accordance with the results obtained with strain ρΔCO (Figure 3) and strengthens the assumption that sequences between positions −949 and −696 must be involved in the drop in apparent transcription rates. Elimination of this sequence in the mitochondrial mutant ρM9410 resulted in run-on transcripts hybridizing to oligonucleotide −29 much more strongly than transcripts of the wild-type strain (Figure 4B). As expected, ρM9410 transcripts did not hybridize to the three oligonucleotides −949, −562 and −319 located within the deleted region. Hybridization of ρM9410 transcripts to oligonucleotide −1079 was similar to hybridization to −29 and was diminished relative to wild type. Whether there is a new, weaker, attenuation site in the tRNA to −1079 interval or transcripts downstream of the tRNA are subject to degradation that is fast enough to have an effect during the 4 min transcription pulse cannot be distinguished.
Figure 4.
Oligonucleotide-based dot-blot analysis of the sequence between tRNAE and COB. Oligonucleotides complementary to the non-coding region of the primary transcript were dotted onto a nylon filter and hybridized to labeled mitochondrial run-on transcripts. (A) Hybridization pattern obtained with labeled run-on transcripts from strain S150-Bio1 carrying a wild-type mitochondrial genome (ρ+). (B) Hybridization pattern obtained with labeled run-on transcripts from strain S150M9410 harboring a 910 bp deletion (Δ) in the intergenic and 5′-UTR region (ρM9410). The relative lengths of the non-coding regions in both strains as well as the position of the oligonucleotides are indicated below each autoradiogram.
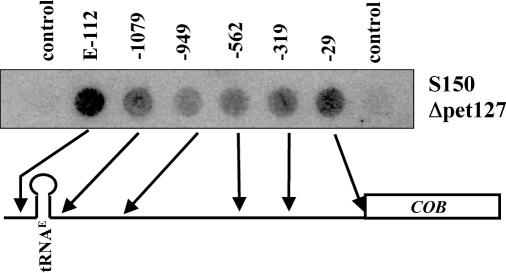
Deletion of PET127 reduces the transcriptional changes within the tRNAE-COB operon
The analysis of mitochondrial mutants revealed that transcriptional changes within the tRNAE-COB operon is, to some extent, based on sequences between the two coding regions. This 5′-UTR region is the target for several proteins involved in COB RNA processing, protection and translation. Thus, mitochondrial run-on transcripts were analyzed from several nuclear mutants. Cbp1 protects COB mRNA through sequences between −961 and −898 of the 5′-UTR (4). Cbs2 promotes translation of COB mRNA through sequences between −170 and −33 of the 5′-UTR (29). Pet127 governs the shortening of the COB mRNA precursor from position −1098 to positions −955/−954. Deletion mutations in CBP1 and CBS2 did not significantly change the transcription pattern of the tRNAE-COB operon (data not shown). However, in cells lacking the Pet 127 protein, the oligonucleotides −562, −319 and −29 gave significantly stronger signals (Figure 5) than the wild-type strain S150 (see Figure 4). Similar to the S150M9410 strain, the Δpet127 strain showed a weaker drop in hybridization intensity in the tRNA to −1079 interval.
Figure 5.
Deletion of PET127 affects hybridization along the tRNAE-COB operon. Mitochondrial run-on transcription was carried out with a strain carrying a deletion of the nuclear gene PET127 in the S150-2B background (see Table 1). The transcripts were hybridized to oligonucleotides representing the non-coding region between tRNAE and COB.
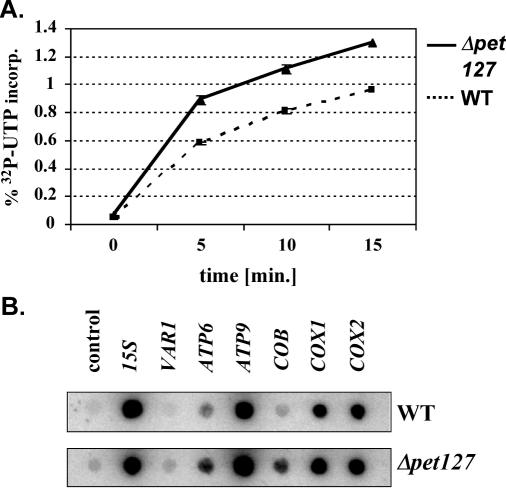
A comparison of overall transcription rates of the entire mitochondrial genome showed that the incorporation of [α-32P]UTP into RNA after 5 min was ∼30% higher in mitochondria of the Δpet127 mutant than in the wild-type strain (Figure 6A). Hybridization of run-on transcripts with gene-specific PCR-products of seven mitochondrial genes revealed that this increase was, however, not distributed evenly across the mitochondrial genome. The transcription of the COB, ATP6, ATP9 and VAR1 genes was more affected than that of the 15S rRNA, COX1 and COX2 genes (Figure 6B).
Figure 6.
Characterization of mitochondrial transcription in the Δpet127 strain. (A) Overall transcription rates in mitochondria isolated from S150-2B and Δpet127 as monitored by incorporation of 32P-UTP into RNA. Aliquots of a run-on assay were taken at 0, 5, 10 and 15 min and the amount of incorporated radioactivity was determined using DE81 paper (see Materials and Methods). Each experiment was repeated four times to give the mean values and standard deviations depicted. (B) Transcription profiles of six protein coding genes. Gene-specific PCR-fragments of six mitochondrial mRNA genes (COX 1, COX 2, COB, ATP6, ATP9 and VAR1), the 15S rDNA and the vector pGem-T for a control were immobilized on a nylon filter and hybridized to labeled run-on transcripts of mitochondria from strain S150-2B and strain Δpet127.
DISCUSSION
Run-on transcription in yeast mitochondria
Isolated yeast mitochondria as well as mitochondrial lysates are capable of carrying out RNA synthesis (14,15), which makes them suitable for run-on transcription analysis. With this method, only newly synthesized transcripts are labeled with radioactive nucleotides and thus can be distinguished from steady-state RNAs. The analysis of run-on kinetics showed that transcription rates are constant for ∼10 min (Figure 1). Measurable RNA degradation occurred after 20 min. The addition of heparin and RNasin had no significant effect on nucleotide incorporation during the first 4 min, while longer reaction times in the presence of heparin resulted in a slight inhibition of transcription. Based on these data, we used short labeling times of not more than 4 min. The observation that incorporation rates were susceptible to degradation by RNase A, as well as to protection by RNase inhibitors, suggests that the membranes of the isolated mitochondria are either permeable to macromolecular substances, such as RNases and their inhibitors, or that a significant percentage of organelles was lysed before or during the reaction.
Asymmetries in transcription of operons
Mitochondrial genes are organized in transcription units preceded by simple promoters. Assuming that transcription proceeds with a constant rate of nucleotide incorporation, co-transcribed genes should be represented by equally strong hybridization signals when analyzed by a combination of run-on transcription and hybrid selection techniques. We found that oligonucleotides that represent different parts of the same coding sequence gave similar hybridization signals, even when the distance between the oligonucleotides was rather large, due to the presence of introns (Figure 2). This result indicates that transcription rates can be assessed faithfully, independent of the location of the probes used for hybridization. In addition, we conclude that transcriptional rates are constant within the coding region of a gene.
In several cases, however, genes located further downstream in an operon showed a significant decrease in transcription rates when compared to the first gene in the operon. Operons exhibiting this transcriptional asymmetry are the tRNAE-COB operon (Figure 2), the ATP9-tRNAS-VAR1 operon (Figure 3) and the tRNAF-COX3 operon (data not shown). It is conspicuous that all operons exhibiting polarity of transcription contain combinations of tRNA genes and protein-encoding genes (COB, COX3 and ATP9). The COX1-ATP8-ATP6 operon, in contrast, consists entirely of protein-coding genes and has constant transcription rates throughout the operon (Figure 2). The co-transcription of genes of different function may be a reason for evolution of the observed transcriptional asymmetries, which results in differences in steady-state values of RNAs transcribed together (30). This mechanism of regulating RNA abundance may be important because regulation via transcriptional initiation is limited in yeast mitochondria, due to the relatively simple and uniform structure of the promoters (10).
Transcription attenuation or very fast RNA degradation?
There are two possible mechanisms that could explain the decrease of run-on values from 5′ to 3′ in gene clusters: (i) the presence of leaky transcription stops leading to partial transcription attenuation or (ii) very rapid processing/degradation events of the primary transcripts. The assessment of in vivo pulse-labeled mRNAs showed fewer transcripts of promoter-distal genes than of promoter-proximal genes within the same transcription unit (19). In some cases (e.g. the tRNAE-COB operon), this difference was as much as 17-fold, a value that is in the same order of magnitude as we observed with the hybridization of run-on transcripts. Mueller and Getz (19) ascribed this phenomenon to the attenuation of transcript elongation, although they could not rule out a selective degradation of downstream transcripts during the pulse-labeling period. In the present work, we showed that the decline in hybridization intensities is not linear but occurs as a sharp drop within a three hundred nucleotide region in wild-type strains. In addition, an experiment with a mutant strain deleted in the region between tRNAE and COB coding sequences showed that the drop in hybridization signals can be reduced in strength (Figure 4). Both observations suggest that transcription is attenuated at a site within the sequence encoding the COB 5′-UTR.
However, lack of the mitochondrially located Pet127 protein was also found to diminish the asymmetry of tRNAE-COB operon transcription significantly (Figure 5). In a Δpet127 null strain, COB precursor RNAs with 5′ ends mapping at the 3′ end of tRNAE (position −1098) accumulate to levels equivalent to those of the processed mature transcript in wild-type strains (5′ ends at positions −955 and −954) (6,7). Precursor RNAs for the 15S rRNA, VAR1 and ATP8/6 mRNAs are also not processed at the 5′ end. Pet127 must then be the nuclease that processes these transcripts at the 5′ end, or it must regulate the nuclease that does. The fact that Pet127 degrades RNAs in addition to processing them to mature forms is suggested by two lines of evidence. First, pet127 null mutations were recovered as suppressors of mutations in the 5′-UTR of COX3 mRNA that make the mRNA unstable (6). COX3 mRNA is cleaved from a precursor RNA containing upstream tRNAs, but the mRNA is not processed further at the 5′ end. Pet127 must be responsible for the turnover of the unstable COX3 mRNAs. Second, overexpression of Pet127 can rescue mutant strains that are null for the mitochondrial 3′–5′ exonuclease activity of the degradosome, which is composed of the Suv3 helicase and the Dss1 nuclease (31–34). The 5′ end-directed activity of Pet127 must be able to compensate for the lack of the 3′–5′ exonuclease activity. Therefore, our hypothesis is that, very rapid RNA degradation events governed by Pet127 are responsible in part for the observed drop in hybridization intensities of the downstream COB gene in the tRNAE-COB operon. The most likely sites for the initiation of degradation dependent on Pet127 are 5′ ends of precursor COB RNA at position −1098 and mature COB mRNA at positions −955/−954. We do not favor the alternative hypothesis wherein Pet127 has a direct role in the attenuation of transcription. The fact that differences in hybridization are observed in the 4 min interval, during which transcription is measured, implies that processing and degradation of transcripts must occur within this time frame. Taken together, it is likely that asymmetries in mitochondrial transcription are a consequence of a combination of transcription attenuation at specific sites between coding sequences and of very rapid nucleolytic degradation processes, which are insensitive to RNasin and heparin.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a postdoctoral grant from the Deutscher Akademischer Austauschdienst (DAAD) to K.K. as well as by grant NIH GM34893 to C.L.D.
REFERENCES
- 1.Dieckmann C.L. and Staples,R.R. (1994) Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. In Jeon,K.W. and Jarvik,J.W. (eds), International Review of Cytology. Academic Press Inc., San Diego, CA, pp. 145–181. [DOI] [PubMed] [Google Scholar]
- 2.Greenleaf A.L., Kelly,J.L. and Lehman,I.R. (1986) Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc. Natl Acad. Sci.USA, 83, 3391–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang S.H. and Jaehning,J.A. (1991) The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial σ factors. J. Biol. Chem., 266, 22671–22677. [PubMed] [Google Scholar]
- 4.Mittelmeier T.M. and Dieckmann,C.L. (1993) In vivo analysis of sequences necessary for Cbp1-dependent accumulation of cytochrome b transcripts in yeast mitochondria. Mol. Cell. Biol., 13, 4203–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W. and Dieckmann,C.L. (1994) Cbp1p is required for message stability following 5′-processing of COB mRNA. J. Biol. Chem., 269, 16574–16578. [PubMed] [Google Scholar]
- 6.Wiesenberger G. and Fox,T.D. (1997) Pet127p, a membrane-associated protein involved in stability and processing of Saccharomyces cerevisiae mitochondrial RNAs. Mol. Cell. Biol., 17, 2816–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islas-Osuna M.A., Ellis,T.P., Marnell,L.L., Mittelmeier,T.M. and Dieckmann,C.L. (2002) Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J. Biol. Chem., 277, 37987–37990. [DOI] [PubMed] [Google Scholar]
- 8.Foury F., Roganti,T., Lecrenier,N. and Prunelle,B. (1998) The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett., 440, 325–331. [DOI] [PubMed] [Google Scholar]
- 9.Christianson T., Edwards,J.C., Mueller,D.M and Rabinowitz,M. (1983) Identification of a single transcriptional initiation site for the glutamic tRNA and COB genes in yeast mitochondria. Proc. Natl Acad. Sci. USA, 80, 5564–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas T.K. (1990) Control of mitochondrial gene expression in the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 87, 9338–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards J.C., Levens,D. and Rabinowitz,M. (1982) Analysis of transcriptional initiation of yeast mitochondrial DNA in a homologous in vitro transcription system. Cell, 31, 337–346. [DOI] [PubMed] [Google Scholar]
- 12.Shadel G.S. and Clayton,D.A. (1995) A Saccharomyces cerevisiae mitochondrial transcription factor, sc-mtTFB, shares features with sigma factors but is functionally distinct. Mol. Cell. Biol., 15, 2101–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangus D.A. and Jaehning,J.A. (1996) Transcription in vitro with Saccharomyces cerevisiae mitochondrial RNA polymerase. Methods Enzymol., 264, 57–66. [DOI] [PubMed] [Google Scholar]
- 14.Iqbal J. and Hudson,A.P. (1996) An in vitro transcription assay for yeast mitochondria using organellar lysates. Anal. Biochem., 243, 270–276. [DOI] [PubMed] [Google Scholar]
- 15.Groot G.S.P., van Harten-Loosbroek,N., van Ommen,G.-J.B. and Pijst,H.L.A. (1981) RNA synthesis in isolated yeast mitochondria. Nucleic Acids Res., 9, 6369–6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McEntee C.M., Cantwell,R., Thomas,L.C. and Hudson,A.P. (1989) Mitochondrial rRNA-containing petite strains of yeast (Saccharomyces cerevisiae) show a normal nuclear-mitochondrial stringent response. Biochem. Biophys. Res. Commun., 164, 362–369. [DOI] [PubMed] [Google Scholar]
- 17.McEntee C.M., Cantwell,R., Thomas,L.C. and Hudson,A.P. (1993) Transcription of the yeast mitochondrial genome requires cyclic AMP. Mol. Gen. Genet., 241, 213–224. [DOI] [PubMed] [Google Scholar]
- 18.Poyton R.O., Bellus,G., McKee,E.E., Sevarino,K.A. and Goehring,B. (1996) In organello mitochondrial protein and RNA synthesis systems from Saccharomyces cerevisiae. Methods Enzymol., 264, 36–42. [DOI] [PubMed] [Google Scholar]
- 19.Mueller D.M. and Getz,G.S. (1986) Transcriptional regulation of the mitochondrial genome of yeast Saccharomyces cerevisiae. J. Biol. Chem., 261, 11756–11764. [PubMed] [Google Scholar]
- 20.Conde J. and Fink,G.G. (1976) A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl Acad. Sci. USA, 73, 3651–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dieckmann C.L. and Mittelmeier,T.M. (1987) Nuclearly encoded Cbp1 interacts with the 5′ end of mitochondrial cytochrome b pre-mRNA. Curr. Genet., 12, 391–397. [DOI] [PubMed] [Google Scholar]
- 22.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Faye G.C., Kujawa,C. and Fukuhara,H. (1974) Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23S ribosomal RNA in petite mutants of Saccharomyces cerevisiae. J. Mol. Biol., 88, 185–203. [DOI] [PubMed] [Google Scholar]
- 24.Hallick R.B., Lipper,C., Richards,O.C. and Rutter,W.J. (1976) Isolation of a transcriptionally active chromosome from Euglena gracilis. Biochemistry, 15, 3039–3045. [DOI] [PubMed] [Google Scholar]
- 25.Krause K., Falk,J., Humbeck,K. and Krupinska,K. (1998) Responses of the transcriptional apparatus of barley chloroplasts to a prolonged dark period and to subsequent reillumination. Physiol. Plant., 104, 143–152. [Google Scholar]
- 26.Krause K., Lopes De Souza,R., Roberts,D.G. and Dieckmann,C.L. (2004) The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell., 15, 2674–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osinga K.A., De Vries,E., Van der Horst,G. and Tabak,H.F. (1984) Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J., 3, 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittelmeier T.M. and Dieckmann,C.L. (1990) CBP1 function is required for stability of a hybrid cob-oli1 transcript in yeast mitochondria. Curr. Genet., 18, 421–428. [DOI] [PubMed] [Google Scholar]
- 29.Mittelmeier T.M. and Dieckmann,C.L. (1995) In vivo analysis of sequences required for translation of cytochrome b transcripts in yeast mitochondria. Mol. Cell. Biol., 15, 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller D.M. and Getz,G.S. (1986) Steady state analysis of mitochondrial RNA after growth of yeast Saccharomyces cerevisae under catabolite repression and derepression. J. Biol. Chem., 261, 11816–11822. [PubMed] [Google Scholar]
- 31.Margossian S.P., Li,H., Zassenhaus,H.P. and Butow,R.A. (1996) The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell, 84, 199–209. [DOI] [PubMed] [Google Scholar]
- 32.Dziembowski A. and Stepien,P.P. (2001) Genetic and biochemical approaches for analysis of mitochondrial degradosome from Saccharomyces cerevisiae. Methods Enzymol., 342, 367–378. [DOI] [PubMed] [Google Scholar]
- 33.Dziembowski A., Piwowarski,J., Hoser,R., Minczuk,M., Dmochowska,A., Siep,M., van der Spek,H., Grivell,L. and Stepien,P.P. (2003) The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem., 278, 1603–1611. [DOI] [PubMed] [Google Scholar]
- 34.Wegierski T., Dmochowska,A., Jablonowska,A., Dziembowski,A., Bartnik,E. and Stepien,P.P. (1998) Yeast nuclear PET127 gene can suppress deletions of SUV3 or DSS1 genes: an indication of a functional interaction between 3′ and 5′ ends of mitochondrial mRNAs. Acta Biochim. Pol., 45, 935–940. [PubMed] [Google Scholar]
- 35.Mayer S.A. and Dieckmann,C.L. (1991) Yeast CBP1 mRNA 3′ end formation is regulated during the induction of mitochondrial function. Mol. Cell. Biol., 11, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]